In Arabidopsis, stomatal patterning is specified by the ERECTA family (ERf) receptor kinases (RKs), the receptor-like protein (RLP) TOO MANY MOUTHS (TMM), and EPIDERMAL PATTERNING FACTOR (EPF) peptides. Here, Lin et al. show that TMM dictates the specificity of ERfs for perception of different EPFs, thus functioning as a specificity switch for the regulation of the activities of ERfs.
Keywords: biochemical interaction, peptide hormones, receptor kinases, receptor-like protein, stomatal development, structure
Abstract
Stomata are microscopic openings that allow for the exchange of gases between plants and the environment. In Arabidopsis, stomatal patterning is specified by the ERECTA family (ERf) receptor kinases (RKs), the receptor-like protein (RLP) TOO MANY MOUTHS (TMM), and EPIDERMAL PATTERNING FACTOR (EPF) peptides. Here we show that TMM and ER or ER-LIKE1 (ERL1) form constitutive complexes, which recognize EPF1 and EPF2, but the single ERfs do not. TMM interaction with ERL1 creates a binding pocket for recognition of EPF1 and EPF2, indicating that the constitutive TMM–ERf complexes function as the receptors of EPF1 and EPF2. EPFL9 competes with EPF1 and EPF2 for binding to the ERf–TMM complex. EPFL4 and EPFL6, however, are recognized by the single ERfs without the requirement of TMM. In contrast to EPF1,2, the interaction of EPFL4,6 with an ERf is greatly reduced in the presence of TMM. Taken together, our data demonstrate that TMM dictates the specificity of ERfs for the perception of different EPFs, thus functioning as a specificity switch for the regulation of the activities of ERfs.
In animals, cell–cell communication via peptide hormones is crucial for development and tissue patterning (MacDonald et al. 2009). Similarly, plants also rely on numerous secreted peptide signals to coordinate growth and development (De Smet et al. 2009; Motomitsu et al. 2015). The signaling pathways enabling cell–cell communications often can generate distinct responses in different tissues. This can be exemplified by the ERECTA family (ERf) signaling pathway in Arabidopsis that regulates different developmental processes in the epidermis and vasculature.
In the epidermis, the ERf signaling pathway regulates the development of stomata, which are microscopic pores ubiquitously used by land plants for gas exchange. Several secreted peptides from the EPIDERMAL PATTERNING FACTOR (EPF) family, LRR receptor kinases (LRR-RKs) from the ERf (ER, ER-LIKE1 [ERL1], and ERL2), and the LRR receptor-like protein (LRR-RLP) TOO MANY MOUTHS (TMM) enforce stomatal patterning (Nadeau and Sack 2002; Shpak et al. 2005; Hara et al. 2007, 2009; Hunt and Gray 2009; Abrash and Bergmann 2010; Hunt et al. 2010; Kondo et al. 2010; Sugano et al. 2010; Abrash et al. 2011; Lee et al. 2012). The EPF family consists of 11 cysteine-rich peptides, with EPF1,2 and EPFL2,4,5,6,9 having been shown to interact with ERf receptors in Arabidopsis (Abrash et al. 2011; Lee et al. 2012, 2015; Uchida et al. 2012; Tameshige et al. 2016). Three peptides—EPF1, EPF2, and EPFL9 (STOMAGEN)—regulate stomata development (Supplemental Fig. S1). EPF2 is expressed during early epidermis development and is perceived primarily by ER to restrict the initiation of stomatal cell lineages, while EPF1 is expressed later than EPF2 and is perceived mainly by ERL1 to enforce stomatal spacing (Shpak et al. 2005; Hara et al. 2007, 2009; Hunt and Gray 2009; Lee et al. 2012). Expressed in mesophyll cells, EPFL9 competes with EPF2 for ER receptor binding, which acts to inhibit receptor function and promote stomatal development (Hunt et al. 2010; Kondo et al. 2010; Sugano et al. 2010; Ohki et al. 2011; Lee et al. 2015).
Another process involving ERf signaling is the elongation of aboveground plant organs, which is regulated primarily by vasculature-localized ERfs. A different subset of EPF family ligands, EPFL6/CHALLAH (CHAL) and EPFL4/CHALLAH-LIKE2 (CLL2), is expressed in endodermal cells and is perceived by vasculature-localized ERfs (Abrash et al. 2011; Uchida et al. 2012). Based on the phenotype of the tmm-1 mutant and the absence of its expression in vasculature, TMM does not regulate the elongation of aboveground plant organs (Nadeau and Sack 2002; Shpak et al. 2005; Abrash et al. 2011).
The precise role of TMM in stomata development has remained ambiguous. TMM is an indispensable component of the ERf signaling pathway in stomatal development (Shpak et al. 2005; Lee et al. 2012), but the phenotype of tmm-1 is tissue-dependent. In stems, the tmm-1 mutant has no stomata, while leaves exhibit a profusion of stomata in clusters (Yang and Sack 1995; Geisler et al. 1998, 2000; Shpak et al. 2005). Interestingly, while mutations in the EPFL4 and EPFL6 genes have no impact on stomata development, their overexpression inhibits stomata formation, especially in the tmm-1 background (Abrash and Bergmann 2010; Abrash et al. 2011). Genetic evidence suggests that, in the epidermis, TMM can act as a specificity factor for ERf signaling by promoting ERf perception of EPF1,2 and EPFL9 but preventing perception of EPFL4,6 (Abrash et al. 2011; Lee et al. 2012, 2015). However, the mechanism of how TMM modulates different signaling with specific ligands has remained elusive.
In the present study, we provide multiple lines of evidence showing that only in complex with TMM can ERfs function as the receptor of EPF1, EPF2, or EPFL9. In contrast, an ERf alone is sufficient for recognition of EPFL4 or EPFL6, and, in fact, TMM interferes with their interaction with ERfs. EPFL9 competes with EPF1 and EPF2 to bind ERfLRR–TMMLRR in vitro. Collectively, these data demonstrate that TMM enables ERf to discriminate between a variety of ligands, and this capability, in concert with tissue-specific expression of TMM, provides a mechanism for tissue-specific function of ERf receptors and their ligands.
Results
TMM forms a ligand-independent complex with ER, ERL1, or ERL2
Genetic and biochemical data show that TMM interacts with ERfs in vivo (Shpak et al. 2005; Lee et al. 2012). Ligand-induced interaction with coreceptors has been demonstrated for many LRR-RKs (Song et al. 2016), but whether TMM–ERf complexes are EPF-promoted has remained unknown. To address this question, we purified the extracellular LRR domain proteins of ER, ERL1, and ERL2 (ERLRR, ERL1LRR, and ERL2LRR) from insect cells. However, purification of the TMMLRR protein was unsuccessful using the same strategy. Given the interaction of TMM and ERfs in plants (Shpak et al. 2005; Lee et al. 2012), we coexpressed TMMLRR individually with the three ERfs in the hope of facilitating purification of the RLP. Indeed, TMMLRR was well expressed when coexpressed with ERLRR or ERL1LRR in insect cells (Supplemental Fig. S2). Gel filtration assays showed that TMMLRR formed a stable complex with ERLRR or ERL1LRR with a stoichiometry of ∼1:1, as indicated by their intensity on SDS-PAGE (Supplemental Fig. S2). However, for unknown reasons, we failed to obtain the ERL2LRR–TMMLRR complex using the coexpression strategy. Nonetheless, these results indicate that, in vitro, both ER and ERL1 form constitutive complexes with TMM through their extracellular domains.
