Abstract
Introduction
Preeclampsia is a leading cause of maternal and fetal mortality worldwide, yet its exact pathogenesis remains elusive.
Objectives
This study, nested within the Vitamin D Antenatal Asthma Reduction Trial (VDAART), aimed to develop integrated omics models of preeclampsia that have utility in both prediction and in the elucidation of underlying biological mechanisms.
Methods
Metabolomic profiling was performed on first trimester plasma samples of 47 pregnant women from VDAART who subsequently developed preeclampsia and 62 controls with healthy pregnancies, using liquid-chromatography tandem mass-spectrometry. Metabolomic profiles were generated based on logistic regression models and assessed using Received Operator Characteristic Curve analysis. These profiles were compared to profiles from generated using third trimester samples. The first trimester metabolite profile was then integrated with a pre-existing transcriptomic profile using network methods.
Results
In total, 72 (0.9%) metabolite features were associated (p<0.01) with preeclampsia after adjustment for maternal age, race, and gestational age. These features had moderate to good discriminatory ability; in ROC curve analyses a summary score based on these features displayed an area under the curve (AUC) of 0.794 (95%CI 0.700, 0.888). This profile retained the ability to distinguish preeclamptic from healthy pregnancies in the third trimester (AUC:0.762 (95% CI 0.663, 0.860)). Additionally, metabolite set enrichment analysis identified common pathways, including glycerophospholipid metabolism, at the two time-points. Integration with the transcriptomic signature refined these results suggesting a particular role for lipid imbalance, immune function and the circulatory system.
Conclusions
These findings suggest it is possible to develop a predictive metabolomic profile of preeclampsia. This profile is characterized by changes in lipid and amino acid metabolism and dysregulation of immune response and can be refined through interaction with transcriptomic data. However validation in larger and more diverse populations is required.
Keywords: Preeclampsia, VDAART, metabolomics, Integrative omics
INTRODUCTION
Preeclampsia is a severe, pregnancy specific disorder characterized by new-onset hypertension and urinary protein after 20 weeks’ gestation. In the United States between 3–7% of all pregnancies are affected, and worldwide, it remains a leading cause of maternal and fetal morbidity and mortality (Kuklina et al., 2009b, Kuklina et al., 2009a, Duley, 2009, Berg et al., 2009). Left untreated, preeclampsia can lead to maternal seizures, multi-organ failure, and death. It cannot be predicted and delivery is the only cure, yet the exact pathogenesis of preeclampsia remains elusive and no early-stage screening tests are available (Ahn et al., 2011).
Advancement of high-throughput technologies has enabled the investigation of the epigenetic, genomic, transcriptomic, proteomic and metabolomic variability underlying complex diseases and represents a novel opportunity for elucidating the biological underpinnings of preeclampsia. However, metabolomics is a largely untapped resource in this area (Allsworth et al., 2013). To date, metabolomic studies of preeclampsia have been limited in scope and size (Austdal et al., 2014, Bahado-Singh et al., 2013, Kenny et al., 2010, Kuc et al., 2014, Odibo et al., 2011, Turner et al., 2008, Schott et al., 2012, Austdal et al., 2015, Bahado-Singh et al., 2015), and clinically translatable biomarkers remain elusive. Metabolomics provides perhaps the most integrated profile of biological status, as it reflects all of the preceding ‘omes’ and is indicative of phenotype and disease state (Bictash et al., 2010). Therefore the metabolic profile of individuals who will develop preeclampsia is likely to have utility both in the identification of early preeclampsia biomarkers, and in furthering understanding of the early pathogenesis of the condition.
Although individual omics platforms are powerful tools, a more global view of the multi-stage disease process is available through integration of multiple ‘omics’, which are intrinsically hierarchical, from the genome to the metabolome. This integrative strategy has already successfully led to the discovery of novel disease pathways and biomarkers for complex diseases that would not have been identified through investigation of a single ‘omics’ platform (Rhee and Gerszten, 2012, Wang et al., 2011, Rhee and Thadhani, 2011).
The aim of this study, nested within the Vitamin D Antenatal Asthma Reduction Trial (VDAART), was to develop predictive models of preeclampsia through metabolic profiling of prospectively-collected blood samples from early pregnancy in order to 1) identify women at risk of developing the preeclampsia prior to any clinical manifestations of the disease and 2) elucidate the biological mechanisms and pathways underlying preeclampsia development. The predictive profiles were tested in third trimester blood samples. They were then refined through integrating transcriptomic preeclampsia profiles with the metabolomic profiles to further explore pathogenesis. To the best of our knowledge this is the first study to integrate both metabolomics and transcriptomics in the study of preeclampsia, providing a novel systems biology perspective on this disorder.
MATERIALS AND METHODS
Study Population
VDAART is a multicenter, randomized, double-blind, placebo-controlled clinical trial examining whether vitamin D supplementation in pregnant women could prevent the development of pregnancy complications, such as preeclampsia, and diseases in their offspring (Litonjua et al., 2016). Briefly, women aged 18–40 years in their first trimester of pregnancy (10–18 weeks pregnant) were recruited from obstetric clinics from three centers in the United States: Boston University Medical Center in Boston, MA, Washington University in St. Louis, MO, and Kaiser Permanente Southern Region in San Diego, CA. Selection criteria included personal history of asthma and/or allergies or a partner with a history of asthma/allergies. The trial was approved by the Institutional Review Boards of the participating institutions and at Brigham and Women’s Hospital and is registered with ClinicalTrials.gov (NCT00920621). Written informed consent was obtained from all women at recruitment.
In total, 882 women were recruited and randomized to either the treatment arm (4000 IU/day of vitamin D3 plus a 400 IU vitamin D3 multivitamin) or to the placebo arm (multivitamin only). At trial entry baseline pre-supplementation vitamin D levels were measured, and blood serum, plasma, and RNA were obtained. The women were then followed throughout pregnancy for the development of complications. Preeclampsia was defined according to ACOG guidelines (ACOG, 2013) followed by an independent review. Controls were chosen from VDAART subjects matched on age (optimally within 5 years), gestational age at entrance into the study (optimally within 2 weeks), race, and center such that each case had 1 or 2 matched controls; additional controls that were similar to the case population with respect to the matching factors were incorporated to enhance the range of vitamin D concentrations. Detailed data on possible confounding variables were also available.
Metabolomic Profiling
Profiling methods are described in detail in Online Resource 1. Briefly, profiling of metabolites was conducted at the Broad Institute (Massachusetts Institute of Technology, Cambridge, MA, USA) using four liquid chromatography-tandem mass spectrometry (LC-MS) methods to measure complementary sets of metabolite classes: (1) HILIC-positive platform: Amines and polar metabolites that ionize in the positive ion mode using hydrophilic interaction liquid chromatography (HILIC) and MS analyses; (2) HILIC-negative platform: Central metabolites (i.e. metabolites directly involved in the maintenance of the essential normal physiological processes of a biological system) and polar metabolites that ionize in the negative ion mode using HILIC chromatography with an amine column and targeted MS; (3) C8-positive platform: Polar and non-polar lipids using reversed phase chromatography and full scan MS; (4) C18-negative platform: Free fatty acids, bile acids, and metabolites of intermediate polarity using reversed chromatography with a T3 UPLC column (C18 chromatography) and MS analyses in the negative ion mode.
LC-MS system sensitivity and chromatography quality were checked prior to analysis by analyzing reference samples. Internal standard peak areas were monitored for quality control during the analyses. A pooled reference sample was analyzed throughout the analytical run as an additional quality control measure and to serve as reference for scaling raw LC–MS peak areas across sample batches.
Features were indexed by their mass-to-charge ratio (m/z) and retention time (rt) and metabolite identities were confirmed using known standards. Metabolite features with a signal-to-noise ratio <10 were considered unquantifiable and excluded, as were features with undetectable/missing levels for >10% of the samples. All remaining missing values were imputed with the median peak intensity for that feature. Features with a coefficient of variance in the quality-control samples greater than 25% across all batches were excluded to ensure good technical reproducibility.
