Abstract
Diverse cellular processes such as autophagic protein degradation require phosphoinositide signaling in eukaryotic cells. In the methylotrophic yeast Pichia pastoris, peroxisomes can be selectively degraded via two types of pexophagic pathways, macropexophagy and micropexophagy. Both involve membrane fusion events at the vacuolar surface that are characterized by internalization of the boundary domain of the fusion complex, indicating that fusion occurs at the vertex. Here, we show that PpAtg24, a molecule with a phosphatidylinositol 3-phosphate-binding module (PX domain) that is indispensable for pexophagy, functions in membrane fusion at the vacuolar surface. CFP-tagged PpAtg24 localized to the vertex and boundary region of the pexophagosome-vacuole fusion complex during macropexophagy. Depletion of PpAtg24 resulted in the blockage of macropexophagy after pexophagosome formation and before the fusion stage. These and other results suggest that PpAtg24 is involved in the spatiotemporal regulation of membrane fusion at the vacuolar surface during pexophagy via binding to phosphatidylinositol 3-phosphate, rather than the previously suggested function in formation of the pexophagosome.
INTRODUCTION
Autophagy is a nonselective protein degradation pathway observed in all eukaryotes that is involved in cellular maintenance and development. During this process, cytoplasmic proteins and organelles are delivered to vacuoles or lysosomes for recycling of cellular proteins. A special type of autophagy called pexophagy regulates the selective degradation of peroxisomes via one of the two autophagic pathways, macropexophagy or micropexophagy.
The methylotrophic yeast Pichia pastoris can undergo either pexophagic pathway depending on the carbon source used for pexophagy induction (macropexophagy with ethanol and micropexophagy with glucose) and develops large peroxisomes when grown on methanol as the sole carbon source (Figure 1; Tuttle and Dunn, 1995). During macropexophagy, peroxisomes are selectively sequestered by the newly formed isolation membrane, which wraps around the peroxisome to form a double-membraned pexophagosome. The pexophagosome is then delivered to a vacuole, where its outer membrane fuses with the vacuolar membrane. During micropexophagy, a rounded vacuole in methanol-grown cells septates, forming a new compartment proximal to the peroxisomal cluster. Vacuolar septation is repeated until the entire peroxisome cluster is almost enclosed. A membrane-bound flattened sac, the micropexophagy-specific membrane apparatus (MIPA), is formed between the membrane tips of an engulfing vacuole to complete the engulfment and sequestration of peroxisomes from the cytosol, forming a micropexophagic body within the lumen of the vacuole. At the last stage of micropexophagy, the incorporated peroxisomes and vacuolar membranes are destroyed to produce the amino acids and lipids for recycling (Figure 1; Sakai et al., 1998; Mukaiyama et al., 2002, 2004; Oku et al., 2003).
Figure 1.
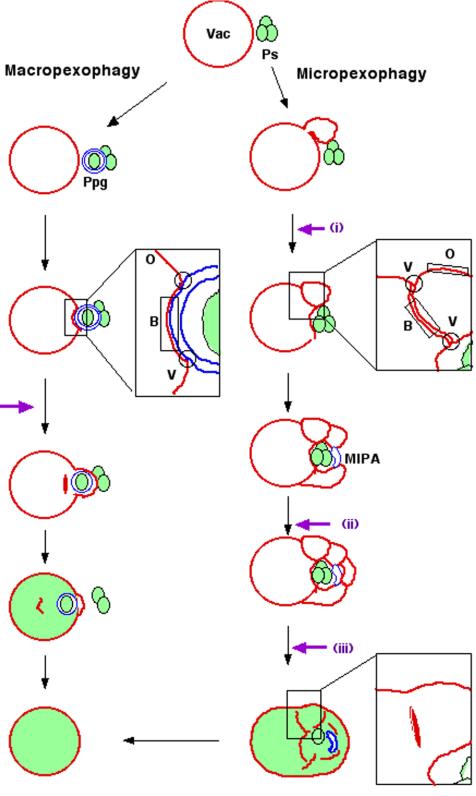
Schematic model for vacuolar membrane dynamics of two distinct pexophagic pathways and subdomains on the vacuolar membrane. Violet arrows represent possible fusion events occurring at the vacuolar membrane surface. Ps, peroxisome; Vac, vacuole; MIPA, micropexophagic apparatus; Ppg, pexophagosome; V, vertex domain; B, boundary domain; O, outside edge domain. Left: macropexophagy. A newly synthesized pexophagosome envelops a single peroxisome within a cluster, and subsequently its outer membrane fuses with vacuolar membrane. This fusion event could occur in two different ways: fusion at a contact point or fusion at a vertex. This figure represents the fusion at a vertex. Although fusion at the vertex involves internalization of the boundary region (as described in this figure), fusion at the contact point does not. Right: micropexophagy. Membrane fusion (or scission) at the vacuolar surface should occur at three different steps (indicated by violet arrows): i) septation of the vacuole; ii) completion of peroxisome engulfment; and iii) degradation of the septum.
Both pexophagic pathways involve membrane fusion events at the vacuolar membrane surface. In macropexophagy, a heterotypic membrane fusion occurs between the pexophagosome and vacuole, delivering peroxisomes into the vacuolar lumen. On the other hand, during micropexophagy, homotypic vacuolar membrane fusion events occur at three distinct steps: i) at septation of the vacuole, ii) at fusion between the membrane tips of the invaginating vacuole to complete peroxisome engulfment mediated by MIPA, and iii) at the internalization of septated vacuoles into the vacuolar lumen (Figure 1).