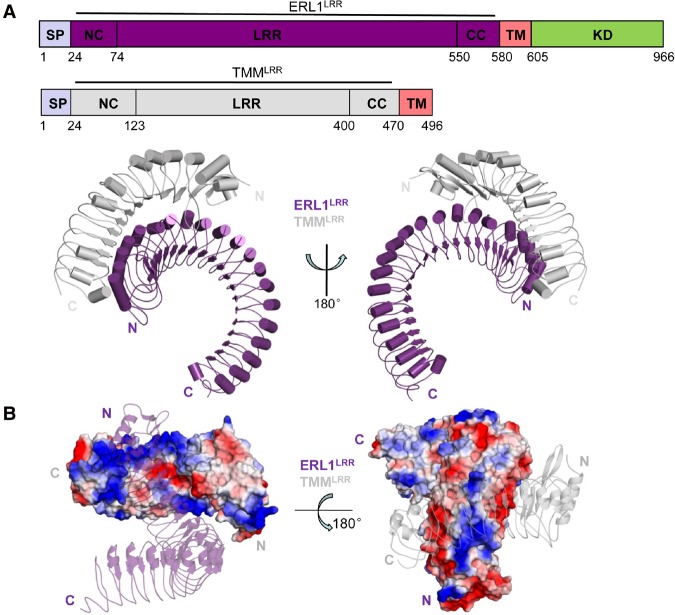
To understand the molecular mechanism underlying TMM–ERf interaction, we solved the crystal structure of the ERL1LRR–TMMLRR complex at a resolution of 3.05 Å (Fig. 1A; Supplemental Table S1). Nearly the whole concave surface of TMMLRR interacts with the N-terminal half convex side of ERL1LRR but with a distortion of ∼30°. TMMLRR and ERL1LRR have extensive interactions, burying a surface area of 1614 Å2. The concave surface of TMMLRR features neutral charges at the center and positive charges at the periphery (Fig. 1B). The charged surfaces of TMMLRR are complementary to those of ERL1LRR at the N-terminal side, whereas the neutral surface forms van der Waals contacts with ERL1LRR (Fig. 1B). TMMR167 is located at the center of the charged interface and forms a pair of hydrogen bonds with the carbonyl oxygen of ERL1P167, whereas a hydrogen bond is established between ERL1N168 and TMMR141 (Fig. 2A). At the periphery of the interface, several polar interactions involving TMMS109, D110, K257 are made with their respective neighboring residues of ERL1. Additionally, hydrophobic contacts made by TMML108, F138, V233 with ERL1 further fortify interactions of this interface (Fig. 2A). At the center of the other interface are TMML281, M307, M330, I331, V355, which form hydrophobic contacts with ERL1LRR (Fig. 2A). However, hydrogen and salting bonds appear to dominate the periphery interactions of this interface (Fig. 2A). Structure-based sequence alignment indicated that the TMM-interacting residues of ERL1 are highly conserved among the ecto-LRR domains of ERfs (Supplemental Fig. S3A), suggesting that they all can constitutively interact with TMM.
Figure 1.
Overall structure of the ERL1LRR–TMMLRR complex. (A) Overall structure of the ERL1LRR–TMMLRR complex. “N” and “C” represent the N terminus and C terminus, respectively. Color codes are indicated. Schematic representations of the domain structures of ERL1LRR and TMMLRR are shown at the top. (SP) Signal peptide; (NC) N-terminal capping; (CC) C-terminal capping; (TM) transmembrane; (KD) kinase domain. (B) Surface representation of the electrostatic potential of ERL1LRR–TMMLRR. The electrostatic potential of TMMLRR and a cartoon representation of ERL1LRR in transparency are shown at the left. The right panel shows the electrostatic potential of ERL1LRR and a cartoon representation of TMMLRR in transparency. (White) Neutral surface; (blue) positive surface; (red) negative surface; (N) N terminus; (C) C terminus. Color codes are indicated.
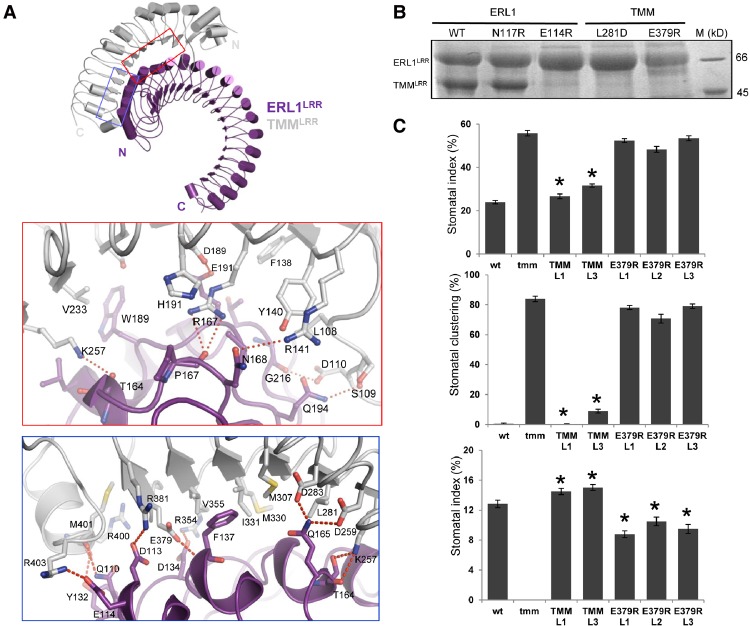
Figure 2.
Mechanism of ERL1LRR–TMMLRR interaction. (A) Detailed interactions of ERL1LRR with TMMLRR. Interaction regions are indicated by red and blue squares in the top panel. Red dashed lines indicate hydrogen bonds or salt bridges. (B) Mutagenesis analysis of the ERL1–TMM interaction in vitro. Mutant ERL1 or TMM was coexpressed with its wild-type partners in insect cells. C-terminally 6xHis-tagged ERL1LRR was pulled down by Ni+ beads. The bound proteins were eluted and visualized by Coomassie blue staining following SDS-PAGE. (C) The TMM E379R reduces the functionality of TMM in plants. The stomatal index (top) and stomatal clustering (middle) were analyzed on the abaxial side of 14-d-old cotyledons of wild-type (wt), tmm, and T2 generation seedlings expressing functional TMM (two independent lines) or TMM with E379R substitution in the tmm background (three independent lines). The stomatal index was analyzed on the mature stem (bottom) of wild-type, tmm, and T2 generation plants expressing functional TMM or TMM with E379R substitution in the tmm background. Values are means ± SE. N = 18–30. Values significantly different from the tmm mutant (P < 0.0001) are indicated by asterisks.
