Abstract
Background and Aims Morphological traits in combination with metabolite fingerprinting were used to investigate inter- and intraspecies diversity within the model annual grasses Brachypodium distachyon, Brachypodium stacei and Brachypodium hybridum.
Methods Phenotypic variation of 15 morphological characters and 2219 nominal mass (m/z) signals generated using flow infusion electrospray ionization–mass spectrometry (FIE–MS) were evaluated in individuals from a total of 174 wild populations and six inbred lines, and 12 lines, of the three species, respectively. Basic statistics and multivariate principal component analysis and discriminant analysis were used to differentiate inter- and intraspecific variability of the two types of variable, and their association was assayed with the rcorr function.
Key Results Basic statistics and analysis of variance detected eight phenotypic characters [(stomata) leaf guard cell length, pollen grain length, (plant) height, second leaf width, inflorescence length, number of spikelets per inflorescence, lemma length, awn length] and 434 tentatively annotated metabolite signals that significantly discriminated the three species. Three phenotypic traits (pollen grain length, spikelet length, number of flowers per inflorescence) might be genetically fixed. The three species showed different metabolomic profiles. Discriminant analysis significantly discriminated the three taxa with both morphometric and metabolome traits and the intraspecific phenotypic diversity within B. distachyon and B. stacei. The populations of B. hybridum were considerably less differentiated.
Conclusions Highly explanatory metabolite signals together with morphological characters revealed concordant patterns of differentiation of the three taxa. Intraspecific phenotypic diversity was observed between northern and southern Iberian populations of B. distachyon and between eastern Mediterranean/south-western Asian and western Mediterranean populations of B. stacei. Significant association was found for pollen grain length and lemma length and ten and six metabolomic signals, respectively. These results would guide the selection of new germplasm lines of the three model grasses in ongoing genome-wide association studies.
Keywords: Association studies, Brachypodium distachyon, Brachypodium stacei, Brachypodium hybridum, metabolite fingerprinting, phenotypic traits, statistical analyses
INTRODUCTION
Wild plants and, to a lesser extent, cultivated crop species exhibit large phenotypic and metabolomic diversity as a consequence of their individual responses to developmental growth conditions and adaptation to environmental factors (Fiehn, 2002). Characterization of phenotypic and metabolomic traits, and their biological dynamics, has become a major line of research in the new era of ‘omics’ (Fiehn, 2001, 2002; Hall et al., 2002), due to their importance for genome-wide association studies (GWAS; Matsuda et al., 2015). Thus, phenomic and high-resolution metabolomic approaches have been used in conjunction with genomic, transcriptomic and proteomic data to detect and map genes, regulatory sequences and epigenomic regulators in genomes, and to unravel the plastic and biochemical variation of individuals under different intrinsic and extrinsic scenarios. Phenotypic and metabolomic responses of plants to different development stages and biotic–abiotic conditions have been investigated in crop species such as cereals (wheat, barley, rice), tomato and potato, and in some model plants, such as the dicot Arabidopsis thaliana (Allwood et al., 2006). However, these studies are less developed in Brachypodium distachyon, the annual temperate grass selected as a model plant for the monocots (Draper et al., 2001; International Brachypodium Initiative, 2010).
Over the past decade, B. distachyon has emerged as one of the preeminent model plant species, for which there are tremendous genetic, molecular and genomic resources (International Brachypodium Initiative, 2010; Mur et al., 2011; Catalán et al., 2014; Gordon et al., 2014). Its flagship plant genome now serves as an anchor for genomic studies across the temperate Pooideae grasses and monocots (Lyons and Scholthof, 2015). Its small genome size, compact genome (e.g. low levels of repetitive DNA), diverse ecological tolerances, ready propagation under controlled growth conditions, and considerable existing molecular and genomic resources make this plant an excellent candidate for addressing fundamental questions in comparative genomics and ecological studies and for its transfer to cereal and biofuel crops (Catalán et al., 2014). Analysis of intraspecific diversity in B. distachyon is under way through the nuclear and organellar resequencing of 54 diverse natural accessions (Gordon et al., 2014; and S. P. Gordon, DOE Joint Genome Institute(USA) et al., unpubl. res.). Phenotypic and metabolomics studies conducted to date in B. distachyon have mostly focused on relevant agricultural traits, such as plant height, biomass, flowering time, seed size, seed production and vernalization requirements (Draper et al., 2001; Opanowicz et al., 2008; Filiz et al., 2009; Vogel et al., 2009), and metabolites related to stress tolerance (Allwood et al., 2006; Parker et al., 2008, 2009; Pasquet et al., 2014; Onda et al., 2015; Shi et al., 2015).
Recently, the demonstration that the model plant is not one but three species (Catalán et al., 2012) has opened the way to a thorough comparative genomic study of this diploid–polyploid complex, which includes the model grass plant B. distachyon and its close allies Brachypodium stacei and Brachypodium hybridum, which show, respectively, 2n = 10, 20 and 30 chromosomes (Catalán et al., 2012, 2014). These three cytotypes were previously attributed to different ploidy levels of the same taxon, B. distachyon s.l. (Robertson, 1981); however, phylogenetic, cytogenetic and phenotypic analyses demonstrated that they should be treated as different species. They consist of two diploids, each with a different chromosome base number [B. distachyon (x = 5, 2n = 10); B. stacei (x = 10, 2n = 20)], and their derived allotetraploid, B. hybridum (x = 5 + 10, 2n = 30). Phylogenetic analyses indicated that the more basally diverging B. stacei and the more recently evolved B. distachyon emerged from two independent lineages, confirming their contribution as genome donors for B. hybridum (Catalán et al., 2012, 2014).
Regarding their broad phenotypic and ecological features, individuals of B. distachyon have overall small stature, require vernalization and are distributed at higher elevations, those of B. stacei are morphologically tall, do not require vernalization and grow mostly in coastlands or at lower altitudes, and those of B. hybridum tend to be physically large, generally lack vernalization requirements and grow in places of intermediate altitude (Opanowicz et al., 2008; Catalán et al., 2012; López-Alvarez et al., 2015). Statistical analysis of morphometric traits showed that five characters (stomata leaf guard cell length, pollen grain length, upper glume length, lemma length and awn length) significantly discriminated between the three species when they were grown under controlled greenhouse conditions (12 inbred lines) and showed stability in 24 additional wild populations (Catalán et al., 2012). However, representation of the three circum-Mediterranean species was biased towards the more exhaustively sampled populations of the western Mediterranean region. This was particularly critical for B. stacei, a species that was only known from its type locality at the time of its description (Catalán et al., 2012), and for which no other statistically analysed phenotypic data have been provided to date despite its known distribution in other native circum-Mediterranean localities (López-Alvarez et al., 2012, 2015).
Recently, López-Alvarez et al. (2012) provided an alternative and reliable genetic method to differentiate the individuals of the three species with a DNA barcoding system using three loci, the plastid trnLF region, the nuclear multicopy ribosomal internal transcribed spacer (ITS) and the low-copy Gigantea (GI) gene, which successfully discriminated between the three species. Interestingly, this study also demonstrated the existence of different bidirectional crosses that likely gave rise to the allotetraploid B. hybridum. This was confirmed through the analysis of the maternally inherited plastid haplotypes in this species; the majority of the surveyed B. hybridum individuals had inherited a maternal B. stacei-like plastome but some of the accessions showed a maternal B. distachyon-like plastome (López-Alvarez et al., 2012). The recurrent, polyphyletic and polytopic origin of this allotetraploid species was supported by ITS and GI evolutionary analyses, which revealed distinct relationships of the B. hybridum sequences to different parental geographical haplotypic groups, though all the studied hybrids correspond to what is considered to be the same allopolyploid species (López-Alvarez et al., 2012; Catalán et al., 2016b). Nonetheless, the potential phenotypic differences between these two types of reciprocal hybrids have not been investigated to date.
Metabolomics is considered to represent the ultimate level of ‘omic’ analysis, facilitating the testing of many biological hypotheses based on the statistical support obtained from high-throughput processing of samples (Allwood et al., 2006; Draper et al., 2013 and references therein). Different metabolomics studies have been used to identify different metabolic pathways, wild-type and mutant accessions, and sensitive versus tolerant lines to biotic and abiotic stresses in plants (Allwood et al., 2006). In B. distachyon, metabolomic analyses have been shown to be useful for identifying the main metabolites produced in response to fungal diseases (Allwood et al., 2006; Pasquet et al., 2014) and to temperature–salinity (Onda et al., 2015) and drought (Shi et al., 2015) stresses, and the dynamics of host–pathogen interactions (Parker et al., 2008, 2009).
Non-targeted metabolite fingerprinting is a technique designed to provide a relatively comprehensive view of the metabolome that can be used to test competing hypotheses in organisms (Draper et al., 2013), where prominent changes in the metabolome are revealed without specifically identifying individual metabolites. This method, originally based on Fourier transform infrared (FT–IR) spectroscopy, was successfully applied to discriminate between control and abiotically stressed lines (Johnson et al., 2003) and responses between tolerant and sensitive lines to fungal attack in plants (Allwood et al., 2006). An additional advantage of metabolite fingerprinting is that the robustness of results can be tested by validation of the analysis using different biological replicates as training and test sets (Draper et al., 2013). Nonetheless, the combined use of ‘first-pass’ metabolic fingerprinting and ultra-high-accuracy mass metabolite profiling, based on direct-injection electrospray ionization-mass spectrometry (ESI–MS), which facilitates the identification of particular metabolites, allows a deeper understanding of the biochemical processes involved in the investigated case studies. Currently, the increased accuracy of mass spectrometry (MS) actually promotes ESI–MS from a merely fingerprinting technique to a metabolite profiling tool that, combined with MS/MS capabilities, improves structural information about the molecules (Draper et al., 2013). A preliminary metabolic fingerprinting analysis conducted in different accessions of diploid B. distachyon found different metabolic profiles related to geography (Opanowicz et al., 2008). However, no metabolomic study has been performed to date in B. stacei and B. hybridum.
The new evolutionary and genomic findings within the B. distachyon s.l. complex taxa have set the stage for high-definition research of the unusual genomic diversity between and within the species of this complex. The nuclear and organellar genomes of B. stacei and B. hybridum are being sequenced and will serve, together with B. distachyon, as a model system for investigating the origins and consequences of speciation and polyploidization events (Catalán et al., 2014; and unpubl. res.) that might parallel those of economically important cereals (e.g. wheats; Marcussen et al., 2014). Despite these advances, the phenotypic and metabolomic studies of the three species of the complex are still incomplete or have not been performed yet. Given the importance of these investigations for future GWAS analysis, the objectives of this study were (1) to analyse the phenotypic variation of a large representation of individuals (and populations) of B. distachyon, B. stacei and B. hybridum collected across their respective native distributions in the circum-Mediterranean region and in some non-native sites; (2) to test the value of potentially informative morphological traits to discriminate between the three species and within geographical ranges and biological origins (B. hybridum only) of each species; (3) to comparatively analyse the metabolite profiles of native accessions of B. distachyon, B. stacei and B. hybridum; (4) to test the value of potentially informative metabolomic traits to discriminate between the three species; and (5) to analyse the potential association of phenotypic and metabolomic variation between the three species of the B. distachyon s.l. complex.
