In this study, Ji et al. identify SIAH1/2 (SIAH) as the E3 ligase mediating Wnt-induced Axin degradation. Their results suggest that Wnt-induced dissociation of the Axin/GSK3 complex allows SIAH to interact with Axin and promote its degradation, which represents an important feed-forward mechanism to achieve sustained Wnt/β-catenin signaling.
Keywords: Axin, SIAH, Wnt
Abstract
The Wnt/β-catenin signaling pathway plays essential roles in embryonic development and adult tissue homeostasis. Axin is a concentration-limiting factor responsible for the formation of the β-catenin destruction complex. Wnt signaling itself promotes the degradation of Axin. However, the underlying molecular mechanism and biological relevance of this targeting of Axin have not been elucidated. Here, we identify SIAH1/2 (SIAH) as the E3 ligase mediating Wnt-induced Axin degradation. SIAH proteins promote the ubiquitination and proteasomal degradation of Axin through interacting with a VxP motif in the GSK3-binding domain of Axin, and this function of SIAH is counteracted by GSK3 binding to Axin. Structural analysis reveals that the Axin segment responsible for SIAH binding is also involved in GSK3 binding but adopts distinct conformations in Axin/SIAH and Axin/GSK3 complexes. Knockout of SIAH1 blocks Wnt-induced Axin ubiquitination and attenuates Wnt-induced β-catenin stabilization. Our data suggest that Wnt-induced dissociation of the Axin/GSK3 complex allows SIAH to interact with Axin not associated with GSK3 and promote its degradation and that SIAH-mediated Axin degradation represents an important feed-forward mechanism to achieve sustained Wnt/β-catenin signaling.
The Wnt/β-catenin signaling pathway plays essential roles during embryonic development and tissue homeostasis through controlling the stability of the transcription cofactor β-catenin (Logan and Nusse 2004; Clevers 2006; MacDonald et al. 2009; Loh et al. 2016). In the absence of Wnt ligands, β-catenin is associated with the β-catenin destruction complex (which contains β-catenin, Axin, GSK3, and APC), phosphorylated by GSK3, and degraded by the ubiquitin proteasome pathway. In a Frizzled- and Dishevelled-dependent manner, Wnt induces phosphorylation of its coreceptor, LRP6, and recruits the Axin-containing β-catenin destruction complex to the plasma membrane to form the signalosome. Within the signalosome, GSK3 is inhibited by phospho-LRP6, which leads to the destabilization of the β-catenin destruction complex and accumulation of β-catenin. Various mechanisms are evolved to control Wnt/β-catenin signaling output, and deregulation of the Wnt pathway is associated with many diseases, including cancer.
Through direct interaction with β-catenin, GSK3, and APC, the scaffolding protein Axin is responsible for the formation of the β-catenin destruction complex. The physical interaction between Axin and GSK3 is regulated. Association between Axin and GSK3 requires the GSK3 kinase activity (Ikeda et al. 1998). Wnt-induced phospho-LRP6 directly inhibits the kinase activity of GSK3 bound to Axin (Cselenyi et al. 2008; Piao et al. 2008; Wu et al. 2009), which might be responsible for decreased Axin–GSK3 association upon Wnt treatment (Liu et al. 2005). The Axin–GSK3 interaction is also regulated by phosphorylation; CK1 and PP1 can reciprocally regulate the interaction between Axin and GSK3 (Luo et al. 2007). Importantly, Axin is a concentration-limiting factor for the assembly of the β-catenin destruction complex (Lee et al. 2003), so mechanisms that control the protein level of Axin are expected to have major impacts on Wnt/β-catenin signaling. Indeed, stabilization of Axin using TNKS inhibitor strongly inhibits Wnt/β-catenin signaling (Chen et al. 2009; Huang et al. 2009). It has been known for many years that activation of Wnt or inhibition of GSK3 promotes degradation of Axin (Willert et al. 1999; Yamamoto et al. 1999). Wnt-induced stabilization of β-catenin precedes Wnt-induced Axin degradation (Willert et al. 1999; Yamamoto et al. 1999; Liu et al. 2005), so Axin degradation is not required for the initial Wnt-induced β-catenin stabilization. Despite the strong interest in studying Axin regulation, the molecular mechanism and biological relevance of Wnt-induced Axin degradation have been elusive.
SIAH1 and SIAH2 are highly homologous RING domain E3 ligases that often target common proteins for degradation (House et al. 2009; Qi et al. 2013). There are two SIAH homologs in humans (SIAH1 and SIAH2) and three in mice (Siah1a, Siah1b, and Siah2) (Carthew and Rubin 1990; Della et al. 1993; Hu et al. 1997). SIAH proteins contain an N-terminal RING domain and a C-terminal substrate-binding domain (SBD) (Hu et al. 1997; House et al. 2003). SIAH proteins are involved in ubiquitination of diverse substrates and are implicated in many biological processes such as RAS signaling (Nadeau et al. 2007), DNA damage (Winter et al. 2008), hypoxia (Nakayama et al. 2004, 2009), p38/JNK/NFкB pathways (Habelhah et al. 2002; Xu et al. 2006), and transcription (Zhang et al. 1998; Frasor et al. 2005).
Here, we identify SIAH as the E3 ligase mediating Wnt-induced ubiquitination and degradation of Axin. SIAH interacts with a VxP motif in the GSK3-binding domain of Axin. Importantly, this binding is counteracted by GSK3 interaction, which is explained by crystal structure analysis. Our work provides mechanistic insights into Wnt-induced Axin degradation and reveals SIAH-dependent degradation of Axin as a powerful feed-forward mechanism to maintain Wnt/β-catenin signaling.
Results
Ubiquitin E3 ligases SIAH1/2 are positive regulators of Wnt signaling
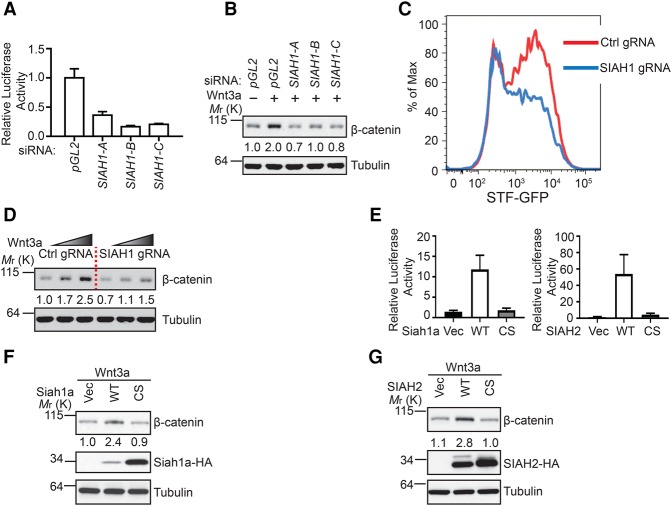
In order to identify novel regulators of the Wnt/β-catenin signaling pathway, we carried out a genome-wide siRNA screen using Wnt3a-induced Super TopFlash (STF) luciferase reporter assay in HEK293 cells. In this screen, SIAH1 scored as a strong hit, and multiple siRNAs against SIAH1 inhibited the STF reporter. We validated the screening results by showing that independent siRNAs against SIAH1 decreased Wnt3a-induced STF reporter and Wnt3a-induced accumulation of cytosolic β-catenin in HEK293 cells (Fig. 1A,B; Supplemental Fig. S1A). Similar findings were made in YAPC cells (Supplemental Fig. S1B). We further validated these results using CRISPR/Cas9-based loss-of-function experiments (Hsu et al. 2014). HEK293 STF-GFP Cas9 cells were infected with virus encoding guide RNA (gRNA) targeting SIAH1, and a pool of SIAH1 knockout cells was used for the further studies to preclude clonal variation. We found that knockout of SIAH1 reduced Wnt3a-induced STF-GFP reporter (Fig. 1C) and attenuated Wnt3a-induced accumulation of cytosolic β-catenin (Fig. 1D). Together, these results demonstrate that inhibition of SIAH1 inhibits Wnt/β-catenin signaling.