During homotypic vacuolar fusion, three distinct membrane domains are identifiable on clusters of vacuoles: the vertex, boundary edge, and outside edge (Wang et al., 2002). Recently, homotypic vacuolar fusion was reported to occur at the vertex of the vacuole, where many known components of the fusion machinery are enriched, resulting in the release of the boundary membrane into the vacuolar lumen (Wickner and Haas, 2000; Wickner, 2002; Wang et al., 2003; Weisman, 2003). However, it remained unknown whether heterotypic fusion occurs also at the vertex ring.
We and others have identified genes that are essential for pexophagy in P. pastoris. The nomenclature for these yeast autophagy-related genes was recently standardized, resulting in their designation as ATG genes (Klionsky et al., 2003). Through biochemical and morphological analysis of P. pastoris Atg8 (PpAtg8) and its modification pathway, many ATG genes have been implicated in MIPA formation (Mukaiyama et al., 2004). This explains the overlapping function of these ATG genes in both pexophagic processes because, in both cases, the membrane structures attached to the peroxisomal surface (MIPA and pexophagosomes) are formed de novo. PpAtg26 is another known molecule that has been shown to be associated with MIPA during micropexophagy (Oku et al., 2003).
The second class of the genes involved in both micro- and macro-autophagic processes is genes related to phosphoinoside(s) (PtdIns). Macroautophagy requires the PtdIns 3-kinase Vps34, which functions as a part of the core complex consisting of Vps34, Vps15, and Vps30/Atg6 (Kihara et al., 2001). PpVPS15 and Hansenula polymopha VPS34 (HpVPS34) were also identified as yeast genes essential for micropexophagy and macropexophagy, respectively (Kiel et al., 1999; Stasyk et al., 1999; Mukaiyama et al., 2002). Yeast VPS15 encodes a protein that belongs to the serine/threonine protein kinase family and activates Vps34 (Stack et al., 1993). Vps34 in turn phosphorylates PtdIns at the D-3 position of the inositol and represents the only detectable PtdIns 3-kinase activity in Saccharomyces cerevisiae (Schu et al., 1993). The Vps15-Vps34 interaction contributes not only to localization of Vps34 to the correct membrane compartment, but also to stimulation of its PtdIns 3-kinase activity (Stack et al., 1995).
Here, we first examined whether or not the heterotypic membrane fusion between the vacuole and pexophagosome during macropexophagy occurs at the vertex ring. Next, we identified a gene that is essential for both modes of pexophagy. PpATG24 (previously designated PAZ16; Klionsky et al., 2003) contains a Phox homology (PX) domain, which belongs to a class of PtdIns(3)P-binding modules that includes the FYVE-(Fab1, YOTB, Vac1, EEA1 homology) domain and the PH (pleckstirn homology) domain. Our results show that PpAtg24 functions in the fusion events at the vacuolar membrane surface during pexophagy rather than in the formation of the pexophagosome and MIPA.
MATERIALS AND METHODS
Strains and Media
The P. pastoris strains used in this study are listed in Table 1. P. pastoris cells were grown in YPD medium (containing 1% yeast extract, 2% bactopeptone, and 2% glucose), methanol medium (containing 0.67% yeast nitrogen base without amino acid, 0.5% [vol/vol] methanol), and glucose medium (containing 0.67% yeast nitrogen base without amino acid, 2% glucose) or ethanol medium (containing 0.67% yeast nitrogen base without amino acid, 0.5% [vol/vol] ethanol), supplemented with appropriate amino acid(s) (100 μg/ml for arginine, 100 μg/ml for histidine). All the components used in these media were purchased from Difco Becton Dickinson (Lincoln Park, NJ).
Table 1.
Pichia pastoris strains used in this study
| Strain | Designation | Genotype (explanation, plasmid used for transformation) | Reference |
|---|---|---|---|
| PPY12 | Wild-type | arg4 his4 | (Sakai et al., 1998) |
| STW1 | GEP-PTS1 | PPY12 his4::PAOX1-GFP-PTS1-HIS4 (pTW51) | (Sakai et al., 1998) |
| SA1017 | YFP-PpAtg8 | PPY12 arg4:: PATG8-YFP-PpATG8 (pSAP115) | Mukaiyama et al. (2003) |
| YAP2401 | Ppatg24Δ[ρ] | PPY12 atg24::ZeocinR (pYA101) | This study |
| YAP2402 | Ppatg24Δ[ρ] | STW1 atg24::ZeocinR (pYA101) | This study |
| YAP2403 | PpATG24/Ppatg24Δ[ρ] | YAP2401 his4::PATG24-PpATG24-HIS4 (pYA101) | This study |
| YAP2404 | PpAtg24-CFP | YAP2401 his4::PATG24-PpATG24-CFP-HIS4 (pYA103) | This study |
| YAP2405 | PpAtg24-CFP/YFP-PTS1 | YAP2403 arg4::PAOX1-YFP-PTS1-ARG4 (pYA005) | This study |
| YAP2406 | PpAtg24-CFP/YFP-2ξFYVE | YAP2403 arg4::PACT1-2 × FYVEHrs-ARG4 (pYA201) | This study |
| YAP2407 | PpAtg24-CFP-PXΔ | YAP2401 his4::PATG24-PpATG24-CFP-PXΔ-HIS4 (pYA104) | This study |
| YAP2410 | YFP-PpAtg8/Ppatg24Δ[ρ] | YAP2401 arg4:: PATG8-YFP-PpATG8-ARG4 (pSAP115) | This study |
| YAP2411 | PpAtg24-CFP/YFP-PpAtg8 | YAP2404 arg4 :: PATG8-YFP-PpATG8-ARG4 (pSAP115) | This study |
| YAP2412 | PpAtg24-CFP/PpVac8-YFP | YAP2404 arg4 :: PVAC8-PpVAC8-YFP-ARG4 (pTN204) | This study |
| YAP2413 | PpAtg24-CFP/YFP-PpAtg17 | YAP2404 arg4 :: PGAP-YFP-PpVAC8-ARG4 (pYA405) | This study |
| YAP2414 | PpAtg24-CFP/Ppvps15 | Ppvps15 his4:: PATG24-PpATG24-CFP-HIS4 (pYA103) | This study |
| YAP2415 | YFP-PpAtg8/CFP-PTS1/Ppatg24Δ[ρ] | YAP2410 his4::PAOX1-CFP-PTS1-HIS4 (pYA006) | This study |
| YAP0004 | YFP-PpAtg8/CFP-PTS1 | SA1017 his4:: PAOX1-CFP-PTS1-HIS4 (pYA006) | This study |
Isolation of Micropexophagy-defective Mutants
Micropexophagy-defective mutants were isolated by alcohol oxidase (Aox) screening after a glucose shift from the methanol condition, from among Zeocin-resistance mutants as previously described (Mukaiyama et al., 2002).