MATERIALS AND METHODS
Sampling
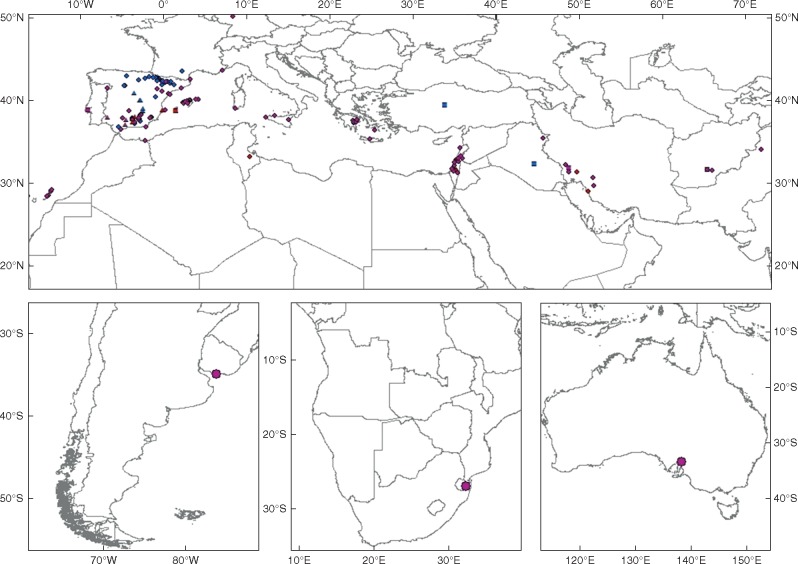
An enlarged sampling was performed in order to increase the representation of populations and individuals of the three B. distachyon s.l. complex species in their respective native circum-Mediterranean regions and in few non-native areas of B. hybridum (Fig. 1, Supplementary Data Table S1). The aim was to cover as much intraspecific phenetic and environmental variability as possible, considering that geographical distribution and environmental variability might affect phenetic variability. Infraspecific phenetic diversity for each of the three species under study has been evidenced in previous studies (Vogel et al., 2009; Catalán et al., 2016b). Additionally, an environmental niche model study indicated that B. distachyon, B. stacei and B. hybridum show overlapping but different environmental niches in their native circum-Mediterranean region, where each species presents a different range of variation for different sets of environmental parameters (López-Alvarez et al., 2015). Also, intraspecific environmental variability has been recorded in the three taxa (Manzaneda et al., 2012, 2015; Shiposha et al., 2016). A total of 1050 individuals, of which 870 were newly collected, from 174 wild populations of B. distachyon (227 individuals, 44 populations), B. stacei (146 individuals, 30 populations) and B. hybridum (497 individuals, 100 populations), plus 180 individual samples from the six inbred lines of B. distachyon [Bd21 (type), ABR1], B. stacei [ABR114 (type)] and B. hybridum [ABR113 (type), ABR110, ABR117] employed in the study of Catalán et al. (2012), were used in the phenotypic analysis. The new samples were collected in the field or were obtained from herbaria and germplasm banks. Seeds from the inbred lines (several generations of selfing) and from new wild germplasm collections (first generation individuals mostly derived from different mother plants) were germinated and grown under standard greenhouse conditions following Catalán et al. (2012). All the studied materials were analysed phenotypically when they reached maturity (e.g. flowering and fruiting stages). The geographical origins and nature [wild (W), herbaria (H), seed bank (S) or inbred (I) plants] of all the studied samples are indicated in Table S1. The taxonomic identity of all the new samples was corroborated by counting DAPI (4′,6-diamidino-2-phenylindole)-stained chromosomes and/or DNA barcoding following the procedures indicated in López-Alvarez et al. (2012). Herbarium vouchers of the newly collected materials have been deposited in the JACA and Unizar (University of Zaragoza) herbaria.
Fig. 1.
Geographical distribution of B. distachyon (blue), B. stacei (red) and B. hybridum (purple) samples used in the phenotypic and metabolomic study. Circles, wild populations; squares, inbred lines; triangles, metabolomic samples.
Phenotypic analysis
Phenotypic analysis was performed using the same 15 potentially informative morphoanatomical characters that were employed to separate and identify the three species of the B. distachyon s.l. complex in a previous study (Catalán et al., 2012). Twelve of the characters were quantitative [(plant) height (H); second leaf length (SLL); second leaf width (SLW); (stomata) leaf guard cell length (LGCL); inflorescence length (IL); spikelet length (total, without awns; SLa); spikelet length (from base to fourth lemma, without awns; SLb); upper glume length (UGL); lemma length (LL); awn length (AL); caryopsis length (CL); pollen grain length (PGL)], and three were discrete characters [number of nodes of tallest culm (NNTC); number of spikelets per inflorescence (NSI); number of flowers per inflorescence (NFI)] (Table 1 and Table S2). Macromorphological characters were measured with a hard ruler under a dissecting microscope. Microanatomical characters (LGCL, PGL) were measured under a microscope using an ocular micrometer. For measurements of stomata LGCL, abaxial epidermises were peeled off from dried leaves that had been pretreated in 90 % lactic acid solution for 8 h. Up to ten individuals (specimens) per population were used for assessment of morphological characters. When possible, five measurements were taken for each character in each individual and the corresponding averaged values were used in the analyses. Statistical analyses were performed separately for wild versus inbred + wild individuals (data from inbred individuals were retrieved from Catalán et al., 2012). Discriminant analysis was also performed for wild versus inbred + wild individuals (see below).
Table 1.
Statistics of 15 phenotypic traits and significance tests of their mean values analysed in individuals from 174 wild populations of Brachypodium distachyon, B. stacei and B. hybridum. Underlined variables are those that significantly discriminate between the three species. N, number of wild individuals analysed. ANOVA (F; d.f. 2) or Kruskal–Wallis (χ2; d.f. 2) tests of variables used for comparisons between species. Superscripts denote Tukey pairwise comparisons between species; means with the same letter do not differ significantly (P<0.05). See text and Supplementary Data Table S2 for abbreviations of variables
| Species | LGCL (μm) | PGL (μm) | H (cm) | NNTC | SLL (cm) | SLW (mm) | IL (cm) | NSI | SLA (cm) | SLB (cm) | NFI | UGL (mm) | LL (mm) | AL (mm) | CL (mm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. distachyon | |||||||||||||||
| N | 227 | 211 | 191 | 190 | 171 | 174 | 184 | 184 | 187 | 183 | 188 | 191 | 188 | 190 | 184 |
| Minimum | 17 | 21 | 5·5 | 1 | 1·1 | 0·5 | 0·92 | 1 | 0·92 | 0·5 | 4 | 2·2 | 5·2 | 6·3 | 3·8 |
| Maximum | 28 | 42 | 56 | 12 | 8·2 | 3·5 | 5 | 4 | 2·4 | 1·82 | 13 | 9·1 | 11 | 15·2 | 7·2 |
| Mean | 22·5a | 30·4a | 19·8a | 4·0a | 3·0a | 1·7a | 2·3a | 2·1a | 1·6a | 1·1a | 7·9a | 6·0a | 7·2a | 10·7a | 5·5a |
| Standard deviation | 2·2 | 3·8 | 11·9 | 2·4 | 1·5 | 0·5 | 0·8 | 0·8 | 0·3 | 0·2 | 2·1 | 1·5 | 0·9 | 1·9 | 0·7 |
| Variance | 5·0 | 14·4 | 141·8 | 5·9 | 2·2 | 0·3 | 0·6 | 0·7 | 0·1 | 0·1 | 4·4 | 2·3 | 0·8 | 3·7 | 0·4 |
| B. stacei | |||||||||||||||
| N | 146 | 154 | 86 | 90 | 75 | 82 | 88 | 91 | 90 | 91 | 88 | 90 | 91 | 88 | 123 |
| Minimum | 16 | 22 | 6·1 | 1 | 1·6 | 1·1 | 2·3 | 2 | 1·3 | 0·61 | 4 | 2·9 | 6·1 | 7·5 | 5·4 |
| Maximum | 36 | 52 | 76 | 9 | 15·1 | 5 | 10 | 5 | 3·3 | 2·82 | 14 | 8·63 | 12·6 | 18·2 | 8·4 |
| Mean | 25·2b | 33·9b | 43·6c | 4·4a | 7·4b | 2·7c | 5·3c | 3·3c | 2·2b | 1·6b | 8·5a | 5·8a | 9·4c | 12·4c | 6·9b |
| Standard deviation | 4·4 | 5·9 | 17·4 | 1·7 | 3·3 | 0·9 | 1·5 | 0·8 | 0·4 | 0·4 | 2·3 | 1·2 | 1·3 | 2·7 | 0·7 |
| Variance | 19·5 | 35·2 | 301·7 | 3·0 | 11·0 | 0·7 | 2·2 | 0·6 | 0·1 | 0·2 | 5·5 | 1·4 | 1·7 | 7·5 | 0·5 |
| B. hybridum | |||||||||||||||
| N | 497 | 518 | 330 | 340 | 320 | 327 | 347 | 350 | 343 | 340 | 343 | 352 | 349 | 349 | 413 |
| Minimum | 20 | 25 | 3·5 | 1 | 1 | 0·7 | 1·2 | 1 | 1 | 0·5 | 2·33 | 2·3 | 3 | 6 | 5 |
| Maximum | 37 | 57 | 78 | 11 | 15·2 | 4·3 | 8 | 6 | 4·1 | 2·96 | 16 | 9·8 | 12·9 | 18·9 | 8·9 |
| Mean | 29·3c | 38·7c | 35·9b | 5·7b | 7·5b | 2·4b | 3·5b | 2·7b | 2·1b | 1·5b | 8·1a | 5·9a | 8·9b | 11·6b | 6·7b |
| Standard deviation | 3·3 | 5·6 | 15·3 | 2·4 | 3·3 | 0·7 | 1·2 | 1·1 | 0·5 | 0·4 | 2·8 | 1·5 | 1·8 | 2·7 | 0·8 |
| Variance | 10·8 | 31·2 | 235·3 | 5·5 | 11·0 | 0·5 | 1·4 | 1·2 | 0·2 | 0·2 | 7·7 | 2·2 | 3·3 | 7·4 | 0·6 |
| χ2 | 411·6 | 322·1 | 164·5 | 82·1 | 213·2 | 127·2 | 255·9 | 86·3 | 194·6 | 132·2 | 22·9 | ||||
| F | 1·5 | 0·6 | 92·3 | 208·7 | |||||||||||
| P | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | >0·05 | >0·05 | <0·001 | <0·001 | <0·001 |
Simple statistic descriptors of intra- and interspecies phenetic diversity (mean, range, standard deviation, box plots of median, range and percentiles) were calculated from the data. The analysis of the interspecies response variables was performed by one-way analysis of variance (ANOVA) χ2 tests or non-parametric Kruskal–Wallis tests when the variables complied or not, respectively, with requirements of normality (this was tested with Kolmogorov–Smirnov tests; Table S2). Multiple pairwise comparisons of means were based on Tukey’s tests for groups with unequal samples sizes. Interspecies response variables were evaluated by multicollinearity analysis to determine which characters were correlated with each other in the common data set and in each of the species data sets; a matrix of Spearman correlation coefficients was obtained by averaging the values from each individual in each case. Comparative analysis of variables from wild populations versus inbred lines was performed using averaged values and pairwise Mann–Whitney (U) tests. In all cases significant tests were performed for the null hypothesis (H0) of μI=μW, where μI and μW are averaged trait values of inbred lines and wild populations, respectively. All the statistical analyses were conducted with the software SPSS v. 15.
Multivariate analysis of phenotypic traits
Multivariate principal component analysis (PCA) of the 15 variables was performed to examine the structure of the taxa, to assess whether the observed groupings were consistent with the taxonomic circumscriptions proposed for the three species of the B. distachyon s.l. complex, and to evaluate the level of covariation in variables. Averaged values of the 15 morphoanatomical characters were estimated for the 174 wild populations and six inbred lines of B. distachyon, B. stacei and B. hybridum. The contribution of each character to the coordinate axes that accounted for the highest percentages of variance was calculated using covariance and variance matrices of the samples with respect to the new axes using PAST v. 2.17 (Hammer et al., 2001). Further PCAs were conducted in each of the B. distachyon, B. stacei and B. hybridum subgroup samples following the procedure of the previous search. These independent analyses allowed estimation of the intraspecific substructure of the taxa and calculation of the contribution of different morphological characters to the separation of intraspecific groups within each species.