Figure 1.
SIAH proteins positively regulate Wnt/β-catenin signaling. (A) Depletion of SIAH1 by independent siRNAs inhibits Wnt3a-induced STF reporter in HEK293 cells. pGL2 nontargeting siRNA served as a negative control. Error bars denote the SD between four replicates. (B) Depletion of SIAH1 by siRNA attenuates Wnt3a-induced cytosolic β-catenin accumulation. The intensity of the β-catenin band was quantified by ImageJ. (C) Knockout of SIAH1 by CRISPR attenuates Wnt3a-induced STF-GFP reporter in HEK293 cells. The FACS plot is a representative from three independent experiments. (D) Knockout of SIAH1 by CRISPR attenuates Wnt3a-induced cytosolic β-catenin accumulation. (E) Overexpression of wild-type Siah1a and SIAH2, but not their RING domain mutants, enhances Wnt3a-induced STF reporter in HEK293 cells. Error bars denote the SD between four replicates. (F,G) Overexpression of wild-type Siah1a or SIAH2 enhances Wnt3a-induced accumulation of cytosolic β-catenin.
In order to identify novel regulators of Wnt signaling, we also performed a large-scale cDNA overexpression screen in HEK293 cells using STF luciferase reporter. SIAH2, a close homolog of SIAH1, scored as a strong positive regulator of Wnt/β-catenin signaling. To follow this observation, we performed a STF luciferase reporter assay by cotransfecting HEK293 cells with STF reporter together with plasmids encoding mouse Siah1a, human SIAH2, or their RING domain mutants (CS mutant, Siah1a C41 and 44S, and SIAH2 C80 and 83S). Overexpression of wild-type Siah1a or SIAH2, but not their RING domain mutants, promoted the STF reporter (Fig. 1E) and enhanced Wnt3a-induced cytosolic β-catenin accumulation (Fig. 1F,G). In these overexpression experiments, SIAH2 showed stronger effects on Wnt signaling than Siah1a, which is likely related to the higher expression levels of SIAH2 proteins. Together with loss-of-function studies, these results suggest that SIAH1 and SIAH2 positively regulate Wnt signaling through promoting accumulation of β-catenin.
Ubiquitin E3 ligases SIAH1/2 regulate Axin stability
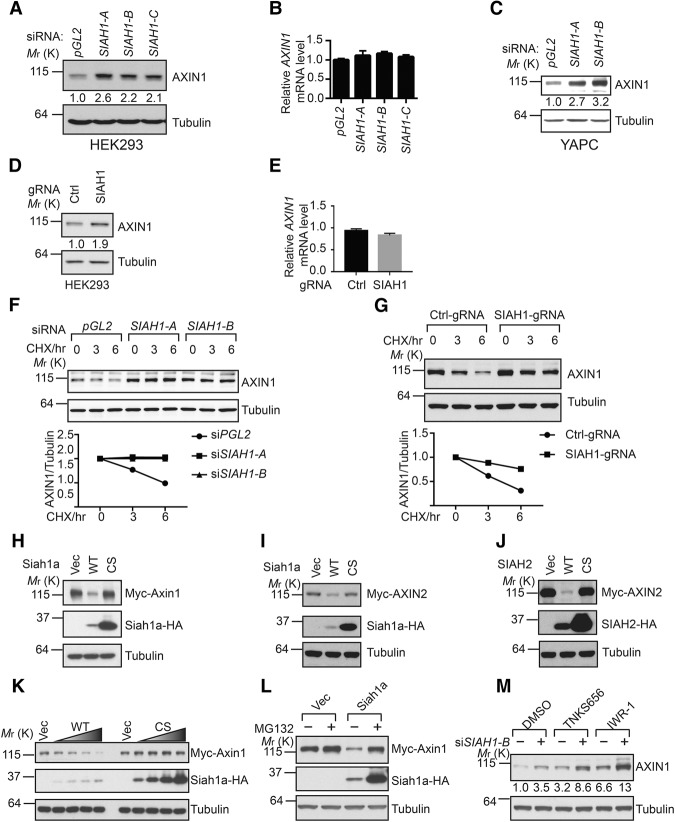
To identify novel binding partners of Axin, we immunoprecipitated AXIN2 from SW480 cells and performed mass spectrometry analysis. In this experiment, both SIAH1 and SIAH2 scored as top hits, suggesting a potential functional link between SIAH1/2 and Axin. To follow this observation, we tested whether SIAH1/2 expression controls Axin protein levels. We found that independent siRNAs against SIAH1 increased the protein level, but not the mRNA level, of AXIN1 in HEK293 cells (Fig. 2A,B). Similar findings were also made in YAPC and U2OS cells (Fig. 2C; Supplemental Fig. S2A,B). Knockout of SIAH1 by CRISPR also increased the protein level, but not mRNA level, of AXIN1 (Fig. 2D,E). We next tested whether inhibition of SIAH1 affects the protein stability of AXIN1 by blocking de novo protein synthesis with cycloheximide (CHX). As seen in Figure 2, F and G, depletion of SIAH1 using siRNA or knockout of SIAH1 using CRISPR increased the protein stability of AXIN1.
Figure 2.
SIAH1/2 regulate the stability of Axin proteins. (A,B) Depletion of SIAH1 increases the protein level, but not the mRNA level, of AXIN1 in HEK293 cells. Error bars denote the SD between quadruplicates. (C) Depletion SIAH1 increases the protein level of AXIN1 in YAPC cells. (D,E) Knockout of SIAH1 using CRISPR increases the protein level, but not mRNA level, of AXIN1 in HEK293 cells. (F,G) Inhibition of SIAH1 by siRNA or CRSIPR increases the protein stability of AXIN1 under CHX treatment. Quantification of normalized AXIN1 band intensity is shown in the bottom panel. (H–J) Overexpression of wild-type Siah1a and SIAH2, but not their RING domain mutants, promotes degradation of coexpressed Axin. (K) Overexpression of Siah1a induces degradation of coexpressed Axin1 in a dose-dependent manner. (L) Proteasome inhibitor MG132 blocks Siah1a-induced degradation of Axin1. (M) Depletion of SIAH1 further enhances the protein level of AXIN1 in cells treated with tankyrase inhibitors.
We next investigated whether overexpression of SIAH proteins affects the stability of coexpressed Axin. Overexpression of wild-type Siah1a or SIAH2, but not their RING domain mutants (CS mutants), decreased expression of coexpressed Axin1 and AXIN2 (Fig. 2H–J). The effect of overexpressed Siah1a on Axin1 protein levels is dose-dependent (Fig. 2K). Notably, the proteasome inhibitor MG132 rescued the effect of Siah1a on Axin1, suggesting that Siah1a promotes proteasomal degradation of Axin1 (Fig. 2L). Combining both loss-of-function and gain-of-function data, these results strongly suggest that SIAH1 and SIAH2 promote proteasomal degradation of Axin.
Tankyrase PARsylates Axin, and PARsylated Axin is recognized by E3 ligase RNF146, causing the PARsylated form to be targeted for proteasomal degradation (Huang et al. 2009; Zhang et al. 2011). We sought to understand whether tankyrase activity is necessary for SIAH-induced Axin degradation. Tanyrase inhibitors TNKS656 (Shultz et al. 2013) and IWR-1 (Chen et al. 2009) increased the protein expression of AXIN1 (Fig. 2M). Interestingly, both tankyrase inhibitors further increased the protein levels of AXIN1 in cells treated with SIAH1 siRNA (Fig. 2M). These results suggest that tankyase/RNF146 and SIAH1/2 represent independent mechanisms that promote Axin degradation.