Data Analysis
Metabolite features were analyzed as measured LC-MS peak areas, which are proportional to feature concentration. All features were log transformed to normalize them and pareto-scaled to reduce the variation in fold-change differences between the features, and to make the effect estimates for each feature comparable. Independent unconditional logistic regression models adjusting for maternal age and race, study site, and gestational age (weeks) were run for each feature using the blood samples extracted at 10–18 weeks’ gestation. Additional models including pre-pregnancy body mass index (BMI) as a further covariate were also explored.
The ability of the significant metabolite features from each platform to prospectively distinguish women who developed preeclampsia from those who did not was assessed using a Receiver Operator Characteristic (ROC) curve. The area under the curve (AUC) of a baseline model including the confounding variables was compared to models additionally including baseline serum vitamin D level and pre-pregnancy BMI, as suspected predictors of preeclampsia, and those including a summary score based on principal component analysis of the selected features.
Metabolomic profiling was additionally performed on plasma samples taken at 32–38 weeks gestation to identify metabolites that distinguished pregnant women who developed preeclampsia from those who remained healthy in the third trimester. The crossover between the significant features at the two time-points was computed to determine whether those metabolites identified in the first trimester retained their predictive ability for preeclampsia in the third. Metabolite set enrichment analyses using the known metabolites and performed with MetaboAnalyst v.2.5 (Xia et al., 2015) was used to explore whether the same pathways were dysregulated at the two time-points. The hypergeometric test was specified for the over-representation analysis and relative-betweenness centrality for the pathway topology analysis.
Integrative Omics
A subset of the metabolomics population also had genome-wide gene expression profiles available. Previous analyses of these data identified 2232 probes corresponding to 1632 genes that were differentially expressed in the first trimester blood samples of women who went on to develop preeclampsia compared to healthy controls (Mirzakhani et al., 2016) (Online Resource 1).. To better understand the biology underlying the predictive metabolomic profiles and to provide a more holistic view of the pathogenesis of preeclampsia, the significant genes and metabolite features were integrated using weighted gene correlation network analysis (WGCNA) (Langfelder and Horvath, 2008). WGCNA employs a systems biology approach to describe correlation patterns in high dimensional datasets and to generate modules based on these correlations. By interrogating the genes and metabolites comprising these modules, the interplay between the transcriptome and the metabolome in the development of preeclampsia can be explored. To account for fold-differences between the genes and the metabolites, normalized but unscaled metabolomic intensity levels and normalized transcriptomic expression levels were pareto-scaled together to form the analytical dataset. Using hierarchical clustering based on topological overlap and applying a soft thresholding power of 2 (chosen to achieve a scale-free topology fitting index >0.9), the interconnectedness between the genes and metabolites was quantified (Online Resource 1).
RESULTS
Baseline Cohort Characteristics
During follow up of the VDAART trial, 67 eligible cases of preeclampsia were diagnosed and 110 matched controls selected. Of these, 47 cases (70%) and 62 controls (56%) had metabolomic profiling on plasma samples extracted at baseline (first trimester), and in the third trimester (five cases did not have a blood sample available at the third trimester). Five (11%) cases were classified as early onset preeclampsia (diagnosed <34 weeks gestation) and 42 (89%) as late onset preeclampsia (diagnosed ≥34 weeks gestation).
Within this ‘metabolomics population’, 44 cases and 57 controls also had transcriptomic profiling performed. These 101 women were termed the ‘integrative omics population’. Baseline characteristics were compared between women who went on to develop preeclampsia and those with healthy pregnancies, in both the ‘metabolomics population’ and the ‘integrative omics’ population (Table 1). Women who developed preeclampsia had a higher BMI prior to pregnancy than controls (metabolomics population; p=0.01, integrative omics population; p=0.027); however there were no other significant differences between the preeclamptic and healthy pregnancies.
Table 1.
Baseline characteristics of the study populations
| Variable | Metabolomics population | Integrative omics population | |||||
|---|---|---|---|---|---|---|---|
| Controls n=62 | Preeclampsia cases n=47 | P-value | Controls n=57 | Preeclampsia cases n=44 | P-value | ||
| Study Site | California | 17 (27.4%) | 15 (31.9%) | 16 (28.1%) | 14 (31.8%) | ||
| Massachusetts | 13 (21.0%) | 11 (23.4%) | 11 (19.3%) | 10 (22.7%) | |||
| Missouri | 32 (51.6%) | 21 (44.7%) | 0.771 | 30 (52.6%) | 20 (45.5%) | 0.773 | |
| Age at enrollment | years (SD) | 26.3 (4.9) | 26.0 (4.8) | 0.81 | 26.4 (4.8) | 26.0 (4.8) | 0.621 |
| Study arm | Vitamin D supplementation | 34 (54.8%) | 24 (51.1%) | 32 (56.1%) | 23 (52.3%) | ||
| Placebo | 28 (45.2%) | 23 (48.9%) | 0.844 | 25 (43.9%) | 21 (47.7%) | 0.853 | |
| Baseline serum vitamin D status | Deficient (<30ng/ml ) | 47 (75.8%) | 41 (87.2%) | 44 (77.2%) | 38 (86.4%) | ||
| Sufficient (≥30ng/ml) | 15 (24.2%) | 6 (12.8%) | 0.21 | 13 (22.8%) | 6 (13.6%) | 0.361 | |
| Race | American Indian/Alaskan | 0 (0%) | 3 (6.4%) | 0 (0%) | 3 (6.8%) | ||
| Asian | 2 (3.2%) | 2 (4.3%) | 2 (3.5%) | 2 (4.5%) | |||
| Black/African American | 34 (54.8%) | 22 (46.8%) | 31 (54.4%) | 20 (45.5%) | |||
| White | 22 (35.5%) | 17 (36.2%) | 20 (35.2%) | 16 (36.4%) | |||
| Other | 4 (6.5%) | 3 (6.4%) | 0.358 | 4 (7.0%) | 3 (6.8%) | 0.360 | |
| Gestational age at enrolment | weeks (SD) | 14.2 (2.7) | 13.9 (2.7) | 0.579 | 14.1 (2.7) | 13.8 (2.7) | 0.584 |
| Pre-pregnancy BMI | kg/m2 (SD) | 26.4 (7.6) | 30.7 (7.6) | 0.010* | 26.6 (7.8) | 30.4 (7.8) | 0.027* |
| History of asthma | No | 38 (61.3%) | 27 (57.4%) | 35 61.4%) | 25 (56.8%) | ||
| Yes | 24 (38.7%) | 20 (42.6%) | 0.835 | 22 (38.6%) | 19 (43.2%) | 0.794 | |
| N. Total pregnancies (n) | 1 | 26 (41.9%) | 22 (46.8%) | 22 (38.6%) | 20 (45.5%) | ||
| 2 | 16 (25.8%) | 11 (23.4%) | 16 (28.1%) | 10 (22.7%) | |||
| 3 | 11 (17.7%) | 9 (19.1%) | 10 (17.5%) | 9 (20.5%) | |||
| >4 | 9 (14.5%) | 5 (10.6%) | 0.908 | 9 (15.8%) | 5 (11.4%) | 0.797 | |
| Gestational diabetes | No | 60 (96.8%) | 43 (91.5%) | 55 (96.5%) | 41 (93.2%) | ||
| Yes | 2 (3.2%) | 3 (6.4%) | 2 (3.5%) | 2 (4.5%) | |||
| Unsure/NA | 0 (0.0%) | 1 (2.1%) | 0.372 | 0 (0%) | 1 (2.3%) | 0.499 | |
| Gender of childa | Female | 33 (53.2%) | 21 (45.7%) | 30 (52.6%) | 19 (44.2%) | ||
| Male | 29 (46.8%) | 25 (54.3%) | 0.559 | 27 (47.4%) | 24 (55.8%) | 0.526 | |
Significant at the 95% confidence interval
child’s gender unknown for one preeclamptic pregnancy
SD – Standard Deviation
Metabolomic Profiling
Development of Predictive Models for Preeclampsia Using First Trimester Samples
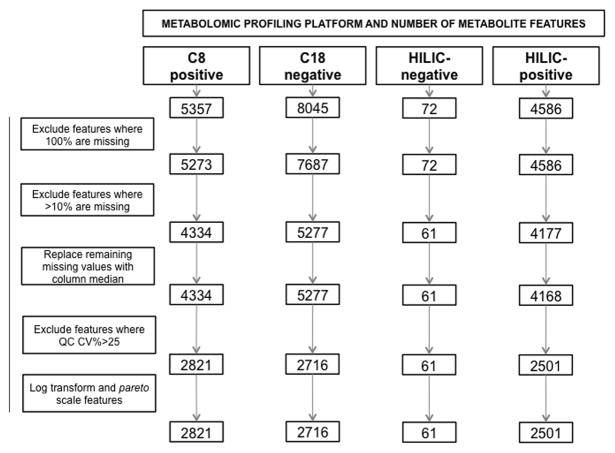
Metabolic profiling yielded a total of 18,060 LC-MS peaks, distinguished by their m/z ratio and rt and hereafter referred to as metabolite features. After data processing and quality control filtering, 8099 metabolite features remained (Fig 1). In total, 484 features (6.0%) were significantly associated with the risk of preeclampsia at a nominal 95% confidence level adjusting for maternal age and race, study site, and gestational age (Table 2). Of these, 377 were upregulated in preeclampsia cases and 107 were downregulated.