Detection of Alcohol Oxidase Activity and Alcohol Oxidase Degradation
The Aox assay was done to assess micropexophagy competence. The YPD-plated P. pastoris colonies were replica plated onto a nylon membrane filter that had been placed on a methanol plate. The plates were incubated at 28°C for 24 h, during which time the peroxisomal enzyme, Aox, was induced. The filter membranes were then transferred to a glucose plate to induce micropexophagic degradation of peroxisomes and, thereby, the degradation of Aox. After 12 h on glucose plates, colonies on the filter were visualized for Aox activity in situ as previously described (Mukaiyama et al., 2002; Oku et al., 2003). For macropexophagy competence, a modified method was applied in which Aox degradation was induced on an ethanol plate and the ethanol shift was prolonged to 48 h.
Morphological Analysis of Micropexophagy
The observation of peroxisome-vacuole dynamics was performed as previously described (Mukaiyama et al., 2002; Oku et al., 2003). To visualize the localization of proteins fused with fluorescence proteins, cells were labeled with 0.93 μg/ml FM4-64 (Molecular Probes, Eugene, OR) during a 12-h incubation in methanol medium with a starting OD600 of 0.5. Samples for the live-cell monitoring experiment were prepared as follows: cells adapted in ethanol medium for 30 min were collected and immediately attached onto a bottom-holed Petri dish precoated with concanavalin A (Wako, Osaka, Japan). Nonattached cells were subsequently aspirated off, and the dish was filled with ethanol medium. Images were acquired with a Sensys CCD (Charged Coupled Device) camera (Photometrics, Tucson, AZ) and analyzed on MetaMorph imaging software (Universal Imaging, West Chester, PA).
Plasmid Construction
The oligonucleotide primers used in this study are listed in Table 2. The flanking DNA of PpATG24 was prepared from two mutant alleles, in which pREMI-Z was inserted in the amino-terminal and carboxy-terminal halves. To obtain the full-length open reading frame (ORF) of PpATG24, plasmids containing pREMI-Z were rescued from each genomic DNA fragment by digestion with KpnI. The nucleotide sequence of PpATG24 has been submitted to DDBJ/EMBL/GenBank databases under Accession No. AB191168. The plasmids with the amino-terminal and carboxy-terminal insertions were digested with XbaI/HindIII and HindIII/StuI, respectively, and their fragments, 1.2 and 1.4 kb, were ligated into the XbaI/SmaI sites of pBluescript II SK+. Using a NotI/XhoI digestion, 2.5 kb of the PpATG24 cassette was ligated into pHM100 (Mukaiyama et al., 2004). The resulting plasmid, pYA101, was digested with BspEI and electoporated into Pichia cells.
Table 2.