SIAH1/2 bind to the GSK3-binding domain of Axin and promote its ubiquitination
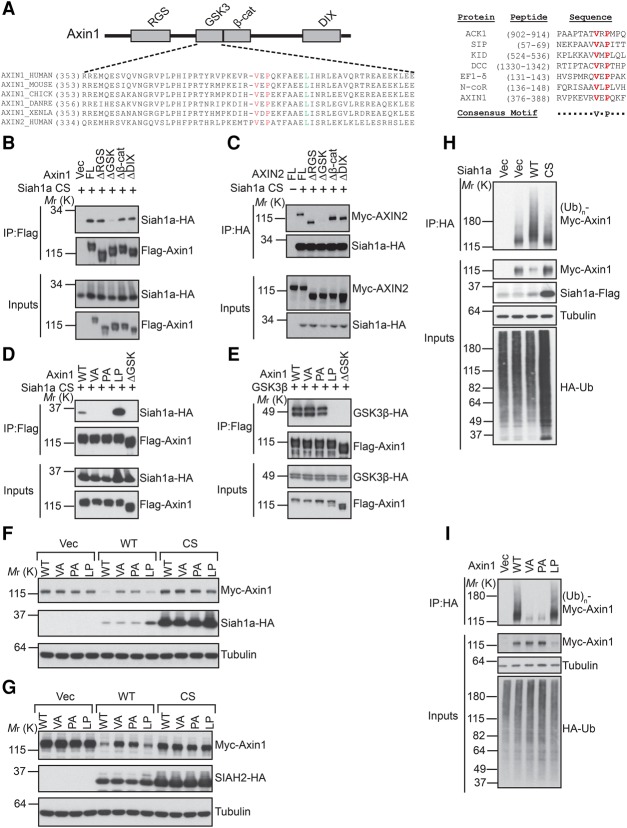
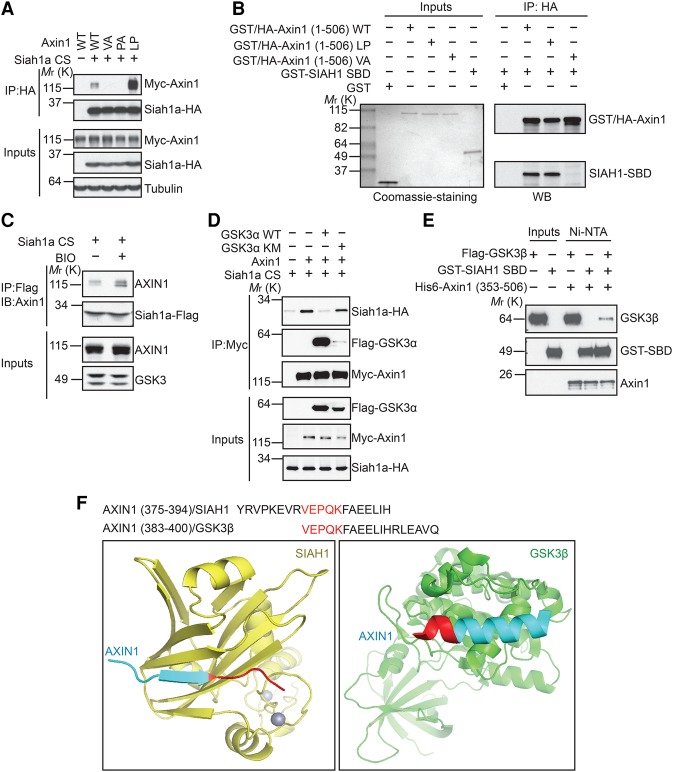
SIAH1 and SIAH2 were detected as top hits of mass spectrometry analysis of AXIN2 immunoprecipitates. We confirmed that Siah1a and SIAH2 interact with Axin1 and AXIN2 in coimmunoprecipitation assays (Fig. 3B,C; Supplemental Fig. S3). Previous studies have identified several critical domains of Axin that allow Axin to interact with its different binding partners (Fig. 3A, top panel). We next sought to determine the domain of Axin that is required for interaction with SIAH. As seen in Figure 3, B and C, deletion of the GSK3-binding domain, but not other domains, of Axin abolished the interaction between Siah1a and Axin (Fig. 3B,C). These results suggest that the GSK3-binding domain of Axin mediates the interaction with SIAH. SIAH proteins have an N-terminal RING domain and a C-terminal SBD. Structural–functional analysis of various SIAH substrates has identified a consensus SIAH-binding motif (VxP) among SIAH substrates (House et al. 2003). Close examination of the GSK3-binding domain of Axin reveals a potential SIAH substrate-binding motif (VxP), which is highly conserved among Axin proteins from different species (Fig. 3A, bottom panel and right panel). To evaluate the importance of this potential SIAH-binding motif in Axin, we mutated Val383 or Pro385 of Axin1 to Ala and tested these mutants in functional assays. Axin1 VA and PA mutations abolished Siah1a inhibition without affecting GSK3β interaction (Fig. 3D,E). As a control, L392P mutation of Axin1, which is known to disrupt Axin–GSK3 interaction (Smalley et al. 1999; Dajani et al. 2003), abolished Axin1–GSK3β interaction without affecting the Axin1–Siah1a interaction (Fig. 3D,E). These results suggest that even though the same general region of Axin binds to both SIAH and GSK3, different key residues of Axin mediate these interactions.
Figure 3.
SIAH1/2 interact with the GSK3-binding domain of Axin and promote its ubiquitination. (A, left panel) A schematic diagram of the domain structure of Axin with the alignment of the GSK3-binding domain of Axin proteins from different species. Val and Pro residues involved in SIAH interaction are highlighted in red, and the Leu residue critical for GSK3 interaction is highlighted in green. (Right panel) Alignment of SIAH1-binding motifs of various SIAH1 substrates. (B,C) The GSK3-binding domain of Axin is required for the interaction with Siah1a. Flag-Axin1 or Myc-AXIN2 was coexpressed with the HA-Siah1a CS mutant in HEK293 cells and subjected to coimmunoprecipitation assay. (D,E) V383A or P385A mutation of Axin1 disrupts Axin1–Siah1a interaction without affecting Axin1–GSK3β interaction. (F,G) Mutation of the SIAH-binding motif attenuates Siah1a-induced Axin1 degradation. (H) Overexpression of wild-type Siah1a, but not its RING domain mutant, promotes Axin1 ubiquitination in vivo. (I) Mutation of the SIAH-binding motif blocks Siah1a-induced ubiquitination of Axin1.
Since overexpression of wild-type SIAH1/2 triggers degradation of Axin, we sought to understand whether SIAH-binding-deficient mutants of Axin are still sensitive to SIAH-induced degradation. Different mutants of Axin1 were coexpressed with SIAH1/2 in HEK293 cells. As seen in Figure 3, F and G, both VA and PA mutations attenuated SIAH1/2 overexpression-induced degradation of Axin1. These results suggest that the SIAH–Axin interaction is required for SIAH-induced degradation of Axin.