Fig 1.
Overview of metabolomic profiling quality control
QC – Quality control; CV – Coefficient of Variance
Table 2.
Summary of the metabolomic profiling analyses of early pregnancy samples based on four profiling platforms
| Analysis | Metabolite features (n=8099) | |
|---|---|---|
| Logistic regression modela | n (%) significant features | |
| p<0.05 | 484 (6.0%) | |
| p<0.01 | 72 (0.9%) | |
| p<0.001 | 2 (0.02%) | |
| ROC analysisb | AUC Model 1 (95% CI) | 0.573 (0.463, 0.682) |
| AUC Model 2 (95% CI) | 0.731 (0.624, 0.838)¥ | |
| AUC Model 3 (95% CI) | 0.752 (0.658, 0.846)¥ | |
| AUC Model 4 (95% CI) | 0.794 (0.700, 0.888)¥ | |
Logistic regression model adjusting for maternal age and race, study site, and gestational age (wks)
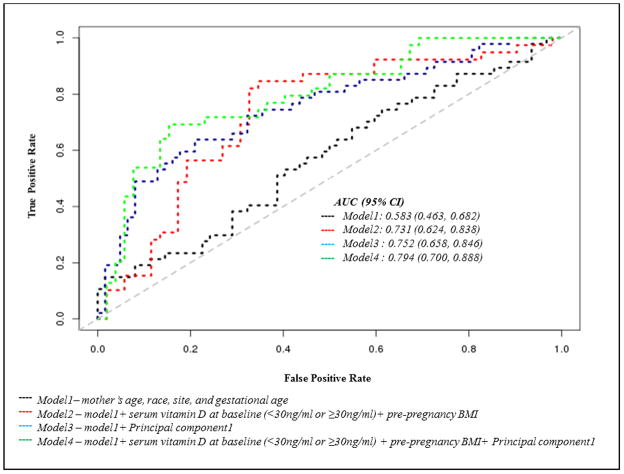
Model1– mother’s age, race, site, and gestational age
Model2 – model1+ serum vitamin D at baseline (<30ng/ml or ≥30ng/ml)+ pre-pregnancy BMI
Model3 – model1+ Principal component1
Model4 – model1+ serum vitamin D at baseline (<30ng/ml or ge;30ng/ml) + pre-pregnancy BMI+ Principal component1
Significantly (p<0.05) different to model 1
∞ Significantly (p<0.05) different to model 2
Ω Significantly (p<0.05) different to model 3
ROC; Receiver Operator Characteristic; AUC – Area Under the Curve; CI – Confidence Interval
The C8-positive platform (polar and non-polar lipids), included the greatest percentage of features (n=215 (7.6%)) significantly associated with preeclampsia risk (Online Resource2: Table S1). The m/z ratio and retention time of all 484 features are shown in Online Resource 3: Fig S1. Within the platforms, a number of features had almost identical m/z ratios and retention times, and it can be speculated that such features may in fact represent ions or adducts of the same metabolites. The top annotated features included Cohibin A (OR [p-value]: 4.52 [6.7×10−4]), 16-alpha-Hydroxypregnenolone (OR [p-value]: 11.89 [9.5×10−4]), and a number of phosphocholines. Seventy-two features (0.9%) (58 upregulated, 14 downregulated) retained significance at the more stringent threshold of p<0.01 (Online Resource2: Table S2) and were used to create a predictive model for preeclampsia, which was assessed using ROC curve analyses (Table 2). The first principal component of these 72 features explained 37% of the variance in the data. When this principal component was included as a model predictor, ROC analysis (model3 AUC: 0.752 (95%CI 0.658, 0.846)) showed that it significantly (p=0.006) outperformed a baseline model including mother’s age, race, site, and gestational age (model 1 AUC: 0.573 (95%CI 0.463, 0.682)) (Fig 2). Pre-pregnancy BMI, which was significantly associated with risk in this population, and serum vitamin D levels have both previously been shown to predict preeclampsia risk (Achkar et al., 2015, Hyppönen et al., 2013, Bodnar et al., 2014, O’Brien et al., 2003). Therefore models that additionally included pre-pregnancy BMI and baseline serum vitamin D (deficient at <30ng/ml versus sufficient at ≥30ng/ml) (model 2 AUC: 0.731 (95%CI 0.624, 0.838) were compared. Although the inclusion of these predictors increased the AUC relative to model1, it was to a lesser extent than the metabolite model3. The highest AUC for the prediction of preeclampsia was achieved when including pre-pregnancy BMI, baseline serum vitamin D levels and the metabolite principal component (model4): AUC: 0.794 (95% CI: 0.700, 0.888), and this model had a significantly higher AUC than both model1 (p difference=0.006) and model2 (p difference=0.048)..
Fig 2.
Receiver operating characteristic curves evaluating metabolomic profiling models ability to distinguish preeclampsia cases and controls
AUC – Area under the Curve
Pre-pregnancy BMI adjusted models
When pre-pregnancy BMI was additionally included in the logistic regression model a total of 265 features (3.3%) were significantly associated with the risk of preeclampsia at a nominal 95% confidence level; 179 were upregulated in preeclampsia cases and 86 were downregulated. The largest proportion of features (n=97 features) were from the C18-negative platform: free fatty acids, bile acids, and metabolites of intermediate polarity. In total 178 of the 484 features from the initial analyses were robust to additional adjustment for pre-pregnancy BMI. Those that did not retain significance included a large number of mono-, di- and tri- acylglycerols. Crucially, among the 72 most significant features from the initial model, 58 (81%) were robust to additional adjustment, including Cohibin A, 16-alpha-Hydroxypregnenolone, phosphocholines, tocopherols and amines (Online Resource2: Table S2). A predictive model based on these 58 metabolites displayed similar predictive power to the 72-metabolite model. It outperformed the baseline model and had a higher AUC than the model including pre-pregnancy BMI and serum vitamin D (Online Resource2: Table S3).
The results were also similar when baseline vitamin D status (<30ng/ml versus ≥30 ng/ml) and the study arm (intervention or placebo) were included as covariates in the logistic regression model (results not shown).