Oligonucleotide primers used in this study
| Primer name | DNA sequence |
|---|---|
| PAZ16-delta-Fw | GCTGTTTCCGATCCCCCACA |
| PAZ16-delta-RV | TGTGAATTGGTCACTCATGCTAGTATTGTC |
| PAZ16-C-SseI-Fw | AAGGTCTGGTTGATGCCTGCAGGATAGAAGGCCTTGAGC |
| PAZ16-C-SseI-Rv | GCTCAAGGCCTTCTATCCTGCAGGCATCAACCAGACCTT |
| SseY/C/BFPFw | CCTGCAGGCGGTGAGCAAGGGCGAGGA |
| SseY/C/BFPRv | CCTGCAGGAACTTGTACAGCTCGTCCATGC |
| PAZ16-PX-delta-Fw | TTACGACGTCTCGGGGAGGTATTTGCTCAT |
| PAZ16-PX-delta-Rv | CCGAGACGTCGTAAATTGGAATACGTTCAAACA |
| BamARG4-Fw | ACGGGATCCTACCTGCCCTCACGGTGGTTA |
| BamARG4-Rv | CGGGATCCTTCTGTACCGGTTTACAGAAGG |
| PAox-5′-SacII | TCCCCGCGGAGATCTAACATCCAAAGACGA |
| PAox-3′-PstI | AACTGCAGCTCGTTTCGAATAATTAGTTGT |
| PAct1-5′-SacII | TCCCCGCGGTCGCTGGTAATCCCGGCTTTT |
| PAct1-3′-PstI | AACTGCAGCATTGTATTGATGAATTTCTTTTACT |
| CFP-SKL-5′ | AAAACTGCAGAATGGTGAGCAAGGGCGAGGA |
| CFP-SKL-3′ | AACTGCAGTTATAATTTGGACTTGTACAGCTCGTCCATGC |
| YFP-2xFYVE-5′-SseI | AGCTCCTGCAGGGGATCCATGGTGAGCAAG |
| YFP-2xFYVE-3′-SalI | ACGCGTCGACTTATGCCTTCTTGTTCAGCT |
| PX-5′-BamHI | CGCGGATCCCCTCCCCGAGATGTTTATATA |
| PX-3′-SalI | TGCGGTCGACATTTACGCTGTCTTCCAAAA |
| BamHI-ATG17-Fw | CGGGATCCGATCAATTAAAGGACTGGACAG |
| ATG17-XhoI-Rv | CCGCTCGAGTTACTTCCTTCCCAGTATTGC |
| PpVac8Fw-KpnI | GGGGTACCCGTCGGGTTGCCATTAAACGGA |
| PpVac8C′SseIRv | AGCTTTCCTGCAGGGCTTTGATCATCTCCAAGATTTGTTG |
The PpATG24 disruption plasmid, pYA102 was constructed as follows from a plasmid recovered from the mutant with a Zeocin-resistance cassette inserted in the carboxy-terminal half: Inverse PCR was performed with the primers paz16-delta-Fw and paz16-delta-Rv to delete the PpATG24 ORF, and this amplified fragment was self-ligated to yield pYA102. To perform homologous recombination, pYA102 was digested with KpnI and introduced into Pichia cells.
For expression of CFP-fusion PpAtg24, pYA103 was constructed. pYA101, a plasmid containing the PpATG24 ORF, was PCR amplified with the primers Paz16-C-SseI-Fw and Paz16-C-SseI-Rv, and the 0.7-kb Sse8387I-Sse8387I. The CFP-coding fragment was PCR-amplified with the primers SseY/C/BFPFw and SseY/C/BFPRv, using the pECFP vector (Clontech Laboratories, Palo Alto, CA) as a template. These PCR fragments were digested with Sse8387I and ligated with T4 DNA ligase to yield pYA103. For deletion of the PX domain, the primers PAZ16-PX-delta-Fw/PAZ16-PX-delta-Rv, were used in inverse PCR using pYA103 as a template, and the PCR fragments were digested with AatII and self-ligated, yielding pYA104. pYA104 was linearized with BspEI and introduced via the PpHIS4 marker for chromosomal integration.
To coexpress various fusion proteins, expression vectors for Pichia cells were constructed. For PpARG4 locus chromosomal integration, pSAP500 was constructed based on pHM100. The 2.0-kb BamHI-BamHI PpARG4-coding fragment was amplified by PCR using P. pastoris genomic DNA as a template and the primers BamARG4-Fw and BamARG4-Rv. The PCR fragment was replaced with the PpHIS4 gene in pHM100. These plasmids, pHM100 and pSAP500 were used as the bone plasmids for other expression vectors, pYA001, pYA002, pYA003, and pYA004. The PCR-amplified fragments encoding the AOX1 and ACT1 promoter from P. pastoris genomic DNA, PAox1-5′-SacII/PAox1-3′-PstI and PAct1-5′-SacII/PAct1-3′-PstI, were ligated to pHM100 and pSAP500, respectively, to overexpress various DNA fragments. pYA006 or pYA005, and pYA201, were prepared for labeling of intracellular peroxisomes and PtdIns(3)P, respectively. The CFP-PTS1 and YFP-PTS1 fragments were prepared by PCR using the primers CFP-SKL-5′ and CFP-SKL-3′, and was ligated into pGEM T-Easy (Promega, Madison, WI). Subsequently the CFP-PTS1 and YFP-PTS1 0.7-kb fragments were EcoRI digested and ligated into pIB4 and pYA002 to yield pYA006 and pYA005, respectively. The mouse Hrs FYVE domain construct YFP-2xFYVEHrs domain was kindly provided by Dr. T. Yoshimori (National Institute of Genetics). Fragments amplified by PCR using the primers YFP-2xFYVE-5′-SseI and YFP-2xFYVE-3′-SalI were ligated to pYA005 to yield pYA201. These plasmids were linearized by AatII digestion and introduction into Pichia cells in the PpARG4 locus for chromosomal integration.
PpATG17 and PpVAC8 were cloned from Pichia genomic DNA by PCR using the primer sets BamHI-ATG17-Fw/ATG17-XhoI-Rv and PpVac8Fw-KpnI/PpVac8C′SseIRv, respectively. After construction of the YFP-fusion plasmid, these were introduced into Pichia cells.
Immunoblot Analysis
SDS-PAGE and immunoblotting were performed using 1:3000 diluted rabbit anti-GFP antiserum (Molecular Probes) and 1:10000 diluted goat anti-rabbit IgG antibody conjugated to horseradish peroxidase (Amersham Pharmacia Biotech, Piscataway, NJ) for detection of PpAtg24-CFP. The detection was performed using the Enhanced Chemiluminescent detection system (Amersham Pharmacia Biotech).