Proteasome-dependent degradation is often induced by polyubiquitination. To determine whether SIAH facilitates Axin ubiquitination, we performed an in vivo ubiquitination assay by coexpressing HA-ubiquitin and Myc-Axin1 with wild-type or RING domain mutated Siah1a. Overexpression of wild-type Siah1a, but not its RING domain mutant, increased polyubiquitination of Axin1 (Fig. 3H). Importantly, Axin1 VA and PA mutants are resistant to Siah1a overexpression-induced ubiquitination (Fig. 3I). Taken together, these results suggest that SIAH proteins promote ubiquitination and proteasome-dependent degradation of Axin through interacting with a VxP motif in the GSK3-binding domain of Axin.
Crystal structure of an AXIN1/SIAH1 complex
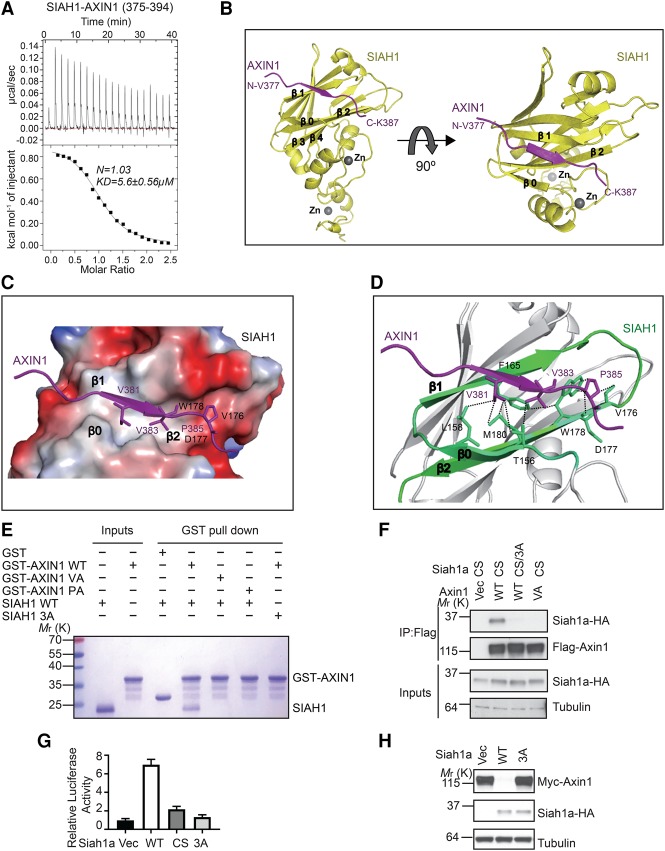
Our isothermal titration calorimetry (ITC) analysis demonstrated that SIAH1-SBD can directly interact with a 20-residue human AXIN1 peptide (residues 375–394) that contains the VxP motif, with a Kd of 5.6 µM (Fig. 4A). To understand the structural basis of the SIAH–Axin interaction, we determined the crystal structure, at 2.1 Å resolution, of the SBD of human SIAH1 (residues 93–282) bound to this AXIN1 peptide that contains the VxP motif (Fig. 4B; Supplemental Table S1; Supplemental Fig. S4A). In the structure, the AXIN1 peptide (residues 377–387) formed a short β strand that tightly packed against the SIAH1 β0 (154–159), β1 (162–169), and β2 (175–182) sheets, largely through hydrophobic interactions (Fig. 4C; Supplemental Fig. S4B). The AXIN1 hydrophobic residues V381, V383, and P385 interacted with SIAH1 residues T156 and L158 in the β0 sheet; V164, F165, and T168 in the β1 sheet; and V176, D177, W178, and M180 in the β2 sheet, respectively (Fig. 4D). This extended β-sheet-binding mode of AXIN1 is used similarly by other SIAH ligands (House et al. 2006).
Figure 4.
Crystal structure of AXIN1 in complex with SIAH1. (A) ITC analysis of the interaction between SIAH1-SBD and AXIN1 peptide. (B) The overall structure of the AXIN1/SIAH1 complex. The sequence of the observable part of AXIN1 in the structure is 377VPKEVRVEPQK387. (C) The surface electrostatic view between AXIN1 (377–387) and SIAH1 β0, β1, and β2 sheets is shown. (D) Major interactions between AXIN1 (377–387) and SIAH1. (E) GST pull-down analysis of the interaction between SIAH1 mutants and GST-tagged AXIN1. (F) The Siah1a 3A mutant (L176A/D177A/W178A) blocks the interaction with Axin1 in the coimmunoprecipitation assay. (G) Overexpression of Siah1a, but not the 3A mutant, enhances Wnt3a-induced STF reporter. Error bars denote the SD between four replicates. (H) Overexpression of Siah1a, but not the 3A mutant, promotes degradation of Axin1.
We next mutated the key residues in AXIN1 377–387 to alanine and investigated their interactions with SIAH1-SBD. The in vitro GST pull-down results show that the AXIN1 P385A mutant and V383A mutant completely abolished the interaction between AXIN1 and SIAH1 (Fig. 4E), which is consistent with coimmunoprecipitation data (Fig. 3D). In our structure, SIAH1 V176, D177, and W178 form the binding site for P385 of AXIN1 (Fig. 4C,D). Consistently, the SIAH1 V176A/D177A/W178A mutant abolished the interaction with AXIN1 in our GST pull-down assay (Fig. 4E). Furthermore, the corresponding Siah1a mutant did not interact with Axin1 in a coimmunoprecipitation assay (Fig. 4F) and did not promote Wnt signaling (Fig. 4G) or enhance Axin1 degradation (Fig. 4H) upon overexpression. Taken together, our crystal structure and mutagenesis analysis clearly demonstrate that the VxP motif in AXIN1 displayed in the extended conformation is critical for the AXIN1–SIAH1 interaction and that this interaction is critical for the Wnt-promoting activity of SIAH.
SIAH1/2 compete with GSK3 over the GSK3-binding domain of Axin
The finding that both SIAH1/2 and GSK3 bind to the same domain of Axin raises the question of whether these two bindings are mutually exclusive. Interestingly, the Axin1 LP mutant, which does not bind to GSK3, showed stronger binding to Siah1a as compared with wild-type Axin1 in the coimmunoprecipitation assay (Figs. 3D, 5A). Notably, enhanced interaction between the Axin1 LP mutant and SIAH1 was observed only in the coimmunoprecipitation assay. In an in vitro binding assay using purified recombinant proteins, wild-type Axin1 and the Axin1 LP mutant demonstrated similar binding to the SBD of SIAH1, while the Axin1 VA mutant lost its binding to SIAH1-SBD as expected (Fig. 5B). The discrepancy between coimmunoprecipitation data and in vitro binding data hints at the possibility that endogenous GSK3 competes with SIAH1 for Axin binding. It has been shown previously that purified wild-type GSK3, but not its catalytic inactive mutant, interacts with Axin in an in vitro binding assay in the absence of ATP (Ikeda et al. 1998), which strongly suggests that only enzymatically active GSK3 can bind to Axin. Consistent with the hypothesis that the endogenous active form of GSK3 inhibits SIAH–Axin interactions, treatment of cells with GSK3 inhibitor BIO enhanced the interaction between AXIN1 and the Siah1a CS mutant (Fig. 5C). In addition, overexpression of wild-type GSK3α blocked the interaction between Siah1a and Axin1 (Fig. 5D). As a control, the catalytic inactive mutant of GSK3α (KM mutant) did not bind to Axin1 and did not affect the interaction between Siah1a and Axin1 (Fig. 5D). Consistently, in an in vitro binding assay using purified recombinant proteins, GST-SIAH1-SBD inhibited the interaction between the Flag-GSK3β and His6-AXIN1 fragments containing the GSK3-binding domain and the β-catenin-binding domain (residues 353–506) (Fig. 5E). Together, these results suggest that GSK3 and SIAH1 compete with each other for interaction with Axin.
Figure 5.