A classification discriminant analysis (DA, cross-validation) was conducted with all the variables (15) and samples (174 wild populations and six inbred lines) to determine the highest-probability membership group of the samples (Legendre and Legendre, 1998). The reference samples for each species’s group were the respective type specimens (B. distachyon, Bd21; B. stacei, ABR114; B. hybridum, ABR113). The more discriminating variables were identified by means of Fisher’s coefficient (Fisher, 1936; Anderson, 1996) at the significance threshold value of 0·05. The posterior probability of classification of each sample and the Wilks’s λ value of each discriminant function were calculated. A Wilks’s λ value closer to zero indicated better discrimination between the predefined groups. The DA was run in SPSS v. 15. Further DAs were also performed following the same procedure as that indicated above within each of the B. distachyon, B. stacei and B. hybridum groups, aiming to classify the samples into intraspecific groups, using as references for each subgroup flag samples from the intraspecific PCA analyses (see the Results section).
Metabolomic analysis
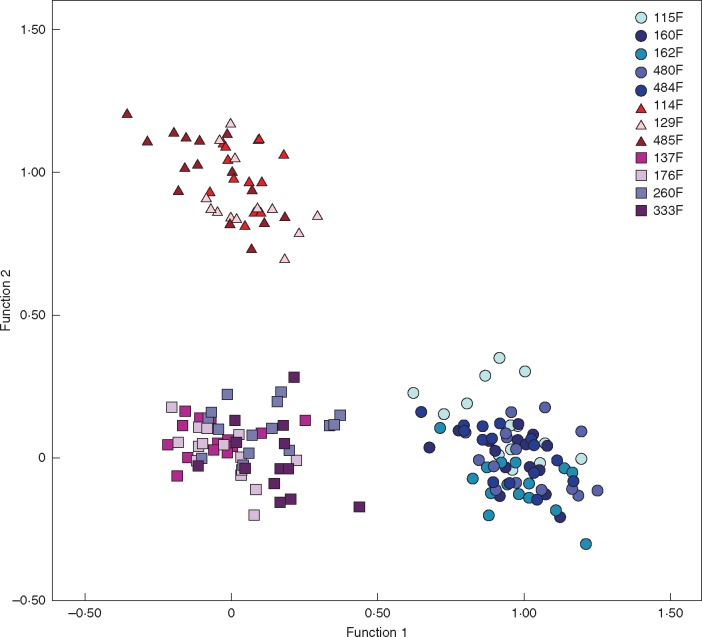
Sampling for metabolomic analysis was performed in a subset of populations from the three studied species (Table S1). Individuals from 12 populations or inbred lines (five of B. distachyon: 115F, 160F, 162F, 480F, 484F; three of B. stacei: 114F, 129F, 485F; and four of B. hybridum: 137F, 176F, 260F, 333F), representing 12 different ecotypes, were grown under standard greenhouse conditions and used in the metabolomic study. A hierarchical metabolomics approach was used to identify and annotate metabolites discriminating the three species. Non-targeted nominal mass metabolite fingerprinting was followed by in-depth data mining and ultra-high-accuracy mass metabolite profiling of explanatory m/z bins. Database searches, MSn fragmentation analysis and comparison with available standards aided signal annotation at different Metabolomics Standards Initiative (MSI) levels of identification (Sumner et al., 2007).
Metabolite fingerprinting was performed using flow infusion electrospray ionization–MS (FIE–MS). For this, sample extraction and mass spectrometric analysis were done following Parker et al. (2009) and Draper et al. (2013). This process involved the use of a single-phase extraction solvent (chloroform:methanol:water, 1:2.5:1, v:v:v) optimized for recovery of a wide range of metabolites, offering relatively comprehensive coverage of the metabolome. Nominal mass FIE–MS analysis was performed using a linear trap quadrupole (LTQ) mass analyser (Finnigan LTQ; Thermo-Finnigan, San Jose, CA), which generated metabolite fingerprints in both positive and negative ionization mode. Ion intensities were detected in the scan range between m/z 50 and 1150, which was subdivided into four small mass ranges for better signal acquisition (low range, m/z 15–200; high1 range, m/z 180–620; high2 range, m/z 600–880; high3 range, m/z 860–1150), and raw data dimensionality was reduced by electronically extracting signals with ±0.1 Da mass accuracy. Mass spectra were combined in a single intensity matrix (runs × m/z ratios) for each ion mode. Data from the intensity matrix were log-transformed and normalized to the total ion count (TIC) before further statistical analysis. Initially, data mining and feature selection was performed using randomForest in the R package FIEms-pro, as reported previously (Enot et al., 2008; RDC Team, 2010).
Ionization products (m/z) identified after data mining were annotated by searching accurate m/z through the MZedDB database (Aberystwyth University) (http://maltese.dbs.aber.ac.uk:8888/hrmet/index.html) at < 5 p.p.m. mass accuracy. Accurate masses from species-representative extracted samples were acquired using the Exactive LC–MS system (Thermo-Scientific) operating in flow-infusion mode. As several overlapping solutions predicting the presence of different metabolites were often possible, the most likely combination of ions putatively identifying a specific metabolite were confirmed by comparing their MSn fragmentation patterns with available standards. MSn fragmentation was carried out with a Finnigan LTQ instrument (Thermo-Finnigan, San Jose, CA) using ion-trap, full MS mode by using a normalized collision energy of 30.0 V. Samples were injected at a flow rate of 3·0 µL min−1 with activation (Q) of 0·250 and activation time 30·0 ms. Thirty scans were used to acquire fragmentation data with an isolation width of 1·0 m/z.
Statistical and multivariate analysis of metabolomic traits
Metabolomic data were subjected to simple statistical analysis of the interspecific diversity (mean, range, standard deviation, box plots of median, range, intervals) and the response variables were estimated with one-way ANOVA χ2 tests for parametric variables and non-parametric Kruskal–Wallis tests, to determine which metabolites were significantly different between the B. distachyon, B. stacei and B. hybridum ecotypes. Only metabolites that were annotated at different MSI levels of identification were used in the multivariate analysis. Discriminant analysis was conducted with the metabolomic data; this consisted of a supervised projection method in which discrimination between groups was based on the spatial classification of ecotypic samples in two-dimensional projections using a priori knowledge of species projections. Fisher’s coefficient (P = 0·05) was used to identify the more discriminating variables, and the posterior probability of classification of each sample and Wilks’s λ value of each discriminant function were also calculated. All statistical analyses were conducted in SPSS v. 15.
Associated phenotypic and metabolomic variation
The potential association of phenotypic and metabolic variation between the three species of the B. distachyon s.l. complex was estimated by correlation analysis using Pearson coefficients. Class means (at ecotype level) of both the 15 phenotypic measurements and the 434 metabolic variables (m/z intensity values) were used for correlation analysis as individual phenotypic measurements could not be attributed to the metabolomic data. Correlation analysis was performed using rcorr in the R package Hmisc and the resulting P values were corrected using the Bonferroni method.
RESULTS
Interspecific phenotypic variation
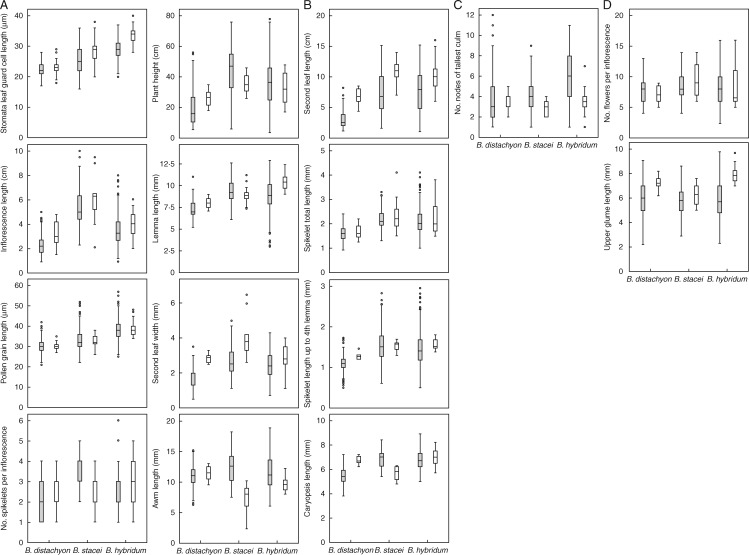
Statistical descriptors and box and whisker plots (Table 1, Fig. 2) summarize the inter- and intraspecific phenotypic diversity detected by the 15 analysed morphological characters across the studied wild populations of the three species. Significant differences (P < 0·001) were found in 13 variables for different combinations of species (Table 1): eight characters discriminate all three species from each other (LGCL, PGL, H, SLW, IL, NSI, LL, AL) (Fig. 2A), four characters discriminate between B. distachyon versus B. stacei and B. hybridum (SLL, SLa, SLb, CL) (Fig. 2B) and one character discriminates B. hybridum from the diploid species (NNTC) (Fig. 2C). Only two characters did not significantly discriminate the species (NFI, UGL) (Table 1, Fig. 2D). Individuals of B. hybridum showed mean values significantly higher for three characters (LGCL, = 29·3 ± 3·3 μm; PGL, = 38·7 ± 0·121μm; NNTC, = 5·7 ± 2·4) than individuals from the diploid species (B. distachyon, = 22·5, = 30·4, = 4·0; B. stacei, = 25·2, = 33·9, = 4·4), while individuals of B. stacei showed mean values significantly higher for six characters (H, = 43·6 ±17·4; SLW, = 2·7 ± 0·9; IL, = 5·3 ± 1·5; LL, = 9·4 ± 1·3; AL, = 12·4 ± 2·7; NSI, = 3·3 ± 0·8) than individuals of B. hybridum and B. distachyon (Table 1, Fig. 2A–C). Individuals of B. distachyon were characterized by smaller and shorter leaf, inflorescence, spikelet, lemma, awn and caryopsis than those of the other species; they also showed fewer nodes in the stem and fewer spikelets and flowers per inflorescence (Table 1, Fig 2). Individuals of B. hybridum showed dimensions intermediate between those of individuals of its parental species in seven characters (H, SLW, IL, NSI, LL, AL, CL) (Table 1, Fig. 2).
Fig. 2.
Box and whisker plots (median, percentiles, range) of 15 morphoanatomical characters analysed in 1050 wild (grey) and inbred (white) individuals of the three species of B. distachyon s.l. complex. (A) Variables that significantly discriminate B. distachyon versus B. stacei versus B. hybridum. (B) Variables that significantly discriminate B. distachyon versus B. stacei + B. hybridum. (C) Variables that significantly discriminate B. hybridum versus B. distachyon + B. stacei. (D) Variables that do not discriminate between species.
When mean value data obtained from the analysis of wild individuals (W; wild versus wild) were compared with those obtained from inbred individuals (I; inbred versus inbred) (Table 2; W-I Mann–Whitney test: wild versus inbred), three characters (PGL, SLA, NFI) did not show significant differences between the two groups in the three species, two (H, NSI) in B. stacei and B. hybridum, and two (LGCL, AL), four (IL, SLB, UGL, LL) and one (SLE) in B. distachyon, B. stacei and B. hybridum, respectively. Among the three common stable traits in both data sets, PGL discriminated between all three species and SLA between B. distachyon and B. stacei or B. hybridum; NFI did not discriminate between species (Table 2). LGCL, LL and AL also discriminated between all three species, and SLL and SLB between B. distachyon and B. stacei or B. hybridum in both data sets but with different means in W and I for some species, SLW between the three species in W (but only between B. distachyon and B. stacei or B. hybridum in I), and CL between B. distachyon and B. stacei or B. hybridum in W (and between B. stacei and B. distachyon or B. hybridum in I) (Table 2). Two traits that significantly discriminated between the three species within the inbred lines (I) discriminated only between B. hybridum and B. distachyon or B. stacei (NNTC), or did not discriminate between them (UGL) in the larger data set of wild individuals (W) (Table 2). The Spearman correlation analysis conducted with the phenotypic traits studied in the wild individuals of B. distachyon, B. stacei and B. hybridum revealed significant correlations (P < 0·001) between several characters (SLA/SLB, 0·89; IL/NSI, 0·75; H/IL, 0·70; SL/SLL, 0·69; H/SLL, 0·66; H/NSI, 0·65; IL/SLA, 0·65; NFI/SLA, 0·62; LL/SLA, 0·59; NFI/SLB, 0·59; LL/SLB, 0·56; NNTC/SLL, 0·56; LGCL/PGL, 0·54; H/SLW, 0·53; H/NNTC, 0·52) (Supplementary Data Table S3).