Electron Microscopy
P. pastoris cells were subjected to rapid freezing and freeze-substitution fixation and observed as previously described (Baba et al., 1997).
Subcellular Fractionation
P. pastoris cells were grown on methanol medium for 15 h with a starting OD600 of 0.5 and shifted to SD medium for 1 h. One thousand OD600 units of the cells were harvested and spheroplasted as previously reported (Mukaiyama et al., 2004). The cells were then osomotically lysed at 4°C in 10 ml of Buffer A (0.2 M sorbitol, 50 mM potassium acetate, 2 mM EDTA, 20 mM HEPES-KOH, pH 6.8, 1 mM dithiothreitol, 20 μg/ml phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 0.7 μg/ml pepstain A). After 10-min centrifugation at 3000 × g in a Hitachi 50F6A rotor (Tokyo, Japan), the supernatant was retrieved and further centrifuged at 100,000 × g for 1 h in a Beckman TLA110 rotor (Fullerton, CA). The precipitate equivalent to 50 OD600 unit cells was mixed with 1 ml of each buffer as follows: Buffer A alone, high-salt solution consisting of 0.67 M potassium acetate and 0.3 M KCl, or 1% Triton X-100 in Buffer A. The mixtures were kept at 25°C for 10 min and were then centrifuged at 100,000 × g for 1 h. Next, the supernatant fractions were trichloroacetic acid-precipitated and resuspended in Laemmli sample buffer (Ausubel et al., 1987). The persistent pellet fractions were also solubilized in Laemmli sample buffer. All samples were subjected to SDS-PAGE and immunoblot analysis, as described above.
Protein Purification and Protein-Lipid Overlay Assay
To construct the GST-PpAtg24 PX domain fusion plasmid, pYA107, the DNA sequence encoding the PX domain of PpAtg24 (amino acids 64-188) was amplified by PCR using the primers PX-5′-BamHI and PX-3′-SalI. The PCR product and vector pGEX6P-1 (Amersham Biosciences, Piscataway, NJ) were digested with BamHI and SalI and subsequently ligated. The resulting plasmid, pYA107, was transformed into Escherichia coli Rosetta DE3 (Novagen, Madison, WI). Simultaneously, the GST fusion 2xFYVEHrs domain from mouse Hrs was also prepared.
The GST fusion protein was purified using a GSTrap FF column (Amersham Pharmacia Biotech). The protein-lipid overlay assay was done with PIP-Strip (Echelon Biosciences, Salt Lake City, UT) The membrane was incubated with 0.5 μg of purified GST-PpAtg24 PX domain or GST-2xFYVEHrs domain. Binding of purified protein to phosphoinositides was detected by enhanced chemiluminescence. Immunoblot analysis was performed as described above using 1:1000 diluted goat anti-GST antiserum and anti-goat antibody conjugated to horseradish peroxidase. Binding of GST alone to phosphoinositides was not detected.
RESULTS
Membrane Fusion between the Pexophagosome and Vacuole Involves Internalization of the Boundary Membrane
Our previous study revealed that YFP-PpAtg8 sequestered peroxisomes specifically after ethanol-adaptation under our experimental conditions (cf. Figure 6; Mukaiyama et al., 2004). To observe membrane fusion events between the pexophagosome and vacuole, sequential fluorescence images of FM4-64-stained vacuolar membranes and the pexophagosomal membrane marker YFP-PpAtg8 during macropexophagic conditions were taken at 3-min intervals (Figure 2A). After 3 min, membrane fusion between the pexophagosome and vacuole could be detected by the sudden flow of FM4-64 into the outer membrane of the pexophagosome and the simultaneous disappearance of the ring-shaped localization of pexophagosomal YFP-PpAtg8. At this time, we could also observe the boundary edge domain of the fused structure. After another 3 min, the vacuolar membrane at the boundary edge domain disappeared.
Figure 6.
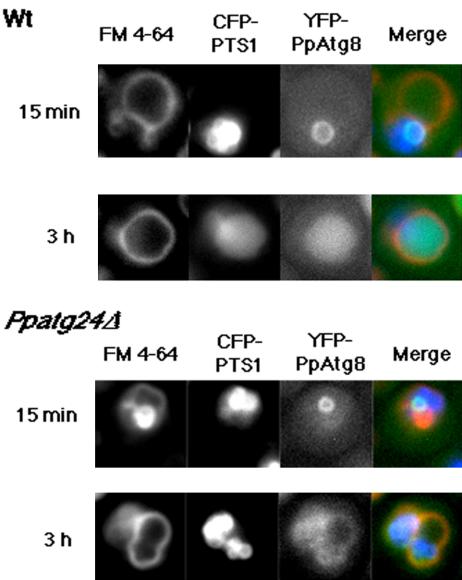
Dynamics of YFP-PpAtg8 localization during macropexophagy in the wild-type and the Ppatg24Δ strain. After ethanol-adaptation for 15 min, both the wild-type (strain YAP0004) and the Ppatg24Δ (YAP2415) strains showed a ring structure representing a pexophagosome. After 3 h, while YFP-PpAtg8 fluorescence could be observed in the vacuolar lumen in wild-type cells, YFP fluorescence in the vacuolar lumen was not observed in the Ppatg24Δ strain.
Figure 2.