GSK3 competes with SIAH1 for Axin binding. (A) The Axin1 L392P mutant shows stronger binding to Siah1a than wild-type Axin1 in the coimmunoprecipitation assay. (B) Wild-type Axin1 and Axin1 L392P have similar binding to the SBD of SIAH1 in an in vitro pull-down assay. GST-HA-Axin1 and GST-SIAH1-SBD were mixed and subjected to immunoprecipitation with anti-HA antibody. (C) GSK3 inhibitor BIO increases the interaction between Axin1 and the Siah1a CS mutant. Cells were treated with MG132 to prevent Axin degradation. (D) Wild-type GSK3α, but not the kinase-dead mutant (K148M), competes with the Siah1a CS mutant for Axin1 binding. (E) The interaction between His6-Axin1 (353–506) and Flag-GSK3β is inhibited by GST-SIAH1-SBD. The indicated proteins were mixed, pulled down using nickel-charged affinity resin, and detected by Western blot. (F) Comparison of the Axin1/Siah1 and Axin1/GSK3β structures.
Our finding that GSK3 and SIAH compete for Axin binding can be better understood by referring to the crystal structures of the AXIN1/SIAH1 and AXIN1/GSK3β complexes (Fig. 5F). The AXIN1 VxP motif adopts distinct conformations in the AXIN1/SIAH1 and AXIN1/GSK3β complexes (Fig. 5F). In the AXIN1/GSK3β complex, the GSK3-binding domain consists of a single α helix (AXIN1 residues 384–399) that packs tightly against the C lobe of GSK3β (Protein Data Bank [PDB] 1O9U) (Dajani et al. 2003). Residues in this helix cover the C-terminal half of the VxP motif, which adopts an extended β-sheet conformation in our AXIN1/SIAH1 complex structure. Thus, the comparison of these two crystal structures demonstrates how SIAH and GSK3 compete for Axin binding (Fig. 5F). In addition, since the GSK3-binding site and SIAH-binding site are right next to each other, competition between GSK3 and SIAH could also result from a steric clash between these two proteins.
SIAH1 is required for Wnt-induced ubiquitination and degradation of Axin
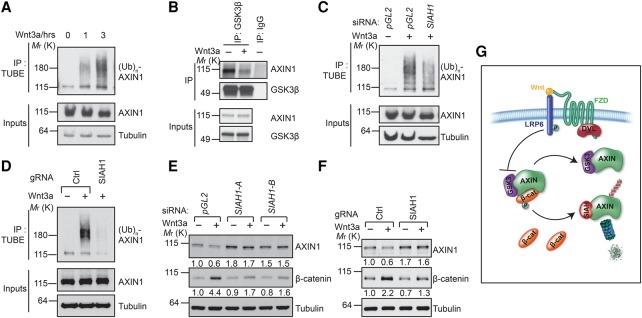
It is well known that Wnt signaling induces proteasome-dependent degradation of Axin (Willert et al. 1999; Yamamoto et al. 1999). However, the underlying molecular mechanism of this signaling event has remained mysterious for many years. Whether Wnt actually induces ubiquitination of Axin is also unknown. Using a tandem repeated ubiquitin-binding entity (TUBE) assay (Hjerpe et al. 2009; Lopitz-Otsoa et al. 2012), we found that Wnt3a strongly induced ubiquitination of endogenous AXIN1 in a time-dependent manner in HEK293 cells (Fig. 6A).
Figure 6.
Inhibition of SIAH1 blocks Wnt-induced Axin ubiquitination and degradation. (A) Wnt3a induces ubiquitination of endogenous AXIN1 in a time-dependent manner. HEK293 cells were pretreated with MG132 to prevent Wnt-induced AXIN1 degradation. (B) Wnt3a decreases the interaction between AXIN1 and GSK3β. Cells were pretreated with MG132 to prevent Wnt-induced Axin degradation and then treated with Wnt3a for 4 h. (C,D) Depletion or knockout of SIAH1 inhibits Wnt3a-induced ubiquitination of AXIN1. (E,F) Depletion or knockout of SIAH1 attenuates Wnt3a-induced AXIN1 degradation and cytosolic β-catenin accumulation. (G) A schematic model of Wnt-induced degradation of Axin through SIAH.
Since Wnt-induced phospho-LRP6 directly binds and inhibits GSK3 (Cselenyi et al. 2008; Piao et al. 2008; Wu et al. 2009) and since only GSK3 in the active conformation can bind to Axin (Ikeda et al. 1998), it is expected that Wnt signaling reduces the physical interaction between GSK3 and Axin. Indeed, a previous report had shown that Wnt signaling decreases the association between Axin and GSK3 (Liu et al. 2005). Consistent with this report, we observed that Wnt treatment led to a modest but consistent decrease of Axin–GSK3 interaction (Fig. 6B; Supplemental Fig. S5). Our finding that SIAH1 can bind only to Axin that is not bound to GSK3 suggests that SIAH1 may play a key role in Wnt-induced Axin ubiquitination and degradation. Indeed, depletion of SIAH1 using siRNA or knockout of SIAH1 using CRISPR inhibited Wnt3a-induced AXIN1 ubiquitination (Fig. 6C,D). Furthermore, depletion of SIAH1 using siRNA or knockout SIAH1 using CRISPR blocked Wnt3a-induced degradation of AXIN1 and attenuated Wnt3a-induced accumulation of β-catenin (Fig. 6E,F). These results suggest that SIAH1 is responsible for Wnt-induced Axin degradation. Taken together, our results suggest a model of Wnt-induced Axin degradation (Fig. 6G). Without Wnt treatment, Axin is part of the β-catenin destruction complex through association with GSK3, APC, and β-catenin. Axin is relatively stable in this complex because GSK3 binding prevents Axin–SIAH association. Wnt stimulation inhibits the kinase activity of Axin-associated GSK3, decreases Axin–GSK3 association, and increases the pool of Axin not associated with GSK3. This allows SIAH to bind to this pool of Axin and promote its ubiquitination and degradation, which enables cells to achieve sustained Wnt/β-catenin signaling.
Discussion
Both negative and positive feedback loops are critical for precise control of cell signaling during development (Freeman 2000). While negative feedback generates stability, positive feedback amplifies the initial small changes and ensures the robustness of a signaling system. Although many negative feedback loops of Wnt signaling have been well described, such as transcriptional activation of negative Wnt regulators AXIN2 (Jho et al. 2002), ZNRF3/RNF43 (Hao et al. 2012; Koo et al. 2012), and NOTUM (Giraldez et al. 2002; Kakugawa et al. 2015; Zhang et al. 2015), it remains a mystery how Wnt signaling is maintained during embryonic development and regeneration processes via counteracting positive feedback. Wnt-induced Axin degradation is well known, but the underlying molecular mechanism of this potential positive feedback loop has been enigmatic. Using genetic and proteomic approaches, we identified SIAH proteins as the key mediators of Wnt-induced Axin degradation. We demonstrated that a consensus VxP motif located in the GSK3-binding domain of Axin is critical for the interaction between SIAH and Axin and that mutating either component of the Axin–SIAH interface attenuates SIAH-dependent degradation of Axin. Thus, Wnt- and SIAH-mediated degradation of Axin can serve as a positive feedback mechanism and enable cells to achieve sustained Wnt/β-catenin signaling. This mechanism may enable degradation of AXIN1 dissociated from the β-catenin destruction complex in response to Wnt signal and likely AXIN2 proteins synthesized by nuclear β-catenin/Tcf complexes upon activation of the Wnt/β-catenin pathway.