Preeclampsia Signatures in the Third Trimester Samples
Matched metabolite profiling of blood samples in the same women, taken during the third trimester of pregnancy (32–38 weeks gestation), identified 502 metabolite features with differential intensity levels by preeclampsia status (p<0.05); 85 of these retained significance at a p<0.01 threshold (Online Resource2: Table S4). At the time of third trimester sampling, one woman had been diagnosed with preeclampsia; the remaining cases were diagnosed subsequently and prior to delivery. Seventy-four (15%) of these 502 features were among those identified in the analysis of the first trimester samples, including 31 annotated metabolites. When a summary score was computed using the 72 metabolite features most strongly associated with subsequent preeclampsia (p<0.01) in the first trimester of pregnancy, there was a significant relationship between the metabolite principal component and preeclampsia status in the third trimester, after adjustment for maternal age, race, gestational age, site and pre-pregnancy BMI (OR: 0.89 95%CI 0.790.99 (p=0.033)). Furthermore, the same metabolites included in the model from the first time-point remained predictive at the later pregnancy time-point. When compared to a baseline model (maternal age, race, site and gestational age - model 1: AUC 0.578 [0.468–0.692]), the inclusion of current vitamin D status and pre-pregnancy BMI increased discriminatory ability (model 2: AUC 0.715 [0.605, 0.825]). But this increase only reached statistical significance when the first trimester metabolite signature was also included (model 4: AUC 0.762 [0.663, 0.860]), p=0.018. (Online Resource 3: Fig S2). These results indicated that a prediagnostic predictive preeclampsia metabolomic signature was present at 10–18 weeks gestation and was also able to distinguish preeclamptic from healthy pregnancies at 32–38 weeks gestation. This provides both some measure of replication as well as helping to elucidate a temporal pattern of the pathogenic changes associated with preeclampsia.
Pathway Analysis of Metabolite Profiles From First and Third Trimesters
To elucidate the biological context and significance of the metabolites, the pathways enriched at 10–18 weeks and those at 32–38 weeks were compared. Only 174 of the 484 significant (p<0.05) features from the first trimester and 53 of the 502 from the third could be annotated to known metabolites, and were available for metabolite set enrichment analysis. Enrichment was determined using Metaboanalyst defaults, all pathways with a nominal p-value<0.5 are reported (Table 3). Twelve pathways were common to metabolite profiles from both first and third trimesters including ‘glycerophospholipid metabolism’, ‘alanine, aspartate and glutamate metabolism’, ‘beta-alanine metabolism’, ‘butanoate metabolism’ and ‘arginine and proline metabolism’.
Table 3.
Metabolite set enrichment analysis of significant metabolite features identified in early and late gestation
| Biological pathway | 10–18 weeks gestation (p-value) | 32–38 weeks gestation (p-value) |
|---|---|---|
| Glycerophospholipid metabolism* | 0.002 | 0.024 |
| Sphingolipid metabolism* | 0.050 | 4.2×10−4 |
| beta-Alanine metabolism* | 0.061 | 0.012 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.156 | |
| Fatty acid biosynthesis | 0.158 | |
| Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | 0.186 | |
| Linoleic acid metabolism* | 0.198 | 0.090 |
| Arachidonic acid metabolism* | 0.227 | 0.325 |
| Taurine and hypotaurine metabolism | 0.255 | |
| Primary bile acid biosynthesis | 0.257 | |
| Alanine, aspartate and glutamate metabolism* | 0.298 | 0.009 |
| Pantothenate and CoA biosynthesis* | 0.328 | 0.156 |
| alpha-Linolenic acid metabolism* | 0.348 | 0.167 |
| Glycolysis or Gluconeogenesis | 0.367 | |
| Glycerolipid metabolism | 0.376 | |
| Pentose phosphate pathway | 0.376 | |
| Butanoate metabolism* | 0.446 | 0.025 |
| Galactose metabolism | 0.454 | |
| Phenylalanine metabolism* | 0.486 | 0.247 |
| Lysine degradation | 0.501 | |
| Glycine, serine and threonine metabolism* | 0.508 | 0.261 |
| Arginine and proline metabolism* | 0.682 | 0.081 |
| Aminoacyl-tRNA biosynthesis | 0.379 | |
| Citrate cycle (TCA cycle) | 0.118 | |
| Nicotinate and nicotinamide metabolism | 0.029 | |
| Nitrogen metabolism | 0.218 | |
| Tryptophan metabolism | 0.395 |
Includes all pathways with a nominal p-value <0.5
Enriched at both time-points
Integrated Omics
A dataset containing 484 significant metabolites and 1632 significant genes was created. WGCNA of this dataset resulted in five gene-metabolite modules; the seven genes and metabolite features that could not be assigned to any of these co-regulated modules were excluded (grey module). The five modules were summarized by their eigenvalues (i.e., the first principal component of variability among all module members) and four (black, brown, green, yellow) were significantly associated with preeclampsia after adjustment for maternal age and race, study site, and gestational age. (Online Resource 2: Table S5).
Co-regulated modules are often enriched for disease-relevant biological functions (Allen et al., 2012); therefore, within each module the genes whose expression levels were significantly correlated (p<0.05) with the intensity levels of the known metabolites were identified (Supplemental Table S6). The significant genes were submitted for GeneOntology (GO) analysis (http://geneontology.org/page/go-enrichment-analysis) to identify related biological processes (Table 4). These analyses implicated distinct biological processes within the modules all of which contribute to the pathophysiology of preeclampsia; including immune processes (brown and green), amino acid metabolism (black), and processes relating to the circulatory system and homeostasis (yellow). These results provided further biological context for the pathways identified in the metabolomics profiling analyses.
Table 4.
Preeclampsia associated WGCNA modules: For each module the annotated metabolites, the number of within module genes they are associated with and the biological processes these genes are enriched for are shown
| Module | Metabolites | Enriched Gene Ontology Processes |
|---|---|---|
| Black | (+/−)-trans- and cis-4,8-Dimethyl-3,7-nonadien-2-ol; (ent-2b,4S,9a)-2,4,9-Trihydroxy-10(14)-oplopen-3-one 2-(2-methylbutanoate) 9-(3-methyl-2E-pentenoate); 1-Methyl-2-pyrrolecarboxaldehyde; 13-Carboxy-gamma-tocopherol; 13-Hydroxy-gamma-tocopherol; 2-Deoxycastasterone; 2-Phenylethyl octanoate; 2,4-Dimethylpyridine; 5,8,11-Eicosatrienoate; 6-Deoxocastasterone; 6alpha-Hydroxycastasterone; 9-POHSA; multiple Sphingomyelins; C38:8 PC; Ganoderiol C; Momordol; Notoginsenoside R10; Pelletierine; Prostaglandin E2; tetradecyl sulfate; Tetrahydrocortisol; Trihydroxycoprostanoate; Tyramine; Uracil; xi-4,5-Dihydro-2,4(5)-dimethyl-1H-imidazole |
n=99 genes detection of chemical stimulus involved in sensory perception of taste negative regulation of viral genome replication benzene-containing compound metabolic process regulation of viral genome replication arginine catabolic process to proline arginine catabolic process to proline via ornithine extracellular amino acid transport extracellular transport TORC1 signaling mitochondrial mRNA 3′-end processing |
| Green | Bergaptol |
n=5 genes regulation of complement activation, alternative pathway negative regulation of complement activation, alternative pathway regulation of complement activation, classical pathway negative regulation of complement activation, classical pathway protein C-linked glycosylation via 2′-alpha-mannosyl-L-tryptophan protein C-linked glycosylation via tryptophan peptidyl-tryptophan modification protein C-linked glycosylation negative regulation of humoral immune response mediated by circulating immunoglobulin regulation of humoral immune response mediated by circulating immunoglobulin |
| Yellow | GABA |
n=43 genes platelet degranulation regulated exocytosis exocytosis secretion by cell regulation of leukocyte migration regulation of hemostasis regulation of blood coagulation regulation of coagulation regulation of cell motility secretion |
| Brown | (2E,6E)-2,6-Nonadienal; (Z)-5-[(5-Methyl-2-thienyl)methylene]-2(5H)-furanone; 1-(10-methylhexadecanyl)-2-(8-[3]-ladderane-octanyl)-sn-glycerophosphocholine; 1-Octene; 1-Stearoylglycerophosphoglycerol; 13-HOTE; 16-alpha-Hydroxypregnenolone; 2-Ethyl-4-methyloxazole; 2-Keto-6-aminocaproate; 2-Keto-6-aminocaproate; 3-Hydroxy-10-apo-b,y-carotenal; 35S-Methylokadaic acid 7-hexadecanoate; 4-oxo-Retinoic acid; 5-(10-Nonadecenyl)-1,3-benzenediol; 5-Hydroxyisourate; 7-Ethoxy-4-methyl-2H-1-benzopyran-2-one; 9-Carboxymethoxymethylguanine; Allitridin; Allodesmosine; Artemoin A; Biliverdin; multiple Lysophosphatidylcholines; multiple choline esters; multiple ceremides; multiple mono-, di- and tri-acylglycerols; multiple phosphatidylcholines; multiple phosphatidylinositol; multiple phosphatidylethanolamines; multiple phosphoserines; Caprate; Caprylic acid; Chloramphenicol; Chlormezanone; Cohibin A/C; Dimethamine; fructose/glucose/galactose; Furaneol; Isovalerylalanine; L-2-Amino-3-methylenehexanoic acid; Lutein; m-Chlorohippurate; Mabioside D; Montecristin; N-Nitroso-3-hydroxypyrrolidine; N-Phenylacetylphenylalanine; PGP(18:0/22:5(4Z,7Z,10Z,13Z,16Z)); Phenylethylamine; Quinoline; Saccharin; Sertaconazole; Sphingosine 1-phosphate (d16:1-P); Taurocyamine; Terpenyl isovalerate; Terpinolene oxide; Terpinyl isobutyrate; Triclosan |
n=792 genes defense response immune response inflammatory response innate immune response response to stress immune system process response to other organism response to external biotic stimulus response to biotic stimulus cellular response to other organism |
WGCNA – weighed gene co-expression correlation analysis
DISCUSSION
In this study a predictive metabolite signature of preeclampsia based on first trimester blood samples was generated. This signature had moderate-to-good discriminatory ability, and retained the ability to discriminate between preeclamptic and normal pregnancies in the third trimester. The integration with transcriptomic data provided a deeper understanding of the biology underlying this signature. To date, the pathophysiology of preeclampsia and its temporal pattern of manifestation in pregnancy are largely unknown, and there are currently no predictive biomarkers or screening tests. The findings from this study will help to address these challenges and to minimize the public health burden of this condition.