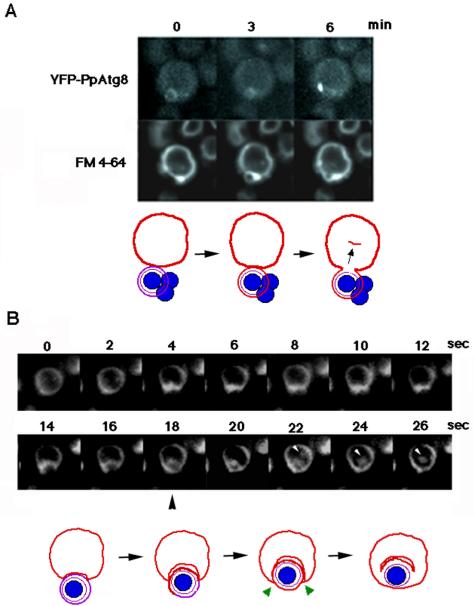
Single-cell observation of FM 4-64-stained vacuolar membrane dynamics during macropexophagy. (A) Sequential images of YFP-PpAtg8 dynamics taken at 3-min intervals. (B) Sequential images taken at 2-s intervals. White arrowheads represent the internalized membrane showing Brownian movement within the vacuolar lumen. FM 4-64 diffuses into the pexophaogome membrane after the fusion (black arrowhead). Bottom panels: a scheme for vacuolar membrane dynamics deduced from the top panels. Red line, vacuolar membrane originally stained with FM 4-64; violet line, pexophagosome membrane; blue circle, peroxisome; green arrow heads, fusion points at the vertex.
Fusion of membranes at the vertex involves internalization of the boundary membrane (Figure 1), whereas fusion at the contact point does not. To see whether or not the heterotypic membrane fusion between the pexophagosome and vacuole involves direct internalization of the vacuolar membrane, we followed in more detail the localization of FM 4-64 in a cell undergoing macropexophagy, capturing images at 2-s intervals. As shown in Figure 2B, vacuolar FM4-64 moved into the pexophaogome membrane, forming a ring of fluorescence after 18 s, indicating that FM 4-64 was distributed throughout the pexophaogome membrane. Thereafter, the boundary edge domain of the FM 4-64-stained membrane was internalized into the vacuolar lumen. The internalized component exhibited Brownian movement (22-26 s) and then disappeared after 1 min (unpublished data). This observation suggests that the membrane fusion between pexophagosome and vacuole occurs at the vertex in the fusion complex.
Although many molecules were identified on the vertex region of homotypic fusion complex between the vacuole (Wang et al., 2003), no Atg molecules were identified as those responsible for the heterotypic fusion between pexophagosome and vacuole. Therefore, we tried to identify an Atg molecule, which localizes to the vertex and is responsible for the heterotypic fusion between the pexophagosome and vacuole.
PpATG24 Is Essential for Micropexophagy and Macropexophagy
Previously, we identified 14 micropexophagy-defective P. pastoris mutants generated by gene-tagging mutagenesis. Further mutant screening identified an ORF sequence whose deduced amino acid sequence showed the highest sequence identity (23% identity and 49% of similarity) to that of ScATG24/SNX4 (YJL036w) from the Saccharomyces Genome Database. The gene was designated as PpATG24 (previously named PAZ16; Klionsky et al., 2003). The full-length fragment of the PpATG24 ORF was cloned from two alleles of PpATG24 mutants, and the coding region was deleted from the P. pastoris genome, with confirmation by Southern analysis (unpublished data). The resultant Ppatg24Δ strain (strain YAP2401) was grown on methanol to induce peroxisome formation and then shifted to ethanol medium and glucose medium to test the competence for macropexophagy and micropexophagy, respectively. The wild-type strain showed Aox degradation after pexophagy (Figure 3, A and B). In contrast, the Ppatg24Δ mutant exhibited strong peroxisomal alcohol oxidase (Aox) activity (Figure 3A) as well as retardation of Aox protein degradation (Figure 3B) after induction of both modes of pexophagy. Diffusion of peroxisomal fluorescent protein (GFP-PTS1) was not detected in the vacuolar lumen after cells were subjected to both pexophagy-inducing conditions (Figure 3C). These mutant phenotypes were complemented by the introduction of plasmid pYA101 containing the full-length PpATG24 ORF under the control of its own promoter. These results indicate that disruption of PpATG24 impaired both types of pexophagy in P. pastoris macropexophagy (after ethanol adaptation) and micropexophagy (after glucose adaptation).
Figure 3.