Table 2.
Comparative Mann–Whitney (U) test of mean values obtained from individuals of 174 wild populations (W) (see Table 1) versus six inbred lines (I) (data retrieved from Catalán et al., 2012) for the 15 analysed phenotypic characters in B. distachyon, B. stacei and B. hybridum. Means with the same letter do not differ significantly (P < 0·05) between species after Tukey’s (wild populations) and Mann–Whitney (inbred lines) pairwise comparison tests in each independent data set). Values that do not differ significantly (P < 0·05) between the two compared data sets (W versus I) for each character and species are underlined. See text and Table S2 for abbreviations of variables
| LGCL | PGL | H | NNTC | SLL | SLW | IL | NSI | SLA | SLB | NFI | UGL | LL | AL | CL | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. distachyon | |||||||||||||||
| W | 22·5a | 30·4a | 19·8a | 4·0a | 3·0a | 1·7a | 2·3a | 2·1a | 1·6a | 1·1a | 7·9a | 6·0a | 7·2a | 10·7a | 5·5a |
| I | 23·26c | 29·87c | 26·13b | 3·30c | 6·68b | 2·84b | 3·25a | 2·69a | 1·63b | 1·27b | 7·00b | 7·23b | 8·05c | 11·45a | 6·75a |
| U | 2820·5 | 3084·5 | 425·5 | 910 | 79 | 46·5 | 559·5 | 791·5 | 1196 | 441·5 | 832 | 538·5 | 425 | 918·5 | 110·5 |
| P | 0·07 | 0·61 | 0·03 | 0·82 | 0 | 0 | 0 | 0·03 | 0·92 | 0 | 0·12 | 0 | 0 | 0·12 | 0 |
| B. stacei | |||||||||||||||
| W | 25·2b | 33·9b | 43·6c | 4·4a | 7·4b | 2·7c | 5·3c | 3·3c | 2·2b | 1·6b | 8·5a | 5·8a | 9·4c | 12·4c | 6·9b |
| I | 28·22b | 32·58b | 35·56c | 2·94b | 11·06a | 4·15a | 6·12b | 2·75a | 2·29a | 1·61a | 9·24a | 6·24c | 8·91b | 7·27c | 5·68b |
| U | 3325 | 2903·5 | 236·5 | 333 | 186 | 171·5 | 423 | 380 | 901 | 897 | 806·5 | 691·5 | 758 | 129 | 165 |
| P | 0 | 0·94 | 0·06 | 0 | 0 | 0 | 0·13 | 0·07 | 0·74 | 0·45 | 0·36 | 0·06 | 0·08 | 0 | 0 |
| B. hybridum | |||||||||||||||
| W | 29·3c | 38·7c | 35·9b | 5·7b | 7·5b | 2·4b | 3·5b | 2·7b | 2·1b | 1·5b | 8·1a | 5·9a | 8·9b | 11·6b | 6·7b |
| I | 33·64a | 38·27a | 33·09a | 3·72a | 10·30a | 2·75b | 3·93a | 2·71a | 2·22a | 1·56a | 8·15ab | 7·92a | 10·49a | 9·68b | 7·02a |
| U | 5090 | 14182 | 1636·5 | 1551·5 | 1377·5 | 2025·5 | 2528 | 3619 | 3321 | 2401·5 | 3310·5 | 919·5 | 1452 | 1784·5 | 8781·5 |
| P | 0 | 0·78 | 0·58 | 0 | 0·01 | 0·06 | 0·04 | 0·9 | 0·81 | 0·03 | 0·79 | 0 | 0 | 0 | 0·01 |
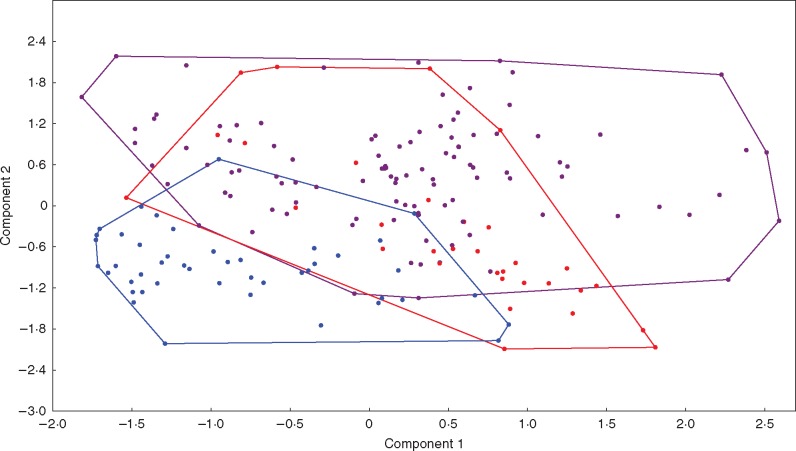
The PCA revealed that 93·3 % of total variation of the 15 analysed characters could be explained by the first three principal components, which accounted for 79·2, 10·4 and 3·6 % of the variance, respectively (Supplementary Data Table S4, Fig. 3). Three characters were identified as the most important contributors to the positive and negative extremes of the first components of the PCA. Plant height (0·98), pollen grain length (0·88) and stomata leaf guard cell length (−0·61) showed the highest contributions to the first, second and third components, respectively (Table S3). The impact of these components on the hierarchical structure of the three species was visualized in a two-dimensional PCA plot (Fig. 3). Population samples of the three species overlapped in the two-dimensional space created by the first two components. The highest overlap was observed between the minimum convex polygons of B. stacei and B. hybridum (Fig. 3).
Fig. 3.
Two-dimensional PCA plot of 46 wild populations of B. distachyon (blue), 31 of B. stacei (red) and 103 of B. hybridum (purple) based on averaged values of individuals analysed for 15 phenotypic traits (see Table 1 and Table S2). The first and second PCA axes explain 79·2 and 10·4 % of the total variation, respectively.
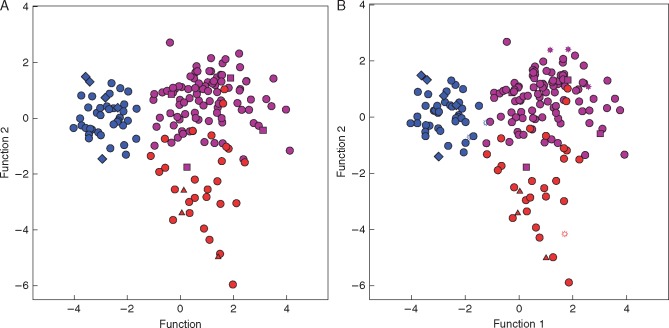
The DA conducted over all 174 wild populations of B. distachyon, B. stacei and B. hybridum using data from the 15 analysed phenotypic traits separated the three species in the two-dimensional plot constructed with the two functions (Fig. 4A). Brachypodium distachyon clustered separately from B. stacei and B. hybridum along the first axis of the plot, which explained 68·2 % of the total variance, whereas the last two species separated along the second axis, which explained 31·8 % of the variance. Catalán et al. (2012) found similar discrimination among the three species when DA was performed only with individuals from the six inbred lines, though sampling of B. stacei was reduced to the type specimen (ABR114). Equivalent results were obtained when DA was performed with the six inbred lines added to the 174 wild populations (Fig. 4B); the three species showed similar clustering patterns in the two-dimensional plot, in which functions 1 and 2 explained 66·8 and 33·2 % of the total variance, respectively. All B. distachyon populations were correctly classified (100 %), both including (46 samples) and excluding (44) the inbred lines (Table 3). However, only 25 (83 %) of 30 wild populations of B. stacei were correctly classified [with the five remaining samples, B891, B921, B621, Bra286 and B100-H141, classified as B. hybridum (17 %)], and only 25 (81 %) of 31 wild + inbred samples were correctly assigned [with six remaining samples classified either as B. hybridum (5; 16 %) or B. distachyon (1 (BGE044241); 3 %)]. Most samples of B. hybridum were correctly classified in both analyses (92 %); 92 out of 100 wild populations and out of 103 wild + inbred samples were correctly assigned, with the eight remaining samples classified either as B. stacei (five samples: B124, 333F, BGE044239, BGE044247, Mog; 5 %) or B. distachyon (three samples: BGE044243, B741, Bra299; 3 %) in both cases (Table 3). Up to ten and five phenotypic characters showed strong correlations to the first and second canonical discriminant functions, respectively (Supplementary Data Table S5), with Wilks’s λ values of 0·12 and 0·44 (P < 0·001), respectively. Of these, three (LGCL, CL, PGL) showed the highest contributions to function 1 and one (IL) to function 2 in the wild populations and the combined inbred lines and wild population data sets (LGCL, 0·54, 0·56; CL, 0·47, 0·46; PGL, 0·42, 0·44; IL, 0·52, 0·53) (Table S5).
Fig. 4.
Two-dimensional DA scatterplots of populations of B. distachyon (blue), B. stacei (red) and B. hybridum (purple) based on averaged values of individuals analysed for 15 phenotypic traits in 174 wild populations (A) and in 174 wild populations plus six inbred lines (B). Wild populations (circles), inbred lines (stars), accessions with metabolites (B. distachyon, diamonds; B. stacei, triangles; B. hybridum, squares).
Table 3.
Assignment probabilities of individuals from 174 wild populations (W) and from six inbred lines plus 147 wild populations (I+W) of B. distachyon, B. stacei and B. hybridum based on discriminant analysis of 15 phenotypic traits. N, number of populations and populations + inbred lines studied
| Data sets | Samples | N | Predicted group membership |
||
|---|---|---|---|---|---|
| B. distachyon | B. stacei | B. hybridum | |||
| W | B. distachyon | 44 | 44 (100 %) | 0 | 0 |
| B. stacei | 30 | 0 | 25 (83·3 %) | 5 (16·7 %) | |
| B. hybridum | 100 | 3 (3 %) | 5 (5 %) | 92 (92 %) | |
| I + W | B. distachyon | 46 | 46 (100 %) | 0 | 0 |
| B. stacei | 31 | 1 (3·2 %) | 25 (80·6 %) | 5 (16·1 %) | |
| B. hybridum | 103 | 3 (3 %) | 5 (5 %) | 95 (92 %) | |
Intraspecific phenotypic variation
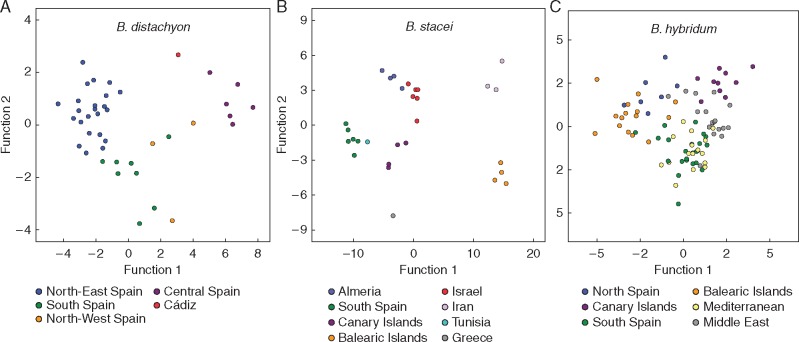
The DAs conducted at the intraspecific level in B. distachyon, B. stacei and B. hybridum using the 15 phenotypic traits showed significant differences for some traits and different geographical groupings of populations in one or other of the studied species. In B. distachyon, the two-dimensional plot constructed with the first two discriminant functions, which explained 79·7 % and 10·8 % of the total variance, indicated the separation of four groups of Iberian populations (Fig. 5A). North-eastern Iberian populations (Aragón, Navarra, Lleida) clustered in the upper left area of the plot, north-western Iberian populations (Valladolid, Palencia) and southern Iberian populations (Andalucía) plus one additional population from southern Spain (Cadiz) in the lower left and middle areas, and central Iberian populations (Albacete, Cuenca, Madrid, Rioja) in the upper right area. The characters that showed higher correlations to functions 1 and 2 were H (0·56) and UGL (0·66), AL (0·46) and NNTC (0·46) (P < 0·001), respectively, with Wilks’s λ values of 0·012 (P < 0·001) and 0·15 (P = 0·058) (Supplementary Data Table S6), respectively. The height of the plant (H) was significantly different between the central ( = 42·2) and north-western ( = 32·2) groups, which in turn showed taller individuals than those of the north-eastern ( = 18·6) and southern ( = 13·8) Iberian groups (Table 4). The length of the upper glume (UGL) significantly separated the southern ( = 4·3) populations from the others ( = 6·4), the number of nodes in the tallest culm (NNTC) the north-eastern populations ( = 2·7) from the rest ( = 6·4), the length of the awn (AL) the north-eastern ( = 11·2) from the southern ( = 9·5) populations, and the number of spikelets per inflorescence (NSI) the central ( = 3·0a) from the north-eastern and southern ( = 1·8) populations (Table 4).