Metabolomic profiling is an underused resource in the study of preeclampsia. Nevertheless, there is great promise for metabolomics in preeclampsia investigations. Metabolites that differ between preeclamptic and healthy pregnancies and between late- and early-onset preeclampsia have been identified in blood and urine (Austdal et al., 2014, Bahado-Singh et al., 2013, Kenny et al., 2008, Koster et al., 2015, Kuc et al., 2014, Turner et al., 2008, Schott et al., 2012). Furthermore, predictive metabolite profiles of preeclampsia with high sensitivity, specificity (Kenny et al., 2005, Bahado-Singh et al., 2012) and predictive power (Koster et al., 2015, Odibo et al., 2011, Kenny et al., 2010), have been reported.
In this study, plasma levels of 72 measured metabolite features in first trimester blood samples were observed to differ significantly between women who remained healthy and those who went on to develop preeclampsia. Previously reported associations between preeclampsia with phosphatidylcholine (Schott et al., 2012), sphingosine-1-phosphate, glycerides (Kenny et al., 2010) and phenylalanine derivatives (Odibo et al., 2011) were validated in this study (with at least nominal significance). Other significant metabolite associations from the literature, including carnitines (Odibo et al., 2011, Koster et al., 2015), taurine (Kuc et al., 2014), arginine (Bahado-Singh et al., 2015) and histidine (Turner et al., 2008) were not replicated. Further previously reported metabolites such as xylitol and 2-hydroxy-3-methy-butanoic acid (Kenny et al., 2008) could not be neither confirmed nor refuted as it is unknown whether they are represented among the significant but unannotated features. The majority of significant features were polar and non-polar lipids (based on platform) however only 40% could be annotated to known metabolites. Pathway analysis was therefore limited but did identify a number of enriched metabolic pathways, which both strengthened the evidence for previously implicated pathways, such as sphingolipid (Melland-Smith et al., 2015) and linoleic acid (Robinson et al., 2009) metabolism, while suggesting novel pathways with biological plausibility.
Overall, the findings implicated altered lipid metabolism in the pathology of preeclampsia; glycerophospholipid, arachidonic acid and glycerolipid metabolism as well as fatty acid biosynthesis were among the top enriched pathways. Abnormalities of lipid metabolism are a recognized feature of preeclampsia and are thought to contribute through endothelial dysfunction leading to vascular remodeling and atherosclerosis (Demir et al., 2011, Robinson et al., 2009). Sphingolipid metabolism may play a particularly important role throughout preeclampsia development, as sphingolipids are the primary components of cell membranes and act as signaling molecules that regulate a diverse range of cellular processes central to immunity and inflammation. They are major components of lipoproteins, which when disrupted have been shown to cause the endothelial dysfunction that can result in hypertension and proteinuria – two of the hallmarks of preeclampsia (Winkler et al., 2003). Lipoproteins were not measured in this study – but these findings suggesting more targeted analysis of the metabolites involved in lipoprotein and specifically sphingolipid metabolism may provide a deeper understanding of preeclampsia pathogenesis. Pathways of carbohydrate metabolism including Glycolysis/Gluconeogenesis, pentose phosphate and butanoate metabolism were also enriched in the first trimester blood samples, suggesting a potential role for these processes in early preeclampsia development. Furthermore, pathways of alanine, aspartate and glutamate metabolism were enriched, which is concordant with previous studies reporting an association of glutamate and alanine (Kenny et al., 2008, Odibo et al., 2011) with preeclampsia. Glutamate and alanine may be released by immune cells, while ischemia (a feature of preeclampsia) results in an increase in glutamate uptake and alanine release. The enrichment of galactose metabolism in these results may relate to the altered metabolic and nutritional status which occurs during pregnancy. Additional studies are needed to further elucidate the roles of all the specific metabolites and pathways identified in this study in the pathogenesis of preeclampsia.
The identified features were found to have the ability to distinguish between women who develop preeclampsia and those who remained healthy, by developing a metabolomic profile based on a summary score. Although an independent replication population was not available to test the validity of this profile, it did retain predictive power to distinguish preeclampsia cases in their third trimester of pregnancy. This could be interpreted to demonstrate that during the multistage evolution of preeclampsia, a number of key metabolites retain their importance. Further, the pathway level replication between the two time-points may provide evidence that, although the exact metabolites differ, key pathways are dysregulated and their downstream products may be identified in the later stages of preeclamptic pregnancies. A number of the metabolites and pathways identified at this later time-point are also among those currently reported in the literature, which likely reflects the fact that many of the existing studies are case-control in design and therefore may be capturing metabolites downstream of the predictive signature identified in this study. However, it must be noted that the two set of blood samples were extracted from the same individuals, therefore similarities in their profiles are to be expected and these results must be interpreted with caution.
The summary score based on these features outperformed serum vitamin D. Vitamin D deficiency is thought to play an important role in the pathophysiology of preeclampsia due to its involvement in immunomodulation and placental development (Novakovic et al., 2009), and has previously been shown to be a predictor of risk (Bodnar et al., 2014, Hyppönen et al., 2013, Achkar et al., 2015). The metabolite score also outperformed pre-pregnancy BMI. In this population a higher BMI prior to pregnancy was associated with an increased risk of preeclampsia, in agreement with the current literature (Demir et al., 2011). However It is of interest that a number of the preeclampsia-metabolite associations observed in this study appeared to be driven by BMI; in particular, the associations with multiple forms of glycerols – the production of which have been shown to increase in obese individuals (Stunff and Bougnères, 1992).
WGCNA was performed to identify modules of genes and metabolites with highly correlated expression and concentration levels suggesting common regulation and biologically relevant associations. Only one of the preeclampsia WCNA modules; the black module, was associated with pre-pregnancy BMI, suggesting that the genes and metabolites of this module may be specifically involved in BMI mediated preeclampsia. mTORC1 signaling, which regulates cellular responses and is implicated in obesity-related conditions, was enriched in this module. This module also contained multiple sphingomyelins, which are involved in both intra- and extracellular transport, one of the other processes enriched among the genes of this module. Further this module contained a number of amino acids, and genes enriched for amino acid transport and metabolism. This points to an additional potential utility of this network based analysis; identifying etiologically relevant associations with suspected preeclampsia risk factors, while disentangling the various mechanism through which preeclampsia may develop.