To confirm the structure of TMMLRR–ERL1LRR, we mutated residues from the interfaces between the two LRRs and individually coexpressed the mutants with their wild-type partners in insect cells. We then used pull-down assays to examine the interaction of the mutant proteins with wild-type TMMLRR or ERL1LRR. As shown in Figure 2B, wild-type His-tagged ERL1LRR failed to pull down the mutant protein of TMMLRRL281D or TMMLRRE379R. To a lesser extent, similar results were obtained for the mutant protein ERL1LRRE114R with wild-type TMMLRR. To further support the biochemical data, we generated transgenic Arabidopsis plants and investigated the effects of ERL1E114R, TMML281D, and TMME379R on stomatal development. As observed previously, clustered stomata and no stomata were observed in cotyledons and stems of tmm mutant plants, respectively (Geisler et al. 1998; Shpak et al. 2005). Expression of wild-type TMM in the tmm-1 mutant restored the stomata patterning of mutant plants to the levels of wild-type plants (Fig. 2C). In contrast, three independent transgenic lines of plants expressing TMM::TMME379R had stomatal patterning similar to the tmm mutant plants, indicating the loss of TMM function with this substitution (Fig. 2C). Intriguingly, stems from the three TMME379R lines only partially phenocopied the tmm mutant, with stomata being present, although fewer than in the wild-type plants. However, a wild-type stomata phenotype was observed in plants expressing ERL1::ERL1E114R in the er erl1 erl2 mutant or TMM::TMML281D in the tmm-1 mutant. We speculate that, in vivo, when coreceptors are properly aligned, due to their attachment to the plasma membrane, these single-point mutations are unable to disrupt their interactions. Overall, our biochemical, structural, and functional data support the idea that TMM and ERf form constitutive complexes.
The constitutive ERL1LRR–TMMLRR complex recognizes EPF1/2
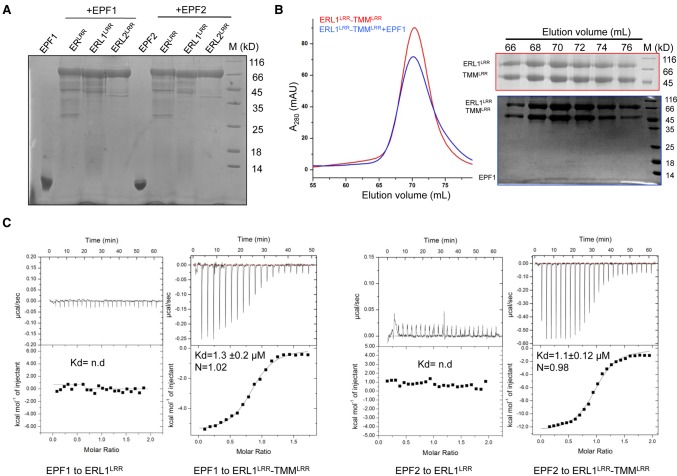
We then investigated whether the constitutive ERLRR–TMMLRR or ERL1LRR–TMMLRR complex is able to interact with EPF1 or EPF2. Our pull-down assays showed that none of the three ERf proteins displayed interaction with these two EPFs (Fig. 3A). These results were further supported by gel filtration (Supplemental Fig. S4A,B). While ERLRR or ERL1LRR alone showed no binding to EPF1/EPF2, complexing with TMMLRR resulted in their strong interaction with these two EPF peptides, as indicated by their comigration under the gel filtration assay (Fig. 3B; Supplemental Fig. S5). To further verify these results, we used isothermal titration calorimetry (ITC) to quantify interaction of the ERL1LRR–TMMLRR complex with EPF1 or EPF2 (Fig. 3C). In support of the data from gel filtration, the ITC results indicated that EPF1 or EPF2 strongly interacted with ERL1LRR–TMMLRR, with dissociation constants of 1.3 µM and 1.1 µM, respectively. In contrast, the ERL1LRR protein alone had no detectable binding to these two EPFs under the ITC assays, further supporting the pull-down data (Fig. 3A). Taken together, our results indicate that ERfs and TMM form constitutive complexes for the recognition of EPF1 and EPF2 in vitro.
Figure 3.
ERL1–TMM complexes function as receptors of EPF1 and EPF2 in vitro. (A) ERfs alone fail to interact with EPF1 or EPF2 in pull-down assays. The 6xHis-tagged ERLRR, ERL1LRR, and ERL2LRR were incubated individually with EPF1 or EPF2, and the mixtures were then flowed through Ni-NTA beads. After extensive washing, the bound proteins were eluted and analyzed with SDS-PAGE followed by Coomassie blue staining. (B) EPF1 directly binds to the ERL1LRR–TMMLRR complex. (Left panel) Size exclusion chromatography analysis of the interaction between EPF1 and ERL1LRR–TMMLRR; the elution volumes and the molecular weight markers are indicated at the top. (Right panel) SDS-PAGE analysis of peak fractions from the left panel. (C) Quantification of the binding affinity of EPF1 (left) or EPF2 (right) to ERL1LRR or ERL1LRR–TMMLRR by ITC. (nd) No detectable binding. EPF1 or EPF2 was titrated into ERL1LRR or ERL1LRR–TMMLRR protein in the ITC cell. Integrated heat measurements from ITC are shown. The calculated stoichiometry (N) and the dissociation constant (Kd) are indicated.
To probe the molecular mechanism of how TMMLRR–ERL1LRR recognizes EPF1 and EPF2, we solved the crystal structures of the tertiary complexes ERL1LRR–TMMLRR–EPF1 (Fig. 4A; Supplemental Table S1) and ERL1LRR–TMMLRR–EPF2 (Supplemental Fig. S6A; Supplemental Table S1) at resolutions of 2.63 Å and 3.5 Å, respectively. Structural comparison showed that the two complexes are highly conserved (Supplemental Fig. S6B). We therefore limit our discussions to the structure of the complex with a higher resolution. In this structure, EPF1 assumes an inverted V-shaped architecture and binds to the groove created by TMMLRR and ERL1LRR (Fig. 4A). This structural observation supports our biochemical data that both TMMLRR and ERL1LRR were required for interaction with EPF1 or EPF2. Structure comparison showed that no striking conformational changes occur to the ERL1LRR–TMMLRR complex following EPF1 binding (Supplemental Fig. S6C). Similar to the solution structure of EPFL9 (Ohki et al. 2011), a long anti-parallel β sheet constitutes the core of the EPF1 structure, with the N-terminal loop region packing against one side of the anti-β sheet, and a short β strand from the C-terminal side pairing with the anti-β sheet (Fig. 4A; Supplemental Fig. S6B). No electron density was found around the hairpin loop of EPF1, consistent with the structural study of EPFL9 showing that multiple conformations were found at the equivalent region of EPFL9 in solution (Ohki et al. 2011). Interestingly, this region of EPFs has been suggested to be important for conferring their functional specificity (Ohki et al. 2011).