Fig. 5.
Two-dimensional DA scatterplots of 41 Spanish populations of B. distachyon (A), 29 circum-Mediterranean populations of B. stacei (B) and 89 circum-Mediterranean populations of B. hybridum (C) based on averaged values of wild individuals analysed for 15 phenotypic traits. Colours for the respective geographical groups of each species are indicated in the charts.
Table 4.
ANOVA test of 15 variables used for comparisons among wild individuals from intraspecific groups within each species (Brachypodium distachyon, B. stacei, B. hybridum). Superscripts denote least significant difference (LSD) pairwise comparisons between geographical groups within species; means with the same letter do not differ significantly (P<0·05). See text and Table S2 for abbreviations of variables. N, number of individuals analysed
| Species | N | LGCL | PGL | H | NNTC | SLL | SLW | IL | NSS | SLA | SLB | NFS | UGL | LL | AL | CL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. distachyon | * | ** | ||||||||||||||
| North-east Spain | 23 | 23·2 | 30·1 | 13·8c | 2·7b | 2·5c | 1·8 | 2·3 | 2·1 | 1·7 | 1·1 | 8·5b | 6·5a | 7·3 | 11·2ab | 5·3 |
| South Spain | 8 | 23·0 | 28·6 | 18·6c | 5·5a | 3·1bc | 1·5 | 1·8 | 1·6 | 1·5 | 1·1 | 6·7c | 4·3b | 7·1 | 9·5c | 5·8 |
| Central Spain | 3 | 22·1 | 28·5 | 32·2b | 7·3a | 4·3ab | 2·1 | 2·6 | 2·4 | 1·6 | 1·0 | 7·8bc | 6·1a | 7·0 | 10·0bc | 5·0 |
| North-west Spain | 6 | 21·4 | 30·9 | 42·3a | 6·5a | 5·5a | 1·9 | 2·8 | 3·0 | 1·5 | 1·2 | 7·3bc | 6·7a | 7·4 | 10·7bc | 5·6 |
| Cádiz | 1 | 21·4 | 34·8 | 36·4ab | 4·0ab | 2·6bc | 1·8 | 4·3 | 3·0 | 2·0 | 1·6 | 12·5a | 6·7a | 7·6 | 14·0a | 6·5 |
| F | 1·2 | 1·1 | 32·2 | 12·9 | 8·6 | 1·4 | 5·2 | 4·9 | 1·6 | 2·0 | 3·7 | 7·1 | 0·5 | 3·5 | 2·2 | |
| P | >0·05 | >0·05 | <0·001 | <0·001 | <0·001 | >0·05 | <0·05 | <0·05 | >0·05 | >0·05 | <0·05 | <0·001 | >0·05 | <0·05 | >0·05 | |
| B. stacei | *** | |||||||||||||||
| Almeria | 4 | 26·9 | 33·2b | 45·7ab | 3·6ab | 6·4 | 2·8 | 5·5ab | 3·1 | 2·7 | 1·9 | 10·0 | 5·0 | 10·1 | 12·1 | 6·7 |
| South Spain | 6 | 22·3 | 30·1b | 55·9a | 6·2a | 9·3 | 2·9 | 6·1a | 3·6 | 2·1 | 1·4 | 7·2 | 6·3 | 9·5 | 14·0 | 6·8 |
| Canary Islands | 4 | 25·0 | 31·3b | 49·1a | 4·0ab | 6·1 | 2·4 | 5·3ab | 3·5 | 2·1 | 1·4 | 8·6 | 5·8 | 9·2 | 10·5 | 6·8 |
| Balearic Islands | 4 | 27·6 | 36·6b | 19·0c | 2·8b | 4·4 | 2·3 | 3·7b | 2·5 | 2·0 | 1·5 | 9·5 | 5·8 | 8·6 | 11·5 | 6·4 |
| Israel | 6 | 25·7 | 35·8b | 42·3ab | 5·8a | 5·0 | 2·5 | 5·3ab | 3·3 | 2·2 | 1·7 | 8·7 | 6·4 | 10·2 | 15·1 | 6·9 |
| Iran | 3 | 28·5 | 47·2a | 27·7bc | 3·6ab | 4·5 | 2·3 | 4·1ab | 2·3 | 2·6 | 2·1 | 11·6 | 5·6 | 9·3 | 14·3 | 7·7 |
| Tunisia | 1 | 28·6 | 30·9b | 42·0abc | 4·0ab | 4·2 | 1·2 | 5·4ab | 3·0 | 2·2 | 1·7 | 10·0 | 5·7 | 7·4 | 11·5 | 6·4 |
| Greece | 1 | 19·7 | 28·6b | 65·0a | 5·8ab | 11·0 | 2·8 | 6·0ab | 4·3 | 1·8 | 0·9 | 7·8 | 5·2 | 10·4 | 14·2 | 6·1 |
| F | 1·8 | 8·1 | 17·0 | 4·6 | 5·4 | 1·4 | 4·1 | 3·7 | 1·7 | 2·0 | 1·9 | 1·0 | 1·9 | 2·0 | 2·3 | |
| P | >0·05 | <0·001 | <0·001 | <0·05 | <0·001 | >0·05 | <0·05 | <0·05 | >0·05 | >0·05 | >0·05 | >0·05 | >0·05 | >0·05 | >0·05 | |
| B. hybridum | ||||||||||||||||
| North Spain | 7 | 30·3 | 35·5b | 21·5c | 2·8c | 3·5c | 1·8c | 4·0b | 2·9abc | 2·2b | 1·3b | 9·9a | 7·2ab | 8·1c | 12·5ab | 6·3bc |
| Canary Islands | 10 | 28·8 | 44·0a | 45·8a | 3·4c | 6·6b | 2·7ab | 5·2a | 3·2ab | 2·8a | 1·9a | 11·1a | 7·6a | 10·2ab | 11·8bc | 7·4a |
| South Spain | 23 | 29·5 | 36·4b | 36·7b | 6·2a | 7·9ab | 2·3b | 3·3c | 2·7bc | 2·1b | 1·5b | 7·3b | 5·4c | 9·4b | 11·6bc | 6·7b |
| Balearic Islands | 15 | 28·5 | 38·8b | 18·2c | 3·8c | 3·5c | 1·6c | 2·7d | 1·9d | 2·1b | 1·4b | 9·6a | 5·7c | 6·7d | 11·0bc | 6·1c |
| Mediterranean | 16 | 30·5 | 37·8b | 46·9a | 6·8a | 9·1c | 2·4ab | 3·7bc | 3·4a | 2·0b | 1·3b | 7·5b | 5·2c | 7·8c | 10·4c | 6·6b |
| Middle East | 18 | 28·7 | 38·3b | 35·3b | 5·2b | 6·7a | 2·7a | 3·3cd | 2·3cd | 2·2b | 1·5b | 7·8b | 6·4b | 10·4a | 13·6a | 7·4a |
| F | 1·2 | 6·0 | 18·5 | 13·7 | 12·1 | 10·0 | 12·2 | 5·5 | 5·2 | 4·7 | 8·0 | 10·1 | 17·2 | 3·9 | 9·5 | |
| P | >0·05 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·001 | <0·05 | <0·001 |
Characters with significant differences only for some comparisons: B. distachyon:
IL, north-east ≠ south; north-west ≠ south; Cádiz ≠ north-east + south + centre + north-west.
NS, north-west ≠ north-east + south; Cádiz ≠ south· B. stacei:
SLL= south ≠ Balearic Islands, Israel, Iran.
In B. stacei the two-dimensional plot constructed with the first two discriminant functions, which accounted for 79·4 and 11·4 % of the total variance, classified three groups of circum-Mediterranean populations (Fig. 5B). The Iranian populations clustered in the upper right area of the plot and the Balearic populations in the lower right area; they were clearly separated from the other populations along function 1. Populations from Israel clustered close to populations from Almeria (southern Spain) in the upper middle area of the plot, those from the Canary Islands in the middle central area, those from southern Spain and Mahgreb (Tunisia) in the leftmost central area, and one population from Greece in the lowest left area (Fig. 5B). The phenotypic traits that showed highest correlations to functions 1 and 2 were IL (−0·11) and SLB (0·18), and SLA (0·17) (P < 0·001) (Supplementary Data Table S7), respectively, with Wilks’s λ values of 0 (P < 0·001) in both cases. The PGL trait significantly discriminated the Iranian populations, which showed longer pollen grains ( = 47·2) than the rest ( = 32·3) (Table 4). Traits IL, NNTC and SLL separated southern Spain ( = 3·7 = 2·8 = 9·2) from Balearic Island ( = 6·1 = 6·2 = 4·3) populations, NNTC and SLL separated southern Spain from the Israel ( = 5·8 = 5) populations, and SLL separated southern Spain from Iran ( = 4·5) populations; the remaining populations showed intermediate measurements for these traits ( = 5·2 = 4·6 = 6·9) (Table 4).
In B. hybridum the first two discriminant functions of the two-dimensional DA plot, which explained 42·7 and 31·4 % of the total variance, did not show a clear-cut geographical clustering of circum-Mediterranean populations, though they separated two main groups along the first axis (Fig. 5C). Populations from northeastern Spain and the Balearic islands clustered in the left area of the plot, whereas those from the Middle East, the Mediterranean basin, southern Spain and the Canary Islands clustered predominantly in the right area. The most influential characters were H (0·51), SLW (0·44) and CL (0·40), and NNTC (0·57), UGL (0·48) and NFI (0·42), which were highly correlated to the first and second functions (P < 0·001), respectively (Supplementary Data Table S8), with Wilks’s λ values of 0·02 and 0·08 (P < 0·001), respectively. The height of the plant separated the taller Mediterranean and Canary Islands individuals ( = 47=45·8) from the shorter Balearic Islands and northern Spain individuals (=18·1=21·5), whereas individuals from other regions (southern Spain =36·7=35·3) showed intermediate statures (Table 4). Caryopsis length (CL) and width of the second leaf (SLW) differentiated the individuals from Middle East and Canary Islands (=7·4; =2·7) from those of northern Spain and Balearic Islands (=6·2; =1·7), whereas the Mediterranean and southern Spain individuals showed intermediate values (=6·6; 2·4) (Table 4). The number of nodes in the tallest culm was higher in Mediterranean and southern Spain individuals (=6·8=6·2 respectively; Table 4) with respect to those of Middle East (=5·2) and Balearic Island, Canary Islands and northern Spain (=3·8=3·3;=2·8) individuals.