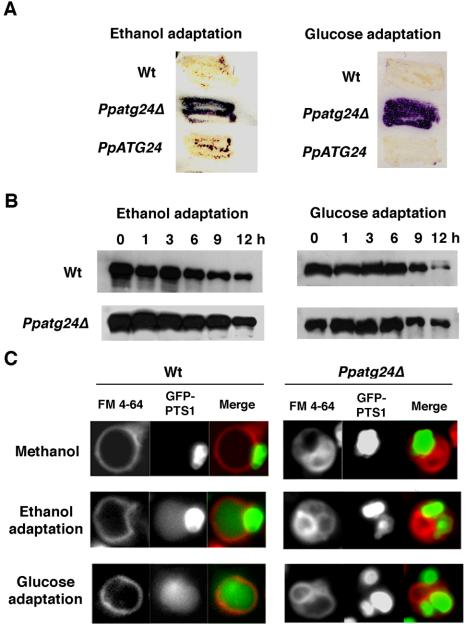
PpATG24 disruption leads to deficient degradation of the peroxisomal enzyme Aox. (A) Remaining Aox activity after pexophagy induction. Wild-type (Wt; strain PPY12), Ppatg24Δ (strain YAP2401), and PpATG24 (strain YAP2403) cells grown on a methanol plate were adapted to glucose or ethanol for 12 or 48 h, respectively. Purple represents the persistence of the peroxisomal protein Aox, as detected by its activity. In wild-type cells (Wt), Aox was degraded after the ethanol adaptation (macropexophagy) and glucose adaptation (micropexophagy), whereas both pathways were impaired in the Ppatg24Δ mutant. By introduction of the PpATG24 gene into Ppatg24Δ, peroxisome degradation was restored completely (PpATG24). (B) Aox degradation during ethanol or glucose adaptation. The methanol-grown wild-type and Ppatg24Δ cells were transferred to glucose or ethanol medium and harvested after the indicated times. Cell lysates were then subjected to immunoblot analysis using anti-Aox antibody. A decrease in the intensity of the Aox signal was retarded in the Ppatg24Δ strain. The decrease of signal in Ppatg24Δ strain was due to the dilution of Aox-protein by cell growth after the medium shift. (C) Fluorescent images of wild-type (STWI) and Ppatg24Δ (YAP2402) cells labeled with GFP-PTS1 and FM 4-64. Pexophagy was detected by diffusion of GFP-PTS1 in the vacuolar lumen in the wild-type cells after ethanol- or glucose-adaptation for 3 h. Diffusion of GFP-PTS1 in the vacuolar lumen was not observed in Ppatg24Δ strain.
Yeast mutants impaired in starvation-induced macroautophagy showed decreased viability after nitrogen-starvation conditions (Tsukada and Ohsumi, 1993). The Ppatg24Δ strain is distinct from Ppatg7 mutant (Oku et al., 2003) in that it did not show decreased viability (unpublished data). As reported for ScAtg24, it was previously suggested that PpATG24 is dispensable for macroautophagy (Nice et al., 2002).
PpATG24 Has a PX Domain That Function as a Phosphatidylinositol-binding Module
Atg24 belongs to a family of sorting nexins (Snx), which are defined partly by the presence of a p40 phox homology (PX) domain that has been shown to bind PtdIns(s) (Ago et al., 2001; Cheever et al., 2001; Kanai et al., 2001; Song et al., 2001; Yu and Lemmon, 2001; Zhou et al., 2003). To study the binding specificity of PpAtg24 PX domain (a.a. 75-195) to PtdIns, we conducted a protein-lipid overlay assay using PIP-Strips (Echelon Biosciences) and the purified GST-PpAtg24 PX domain-fusion protein. The purified protein gave a single band as judged by SDS-PAGE (Figure 4A). Of the PtdIns proteins present in yeast cells (PtdIns(3)P, PtdIns(3,5)P2, PtdIns(4)P, and PtdIns(4,5)P), the PpAtg24 PX domain binds to PtdIns(3)P with the 2highest affinity (Figure 4B).
Figure 4.
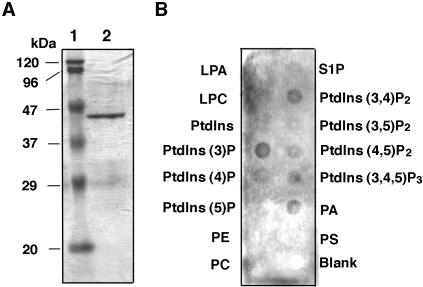
Specific binding of PpAtg24 phox homology (PX) domain to phosphatidylinositol-3-phosphate (PtdIns(3)P). (A) Purity of the GST-PpAtg24 PX domain detected on SDS-PAGE. The GST-fused PpAtg24 PX domain was purified using an E. coli Rosetta DE3 by GSTrap FF column. The purity of the purified protein was determined by Coomassie staining. Lane 1, molecular weight markers; lane 2, purified GST-PpAtg24 PX domain protein. (B) Protein-lipid overlay assay. Purified GST-PpAtg24 PX domain was subjected to PIP-Strip using anti-GST antiserum for immunoblot analysis. LPA, lysophosphatidic acid; LPC, lysophosphocholine; PtdIns, phsophatidylinositol; PE, phosphatidylethanolamine; PC, phsophatidylcholine; S1P, sphingosine-1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine.
Next, we introduced the full-length PpAtg24-CFP fusion protein or the PX-domain (a.a. 75-195)-deleted PpAtg24-CFP construct into the Ppatg24Δ strain under its own promoter. Although the full-length PpAtg24-CFP could rescue the pexophagy defect of the Ppatg24Δ strain, resulting in perivacuolar spots of fluorescence (see below), the PX-deleted construct could not and lost perivacuolar fluorescence.
Subcellular Fractionation of PpAtg24-CFP
Pichia cells expressing functional PpAtg24-CFP were grown on methanol medium, spheroplasted, and lysed osmotically. The obtained cell lysates were centrifuged at 13,000 × g and separated into low-speed pellet (P13) and supernatant (S13) fractions. The S13 fraction was further centrifuged at 100,000 × g and fractionated into high-speed pellet (P100) and supernatant (S100) fractions. The PpAtg24-CFP was detected as a 105-kDa band by immunoblotting using rabbit anti-GFP antiserum. Most of the signal was detected in soluble fractions (S13 and S100) and was also detected in an organellar fraction (P13) containing peroxisomes, vacuoles, and the nucleus (Supplementary Figure 1A). To investigate the properties of PpAtg24 membrane association, cell lysates were solubilized with Triton X-100 or high-salt conditions (0.67 M potassium acetate plus 0.3 M KCl). The treated cell lysates were separated into membrane and soluble fractions by high-speed centrifugation at 100,000 × g. The Triton X-100-treated cell lysate showed a signal only in the soluble fraction, whereas high-salt treatment did not affect membrane association of PpAtg24-CFP (Supplementary Figure 1B).