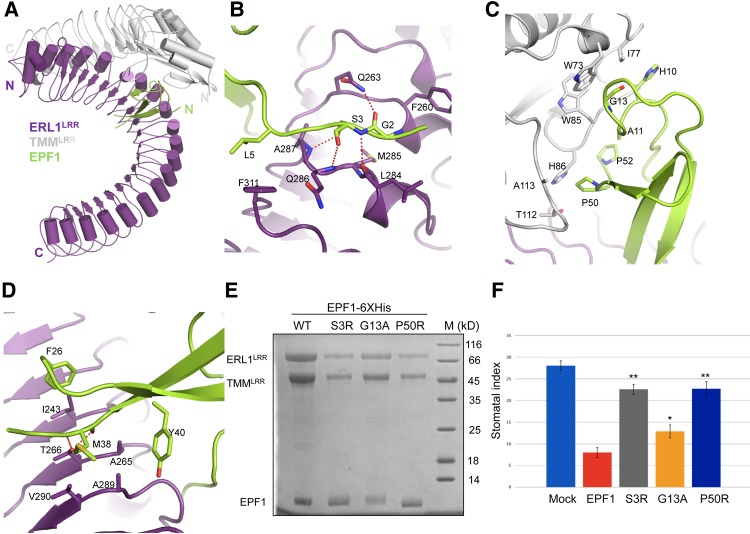
Figure 4.
Recognition mechanism of EPF1 by ERL1LRR–TMMLRR. (A) Overall structure of the EPF1–ERL1LRR–TMMLRR complex. (N) N terminus; (C) C terminus. Color codes are indicated. (B) Detailed interactions of the N-terminal side of EPF1 with ERL1LRR–TMMLRR. (C) Detailed interactions of the central region of EPF1 with ERL1LRR–TMMLRR. (D) Detailed interactions of EPF1 with ERL1LRR–TMMLRR. (E) Mutagenesis analysis of EPF1-6xHis's interaction with ERL1LRR–TMMLRR in vitro. The pull-down assays were performed as described in Figure 3A. (F) Effects of the mutations of EPF1 on the stomatal density of Arabidopsis cotyledons; the concentration of peptide was 10 µM. The error bars show SD. n > 8. (*) P < 0.05; (**) P < 0.01, versus the controls by Student's t-test.
The three parts of EPF1 interact with the ERL1LRR–TMMLRR complex (Fig. 4A). The N-terminal loop of EPF1 is sandwiched between two consecutive LRRs of ERL1LRR, and the interactions around this region are mediated mainly by polar contacts (Fig. 4B). At the center of this interface is EPF1Ser3, which hydrogen-bonds with ERL1Leu284, Gln286, and Ala287. In addition to making Van der Waals contacts with ERL1Leu284, EPF1Gly2 establishes a hydrogen bond with ERL1Gln263. These two EPF1 residues (called the “GS” motif here) are highly conserved among all of the EPF members (Supplemental Fig. S3B). Additionally, EPF1Leu5 hydrophobically packs against ERL1Phe311. The N-terminal loop of EPF1 also interacts with TMMLRR through His10 (aromatic), Ala11, and Gly13, which stack tightly against Ile77, Trp85, and Trp73 of TMM, respectively (Fig. 4C). The TMMLRR–EPF1 interaction is further strengthened by Pro50 and Pro52 from the C-terminal side of EPF1, with the former residue packing against T112 and Ala113 of TMMLRR, and the latter residue stacking against Trp85 and His86 of TMMLRR (Fig. 4C). It is of interest to note that some of the TMM-interacting residues of EPF1 are largely conserved in EPF2 but not in EPFL4,5,6 (Supplemental Figs. S3B, S6D). The inner surface of ERL1LRR interacts with one β strand of EPF1 (Fig. 4D). ERL1Thr266 from underneath forms a pair of hydrogen bonds with EPF1Met38, which also packs against ERL1Val290. Hydrophobic contacts made by Ala265 and Ala289 of ELR1 with EPF1Tyr40 further fortify the interaction around this region.
To further verify the structure of the TMMLRR–ERL1LRR–EPF1 complex, we mutated some of the critical residues in EPF1 that interact with TMMLRR or ERL1LRR and then tested the interaction of these mutant EPF1 proteins with the TMMLRR–ERL1LRR complex. In support of the structural observation, mutation of Ser3 from the “GS” motif compromised the interaction of EPF1 with the TMMLRR–ERL1LRR complex, as indicated by pull-down assays using tagged EPF1 (Fig. 4E). Similar observations were made with mutations of EPF1G13 and EPF1P50 that pack against TMMLRR. Consistent with previous data (Lee et al. 2012), treatment of Arabidopsis with wild-type EPF1 strikingly reduced the number of stomata in cotyledons (Fig. 4F). In contrast, the activity of EPF1 was reduced substantially by the three EPF1 mutations, in particular EPF1S3R and EPF1P50R (Fig. 4F).
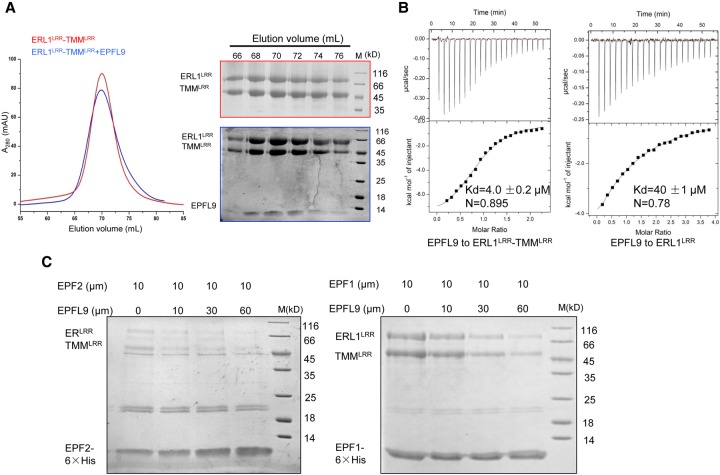
EPFL9 competes with EPF1 and EPF2 to bind ERfLRR–TMMLRR
EPFL9 was shown recently to compete with EPF2 for interaction with ER, thus promoting stomatal development (Lee et al. 2015). Our data demonstrate that ERfs and TMM form constitutive complexes for the recognition of EPF1 and EPF2. We then investigated whether the ERL1LRR protein alone or the constitutive ERL1LRR–TMMLRR complex is able to interact with EPFL9 using gel filtration. The assay showed that a tiny amount of EPFL9 comigrated with ERL1LRR, indicating a weak interaction between them (Supplemental Fig. S4C). In contrast, when the ERL1LRR–TMMLRR complex protein was used for the assay, much more EPFL9 was shifted to a higher-molecular-weight species and comigrated with the complex (Fig. 5A). This result showed that complexing with TMMLRR greatly promotes ERL1LRR recognition of EPFL9. These gel filtration data were confirmed further by our ITC assays showing that EPFL9 bound to the ERL1LRR–TMMLRR complex with an affinity of 4 µM, ∼10 times higher than that of its binding to ERL1LRR alone (Fig. 5B). Next, we tested whether EPFL9 is able to compete with EPF2 or EPF1 for binding to the ERLRR–TMMLRR or ERL1LRR–TMMLRR complex. To this end, we individually incubated the complexes with the C-terminally His-tagged EPF1 or EPF2. Purification of the mixture using Ni-resin showed that increasing concentrations of EPFL9 gradually decreased the amount of ERLRR–TMMLRR or ERL1LRR–TMMLRR pulled down by EPF2 or EPF1, indicating that EPFL9 and these two EPFs compete with each other for binding to the ERfLRR–TMMLRR complex (Fig. 5C).