The brown module identified co-regulation between genes involved in the immune response with triglycerides, diglycerides, phosphotidylcholines and myoinsitol. This is supported by a known link between glyceride levels and immune function (Barcia and Harris, 2005). In particular there is evidence for biologically important relationships between inositol, diacylglycerols and triacylglycerols with cytokines (Tsuchiya et al., 2015, Kiely et al., 2007, Smith et al., 2007) that may explain their role in preeclampsia. While phosphatidylcholine has been found to have anti-inflammatory effects in other disease systems (Treede et al., 2007). Further support for a meaningful interaction between the genes and metabolites of this module comes from the fact that several of the identified genes, such as AOC3 and CDS2, have been shown to directly regulate triglyceride and phosphotidyl levels.
The yellow module included a single annotated metabolite; gamma-Aminobutyric acid (GABA). This neurotransmitter has been linked to a number of the key pathogenic processes in preeclampsia (Konijnenberg et al., 1997, Brown, 1995) including hemostasis, coagulation and platelet aggregation (Tyurenkov et al., 2014, Kaneez and Saeed, 2009), and homeostasis (Fregoneze, 2014). These processes were enriched among the identified genes. These findings again point to a mechanistically meaningful module with a biologically plausible link to preeclampsia risk.
Overall these findings suggest the importance of immune function, amino acid metabolism and the regulation and control of cardiovascular system in the pathology of preeclampsia. Crucially, the integration with transcriptomic data allowed the refinement of the set of significant metabolites, differentiating the mechanisms of preeclampsia pathogenesis into distinct modules. This provided a deeper understanding of the biology underlying the identified predictive signature than could be achieved with the metabolome alone. On a broader level, the identification of biologically meaningful and functionally relevant modules containing both genes and metabolites provides definitive evidence of a relationship between the metabolome and transcriptome that can be captured using systems biology approaches, and supports the utility of integrative omics in the study of complex disorders.
There were a number of limitations to these analyses. Due to sample size, findings were not stratified by parity or preeclampsia onset. As the ultimate aim of such analyses is to develop predictive clinically-translatable and minimally invasive biomarkers of preeclampsia; blood was chosen as the biological medium rather than placental tissue. The use of non-fasting plasma samples may have affected the measured metabolite features, and only a small proportion of these features could be annotated. However the ROC analyses including only the known metabolites demonstrated comparable discriminatory ability, and all pathway analyses were performed based on the known metabolites alone. Finally, a number of the reported results would not be robust to correction for multiple testing, rendering false positive findings a possibility. However, many FDR methods are considered too stringent for metabolomics analysis, due to the high correlation and redundancy between metabolite features, and there is a lack of agreed-upon standards in the field (Chadeau-Hyam et al., 2010). In order to minimize the impact of false positive findings on the conclusions, differing nominal p-values and potential confounders were explored, and replication of the findings at two different pregnancy time-points was attempted. It should be noted that a large number of the significant metabolites are likely to be related based on their mass-to-charge ratio and retention times, and given the biological relevance of the findings confidence can be placed in the results.
The generalizability of this population must also be taken into account. Subjects were recruited into VDAART on the basis of a maternal or paternal history of asthma or allergic rhinitis, and almost 60% of participants in this study reported a history of asthma. Maternal asthma has been reported to increase the risk of preeclampsia by more than 50% (Murphy et al., 2011). However there was no significant difference in the proportion of asthmatics between cases and controls, therefore this is unlikely to have induced spurious findings. Nevertheless a number of the metabolites and pathways identified in this study, and the dysregulation in amino acid metabolism, lipid metabolism and in immune processes implicated by these findings have also been observed in metabolomics studies of asthma (Kelly et al., 2016). Interrogation of metabolomics findings offers the potential to gain a better understanding of the common pathogenesis of these disorders. Nevertheless in such high dimensionality the possibility of false potivies and the generalizability of the findings requires further exploration. Ultimately, validation of the reported findings will only be possible through the use of a larger independent cohort.
CONCLUSIONS
The findings from this unique prospective cohort support the role of metabolomics and integrated omics in the study of preeclampsia. A metabolomic profile based on blood samples extracted in early pregnancy was able to predict the risk of subsequent preeclampsia. The interrogation of this signature identified biologically relevant pathways, particularly those involved in lipid imbalance. Through the integration of transcriptomic data, these results were further refined to suggest that the roles of lipids in immune function were among the biggest contributors to the pathogenesis of preeclampsia. The findings require independent replication to eliminate the impact of false positive findings, but those that can be validated have the potential to support clinical translation both through candidate biomarkers and candidate therapeutic targets. Results from transcriptomics both confirm and expand upon the results of the metabolomic analyses; fulfilling the fundamentally important knowledge gap between genetics, genomics and disease, and demonstrate that metabolomic and transcriptomic profiling can be integrated in a biologically meaningful way.
Supplementary Material
Acknowledgments
Funding: This manuscript is supported by an R01 grant from the National Heart, Lung, and Blood Institute of the National Institutes of Health, entitled Integrative metabolomics of asthma severity NHLBI 1R01HL123915-01 (JALS).
The generation of the transcriptomic dataset was funded by the ongoing VDAART grant and the data is currently being prepared for submission to the Gene Expression Omnibus (GEO) database, pending acceptance of the primary gene expression manuscript.
Abbreviations
- AUC
Area Under the Curve
- FDR
False Discovery Rate
- HILIC
Hydrophilic Interaction Liquid Chromatography
- LCMS
Liquid Chromatography Mass Spectroscopy
- UHPLC
Ultra High Performance Liquid Chromatography
- m/z
mass-to-charge ratio
- RT
retention time
- OR
odds ratio
- CI
confidence interval
Footnotes
Conflict of Interest: All authors declare that they have no conflicts of interest.
Ethical approval: The VDAART study was approved by the IRBs of the participating institutions (Washington University in St. Louis, Boston Medical Center, Kaiser Health Care San Diego) and Brigham and Women’s Hospital. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent: Written informed consent was obtained from all individual participants included in the study.
Availability of Data: The metabolomics dataset supporting the conclusions of this article will be made publically available at such a time as a suitable online repository that is approved by the metabolomics society becomes available.