Interspecific metabolomic variation
Metabolite fingerprinting analysis resulted in a total of 2219 nominal m/z signals in combined positive and negative ionization datasets in the 12 studied ecotypes of B. distachyon, B. stacei and B. hybridum. However, only 693 metabolite signals showed significant differences between species based on randomForest feature selection, and of these only 434 nominal m/z signals could be further annotated with a targeted accurate mass m/z search through the MZedDB database (Supplementary Data Table S9). Where more than one m/z signal resulted in the annotation of the same metabolite, e.g. due to the formation of various adducts, the most likely adduct was selected based on the most abundant adduct or by giving preference to the [M+H]1+ and [M+K]1+ adducts in positive ion mode and the [M-H]1− adduct in negative ion mode over the other adducts. The other duplicate adducts were, however, kept in the annotation list of 434 m/z signals (in plain text rather than bold) (Table S9) as they give added confidence in the annotation of m/z signals. These m/z signals with duplicate annotations were, however, later removed from the DA, details of which are provided below. Some of the annotation results were confirmed by comparing MSn fragmentation patterns of m/z signals with those of available standards (see Supplementary Data Table S10 for the list of annotated m/z signals).
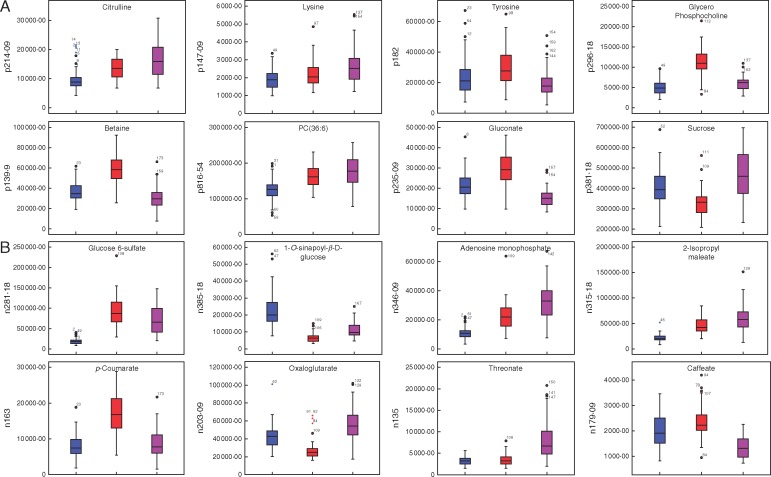
Statistical descriptors and box and whisker plots indicated that 434 fingerprint m/z signals were able to significantly discriminate between the three species (Supplementary Data Tables S11 and S12, Fig. 6). Of these, several positive (e.g. p214·09, p147·09, p182, p296·18, p139·9, p816·54, p235·09, p381·18) (Fig. 6A) and negative (e.g. n281·18, n385·18, n346·09, n315·18, n163, n203·09, n135, n179·09) (Fig. 6B) m/z signals discriminated the ecotypes of B. distachyon, B. stacei and B. hybridum from each other (Table S12).
Fig. 6.
Box and whisker plots (median, percentiles, range) of a subsample of 16 significantly discriminant metabolomic traits analysed in five ecotypes (60 replicates) of B. distachyon (blue), three (36) of B. stacei (red) and four (48) of B. hybridum (purple). (A) Positive mode metabolites. (B) Negative mode metabolites. Assignment of metabolomics signals to putative ionization products is indicated for each trait. The y-axis represents m/z signal intensities.
The DAs conducted on the 12 ecotypes of B. distachyon, B. stacei and B. hybridum using the data from the 434 (positive 217, and negative 217) metabolomic traits discriminated the three species in the two-dimensional plots formed by the two functions (Table S11, Fig. 7). Removing m/z signals of the same annotations showing different adducts did not alter the DA classification results (Tables S9 and S11, Supplementary Data Fig. S1). The five ecotypes of B. distachyon clustered separately from those of B. stacei and B. hybridum along the first axis of the plot, which explained 84·7 % (positive data set) or 80 % (negative data set) of the total variance, whereas the ecotypes of the last two species separated respectively along the opposite extremes of the second axis, which explained 20 % (positive data set) or 15·3 % (negative data set) of the variance. Both positive and negative metabolites were significantly correlated with the first (Wilks’s λ= 0; P < 0·001) and second [Wilks’s λ= 0·01 (positive data set), 0·02 (negative data set); P < 0·001] functions of their respective analysis, though their correlation values were usually low (Table S11). The ecotypes of each species were correctly classified to their respective groups in all cases (100 %).
Fig. 7.
Two-dimensional DA scatterplot of five ecotypes (60 replicates) of B. distachyon (circles), three (36) of B. stacei (triangles) and four (48) of B. hybridum (squares) based on averaged values of individuals analysed for 434 metabolomic traits. Colours for each ecotype are indicated in the charts.
Association of phenotypic traits and metabolites
The Pearson correlation analysis between phenotypic traits and 434 fingerprint m/z signals indicated that the highest significant Bonferroni-corrected P value (<0·05) correlations were for PGL and LL (Table 5) to discriminate the three species. PGL showed significant correlations with ten metabolite signals [hydroxybutyrate, threonate, shikimate, 13C isotope of shikimate, quinate, 13C isotope of quinate, sinapate, sedoheptulose 7-phosphate, ADP-glucose, phosphatidylglycerol (PG)(18:1(11Z)/22:6 (4Z,7Z,10Z,13Z,16Z,19Z)] and LL showed significant correlations with six metabolites [O-phosphohomoserine and lipids tentatively assigned as phosphatidylcholine (PC)(16:0/18:2(2Z,4Z)), sulfoquinovosyl diacylglycerol (SQDG)(16:0/16:1 (11Z)), PC(18:3(8E,10E,12E)/18:3(8E,10E,12E)) (PC(36:6)), PC(18:0/18:3(9Z,12Z,15Z)/0:0), PG(18:0/20:3(5Z,8Z,11Z))] (Table 5, Supplementary Data Fig. S2).
Table 5.
Pearson correlation coefficient between 435 metabolic variables (m/z) and 15 phenotypic traits based on 12 ecotypes of B. distachyon, B. stacei and B. hybridum. See text and Table S2 for abbreviations of variables
| m/z | LGCL | PGL | H | NNTC | SLL | SLW | IL | NSS | SLA | SLB | NFS | UGL | LL | AL | CL | Putative ionization product | Adduct |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n103·09 | 0·707 | 0·906* | 0·656 | 0·834 | 0·488 | 0·300 | 0·439 | 0·537 | 0·539 | 0·500 | 0·533 | 0·702 | 0·555 | 0·468 | 0·474 | Hydroxybutyrate | [M-H]1− |
| n135 | 0·784 | 0·936* | 0·601 | 0·807 | 0·553 | 0·279 | 0·384 | 0·436 | 0·427 | 0·405 | 0·462 | 0·637 | 0·553 | 0·432 | 0·487 | Threonate | [M-H]1− |
| n173·09 | 0·578 | 0·900* | 0·618 | 0·878 | 0·347 | 0·107 | 0·394 | 0·597 | 0·520 | 0·481 | 0·581 | 0·641 | 0·505 | 0·506 | 0·450 | Shikimate | [M-H]1− |
| n174·09 | 0·724 | 0·929* | 0·617 | 0·885 | 0·429 | 0·101 | 0·345 | 0·533 | 0·444 | 0·405 | 0·488 | 0·633 | 0·497 | 0·437 | 0·519 | 13C isotope of shikimate | [M-H]1− |
| n191·09 | 0·605 | 0·919* | 0·610 | 0·856 | 0·429 | 0·189 | 0·423 | 0·608 | 0·530 | 0·492 | 0·588 | 0·682 | 0·534 | 0·496 | 0·463 | Quinate | [M-H]1− |
| n192·09 | 0·590 | 0·897* | 0·646 | 0·856 | 0·394 | 0·218 | 0·460 | 0·628 | 0·587 | 0·552 | 0·633 | 0·707 | 0·547 | 0·528 | 0·474 | 13C isotope of quinate | [M-H]1− |
| n222·09 | 0·528 | 0·873 | 0·650 | 0·890* | 0·343 | 0·076 | 0·460 | 0·699 | 0·608 | 0·556 | 0·640 | 0·655 | 0·551 | 0·561 | 0·534 | N-acetyl tyrosine | [M-H]1− |
| n223·09 | 0·762 | 0·911* | 0·638 | 0·819 | 0·583 | 0·316 | 0·481 | 0·596 | 0·463 | 0·447 | 0·548 | 0·672 | 0·546 | 0·430 | 0·571 | Sinapate | [M-H]1− |
| n236·09 | 0·658 | 0·714 | 0·823 | 0·571 | 0·573 | 0·421 | 0·752 | 0·614 | 0·835 | 0·808 | 0·730 | 0·643 | 0·891* | 0·768 | 0·690 | O-phosphohomoserine | [M+K-2H]1− |
| n289·18 | 0·697 | 0·890* | 0·636 | 0·817 | 0·398 | 0·229 | 0·347 | 0·425 | 0·526 | 0·483 | 0·466 | 0·694 | 0·547 | 0·490 | 0·445 | Sedoheptulose 7-phosphate | [M-H]1− |
| n420·18 | 0·472 | 0·632 | 0·811 | 0·571 | 0·539 | 0·661 | 0·866 | 0·780 | 0·874 | 0·876 | 0·891* | 0·760 | 0·756 | 0·700 | 0·630 | β-1,4-mannose-N-acetyl glucosamine | [M+K-2H]1− |
| n588·09 | 0·736 | 0·892* | 0·658 | 0·803 | 0·663 | 0·500 | 0·628 | 0·572 | 0·554 | 0·546 | 0·583 | 0·631 | 0·648 | 0·515 | 0·578 | ADP-glucose | [M-H]1− |
| n819·63 | 0·598 | 0·907* | 0·697 | 0·811 | 0·532 | 0·309 | 0·576 | 0·588 | 0·659 | 0·626 | 0·673 | 0·651 | 0·723 | 0·666 | 0·553 | PG(18:1(11Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z))** | [M-H]1− |
| n89·09 | 0·666 | 0·869 | 0·521 | 0·892* | 0·318 | 0·030 | 0·237 | 0·478 | 0·311 | 0·284 | 0·426 | 0·558 | 0·311 | 0·290 | 0·416 | Oxalate | [M-H]1− |
| p445·27 | −0·576 | −0·751 | −0·607 | −0·534 | −0·320 | −0·352 | −0·480 | −0·519 | −0·778 | −0·764 | −0·601 | −0·904* | −0·749 | −0·723 | −0·580 | Unknown | – |
| p796·63 | 0·413 | 0·582 | 0·687 | 0·409 | 0·625 | 0·537 | 0·834 | 0·581 | 0·806 | 0·788 | 0·735 | 0·470 | 0·897* | 0·768 | 0·536 | PC(16:0/18:2(2Z,4Z))** | [M+K]1+ |
| p815·54 | 0·555 | 0·724 | 0·758 | 0·530 | 0·730 | 0·594 | 0·787 | 0·620 | 0·804 | 0·770 | 0·716 | 0·663 | 0·910* | 0·746 | 0·633 | SQDG(16:0/16:1(11Z))** | [M+Na]1+ |
| p816·54 | 0·548 | 0·747 | 0·708 | 0·493 | 0·699 | 0·593 | 0·790 | 0·664 | 0·820 | 0·803 | 0·765 | 0·724 | 0·913* | 0·758 | 0·607 | PC(18:3(8E,10E,12E)/18:3(8E,10E,12E))** | [M+K]1+ |
| p822·54 | 0·593 | 0·688 | 0·741 | 0·493 | 0·647 | 0·531 | 0·801 | 0·589 | 0·813 | 0·798 | 0·737 | 0·590 | 0·900* | 0·746 | 0·608 | PC(18:0/18:3(9Z,12Z,15Z)/0:0)** | [M+K]1+ |
| p823·54 | 0·690 | 0·692 | 0·840 | 0·585 | 0·668 | 0·478 | 0·772 | 0·563 | 0·809 | 0·776 | 0·682 | 0·591 | 0·904* | 0·745 | 0·764 | PG(18:0/20:3(5Z,8Z,11Z))** | [M+Na]1+ |
Bonferroni-corrected significant P values (< 0·05);
many possible isomers with this particular accurate mass (see Table S9).