Figure 5.
EPFL9 competes with EPF1 and EPF2 for binding to the ERfLRR–TMMLRR complexes. (A, left panel) Size exclusion chromatography analysis of the interaction between EPFL9 and ERL1LRR–TMMLRR. (Right panel) SDS-PAGE analysis of peak fractions from the left panel. The elution volumes and molecular weight markers are indicated at the top. (B) TMM promotes EPFL9 binding to ERL1LRR. Quantification of the binding affinity of EPF9 with ERL1LRR or ERL1LRR–TMMLRR by ITC. EPFL9 was titrated into ERL1LRR or ERL1LRR–TMMLRR protein in the ITC cell. Raw data (top panel) and integrated heat measurements (bottom panel) from ITC are shown. The calculated stoichiometry (N) and the dissociation constant (Kd) are indicated. (C) EPFL9 competes with EPF1,2 for binding to ERfLRR–TMMLRR. ERLRR–TMMLRR or ERL1LRR–TMMLRR was incubated with 10 µM EPF2-6xHis or EPF1-6xHis with increasing concentrations of EPFL9 and subjected to Ni+ resin. The bound proteins were eluted and further analyzed with SDS-PAGE followed by Coomassie blue staining.
EPFL4/6 interacts with ER, ERL1, or ERL2 with no need of TMM
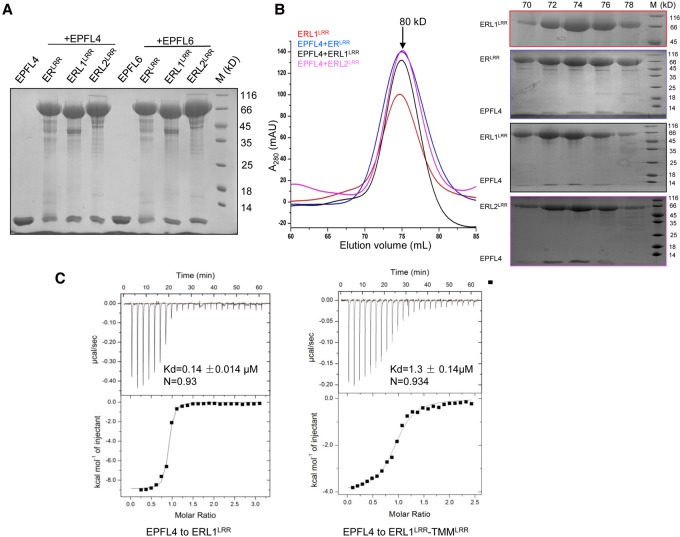
In contrast to EPF1,2, which are specialized for stomatal regulation, CHAL-related family members (EPFL4,5,6) appear to be dedicated to growth regulatory processes. Genetic evidence has suggested that ERfs also function as receptors of EPFL4,5,6 (Abrash et al. 2011; Uchida et al. 2012). To further support this conclusion, we used pull-down and gel filtration to assay the interaction of EPFL4 with ERLRR, ERL1LRR, or ERL2LRR. In contrast to EPF1,2, EPFL4 interacted strongly with all the three ERf proteins in the absence of TMMLRR (Fig. 6A,B). The EPFL4–ERL1LRR interaction was further verified by our ITC assay showing that their binding affinity was ∼0.14 µM (Fig. 6C). Unexpectedly, complexing with TMMLRR significantly compromised the binding affinity of ERL1LRR with EPFL4 (1.3 μM), indicating that TMM blocks ERL1LRR recognition of EPFL4. Similar results were obtained with EPFL6 and the ERL1LRR–TMMLRR complex (Supplemental Fig. S7).
Figure 6.
EPFL4 directly binds to ERfs independent of TMM. (A) ERfs alone are sufficient for interaction with EPFL4 or EPFL6. The 6xHis-tagged ERLRR, ERL1LRR, or ERL2LRR was incubated individually with EPFL4 or EPFL6 and subjected to Ni+ resin. Next, the bound proteins were eluted from Ni-NTA and further analyzed by SDS-PAGE with Coomassie blue staining. (B, left panel) Size exclusion chromatography analysis of the interaction between EPFL4 and ERfLRR. (Right panel) SDS-PAGE analysis of peak fractions from the left panel. (C) TMM dampens EPFL4 binding to ERL1LRR in vitro. Quantification of the binding affinity of EPFL4 with the ERL1LRR or ERL1LRR–TMMLRR by ITC. EPFL4 was titrated into ERL1LRR or ERL1LRR–TMMLRR protein in the ITC cell. Raw data (top panel) and integrated heat measurements (bottom panel) from ITC are shown. The calculated stoichiometry (N) and the dissociation constants (Kd) are indicated.
To elucidate the molecular mechanism underlying ERf recognition of EPFL4, we solved the crystal structure of ERL2LRR bound by EPFL4 (Fig. 7A; Supplemental Table S1). Like EPF1, EPFL4 also adopts an inverted V-shaped structure consisting of an anti-parallel β-sheet and a long N-terminal loop (Fig. 7A). These two structural elements mediate EPFL4's interaction with ERL2LRR by clamping one lateral side of the receptor. EPFL4 binding to ERL2LRR results in a structure resembling that of EPF1–ERL1LRR from the ternary complex ERL1LRR–TMMLRR–EPF1 (Fig. 7B). However, the apical loop region linking the first β strand and the N-terminal side of EPFL4 was not well defined by electron density (Fig. 7A). In contrast, the equivalent regions of both EPF1 and EPF2 make contact with their C-terminal sides and interact with TMMLRR (Supplemental Fig. S8A). Ala11 conserved in EPF1 and EPF2 (Supplemental Fig. S3B) packs tightly against Pro52 and Val48 in the two peptides (Supplemental Fig. S8A). This amino acid is substituted with the bulkier lysine residue at the same position of EPFL4, which is expected to disrupt its local conformation. These structural differences provide an explanation of why TMMLRR reduces the binding affinity of EPFL4 with ERL1LRR or ERL2LRR. To further support this model, we substituted the apical loop region of EPFL4 with that of EPF1 and assayed interaction of the swapped EPFL4 mutant (called EPFL4–EPF1) with ERL1LRR–TMMLRR using ITC. The results from the assay showed that EPFL4–EPF1 bound to the ERL1LRR–TMMLRR complex with an affinity of 0.42 µM, which is approximately three times as high as that of wild-type EPFL4 to the same complex (Supplemental Fig. S8B).
Figure 7.