References
- ACHKAR M, DODDS L, GIGUÈRE Y, FOREST JC, ARMSON BA, WOOLCOTT C, AGELLON S, SPENCER A, WEILER HA. Vitamin D status in early pregnancy and risk of preeclampsia. American Journal of Obstetrics and Gynecology. 2015;212:511.e1–511.e7. doi: 10.1016/j.ajog.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACOG. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet Gynecol. 2013;122:1122–31. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- AHN H, PARK J, GILMAN-SACHS A, KWAK-KIM J. Immunologic characteristics of preeclampsia, a comprehensive review. American journal of reproductive immunology. 2011;65:377–94. doi: 10.1111/j.1600-0897.2010.00913.x. [DOI] [PubMed] [Google Scholar]
- ALLEN JD, XIE Y, CHEN M, GIRARD L, XIAO G. Comparing Statistical Methods for Constructing Large Scale Gene Networks. PLoS ONE. 2012;7:e29348. doi: 10.1371/journal.pone.0029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALLSWORTH J, THEILEN L, SPAIN J. Discussion: ‘Metabolomic prediction of late-onset preeclampsia’, by Bahado-Singh et al. American journal of obstetrics and gynecology. 2013;208:e10–1. doi: 10.1016/j.ajog.2012.11.021. [DOI] [PubMed] [Google Scholar]
- AUSTDAL M, SKRASTAD RB, GUNDERSEN AS, AUSTGULEN R, IV, ERSEN AC, BATHEN TF. Metabolomic biomarkers in serum and urine in women with preeclampsia. PloS one. 2014;9:e91923. doi: 10.1371/journal.pone.0091923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AUSTDAL M, TANGERÅS LH, SKRÅSTAD RB, SALVESEN KÅ, AUSTGULEN R, IV, ERSEN AC, BATHEN TF. First Trimester Urine and Serum Metabolomics for Prediction of Preeclampsia and Gestational Hypertension: A Prospective Screening Study. International Journal of Molecular Sciences. 2015;16:21520–21538. doi: 10.3390/ijms160921520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAHADO-SINGH RO, AKOLEKAR R, MANDAL R, DONG E, XIA J, KRUGER M, WISHART DS, NICOLAIDES K. Metabolomics and first-trimester prediction of early-onset preeclampsia. The Journal of Maternal-Fetal & Neonatal Medicine. 2012;25:1840–1847. doi: 10.3109/14767058.2012.680254. [DOI] [PubMed] [Google Scholar]
- BAHADO-SINGH RO, AKOLEKAR R, MANDAL R, DONG E, XIA J, KRUGER M, WISHART DS, NICOLAIDES K. First-trimester metabolomic detection of late-onset preeclampsia. American journal of obstetrics and gynecology. 2013;208:58.e1–7. doi: 10.1016/j.ajog.2012.11.003. [DOI] [PubMed] [Google Scholar]
- BAHADO-SINGH RO, SYNGELAKI A, AKOLEKAR R, MANDAL R, BJONDAHL TC, HAN B, DONG E, BAUER S, ALPAY-SAVASAN Z, GRAHAM S, TURKOGLU O, WISHART DS, NICOLAIDES KH. Validation of metabolomic models for prediction of early-onset preeclampsia. Am J Obstet Gynecol. 2015;213:530.e1–530 e10. doi: 10.1016/j.ajog.2015.06.044. [DOI] [PubMed] [Google Scholar]
- BARCIA AM, HARRIS HW. Triglyceride-Rich Lipoproteins as Agents of Innate Immunity. Clinical Infectious Diseases. 2005;41:S498–S503. doi: 10.1086/432005. [DOI] [PubMed] [Google Scholar]
- BERG CJ, MACKAY AP, QIN C, CALLAGHAN WM. Overview of maternal morbidity during hospitalization for labor and delivery in the United States: 1993–1997 and 2001–2005. Obstetrics and gynecology. 2009;113:1075–81. doi: 10.1097/AOG.0b013e3181a09fc0. [DOI] [PubMed] [Google Scholar]
- BICTASH M, EBBELS T, CHAN Q, LOO RL, YAP I, BROWN IJ, DE IORIO M, DAVIGLUS M, HOLMES E, STAMLER J, NICHOLSON JK, ELLIOTT P. Metabolic phenotyping in epidemiology and metabolome-wide association studies. Journal of clinical epidemiology. 2010;63:970–979. doi: 10.1016/j.jclinepi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BODNAR LM, SIMHAN HN, CATOV JM, ROBERTS JM, PLATT RW, DIESEL JC, KLEBANOFF MA. Maternal vitamin D status and the risk of mild and severe preeclampsia. Epidemiology (Cambridge, Mass) 2014;25:207–214. doi: 10.1097/EDE.0000000000000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN MA. THE PHYSIOLOGY OF PRE-ECLAMPSIA. Clinical and Experimental Pharmacology and Physiology. 1995;22:781–791. doi: 10.1111/j.1440-1681.1995.tb01937.x. [DOI] [PubMed] [Google Scholar]
- CHADEAU-HYAM M, EBBELS TMD, BROWN IJ, CHAN Q, STAMLER J, HUANG CC, DAVIGLUS ML, UESHIMA H, ZHAO L, HOLMES E, NICHOLSON JK, ELLIOTT P, DE IORIO M. Metabolic Profiling And The Metabolome-Wide Association Study: Significance Level For Biomarker Identification. Journal of proteome research. 2010;9:4620–4627. doi: 10.1021/pr1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEMIR B, DEMIR S, ATAMER Y, GUVEN S, ATAMER A, KOCYIGIT Y, HEKIMOGLU A, TOPRAK G. Serum Levels of Lipids, Lipoproteins and Paraoxonase Activity in Pre-Eclampsia. Journal of International Medical Research. 2011;39:1427–1431. doi: 10.1177/147323001103900430. [DOI] [PubMed] [Google Scholar]
- DULEY L. The Global Impact of Pre-eclampsia and Eclampsia. Seminars in Perinatology. 2009;33:130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- FREGONEZE JF, HS, LUZ CPN. Serotonergic Receptors and control of FluiIntake and Cardiovascular Function in Rats. In: AKDLLMJ, editor. Neurobiology of Body Fluid Homeostasis: Transduction and Integration. Boca Raton (FL): CRC Press/Taylor and Francis; 2014. [Google Scholar]
- HYPPÖNEN E, CAVADINO A, WILLIAMS D, FRASER A, VERECZKEY A, FRASER WD, BÁNHIDY F, LAWLOR D, CZEIZEL AE. Vitamin D and Pre-Eclampsia: Original Data, Systematic Review and Meta-Analysis. Annals of Nutrition and Metabolism. 2013;63:331–340. doi: 10.1159/000358338. [DOI] [PubMed] [Google Scholar]
- KANEEZ FS, SAEED SA. Investigating GABA and its function in platelets as compared to neurons. Platelets. 2009;20:328–333. doi: 10.1080/09537100903047752. [DOI] [PubMed] [Google Scholar]
- KELLY RS, DAHLIN A, MCGEACHIE MJ, QIU W, SORDILLO J, WAN ES, WU AC, LASKY-SU J. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest. 2016 doi: 10.1016/j.chest.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENNY LC, BROADHURST D, BROWN M, DUNN WB, REDMAN CWG, KELL DB, BAKER PN. Detection and Identification of Novel Metabolomic Biomarkers in Preeclampsia. Reproductive Sciences. 2008;15:591–597. doi: 10.1177/1933719108316908. [DOI] [PubMed] [Google Scholar]
- KENNY LC, BROADHURST DI, DUNN W, BROWN M, NORTH RA, MCCOWAN L, ROBERTS C, COOPER GJ, KELL DB, BAKER PN. Robust early pregnancy prediction of later preeclampsia using metabolomic biomarkers. Hypertension. 2010;56:741–9. doi: 10.1161/HYPERTENSIONAHA.110.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENNY LC, DUNN WB, ELLIS DI, MYERS J, BAKER PN, KELL DB. Novel biomarkers for pre-eclampsia detected using metabolomics and machine learning. Metabolomics. 2005;1:227–234. [Google Scholar]
- KIELY A, MCCLENAGHAN NH, FLATT PR, NEWSHOLME P. Pro-inflammatory cytokines increase glucose, alanine and triacylglycerol utilization but inhibit insulin secretion in a clonal pancreatic β-cell line. Journal of Endocrinology. 2007;195:113–123. doi: 10.1677/JOE-07-0306. [DOI] [PubMed] [Google Scholar]
- KONIJNENBERG A, STOKKERS EW, VAN DER POST JAM, SCHAAP MCL, BOER K, BLEKER OP, STURK A. Extensive platelet activation in preeclampsia compared with normal pregnancy: Enhanced expression of cell adhesion molecules. American Journal of Obstetrics and Gynecology. 1997;176:461–469. doi: 10.1016/s0002-9378(97)70516-7. [DOI] [PubMed] [Google Scholar]