DISCUSSION
Phenotypic differentiation is shaped both taxonomically and geographically within the B. distachyon s.l. complex species
This large phenotypic analysis conducted on 1050 individuals from 174 wild populations and six inbred lines of B. distachyon, B. stacei and B. hybridum has provided a wealth of data and strong statistical evidence for disentangling the intraspecific phenotypic plasticity and interspecific differentiation of the three species, once considered cryptic taxa (Garvin et al., 2008; Catalán et al., 2012; López-Alvarez et al., 2012). The considerably increased sampling size of this study with respect to the pioneer work of Catalán et al. (2012) has also made it possible to increase the number of significantly discriminating traits between the species from five to eight (Table 1, Fig. 2). To the five characters (LGCL, PGL, UGL, LL, AL) found previously Catalán et al. (2012) to significantly discriminate between the three taxa when inbred lines were grown under controlled greenhouse conditions, the present study adds four more characters (H, SLW, IL, NSI) and discards one (UGL) for discriminating between wild individuals of the three species. Exhaustive analysis shows that despite an increase in intraspecific phenotypic variability within each species (Tables 1 and 2, Fig. 2) when compared with Catalán et al. (2012), the number of discriminant taxonomic traits also rises (from the same 15 original characters) as a consequence of the more robust statistical inference and the underlying evolutionary phenotypic differentiation of the three members of the complex (Catalán et al., 2012, 2014).
This study has also shown geographically diverse phenotypic data for the poorly known species B. stacei (Tables 1 and 2, Table S1, Fig. 2). This taxon emerges as the tallest plant and with some of the largest features (inflorescence, number of spikelets per inflorescence, lemma and awn) of the three species of the complex, exceeding the measurements of the allotetraploid B. hybridum for these traits (Table 1, Fig. 1). Additionally, this study confirms previous findings of Catalán et al. (2012), such as smaller stature and shorter features (12 out of the 15 studied traits) of B. distachyon with respect to its two congeners. The considerable phenotypic gap observed between the two diploid species B. stacei and B. distachyon (Table 1, Fig. 2) could be a consequence of their distinct evolutionary origins, large divergence and genomic expression (Catalán et al., 2012, 2014). The allotetraploid B. hybridum shows significantly larger measurements for two non-endoreduplicating plant cell types, LGCL and PGL, than its two diploid progenitors (Table 1). Correlations between increasing ploidy level and larger pollen grain and stomata guard cell sizes have been reported in other pooids, like Lolium (Speckmann et al., 1965), Bromus (Tan and Dunn, 1973) and Dactylis (Bretagnolle and Lumaret, 1995). These traits could be also used as proxies to differentiate B. hybridum from its parents.
The use of morphological traits is considered to be limited, because most of the traits are multigenic, quantitative characters that could be influenced by environmental conditions, plant age, phenological stage or cultivation conditions (Smykal et al., 2008). However, the data presented here support the stability of the phenotypic characters in natural populations when compared with the propagated inbred lines (Table 3). Most of the studied characters show the same discriminant value to separate between wild, cultivated, or wild and cultivated individuals of B. distachyon, B. stacei and B. hybridum (Table 3, Table S5). Furthermore, three of the traits (PGL, SL, NFI) have been shown to be similarly discriminant among the three species, both in wild and inbred plants. Results of this study suggest that these traits could be genetically fixed and might constitute a valuable tool to separate and identify individuals of the B. distachyon, B. stacei and B. hybridum. However, the precise phenotypic characterization and identification of all individuals is not always possible, as demonstrated by the failure to correctly classify a few individuals of B. stacei and B. hybridum, erroneously assigned to other species (Table 3, Fig. 4A, B). Because the taxonomic identity of these individuals was confirmed by chromosome counting and/or DNA barcoding, their diverging phenetic features could have resulted from extreme plasticity or could be a consequence of the restricted number of phenotypic traits used in the study. By contrast, the low percentages of failures detected (up to 20 % in B. stacei, 8 % in B. hybridum and 0 % in B. distachyon) suggest that the morphological traits employed here are good diagnostic features to differentiate these taxa. In a recent updated taxonomic description of these species, five additional qualitative phenotypic traits have been found to be useful discriminators: leaf blade colour to separate all three species; occasional production of short rhizomes to discriminate B. stacei and B. hybridum; and leaf blade shape, softness and hairiness to separate B. stacei from the other two species (Catalán et al., 2016a).
The analysis of phenotypic variation within each of the three species of the B. distachyon complex has also untapped the organization of their intraspecific diversity (Fig. 5). The geographical phenotypic structure observed between the northern, central and southern Iberian populations of B. distachyon (Fig. 5A) might reflect different adaptations to environmental conditions (Manzaneda et al., 2015) and could also be indicative of genotypic differences (López-Álvarez and Catalán, unpubl. res.). Also, the observed differences in plant height (H), with central and northern Spanish individuals being significantly taller than southern Spanish individuals (Table 4), has been corroborated in independent phenomic studies of Iberian B. distachyon inbred lines (E. Pérez-Collazos, University of Zaragoza (Spain), and J. Finch, Aberystwyth University (UK), pers. comm.). The broader but less congruent geographical structure of B. stacei phenotypes (Fig. 5B) suggests the isolation of circum-Mediterranean-edge Iranian and continental-island Balearic populations versus the proximity of largely disjunct Israel–southern Spain (Almeria) and southern Spanish–Canary Islands populations. Long-distance dispersal of seeds has been proposed for B. distachyon and B. hybridum (Vogel et al., 2009) and could also operate in B. stacei. The low geographically structured variation within B. hybridum detects, however, a slight differentiation of the Balearic and northern Spanish populations from the rest (Fig. 5C). It is important to stress, however, that the southern Spanish allotetraploid individuals derived from the unusual cross of maternal B. distachyon parent and paternal B. stacei parent (cf. López-Alvarez et al., 2012) are morphologically close to other Mediterranean individuals derived from the common cross of maternal B. stacei and paternal B. distachyon (Fig. 5C). In allopolyploid grasses different bidirectional crosses of parental species might be the origin of the same or different hybrid allopolyploid species (e.g. Aegylops; Meimberg et al., 2009). However, our data corroborate the idea of a unique speciation event in the origin of B. hybridum, even if its individuals could have originated from alternative bidirectional crosses between different ancestral diploid progenitors (Catalán et al., 2016b).
Metabolomic characterization of B. distachyon, B. stacei and B. hybridum and association of phenotypic with metabolite traits
The use of FIE–MS fingerprinting has provided a preliminary metabolite profile for the three species of the B. distachyon complex. This study shows that a large number of metabolites (434) could be used to discriminate between B. distachyon, B. stacei and B. hybridum (Tables S9–S12, Fig. 6). For example, citrulline content is significantly larger in B. hybridum than in its parental species, of which B. distachyon shows the lowest concentration (Fig. 6A). Citrulline has been shown to play an important role in transporting and storing nitrogen, and is reported to be an important biochemical indicator of plant tolerance of saline and drought stresses (Kawasaki et al., 2000; Kusvuran et al., 2013). In this study, citrulline levels reflect climatic variation from warmer and more aridic Iberian places (B. hybridum ecotypes), through warm but shady places (B. stacei ecotypes) to more mesic places (B. distachyon ecotypes) (Fig. 1; Catalán et al., 2016a), supporting the different ecological adaptations of the three species’ ecotypes to their respective environments. Another example is a phospholipid assigned as phosphatidylcholine(36:6) [PC(36:6)], whose levels were significantly higher in B. hybridum than in its parental species (Fig. 6A). Drought stress has been shown to induce changes in leaf lipid composition by increasing the levels of phosphatidylcholine, suggesting specific adaptive alterations in membrane composition to compromise drought stress tolerance (Vigh et al., 1986; Toumi et al., 2008).
Discriminant analysis indicates not only that each species could be significantly separated from each other (Fig. 7, Fig. S1) but that B. distachyon is clearly apart from B. stacei and B. hybridum, followed by metabolomic differentiation of the latter. This pattern parallels what has been observed in the phenotypic analysis (Fig. 4) and in molecular relationships between the species, especially in the plastid genome-based reconstructions (López-Alvarez et al., 2012). Thus, metabolite fingerprinting is concordant with phenotypic analysis within the B. distachyon species complex. The Pearson correlation analysis between phenotypic and metabolomics traits indicated a significant association of PGL and LL with two distinct groups of metabolites, respectively: members of the phenolics biosynthesis and lipids. Pollen grain length showed significant correlations with metabolite signals for shikimate, the 13C isotope of shikimate, sinapate, quinate, the 13C isotope of quinate as well as sedoheptulose-7-phosphate, ADP-glucose and PG(40:7). Lemma length correlated mainly with lipids, including phosphatidylcholines (Table 5). Since the correlation analysis applied here is only preliminary and these phenotypic traits are among the best markers to discriminate the three species, PLG in particular is likely genetically fixed (Table 2, Table S3), one has to consider chance: high, significant correlations found between metabolites and phenotypic traits might reflect their statistical value in separating the three species, rather than a direct association with PGL and LL. However, there is evidence that these results indicate a potentially strong link between PGL and lignin/phenolic biosynthesis, and between LL and lipid metabolism. Pollen grain length can be associated with phenolic composition as it provides structural support to the pollen grain, because mature angiosperm pollen grains are covered by three distinct layers of cell walls: (1) an outer exine coating, composed of a tough, chemically resistant biopolymer (sporopollenin), which is interrupted by openings called apertures; (2) an inner intine coating, made primarily of cellulose; and (3) a pollen coat, composed of lipids, proteins, pigments and aromatic compounds, that fills the sculpted cavities of the pollen exine wall (Edlund et al., 2004). Sporopollenin, which is present in the spore/pollen walls of all land plants, comprises both aliphatic (unsaturated lipids) and aromatic (phenolics) components and is regarded as one of the most recalcitrant biomacromolecules, providing protection against harsh terrestrial environments, including a range of abiotic stresses (de Leeuw et al., 2006; Fraser et al., 2012). Results therefore show that phenolic biosynthesis may be crucial for pollen grain development as phenolics provide structural components for both sporopollenin and pollen coat formation. An alternative explanation is that the levels of accumulation of these metabolites in the different species indicate that some biochemical pathways have or have not been switched on yet. A genome-wide SNP scan study to identify trait-regulatory genomic loci in chickpeas showed that upregulation of a superior gene haplotype correlated with increased transcript expression of the Ca_Kabuli_CesA3 gene in the pollen and pod of an accession with a high pod/seed number, resulting in higher cellulose accumulation for normal pollen and pollen tube growth (Kujur et al., 2015). Environmental triggers like temperature or drought might also influence the availability of metabolites for enzymatic systems regulating pollen grain and lemma length, but gaps in our knowledge of how constraints affect plant survival and seed production are being filled only slowly (Ejsmond et al., 2011).
A number of studies have corroborated the differentiation of the three species of the B. distachyon complex, like those based on seed protein data (Hammami et al., 2011), phenotypic and cytogenetic traits and nuclear and plastid phylogenetic markers (Catalán et al., 2012), nuclear single sequence repeats (SSRs) (Giraldo et al., 2012), DNA barcoding (López-Alvarez et al., 2012), isozymes (Jaaska, 2014) and comparative chromosome painting (Idziak et al., 2011; Betekhtin et al., 2014). This study shows that metabolomics can discriminate the three species. Furthermore, preliminary metabolite discriminant analysis suggests a closer metabolomic affinity of B. hybridum to its maternal B. stacei parent than to its paternal B. distachyon parent for the studied ecotypes (Fig. 7). The closeness of the allotetraploid hybrid to its B. stacei parent, supported by both phenotypic and metabolomic data (Figs 4 and 7) is also in agreement with whole-genome sequence (Vogel, Joint Genome Institute (USA), pers. comm.) and environmental data; environmental niche model analyses demonstrated a larger niche overlap and niche affinity of the B. hybridum niche to that of B. stacei than to the B. distachyon niche (López-Alvarez et al., 2015). Because phenotypic and metabolomic data reflect the summed effects of genotypic composition and environmentally mediated gene expression, larger genomic, phenomic and metabolomic analyses should be conducted on higher numbers of replicates and different ecotypes of the three species to identify the allelic variants and regulatory elements responsible for the observed phenotypic and metabolomics profiles at both species and ecotype level. The preliminary phenotypic and metabolomic analyses performed in this study have set the way for future, more exhaustive, GWAS studies.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Tables S1–S12 and Figures S1 and S2A: detailed description of phenotypic and metabolomics variables analysed in inter and intraspecific statistical studies of Brachypodium distachyon, B. stacei and B. hybridum populations.