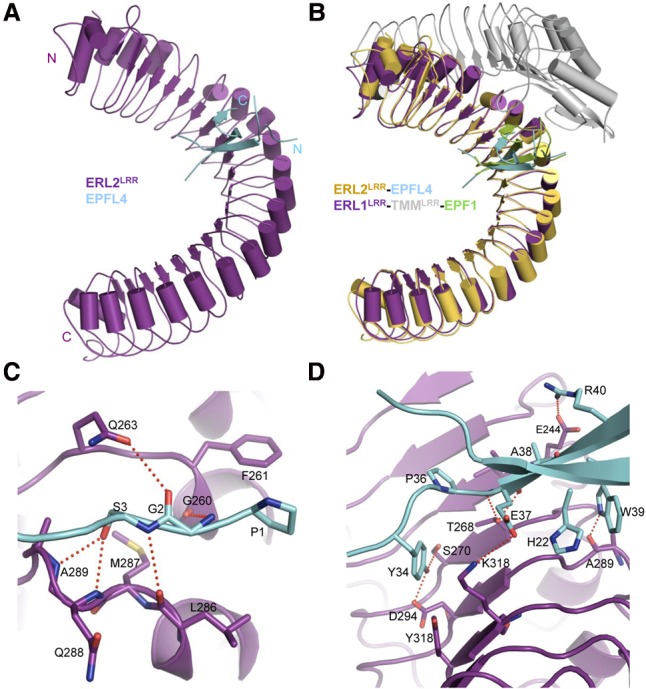
Recognition mechanism of EPFL4 by ERL2LRR. (A) Overall structure of EPFL4–ERL2LRR. (N) N terminus; (C) C terminus. (B) Structural alignment of ERL2LRR–EPFL4 and EPF1–ERL1LRR–TMMLRR complexes. The structure of ERL1LRR was used as the template for the alignment. (C) Detailed interactions of the N-terminal side of EPFL4 with ERL2LRR. (D) Detailed interactions of the central region of EPFL4 with ERL2LRR.
The “GS” motif from the N-terminal loop of EPFL4 forms a set of polar interactions with ERL2LRR similar to those of EPF1 with ERL1LRR (Fig. 7C). ERL2M287 is the equivalent of ERM282, and mutation of this ERL2 residue is expected to perturb the local conformation of the N-terminal loop-binding groove of ERL2 and consequently compromise ERL2's interaction with EPFL4. This is consistent with the reduced functionality of the ERM282I mutant (er-103) (Torii et al. 1996). Interaction of EPFL4 through its one β strand with the inner surface of ERL2LRR buries a total surface area of 468.0 Å2, which is much more extensive than that of EPF1 with the inner surface of ERL1LRR (291.0 Å2) (Fig. 7D). This structural observation affords an explanation of why EPFL4 is recognized by ERL2LRR in the absence of TMM. In addition to hydrogen-bonding with ERL2Thr268, EPFL4Glu37 that is substituted with Met38 in EPF1 forms a salt bridge with ERL2Lys318 (Fig. 7D). Salt bridges are also established between EPFL4Arg40 and ERL2Glu244. Tyr40 of EPF1 is substituted with Trp39 in EPFL4, which makes a hydrogen bond with the carbonyl oxygen of Ala289 from ERL2. Compared with EPF1, EPFL4 has two more residues well defined in electron density from the C-terminal side of the ERL2LRR-interacting β strand. One of these two residues, Tyr34, makes a pair of hydrogen bonds with ERL2Ser270, Asp294. Additionally, this EPFL4 residue also forms hydrophobic contacts with ERL2Tyr318. Structure-based sequence alignment showed that all of the amino acids critical for EPFL4 binding to ERL2LRR are conserved among EPFL4,5,6 (Supplemental Fig. S3B).
Discussion
Here we provide several lines of evidence showing that TMM and ERfs form constitutive complexes. Our structural studies demonstrate that both TMM and ER/ERL1 are directly involved in the recognition of the stomatal regulatory peptides EPF1,2. These results indicate that the constitutive TMM–ERf complexes as a whole function as the receptors of the two peptides. ERfs, however, recognize the growth regulatory peptides EPFL4,6 independent of TMM. In stark contrast to EPF1,2, EPFL4,6 were greatly compromised in interaction with ERL1,2 in the presence of TMM. These results are consistent with previous genetic models (Abrash et al. 2011; Shpak 2013). Overall, our results together with others suggest that TMM endows ERfs with ligand specificity, allowing ERf-mediated signaling to regulate TMM-dependent stomatal development and TMM-independent responses (Supplemental Fig. S9). The requirement of TMM for EPF1,2 interaction with ERfs suggests that TMM might have evolved to specifically regulate stomatal development. This is in line with the genomic studies indicating that TMM is present only in land plants (Caine et al. 2016; Olsen et al. 2016) and the fact that TMM is expressed specifically in the epidermis but not in other tissues (Nadeau and Sack 2002; Shpak et al. 2005). The ligand specificity switch mechanism enabled by TMM offers an explanation for the tissue-dependent phenotypes of tmm knockout plants. In leaves, TMM deletion blocks the negative regulation of stomata development by EPF1,2-induced ERf signaling. In tmm stems where EPFL4,6 are expressed, loss of the EPF1,2-induced ERf signaling activity would be compensated by the two EPFLs, which are recognized by ERfs without TMM, thus maintaining negative regulation of stomatal development. Many RLPs are present in plants (Gust and Felix 2014), and it will be intriguing to investigate whether other RLPs can use TMM-like mechanisms for ligand discrimination.
Binding of EPFs did not induce homodimerization of ERfs in either the presence or absence of TMM (Figs. 2B, 6B; Supplemental Fig. S5), suggesting that coreceptors are required for EPF-induced activation of ERfs. Indeed, a recent study demonstrated that the LRR-RK SERKs function as coreceptors with ERfs and TMM for the regulation of stomatal patterning (Meng et al. 2015). The structural mechanism of this remains unclear, but a clue to this can be gained from structural comparison between EPF1–ERL1LRR–TMMLRR and other SERK-containing protein complexes. When the LRR-RK PXY (phloem intercalated with xylem) (Zhang et al. 2016) and ERL1 are aligned, EPF1 is located at a position similar to the peptide ligand TDIF bound by PXYLRR–SERK2LRR (Supplemental Fig. S10). The hairpin loop of EPF1 is close to SERK2, suggesting that this region of EPF1 could be involved in the interaction with a SERK member or other partners to initiate ERf signaling. Interestingly, the hairpin loop region of the EPFL family members is much more variable as compared with the remaining parts (Supplemental Fig. S3B). Given the unequal redundancy of individual SERK members in stomatal patterning, this could be associated with the functional specificity of EPFL members (Meng et al. 2015). The positive regulation of stomatal development by EPFL9 can be explained by the possibility that its hairpin loop is unable to interact with a SERK member, although recruitment of a different partner by the loop remains formally possible. Consistently, EPF2 but not EPFL9 induced phosphorylation of downstream signaling components in plants (Lee et al. 2015). This possibility also agrees with the observation that substitution of this region of EPFL9 with that of EPF2 switched EPFL9 from positive to negative regulation of stomatal development (Ohki et al. 2011).
Negative regulation of stomata development in the stems of tmm knockout plants (Abrash and Bergmann 2010; Abrash et al. 2011) where EPF1,2 signaling is silenced suggests that the growth regulatory EPFL4,6 can potentiate the transduction of signals in the epidermis. On the other hand, although TMM is required for ERL1 recognition of EPF1, the structure of EPF1–ERL1 is similar to that of EPFL4–ERL2, in particular around the anti-parallel β sheet of EPF1 and EPFL4, indicating that TMM binding has no impact on the relative orientation between ERL1LRR and EPF1. These results collectively suggest that EPF1–ERL1 and EPFL4–ERL2 may have shared coreceptors for signaling. In the future, it will be of interest to investigate whether SERK members have a role in signaling induced by EPFLs such as EPFL4,6.