- KOSTER MPH, VREEKEN RJ, HARMS AC, DANE AD, KUC S, SCHIELEN PCJI, HANKEMEIER T, BERGER R, VISSER GHA, PENNINGS JLA. First-Trimester Serum Acylcarnitine Levels to Predict Preeclampsia: A Metabolomics Approach. Disease Markers. 2015;2015:8. doi: 10.1155/2015/857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUC S, KOSTER MP, PENNINGS JL, HANKEMEIER T, BERGER R, HARMS AC, DANE AD, SCHIELEN PC, VISSER GH, VREEKEN RJ. Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PloS one. 2014;9:e98540. doi: 10.1371/journal.pone.0098540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUKLINA EV, AYALA C, CALLAGHAN WM. Hypertensive disorders and severe obstetric morbidity in the United States. Obstetrics and gynecology. 2009a;113:1299–306. doi: 10.1097/AOG.0b013e3181a45b25. [DOI] [PubMed] [Google Scholar]
- KUKLINA EV, AYALA C, CALLAGHAN WM. Hypertensive disorders and severe obstetric morbidity in the United States. Obstet Gynecol. 2009b;113:1299–306. doi: 10.1097/AOG.0b013e3181a45b25. [DOI] [PubMed] [Google Scholar]
- LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559–559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LITONJUA AA, CAREY VJ, LARANJO N, HARSHFIELD BJ, MCELRATH TF, O’CONNOR GT, SANDEL M, IV, ERSON RE, JR, LEE-PARITZ A, STRUNK RC, BACHARIER LB, MACONES GA, ZEIGER RS, SCHATZ M, HOLLIS BW, HORNSBY E, HAWRYLOWICZ C, WU AC, WEISS ST. Effect of Prenatal Supplementation With Vitamin D on Asthma or Recurrent Wheezing in Offspring by Age 3 Years: The VDAART Randomized Clinical Trial. JAMA. 2016;315:362–70. doi: 10.1001/jama.2015.18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MELLAND-SMITH M, ERMINI L, CHAUVIN S, CRAIG-BARNES H, TAGLIAFERRO A, TODROS T, POST M, CANIGGIA I. Disruption of sphingolipid metabolism augments ceramide-induced autophagy in preeclampsia. Autophagy. 2015;11:653–669. doi: 10.1080/15548627.2015.1034414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIRZAKHANI H, LITONJUA AA, MCELRATH TF, O’CONNOR G, LEE-PARRITZ A, IVERSON R, MACONES G, STRUNK RC, BACHARIER LB, ZEIGER R, HOLLIS BW, HANDY DE, SHARMA A, LARANJO N, CAREY V, QIU W, SANTOLINI M, LIU S, CHHABRA D, ENQUOBAHRIE DA, WILLIAMS MA, LOSCALZO J, WEISS ST. Early pregnancy vitamin D status and risk of preeclampsia. The Journal of Clinical Investigation. 2016:126. doi: 10.1172/JCI89031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURPHY VE, NAMAZY JA, POWELL H, SCHATZ M, CHAMBERS C, ATTIA J, GIBSON PG. A meta-analysis of adverse perinatal outcomes in women with asthma. BJOG: An International Journal of Obstetrics & Gynaecology. 2011;118:1314–1323. doi: 10.1111/j.1471-0528.2011.03055.x. [DOI] [PubMed] [Google Scholar]
- NOVAKOVIC B, SIBSON M, NG HK, MANUELPILLAI U, RAKYAN V, DOWN T, BECK S, FOURNIER T, EVAIN-BRION D, DIMITRIADIS E, CRAIG JM, MORLEY R, SAFFERY R. Placenta-specific Methylation of the Vitamin D 24-Hydroxylase Gene: IMPLICATIONS FOR FEEDBACK AUTOREGULATION OF ACTIVE VITAMIN D LEVELS AT THE FETOMATERNAL INTERFACE. Journal of Biological Chemistry. 2009;284:14838–14848. doi: 10.1074/jbc.M809542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’BRIEN TE, RAY JG, CHAN WS. Maternal body mass index and the risk of preeclampsia: a systematic overview. Epidemiology. 2003;14:368–74. doi: 10.1097/00001648-200305000-00020. [DOI] [PubMed] [Google Scholar]
- ODIBO AO, GOETZINGER KR, ODIBO L, CAHILL AG, MACONES GA, NELSON DM, DIETZEN DJ. First-trimester prediction of preeclampsia using metabolomic biomarkers: a discovery phase study. Prenatal diagnosis. 2011;31:990–4. doi: 10.1002/pd.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RHEE EP, GERSZTEN RE. Metabolomics and cardiovascular biomarker discovery. Clin Chem. 2012;58:139–47. doi: 10.1373/clinchem.2011.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RHEE EP, THADHANI R. New insights into uremia-induced alterations in metabolic pathways. Curr Opin Nephrol Hypertens. 2011;20:593–8. doi: 10.1097/MNH.0b013e32834b8a1d. [DOI] [PubMed] [Google Scholar]
- ROBINSON NJ, MINCHELL LJ, MYERS JE, HUBEL CA, CROCKER IP. A potential role for free fatty acids in the pathogenesis of preeclampsia. J Hypertens. 2009;27:1293–302. doi: 10.1097/hjh.0b013e328329fbfe. [DOI] [PubMed] [Google Scholar]
- SCHOTT S, HAHN J, KURBACHER C, MOKA D. (31)P and (1)h nuclear magnetic resonance spectroscopy of blood plasma in female patients with preeclampsia. International journal of biomedical science : IJBS. 2012;8:258–63. [PMC free article] [PubMed] [Google Scholar]
- SMITH N, BROWNING CA, DUROUDIER N, STEWART C, PEEL S, SWAN C, HALL IP, SAYERS I. Salmeterol and cytokines modulate inositol-phosphate signalling in Human airway smooth muscle cells via regulation at the receptor locus. Respiratory Research. 2007;8:68–68. doi: 10.1186/1465-9921-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUNFF CL, BOUGNÈRES PF. Glycerol Production and Utilization During the Early Phase of Human Obesity. Diabetes. 1992;41:444–450. doi: 10.2337/diab.41.4.444. [DOI] [PubMed] [Google Scholar]
- TREEDE I, BRAUN A, SPARLA R, KÜHNEL M, GIESE T, TURNER JR, ANES E, KULAKSIZ H, FÜLLEKRUG J, STREMMEL W, GRIFFITHS G, EHEHALT R. Anti-inflammatory Effects of Phosphatidylcholine. Journal of Biological Chemistry. 2007;282:27155–27164. doi: 10.1074/jbc.M704408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSUCHIYA R, TANAKA T, HOZUMI Y, NAKANO T, OKADA M, TOPHAM MK, II, NO M, GOTO K. Downregulation of diacylglycerol kinase ζ enhances activation of cytokine-induced NF-κB signaling pathway. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2015;1853:361–369. doi: 10.1016/j.bbamcr.2014.11.011. [DOI] [PubMed] [Google Scholar]
- TURNER E, BREWSTER JA, SIMPSON NA, WALKER JJ, FISHER J. Aromatic amino acid biomarkers of preeclampsia--a nuclear magnetic resonance investigation. Hypertension in pregnancy. 2008;27:225–35. doi: 10.1080/10641950801955725. [DOI] [PubMed] [Google Scholar]
- TYURENKOV IN, PERFILOVA VN, KARAMYSHEVA VI, REZNIKOVA LB, MOKROUSOV IS, MIKHAILOVA LI, BERESTOVITSKAYA VM, VASIL’EVA OS. Effect of GABA derivatives on the rate of thrombus formation, platelet aggregation, and plasma coagulation capacity in rats with experimental gestosis. Bull Exp Biol Med. 2014;158:219–21. doi: 10.1007/s10517-014-2726-3. [DOI] [PubMed] [Google Scholar]
- WANG TJ, LARSON MG, VASAN RS, CHENG S, RHEE EP, MCCABE E, LEWIS GD, FOX CS, JACQUES PF, FERNANDEZ C, O’DONNELL CJ, CARR SA, MOOTHA VK, FLOREZ JC, SOUZA A, MELANDER O, CLISH CB, GERSZTEN RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINKLER K, WETZKA B, HOFFMANN MM, FRIEDRICH I, KINNER M, BAUMSTARK MW, ZAHRADNIK HP, WIELAND H, MÄRZ W. Triglyceride-Rich Lipoproteins Are Associated with Hypertension in Preeclampsia. The Journal of Clinical Endocrinology & Metabolism. 2003;88:1162–1166. doi: 10.1210/jc.2002-021160. [DOI] [PubMed] [Google Scholar]
- XIA J, SINELNIKOV IV, HAN B, WISHART DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.