Our data are consistent with a model in which Wnt induces Axin degradation by affecting the GSK3–Axin interaction. Our structural and biochemical analysis demonstrates that SIAH and GSK3 bind to the same region of Axin in a mutually exclusive manner. In the absence of Wnt signal, Axin is bound to GSK3 and several other proteins to form the β-catenin destruction complex, and SIAH is excluded from this complex (Fig. 6G). Wnt-induced phospho-LRP6 can directly bind and inhibit Axin-bound GSK3 (Cselenyi et al. 2008; Piao et al. 2008; Wu et al. 2009). The previous and current observation that Wnt decreases the Axin–GSK3 interaction might be explained by Axin's preference for interacting with active GSK3 (Ikeda et al. 1998). The separation of Axin and GSK3 upon Wnt stimulation allows SIAH to interact with Axin, leading to its ubiquitination and proteasomal degradation. It should be noted that Drosophila Axin is degraded upon Wnt signaling (Tolwinski et al. 2003), although it does not have a VxP motif. It is possible that another E3 ligase mediates Wnt-induced Axin degradation in Drosophila.
It has been shown previously that Axin is a concentration-limiting factor of the β-catenin destruction complex in Xenopus oocyte extracts (Lee et al. 2003), although another study had suggested that Axin may not be limiting in mammalian cells (Tan et al. 2012). Nevertheless, increasing the concentration of Axin through blocking either tankyrase/RNF146 or SIAH strongly inhibits Wnt signaling in mammalian cells. Tankyrase/RNF146 and SIAH represent independent mechanisms that control Axin stability. Tankyrase binds to the N terminus of Axin and promotes its PARsylation, and PARsylated Axin is degraded by RNF146 (Huang et al. 2009; Zhang et al. 2011; Wang et al. 2012; DaRosa et al. 2015). Tankyrase/RNF146 can potentially target Axin residing in the β-catenin destruction complex. On the contrary, SIAH can bind only to Axin not associated with GSK3, and its activity on Axin is controlled by Wnt. Beyond tankyrase/RNF146 and SIAH, there could be other mechanisms that control Axin stability, which need to be further investigated in the future.
Several previous studies have suggested that SIAH1 represses Wnt signaling by targeting β-catenin for degradation through a phosphorylation-independent mechanism (Liu et al. 2001; Matsuzawa and Reed 2001; Dimitrova et al. 2010; Jumpertz et al. 2014; Shin et al. 2016), which is inconsistent with our conclusion. Underlying causes of discrepancies are currently unclear. It is possible that cell types and experimental conditions confound results. In our hands, overexpression of either SIAH1 or SIAH2 using the minimal amount of SIAH1/2 expression plasmids enhances Wnt/β-catenin signaling and promotes ubiquitination and degradation of Axin in a manner that is dependent on the SIAH–Axin interaction. These results are further supported by RNAi and CRSIPR-based loss-of-function experiments. Through combining loss-of-function, gain-of-function, crystal structure, and mutagenesis experiments, we defined SIAH1 as a positive regulator of Wnt signaling.
SIAH1 and SIAH2 are highly homologous (85% identity), and mouse genetic studies suggest that they have overlapping functions in vivo (Dickins et al. 2002; Frew et al. 2003). We found that both SIAH1 and SIAH2 interact with Axin, and overexpression of either of them promotes ubiquitination and degradation of Axin. In cell lines used in this study, SIAH1 appears to play a dominant role over SIAH2 in regulating Wnt signaling. However, it is possible that SIAH2 also regulates Wnt signaling in certain cell types and biological conditions. SIAH proteins promote ubiquitination and proteasomal degradation of diverse substrates and are implicated in many biological processes. Simple gain-of-function or loss-of-function experiments to study the connection between SIAH proteins and Wnt signaling in model organisms would be difficult since Wnt-related phenotypes are likely obscured by the effect of SIAH on other proteins and biological processes. Nevertheless, the bone phenotype of Siah1a knockout mice is consistent with a positive role of SIAH in Wnt signaling (Frew et al. 2004). The Wnt/β-catenin signaling pathway plays a dominant role in promoting bone formation (Baron and Kneissel 2013). Interestingly, Siah1a knockout mice have striking low bone volume (Frew et al. 2004). Whether this phenotype is associated with decreased Wnt/β-catenin signaling should be examined in future studies.
Accumulating evidence suggests a tumor-promoting function of SIAH proteins (House et al. 2009; Qi et al. 2013). SIAH proteins are overexpressed in various tumors, and depletion of SIAH suppresses tumor growth. Generation of SIAH inhibitors is being explored for treatment of cancer (Stebbins et al. 2013). Since Wnt/β-catenin signaling plays important roles in many aspects of tumorigenesis, overexpression of SIAH proteins could promote tumorigenesis through enhancing Wnt/β-catenin signaling beyond other proposed mechanisms. Whether a SIAH inhibitor could be used to treat tumors with aberrant Wnt/β-catenin signaling, especially those with upstream pathway mutations (Hao et al. 2016), should be investigated.
Materials and methods
Cell culture, transfection, and stable cell lines
HEK293, YAPC, and U2OS cells were obtained from American Type Culture Collection and grown in DMEM or McCoy's 5A supplemented with 10% FBS and penicillin/streptomycin (Invitrogen). Plasmid or siRNA transfection was done by Fugene 6 (Roche) or Lipofectamine RNAiMax (ThermoFisher Scientific), respectively. Lentivirus was produced from HEK293 cells according to the standard virus packaging protocol and used to generate stable cell lines.
STF reporter assay
STF luciferase assays were performed as reported previously (Ettenberg et al. 2010). HEK293 cells were transfected with STF-luciferase, SV40-Renilla luciferase, and the indicated plasmids and treated with Wnt3a conditioned medium. Luciferase assays were done using Dual-Glo Luciferase assay kit (Promega) according to manufacturer's instructions.
Immunoblotting, immunoprecipitation, and in vitro pull-down assay
For immunoblotting, total cell lysates were prepared by direct lysis of cells with RIPA buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, protease inhibitors, phosphatase inhibitors) followed by centrifugation at 13,000 rpm for 15 min at 4°C. Equal amount of proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane, and probed with the indicated antibodies.
For immunoprecipitation experiments, cleared cell lysates were incubated with magnetic beads conjugated with Flag, HA, or Myc tag antibody for 2 h at 4°C. Beads were washed three times with lysis buffer, and bound proteins were eluted in SDS sample buffer for immunoblotting analysis.
For the pull-downs of GST-tagged AXIN1 (353–436) with SIAH1, 30 µL of glutathione Sepharose 4B beads was suspended with 200 µL of binding buffer (50 mM potassium phosphate, 500 mM NaCl at pH 7.2, 1 mM DTT) and mixed with 1 nmol of GST-tagged AXIN1 or AXIN1 mutants for 20 min. Purified SIAH1 (0.5 nmol) was added and incubated for another 3 h. Beads were washed quickly four times with 1 mL of washing buffer (binding buffer, 1% Triton X-100) before addition of 50 µL of sample loading buffer. All samples were analyzed by SDS-PAGE.
AXIN1 (C76H11), Myc tag (71D10), GFP (D5.1), GSK3α (D80E6), GSK3β (27C10), DYKDDDDK (D6W5B), and His (D3I1O) antibodies were purchased from Cell Signaling Technology. β-Catenin antibody was from BD Transduction. HA (3F10) antibody was from Roche. α-Tubulin antibody was from Sigma.
In vivo ubiquitin assay and TUBE ubiquitination assay
To detect the ubiquitination of Axin1 under denaturing conditions, HEK293 cells were transfected with plasmids encoding HA-tagged ubiquitin, Myc-tagged Axin1, and Flag-tagged Siah1a or Siah1a CS mutant. Two days after transfection, cells were treated with 10 µM MG132 for 6 h and lysed in 150 µL of lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% SDS with protease and phosphatase inhibitors). Cell lysates were boiled for 10 min, diluted 10 times in lysis buffer without SDS, and subjected to immunoprecipitation using HA antibody-conjugated beads. Immunoprecipitates were resolved by SDS-PAGE and blotted with Myc tag antibody.