The present study together with others demonstrates that TMM–ERf complexes and ERfs are the receptors of EPF1,2, EPFL9, and EPFL4,6, respectively (Abrash et al. 2011; Lee et al. 2012, 2015; Uchida et al. 2012). Our data also suggest possible ligand–receptor interactions between other EPFLs and ERfs. The “GS” motif is highly conserved among all of the EPFL members and is involved in the interaction with ERfs for both EPF1,2 and EPFL4. Given the conserved structures of the EPFL members as supported by modeling (Ohki et al. 2011) and structural studies, these data suggest that other EPFL members can also interact with ERfs. This possibility is supported by a recent study showing that EPFL2 interacts with ERfs for the regulation of leaf margin morphogenesis in Arabidopsis (Tameshige et al. 2016). Interestingly, in addition to the “GS” motif, the ERL2LRR-interacting residues of EPFL4,5,6 are also highly conserved in EPFL1,2,3,8 (Supplemental Fig. S3B). Furthermore, like EPFL4,5,6, EPFL1,2,3,8 have shorter C-terminal sides than EPF1,2, which interact with TMMLRR for the last two EPF peptides. These results suggest that EPFL1,2,3,8 might also interact with ERfs independent of TMM. Indeed, our biochemical assay showed that EPFL8 interacted with ERL1LRR in the absence of TMMLRR (Supplemental Fig. S11). This hypothesis is also in line with the observations that overexpression of EPFL1,2 has no obvious stomatal phenotypes in a wild-type background but displays noticeably reduced stomatal density in tmm, phenocopying that of EPFL4,5,6 (Abrash et al. 2011). In contrast, both the ERf- and TMM-interacting residues of EPF1,2 are conserved in EPFL7, suggesting that the interaction of this EPFL peptide with ERfs might require TMM. Further studies are needed to investigate whether the function of EPFL7 is TMM-dependent.
Materials and methods
Protein expression and purification
The extracellular LRR domains of ER (residues 24–580), ERL1 (residues 24–580), ERL2 (residues 28–580), TMM (residues 47–470), EPF1 (residues 53–104), EPF2 (residues 69–120), EPFL9 (residues 58–102), EPFL4 (residues 59–109), EPFL6 (residues 106–156), and EPFL8 (residues 46–99) were cloned into modified pFastBac vectors containing an N-terminal hemolin (Hem) signal peptide or a cleavable N-terminal 6xHis-SUMO tag (Hem SUMO) with or without a C-terminal 6xHis tag (3HT or 3T, respectively). All of the proteins were expressed using the Bac-to-Bac baculovirus expression system (Invitrogen) protocols in High Five cells at 22°C. High Five cells were grown in ESF 921 medium (Expression Systems) by shaking at 120 rpm at 28°C until the density reached 2 × 106 cells per milliliter. One liter of cells (2.0 × 106 cells per milliliter) was infected with 20 mL of recombinant baculovirus. For purification of ERfLRR–TMMLRR, baculoviruses of ERf and TMM were added at the same time. The supernatant was harvested via centrifugation after 60 h of infection. The supernatant was flowed through Ni-NTA (Novagen). Bound proteins were eluted by buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, and 250 mM imidazole and further purified by size exclusion chromatography (Hiload 16/60Superdex 200 prep grade, GE Healthcare) in a buffer containing 10 mM Bis-Tris (pH 6.0) and 100 mM NaCl. To obtain the ERfLRR–EPF4 or ERfLRR–TMMLRR–EPF1/2 complexes, the purified ERfLRR or ERfLRR–TMMLRR and EPF proteins were mixed and incubated for 30 min at 4°C. The mixture was subsequently subjected to gel filtration (Hiload 16/60, Superdex 200 prep grade, GE Healthcare) in a buffer containing 10 mM Bis-Tris (pH 6.0) and 100 mM NaCl. Fractions corresponding to the complex were pooled together and subjected to ultrafiltration to ∼10 mg/mL for crystallization.
Crystallization, data collection, structure determination, and refinement
Crystallization experiments were performed by hanging drop vapor diffusion methods by mixing 1.0 µL of protein with 1.0 µL of reservoir solution at 18°C. Diffraction-quality crystals of ERL1LRR–TMMLRR were obtained in the buffer containing 4% (v/v) tacsimate (pH 6.0) and 10% (w/v) polyethylene glycol 3350. Good-quality crystals of ERL1LRR–TMMLRR–EPF1 and ERL1LRR–TMMLRR–EPF2 were obtained in the buffer containing 0.1 mM ammonium citrate tribasic (pH 7.0) and 10% polyethylene glycol 3350. For ERL2–EPF4, high-quality crystals emerged in the buffer containing 0.1 M MES monohydrate (pH 6.5) and 12% polyethylene glycol 20000. All of the crystals were flash-frozen using the reservoir buffer plus 15% glycerol as the cryoprotectant. All of the diffraction data sets were collected at the Shanghai Synchrotron Radiation Facility (SSRF) on beam line BL17U1. All of the data were processed using the HKL2000 software package (Otwinowski and Minor 1997). The crystal structure of ERL1LRR–TMMLRR was determined by molecular replacement (MR) with PHASER (McCoy et al. 2007) using the structure of FLS2 (Protein Data Bank: 4MN8) as the initial searching model. The model from MR was built with the program Coot (Emsley and Cowtan 2004) and subsequently subjected to refinement by the program PHENIX (Adams et al. 2002). The other crystal structures were determined by MR using the structure of ERL1LRR–TMMLRR or ERL1LRR as the initial searching model. The final refinement statistics are summarized in Supplemental Table 1. Structural figures were drawn using the program PyMOL (http://www.pymol.org).
ITC
The binding affinities of ERL1LRR and ERL1LRR–TMMLRR with EPFs were measured using ITC200 (Microcal LLC) at 25°C in a buffer containing 10 mM Bis-Tris (pH 6.0) and 100 mM NaCl. Approximately 0.3 mM EPF peptides was injected into the stirred calorimeter cell (250 µL) containing 0.03 mM ERL1LRR or ERL1LRR–TMMLRR with 24 × 1.5 µL at 150-sec intervals. The stirring speed was 750 rpm. The heat of dilution generated by the titration of peptides into the buffer was subtracted. Measurements of the binding affinity of all of the titration data were analyzed using Origin software (MicroCal Software).
Pull-down assay
The purified 6xHis-tagged ERfLRR protein with excess EPFs was mixed and incubated with 40 µL of Ni+ resin for 30 min on ice. The resin was washed four times with 1 mL of washing buffer containing 25 mM Tris (pH 8.0), 250 mM NaCl, and 15 mM imidazole. All of the resins were added with 100 µL of SDS loading buffer and heated for 5 min to 100°C. Next, the supernatants containing bound proteins were separated by SDS-PAGE, and proteins were detected by Coomassie blue staining. Similar procedures were used to test the interaction of ERL1LRR–TMMLRR with various 6xHis-tagged EPF1 mutants.