Supplementary Material
ACKNOWLEDGEMENTS
We thank several colleagues (Table S1), germplasm and herbaria for providing us with B. distachyon, B. stacei and B. hybridum samples and information. The study was funded by two consecutive Spanish Ministry of Science grant projects (CGL2009-12955-C02-01, CGL2012-39953-C02-01) and one Aragon Government and European Social Fund Bioflora grant to P.C. and D.L.-A., and one European Plant Phenotyping Network (EPPN) BRACHY-DROUGHT grant to P.C. D.L.-A. was funded by a Spanish Ministry of Science and Innovation PhD FPI grant and by a Spanish Instituto de Estudios Altoaragoneses fellowship.
LITERATURE CITED
- Allwood JW, Ellis DI, Heald JK, Goodacre R, Mur LAJ.. 2006. Metabolomic approaches reveal that phosphatidic and phosphatidyl glycerol phospholipids are major discriminatory non-polar metabolites in responses by Brachypodium distachyon to challenge by Magnaporthe grisea. Plant Journal 46: 351–368. [DOI] [PubMed] [Google Scholar]
- Anderson TW. 1996. R.A. Fisher and multivariate analysis. Statistical Science 11: 20–34. [Google Scholar]
- Betekhtin A, Jenkins G, Hasterok R.. 2014. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS One 9: e115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretagnolle F, Lumaret R.. 1995. Bilateral polyploidization in Dactylis glomerata L. subsp. lusitanica: occurrence, morphological and genetic characteristics of first polyploids. Euphytica 84: 197–207. [Google Scholar]
- Catalán P, Muller J, Hasterok R, et al. 2012. Evolution and taxonomic split of the model grass Brachypodium distachyon. Annals of Botany 109: 385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán P, Chalhoub B, Chochois V, et al. 2014. Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI). Trends in Plant Science 19: 414–418. [DOI] [PubMed] [Google Scholar]
- Catalán P, López-Alvarez D, Bellosta C, Villar L.. 2016a. Updated taxonomic description, iconography and habitat preferences of Brachypodium distachyon, B. stacei and B. hybridum (Poacea). Anales Del Jardin Botanico De Madrid 73: e028. [Google Scholar]
- Catalán P, López-Alvarez D, Sancho R, López-Herranz ML, Díaz-Peréz A.. 2016b. Phylogeny, evolution and environmental niches of Brachypodium In: Vogel J, ed. Genetics and genomics of Brachypodium. Springer, New York: Series Plant Genetics and Genomics: Crops Models, 9–38. [Google Scholar]
- Draper J, Mur LAJ, Jenkins G, et al. 2001. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiology 127: 1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Draper J, Lloyd AJ, Goodacre R, Beckmann M.. 2013. Flow infusion electrospray ionisation mass spectrometry for high throughput, non-targeted metabolite fingerprinting: a review. Metabolomics 9: S4–S29. [Google Scholar]
- Edlund AF, Swanson R, Preuss D.. 2004. Pollen and stigma structure and function: the role of diversity in pollination. The Plant Cell 16: S84–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsmond MJ, Wronska-Pilarek D, Ejsmond A, et al. 2011. Does climate affect pollen morphology? Optimal size and shape of pollen grains under various desiccation intensity. Ecosphere 2: art117. doi:10.1890/ES11-00147.1. [Google Scholar]
- Enot DP, Lin W, Beckmann M, Parker D, Overy DP, Draper J.. 2008. Preprocessing, classification modeling and feature selection using flow injection electrospray mass spectrometry metabolite fingerprint data. Nature Protocols 3: 446–470. [DOI] [PubMed] [Google Scholar]
- Fiehn O. 2001. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative and Functional Genomics 2: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. 2002. Metabolomics – the link between genotypes and phenotypes. Plant Molecular Biology 48: 155–171. [PubMed] [Google Scholar]
- Filiz E, Ozdemir BS, Budak F, Vogel JP, Tuna M, Budak H.. 2009. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52: 876–890. [DOI] [PubMed] [Google Scholar]
- Fisher RA. 1936. The use of multiple measurements in taxonomic problems. Annals of Eugenics 7: 179–188. [Google Scholar]
- Fraser WT, Scott AC, Forbes AES, et al. 2012. Evolutionary stasis of sporopollenin biochemistry revealed by unaltered Pennsylvanian spores. New Phytologist 196: 397–401. [DOI] [PubMed] [Google Scholar]
- Garvin DF, Gu YQ, Hasterok R, et al. 2008. Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Science 48: S69–S84. [Google Scholar]
- Giraldo P, Rodriguez-Quijano M, Vazquez JF, Carrillo JM, Benavente E.. 2012. Validation of microsatellite markers for cytotype discrimination in the model grass Brachypodium distachyon. Genome 55: 523–527. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Priest H, Marais DLD, et al. 2014. Genome diversity in Brachypodium distachyon: deep sequencing of highly diverse inbred lines. The Plant Journal 79: 361–374. [DOI] [PubMed] [Google Scholar]
- Hall R, Beale M, Fiehn O, Hardy N, Sumner L, Bino R.. 2002. Plant metabolomics: the missing link in functional genomics strategies. The Plant Cell 14: 1437–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Jouve N, Cuadrado A, Soler C, Gonzalez JM.. 2011. Prolamin storage proteins and alloploidy in wild populations of the small grass Brachypodium distachyon (L.) P. Beauv. Plant Systematics and Evolution 297: 99–111. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD.. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4: 9. [Google Scholar]
- International Brachypodium Initiative. 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Idziak D, Betekhtin A, Wolny E, et al. 2011. Painting the chromosomes of Brachypodium – current status and future prospects. Chromosoma 120: 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaska V. 2014. Isozyme variation and differentiation of morphologically cryptic species in the Brachypodium distachyon complex. Biochemical Systematics and Ecology 56: 185–190. [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ.. 2003. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology 133: 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Miyake C, Kohchi T, Fujii S, Uchida M, Yokota A.. 2000. Responses of wild watermelon to drought stress: accumulation of an ArgE homologue and citrulline in leaves during water deficits. Plant and Cell Physiology 41: 864–873. [DOI] [PubMed] [Google Scholar]
- Kujur A, Bajaj D, Upadhyaya HD, et al. 2015. A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Scientific Reports 5: art11166. doi:10.1038/srep11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusvuran S, Dasgan HY, Abak K.. 2013. Citrulline is an important biochemical indicator in tolerance to saline and drought stresses in melon. Scientific World Journal 2013: 253414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw JW, Versteegh GJM, van Bergen PF.. 2006. Biomacromolecules of algae and plants and their fossil analogues. Plant Ecology 182: 209–233. [Google Scholar]
- Legendre P, Legendre L.. 1998. Numerical ecology. Amsterdam: Elsevier. [Google Scholar]
- López-Alvarez D, López-Herranz ML, Betekhtin A, Catalan P.. 2012. A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Alvarez D, Manzaneda AJ, Rey PJ, et al. 2015. Environmental niche variation and evolutionary diversification of the Brachypodium distachyon grass complex species in their native circumMediterranean range. American Journal of Botany 102: 1–16. [DOI] [PubMed] [Google Scholar]
- Lyons C, Scholthof K.. 2015. Watching grass grow: the emergence of Brachypodium distachyon as a model for the Poaceae. Archimedes 40: 479–501. [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T.. 2012. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytologist 193: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Anderson JT, Raskin E, Weiss-Lehman C, Mitchell-Olds T.. 2015. Natural variation, differentiation, and genetic trade-offs of ecophysiological traits in response to water limitation in Brachypodium distachyon and its descendent allotetraploid B. hybridum (Poaceae). Evolution 69: 2689–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T, Sandve SR, Heier L, et al. International Wheat Genome Sequencing Consortium. 2014. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Nakabayashi R, Yang Z, et al. 2015. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. The Plant Journal 81: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimberg H, Rice KJ, Milan NF, Njoku CC, McKay JK.. 2009. Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae). American Journal of Botany 96: 1262–1273. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Allainguillaume J, Catalan P, et al. 2011. Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytologist 191: 334–347. [DOI] [PubMed] [Google Scholar]
- Onda Y, Hashimoto K, Yoshida T, et al. 2015. Determination of growth stages and metabolic profiles in Brachypodium distachyon for comparison of developmental context with Triticeae crops. Proceedings of the Royal Society. B, Biological Sciences 282: 20150964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opanowicz M, Vain P, Draper J, Parker D, Doonan JH.. 2008. Brachypodium distachyon: making hay with a wild grass. Trends in Plant Science 13: 172–177. [DOI] [PubMed] [Google Scholar]
- Parker D, Beckmann M, Enot DP, et al. 2008. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nature Protocols 3: 435–445. [DOI] [PubMed] [Google Scholar]
- Parker D, Beckmann M, Zubair H, et al. 2009. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant Journal 59: 723–737. [DOI] [PubMed] [Google Scholar]
- Pasquet J-C, Chaouch S, Macadre C, et al. 2014. Differential gene expression and metabolomic analyses of Brachypodium distachyon infected by deoxynivalenol producing and non-producing strains of Fusarium graminearum. BMC Genomics 15: 629. doi:10.1186/1471-2164-15-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson IH. 1981. Chromosome numbers in Brachypodium Beauv. (Gramineae). Genetica 56: 55–60. [Google Scholar]
- Shi H, Ye T, Song B, Qi X, Chan Z.. 2015. Comparative physiological and metabolomic responses of four Brachypodium distachyon varieties contrasting in drought stress resistance. Acta Physiologiae Plantarum 37: 122. doi:10.1007/s11738-015-1873-0. [Google Scholar]
- Shiposha V, Catalán P, Olonova M, Marques I.. 2016. Genetic structure and diversity of the selfing model grass Brachypodium stacei (Poaceae) in Western Mediterranean: out of the Iberian Peninsula and into the islands. PeerJ 4: e2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal P, Horacek J, Dostalova R, Hybl M.. 2008. Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. Journal of Applied Genetics 49: 155–166. [DOI] [PubMed] [Google Scholar]
- Speckmann GJ, Post J Jr, Dijkstra H. 1965. The length of stomata as an indicator for polyploidy in rye-grasses. Euphytica 14: 225–230. [Google Scholar]
- Sumner LW, Amberg A, Barrett D, et al. 2007. Proposed minimum reporting standards for chemical analysis. Metabolomics 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GY, Dunn GM.. 1973. Relationship of stomatal length and frequency and pollen-grain diameter to ploidy level in Bromus inermis Leyss. Crop Science 13: 332–334. [Google Scholar]
- RDC Team. 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Toumi I, Gargouri M, Nouairi I, et al. 2008. Water stress induced changes in the leaf lipid composition of four grapevine genotypes with different drought tolerance. Biologia Plantarum 52: 161–164. [Google Scholar]
- Vigh L, Huitema H, Woltjes J, Vanhasselt PR.. 1986. Drought stress-induced changes in the composition and physical state of phospholipids in wheat. Physiologia Plantarum 67: 92–96. [Google Scholar]
- Vogel JP, Tuna M, Budak H, Huo NX, Gu YQ, Steinwand MA.. 2009. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biology 9: 88. doi:10.1186/1471-2229-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.