Ubiquitination of endogenous AXIN1 protein was determined with the TUBE assay according to the manufacturer's protocol (LifeSensors). Cells were pretreated with 10 µM MG132 for 4 h and treated with Wnt3a conditioned medium before harvesting for the TUBE assay.
RNAi and CRISPR knockout
siRNA duplexes against SIAH1 were obtained from Dharmacon and Qiagen. The information and target sequences used were as follows: SIAH1-A (Smart Pool against human SIAH1 from Dharmacon), SIAH1-B (target sequence, 5′-ACCTAAAACTCTTTCGGTA-3′) (Qiagen), SIAH1-C (target sequence 5′-CAGCAGTTCTTCGCAATCGTA-3′) (Dharmacon), and pGL2(luciferase) (sense, 5′-CGTACGCGGAATACTTCGA-3′) (Dharmacon). For CRISPR knockout, cells stably expressing Cas9 were infected with lentivirus expressing gRNA fused with tracerRNA. gRNA sequences used in this work were as follows: control (ACCGGAACGATCTCGCGTA) and SIAH1 (TAATGCTGTAGCAGTCTGA).
Quantitative RT–PCR
Total RNA was extracted using RNeasy Plus minikit (Qiagen) and reserve transcribed with TaqMan reverse transcription reagents (Applied Biosystem) according to the manufacturers’ instructions. Gene expression levels were detected by TaqMan assay, and all experiments were carried out in quadruplicate. Gene expression analysis was carried out using the comparative cycle threshold method with housekeeping gene GUSB for normalization.
Protein purification, crystallization, and data collection
Both AXIN1 and SIAH1 proteins used in crystallographic analysis and in vitro GST pull-down were expressed in the BL21(DE3) cell strain (Stratagene). Human SIAH1-SBD (91–282, NP_001006611) was cloned into the pET28a vector (Novagen). Human AXIN1 (353–436, NP_003493) was cloned into pGEX-6p-1 with an N-terminal GST tag fusion protein. Cell cultures were grown at 37°C and induced with isopropyl 1-thio-β-D-galactopyranoside. SIAH1 was purified by a Ni-NTA affinity column (GE Healthcare) followed by removal of the histidine tag with TEV protease and further purified on a HiLoad 10/300 Superdex 200 column (GE Healthcare). GST-AXIN1 proteins were purified by glutathione Sepharose columns. The fusion proteins were purified by Resource Q anion exchange chromatography (GE Healthcare). All mutants were generated by standard PCR procedures and expressed and purified as well as native proteins.
Crystals of SIAH1-SBD (91–282) in complex with AXIN1 peptide (375–394) were grown at 4°C using the hanging drop vapor diffusion method. Crystals were obtained with buffer containing 25% (w/v) PEG3350, 200 mM MgCl2, and 100 mM Tris-Hcl (pH 8.0). The crystals were cryoprotected by 15%–20% glycerol and flash-frozen by liquid nitrogen. X-ray diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF) on beamline BL19U1 equipped with a PILATUS6M detector. The diffraction images were indexed and integrated using HKL2000 (Collaborative Computational Project, Number 4 1994). The data collection statistics are in Supplemental Table 1.
Structure determination and refinement
The crystal structure of the SIAH1-SBD/AXIN1 (375–394) complex was solved by the molecular replacement method using PHASER in the PHENIX suite (Adams et al. 2010) with one monomer of SIAH1 (PDB 4CA1) (Rimsa et al. 2013) as the search model. Iterative cycles of refinement and manual model building were carried out with PHENIX refinement programs and COOT (Emsley and Cowtan 2004), respectively, at 2.1 Å resolution. All structural images were drawn using PyMOL (http://www.pymol.org). Detailed crystallographic statistics are in Supplemental Table 1. Coordinates have been deposited in PDB under accession code 5WZZ.
ITC
The binding affinities of SIAH1 with constructs of AXIN1 (375–394) were measured at 25°C using ITC200 calorimeters (GE Life Science and MicroCal). Proteins were dialyzed against ITC buffer (20 mM Tris at pH 8.0, 100 mM NaCl). Protein concentrations were measured based on their respective ultraviolet absorption at 280 nm. Two microliters of 1 mM AXIN1 (375–394) was injected 20 times into 300 µL of 60 µM SIAH1 proteins. Data were analyzed using Origin software (version 7.0). A single binding site model for SIAH/AXIN gave the best fit to the data. Errors are given as SD of the fit from the original data points.
Accession codes
The coordinates and structure factors for the AXIN1/SIAH1 structure have been deposited in the PDB under accession number 5WZZ.
Supplementary Material
Acknowledgments
We thank B. Lu, R. Zamponi, X. Xie, X. Shi, M. Hild, and D. Glass for comments and advice. We appreciate assistance in data collection from the staff of beamline BL19U1/BL17U at the Shanghai Synchrotron Radiation Facility, China. We also thank Zhenwei Yang and Yuanyuan Chen for help with ITC analyses at the Institute of Biophysics, Chinese Academy of Sciences. This work was supported in part by National Natural Science Foundation of China grants 31570794 and 31629002, and Chinese Academy of Sciences Pilot Strategic Science and Technology Projects B grant XDB08010303. L.J. and X.J. performed mechanistic studies. B.J. built constructs and performed protein purification. B.J., W.X., and X.Y. carried out ITC and GST pull-down assays. B.J. and X.Y. obtained crystals of the AXIN1/SIHA1 complex, performed the X-ray diffraction measurements, and determined the crystal structure. A.C. conducted the siRNA screen. C.M. conducted the cDNA overexpression screen. A.B. conducted the proteomics screen. O.C. provided critical technical assistance. L.J., B.J., W.X., X.Y., and F.C. wrote the paper. All authors provided editorial comments.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.300053.117.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R, Kneissel M. 2013. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med 19: 179–192. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Rubin GM. 1990. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63: 561–577. [DOI] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. 2009. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 5: 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2006. Wnt/β-catenin signaling in development and disease. Cell 127: 469–480. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Cselenyi CS, Jernigan KK, Tahinci E, Thorne CA, Lee LA, Lee E. 2008. LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3′s phosphorylation of β-catenin. Proc Natl Acad Sci 105: 8032–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, Dale TC, Pearl LH. 2003. Structural basis for recruitment of glycogen synthase kinase 3β to the axin-APC scaffold complex. EMBO J 22: 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, Xu W. 2015. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della NG, Senior PV, Bowtell DD. 1993. Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117: 1333–1343. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Frew IJ, House CM, O'Bryan MK, Holloway AJ, Haviv I, Traficante N, de Kretser DM, Bowtell DD. 2002. The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol Cell Biol 22: 2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova YN, Li J, Lee YT, Rios-Esteves J, Friedman DB, Choi HJ, Weis WI, Wang CY, Chazin WJ. 2010. Direct ubiquitination of β-catenin by Siah-1 and regulation by the exchange factor TBL1. J Biol Chem 285: 13507–13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Ettenberg SA, Charlat O, Daley MP, Liu S, Vincent KJ, Stuart DD, Schuller AG, Yuan J, Ospina B, Green J, et al. 2010. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proc Natl Acad Sci 107: 15473–15478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Funk CC, Katzenellenbogen BS. 2005. Estrogen down-regulation of the corepressor N-CoR: mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc Natl Acad Sci 102: 13153–13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. 2000. Feedback control of intercellular signalling in development. Nature 408: 313–319. [DOI] [PubMed] [Google Scholar]