Ligand competition assays
The ERLRR–TMMLRR or ERL1LRR–TMMLRR complex was mixed and incubated with 10 µM EPF2-6xHis or EPF1-6xHis peptides in the presence or absence of different concentrations of bioactive EPFL9 peptide for 1 h at 4°C with gentle agitation. The reaction mixture was then flowed through Ni+ beads and washed four times with 1 mL of wash buffer to separate the bound and free ERLRR–TMMLRR or ERL1LRR–TMMLRR. Bound proteins were eluted by elution buffer, added to SDS loading buffer for 5 min at 100°C, and detected on SDS-PAGE gel by Coomassie blue staining.
Gel filtration assay
The ERfLRR, ERfLRR–TMMLRR, and EPFs proteins, purified as described above, were mixed and incubated for 30 min at 4°C before being subjected to gel filtration analysis (Hiload 16/60 Superdex 200 prep grade, GE Healthcare) in a buffer containing 10 mM Bis-Tris (pH 6.0) and 100 mM NaCl. Samples from relevant fractions were applied to SDS-PAGE and visualized by Coomassie blue staining.
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Columbia was used as the wild type. Tmm-1 (CS6140) was acquired from the Arabidopsis Biological Resource Center (Ohio State University). Seedlings were grown on modified Murashige and Skoog medium plates supplemented with 1% (w/v) sucrose. Plants were grown in a soil mixture with a 1:1 ratio of Promix PGX (Premier Horticulture, Inc.) and vermiculite (Pametto Vermiculite Co.) supplemented with Miracle-Gro (Scotts) and ∼3.5 mg/cm3 Osmocoat 15-9-12 (Scotts) at 20°C under long-day conditions (18 h light/6 h dark).
Plasmid construction and plant transformation
To construct a plasmid carrying proTMM:TMM-3XHA (pESH 730), the TMM coding region was cloned behind the 540-base-pair TMM promoter sequence and in-frame with 3× hemagglutinin (HA) tag (YPYDVPDYAGAYPYDVPDYAQLYPYDVPDYA) into the pZP222 vector. A 35S terminator was placed after the stop codon of 3xHA. The L281D and E379R substitutions were introduced into TMM by overlap extension PCR using pESH730 as a template. The amplified fragment was digested with RsrII and PstI, inserted into pESH 730, and sequenced. The plasmids carrying proTMM:TMM(L281D)-3XHA and proTMM:TMM(E379R)-3XHA sequences were named pESH 731 and pESH 732, respectively. The described plasmids were introduced into an Agrobacterium tumefaciens strain (GV3101/pMP90) by electroporation and into tmm-1 mutants by the floral dip method (Clough and Bent 1998). The transgenic plants were selected on Murashige and Skoog medium plates containing gentamicin.
Differential interference contrast (DIC) microscopy
Samples were fixed overnight with ethanol:acetic acid (9:1 [v/v]). After fixation, samples were rehydrated with an ethanol series to 30% (v/v) ethanol and cleared in chloral hydrate solution (chloral hydrate:water:glycerol, 8:1:1 [w/v/v]). Microscopic observations were performed using a Nikon Eclipse 80i microscope with DIC optics; images were obtained with a 12-megapixel cooled color DXM-1200c (Nikon) camera. The stomatal index was calculated using the formula (number of stomata/total number of epidermal cells) × 100. The stomatal clustering fraction was calculated using the formula number of clustered stomata/total number of stomata.
EPF1 bioassay
Wild-type Arabidopsis seeds were germinated on modified Murashige and Skoog medium plates supplemented with 1× Gamborg B5 vitamins and 1% (w/v) sucrose. One day after germination, the seedlings were transferred to a liquid MS-0 medium containing 1% (w/v) sucrose, 40 mg/mL timentin, and 10 mM EPF1 or EPF1 with substitutions. After 5 d of incubation at 20°C under long-day conditions (18 h light/6 h dark), the stomatal index was measured on the abaxial side of cotyledons by DIC microscopy. For microscopy, seedlings were fixed overnight in ethanol:acetic acid (9:1), rehydrated with an ethanol series to 50%(v/v) ethanol, and then cleared in a mixture of 8:1:1 chloral hydrate:water:glycerol.
Supplementary Material
Acknowledgments
We thank S. Huang and J. He at the Shanghai Synchrotron Radiation Facility (SSRF) for assistance with data collection. We thank Dr. Heqiao Zhang and Dr. Zhen Wu for assistance with data collection and ITC assays. This research was funded by Projects of International Cooperation and Exchanges National Natural Science Foundation of China (31420103906), the Chinese Ministry of Science and Technology (2015CB910200), and the National Natural Science Foundation of China (31421001) to J.C., and the National Science Foundation (IOS-0843340) to E.D.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.297580.117.
References
- Abrash EB, Bergmann DC. 2010. Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455. [DOI] [PubMed] [Google Scholar]
- Abrash EB, Davies KA, Bergmann DC. 2011. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954. [DOI] [PubMed] [Google Scholar]
- Caine RS, Chater CC, Kamisugi Y, Cuming AC, Beerling DJ, Gray JE, Fleming AJ. 2016. An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 143: 3306–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- De Smet I, Voss U, Jurgens G, Beeckman T. 2009. Receptor-like kinases shape the plant. Nat Cell Biol 11: 1166–1173. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Geisler M, Yang M, Sack FD. 1998. Divergent regulation of stomatal initiation and patterning in organ and suborgan regions of the Arabidopsis mutants too many mouths and four lips. Planta 205: 522–530. [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD. 2000. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Felix G. 2014. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr Opin Plant Biol 21: 104–111. [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. 2007. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T. 2009. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE. 2009. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869. [DOI] [PubMed] [Google Scholar]
- Hunt L, Bailey KJ, Gray JE. 2010. The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol 186: 609–614. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. 2010. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol 51: 1–8. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU. 2012. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YC, Putarjunan A, Han SK, Avila J, Torii KU. 2015. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, Shan L. 2015. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol 25: 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomitsu A, Sawa S, Ishida T. 2015. Plant peptide hormone signalling. Essays Biochem 58: 115–131. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. 2002. Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Ohki S, Takeuchi M, Mori M. 2011. The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat Commun 2: 512. [DOI] [PubMed] [Google Scholar]
- Olsen JL, Rouze P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Shpak ED. 2013. Diverse roles of ERECTA family genes in plant development. J Integr Plant Biol 55: 1238–1250. [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. 2005. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293. [DOI] [PubMed] [Google Scholar]
- Song W, Han Z, Wang J, Lin G, Chai J. 2016. Structural insights into ligand recognition and activation of plant receptor kinases. Curr Opin Struct Biol 43: 18–27. [DOI] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. 2010. Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244. [DOI] [PubMed] [Google Scholar]
- Tameshige T, Okamoto S, Lee JS, Aida M, Tasaka M, Torii KU, Uchida N. 2016. A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Curr Biol 26: 2478–2485. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. 1996. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, Tasaka M, Torii KU. 2012. Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc Natl Acad Sci 109: 6337–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sack FD. 1995. The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7: 2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin X, Han Z, Wang J, Qu LJ, Chai J. 2016. SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol Plant 9: 1406–1414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.