- Frew IJ, Hammond VE, Dickins RA, Quinn JM, Walkley CR, Sims NA, Schnall R, Della NG, Holloway AJ, Digby MR, et al. 2003. Generation and analysis of Siah2 mutant mice. Mol Cell Biol 23: 9150–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew IJ, Sims NA, Quinn JM, Walkley CR, Purton LE, Bowtell DD, Gillespie MT. 2004. Osteopenia in Siah1a mutant mice. J Biol Chem 279: 29583–29588. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Copley RR, Cohen SM. 2002. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev Cell 2: 667–676. [DOI] [PubMed] [Google Scholar]
- Habelhah H, Frew IJ, Laine A, Janes PW, Relaix F, Sassoon D, Bowtell DD, Ronai Z. 2002. Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J 21: 5756–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al. 2012. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485: 195–200. [DOI] [PubMed] [Google Scholar]
- Hao HX, Jiang X, Cong F. 2016. Control of Wnt receptor turnover by R-spondin–ZNRF3/RNF43 signaling module and its dysregulation in cancer. Cancers (Basel) 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. 2009. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 10: 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House CM, Frew IJ, Huang HL, Wiche G, Traficante N, Nice E, Catimel B, Bowtell DD. 2003. A binding motif for Siah ubiquitin ligase. Proc Natl Acad Sci 100: 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House CM, Hancock NC, Moller A, Cromer BA, Fedorov V, Bowtell DD, Parker MW, Polekhina G. 2006. Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure 14: 695–701. [DOI] [PubMed] [Google Scholar]
- House CM, Moller A, Bowtell DDL. 2009. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res 69: 8835–8838. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Chung YL, Glover T, Valentine V, Look AT, Fearon ER. 1997. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46: 103–111. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461: 614–620. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J 17: 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpertz S, Hennes T, Asare Y, Vervoorts J, Bernhagen J, Schutz AK. 2014. The β-catenin E3 ubiquitin ligase SIAH-1 is regulated by CSN5/JAB1 in CRC cells. Cell Signal 26: 2051–2059. [DOI] [PubMed] [Google Scholar]
- Kakugawa S, Langton PF, Zebisch M, Howell SA, Chang TH, Liu Y, Feizi T, Bineva G, O'Reilly N, Snijders AP, et al. 2015. Notum deacylates Wnt proteins to suppress signalling activity. Nature 519: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, et al. 2012. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488: 665–669. [DOI] [PubMed] [Google Scholar]
- Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. 2003. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 1: E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL, Matsunami N. 2001. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell 7: 927–936. [DOI] [PubMed] [Google Scholar]
- Liu X, Rubin JS, Kimmel AR. 2005. Rapid, Wnt-induced changes in GSK3β associations that regulate β-catenin stabilization are mediated by Gα proteins. Curr Biol 15: 1989–1997. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810. [DOI] [PubMed] [Google Scholar]
- Loh KM, van Amerongen R, Nusse R. 2016. Generating cellular diversity and spatial form: Wnt signaling and the evolution of multicellular animals. Dev Cell 38: 643–655. [DOI] [PubMed] [Google Scholar]
- Lopitz-Otsoa F, Rodriguez-Suarez E, Aillet F, Casado-Vela J, Lang V, Matthiesen R, Elortza F, Rodriguez MS. 2012. Integrative analysis of the ubiquitin proteome isolated using tandem ubiquitin binding entities (TUBEs). J Proteomics 75: 2998–3014. [DOI] [PubMed] [Google Scholar]
- Luo W, Peterson A, Garcia BA, Coombs G, Kofahl B, Heinrich R, Shabanowitz J, Hunt DF, Yost HJ, Virshup DM. 2007. Protein phosphatase 1 regulates assembly and function of the β-catenin degradation complex. EMBO J 26: 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa SI, Reed JC. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol Cell 7: 915–926. [DOI] [PubMed] [Google Scholar]
- Nadeau RJ, Toher JL, Yang X, Kovalenko D, Friesel R. 2007. Regulation of Sprouty2 stability by mammalian Seven-in-Absentia homolog 2. J Cell Biochem 100: 151–160. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, et al. 2004. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell 117: 941–952. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Qi J, Ronai Z. 2009. The ubiquitin ligase Siah2 and the hypoxia response. Mol Cancer Res 7: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Lee SH, Kim H, Yum S, Stamos JL, Xu Y, Lee SJ, Lee J, Oh S, Han JK, et al. 2008. Direct inhibition of GSK3β by the phosphorylated cytoplasmic domain of LRP6 in Wnt/β-catenin signaling. PLoS One 3: e4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Kim H, Scortegagna M, Ronai ZA. 2013. Regulators and effectors of Siah ubiquitin ligases. Cell Biochem Biophys 67: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsa V, Eadsforth TC, Hunter WN. 2013. Two high-resolution structures of the human E3 ubiquitin ligase Siah1. Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Yi EH, Kim BH, Shin JC, Park JY, Cho CH, Park JW, Choi KY, Ye SK. 2016. STAT3 potentiates SIAH-1 mediated proteasomal degradation of β-catenin in human embryonic kidney cells. Mol Cells 39: 821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz MD, Cheung AK, Kirby CA, Firestone B, Fan J, Chen CH, Chen Z, Chin DN, Dipietro L, Fazal A, et al. 2013. Identification of NVP-TNKS656: the use of structure-efficiency relationships to generate a highly potent, selective, and orally active tankyrase inhibitor. J Med Chem 56: 6495–6511. [DOI] [PubMed] [Google Scholar]
- Smalley MJ, Sara E, Paterson H, Naylor S, Cook D, Jayatilake H, Fryer LG, Hutchinson L, Fry MJ, Dale TC. 1999. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO J 18: 2823–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins JL, Santelli E, Feng Y, De SK, Purves A, Motamedchaboki K, Wu B, Ronai ZA, Liddington RC, Pellecchia M. 2013. Structure-based design of covalent Siah inhibitors. Chem Biol 20: 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CW, Gardiner BS, Hirokawa Y, Layton MJ, Smith DW, Burgess AW. 2012. Wnt signalling pathway parameters for mammalian cells. PLoS One 7: e31882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolwinski NS, Wehrli M, Rives A, Erdeniz N, DiNardo S, Wieschaus E. 2003. Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3β activity. Dev Cell 4: 407–418. [DOI] [PubMed] [Google Scholar]
- Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. 2012. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev 26: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K, Shibamoto S, Nusse R. 1999. Wnt-induced dephosphorylation of axin releases β-catenin from the axin complex. Genes Dev 13: 1768–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, Hofmann TG. 2008. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 10: 812–824. [DOI] [PubMed] [Google Scholar]
- Wu G, Huang H, Garcia Abreu J, He X. 2009. Inhibition of GSK3 phosphorylation of β-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One 4: e4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Sproul A, Wang W, Kukekov N, Greene LA. 2006. Siah1 interacts with the scaffold protein POSH to promote JNK activation and apoptosis. J Biol Chem 281: 303–312. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kishida S, Kishida M, Ikeda S, Takada S, Kikuchi A. 1999. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3β regulates its stability. J Biol Chem 274: 10681–10684. [DOI] [PubMed] [Google Scholar]
- Zhang J, Guenther MG, Carthew RW, Lazar MA. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev 12: 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, et al. 2011. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13: 623–629. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cheong SM, Amado NG, Reis AH, MacDonald BT, Zebisch M, Jones EY, Abreu JG, He X. 2015. Notum is required for neural and head induction via Wnt deacylation, oxidation, and inactivation. Dev Cell 32: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.