These biochemical behavior of PpAtg24 in P. pastoris was similar to that of ScAtg24 in S. cerevisiae (Nice et al., 2002). PpAtg24 was found to be expressed constitutively (see below), and glucose- and ethanol-shifted cells also showed a similar biochemical behavior to methanol-grown cells (unpublished data).
Aberrant Vacuole Morphology and Impaired Peroxisomal Clustering in PpATG24 Mutant Strain
When P. pastoris was grown on methanol medium, a single cell contained a cluster of few giant peroxisomes and a single rounded vacuole, which can be easily observed by double-staining for the peroxisome marker GFP-PTS1 and the vacuolar membrane marker FM4-64. In contrast, the disruption of PpATG24 resulted in aberrant vacuole morphology in methanol-grown cells (Figure 5A). In addition, clustering of peroxisomes was observed to be impaired in some methanol-induced cells (Figure 5B). A shift from methanol medium to glucose or ethanol medium did not cause detectable changes in the fluorescent morphology of GFP-PTS1 and FM4-64, and diffusion of GFP-PTS1 to vacuolar lumen was never detected (Figure 3C).
Figure 5.
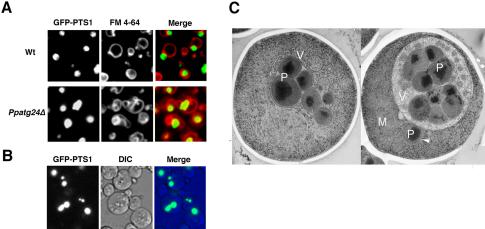
The deletion of PpATG24 causes aberrant vacuole morphology and peroxisome cluster impairment in methanol-grown cells. (A) Methanol-grown wild-type (STWI) and Ppatg24Δ (YAP2402) cells were labeled with GFP-PTS1 and FM 4-64 and were observed by fluorescence microscopy. Wild-type cells formed a large, round vacuole before pexophagy. In contrast, Ppatg24Δ cells showed aberrant vacuole morphology. Wild-type methanol-grown cells always contained one peroxisomal cluster. (B) Peroxisomal clustering was inhibited in some Ppatg24Δ cells (strain YAP2402). (C) Two electron micrographs of methanol-grown Ppatg24Δ cells. Before induction of pexophagy, cells showed extensive engulfment of peroxisomes and cytosol by vacuolar membranes. Note that the mitochondrion was not wrapped in the vacuolar membrane, and one peroxisome was isolated from the other clusters (indicated by an arrowhead). P, peroxisome; V, vacuole; M, mitochondrion; N, nucleus.
To observe these aberrant intracellular structures of the Ppatg24Δ strain in more detail, cells were subjected to rapid freezing and freeze-substitution fixation and observed by electron microscopy. As shown in Figure 5C (left), peroxisomes (and cytosol) were extensively wrapped by vacuolar membranes in methanol-grown Ppatg24Δ cells, although some peroxisomes escaped this vacuolar engulfment (Figure 5C, right). Previously, we reported that PpATG8/PAZ2 disruption caused a similar vacuolar morphology in which vacuole could not complete peroxisome engulfment because of inability to form MIPA (Mukaiyama et al., 2004). However, peroxisome clustering was not impaired in the Ppatg8Δ strain (Mukaiyama et al., 2002, 2004). In Ppatg24Δ cells, the vacuole was observed to initiate engulfment of peroxisomes and cytosol via microautophagic process but could not complete their sequestration suggesting that the fusion between vacuolar tips mediated by MIPA was inhibited.
A similar aberrant vacuolar morphology, as well as a slight growth inhibition, was also observed for glucose- and oleate-grown Ppatg24Δ cells (unpublished data). These results show that PpAtg24 has pleiotropic functions in Pichia cells even before the onset of pexophagy. Moreover, the phenotype of Ppatg24Δ strain suggests that PpAtg24 is involved in the septation or division of vacuoles throughout the entire life cycle of the yeast cells.
Macropexophagy Is Blocked Before the Fusion between the Pexophagosome and Vacuole
Next, we wanted to know the stage at which macropexophagy was blocked in the Ppatg24Δ strain, namely, whether or not the pexophagosome is formed in the Ppatg24Δ strain. In wild-type cells, pexophagosome formation could be followed by using a strain coexpressing YFP-PpAtg8 and CFP-PTS1 after ethanoladaptation (Figure 6). We could see YFP-PpAtg8 ring structures marking pexophagosomes in Ppatg24Δ cells after 15 min of ethanol adaptation, similar to those in the wild-type strain (Figure 6). In contrast, this ring-like structure was not observed in many other Ppatg mutants, including Ppatg7 and Ppatg4 (Mukaiyama et al., 2004).
During macroautophagy, autophagosomal ScAtg8 was eventually transported into the vacuolar lumen (Kirisako et al., 1999, 2000). Similarly, significant YFP-PpAtg8 fluorescence was observed in the vacuolar lumen of wild-type yeast cells after 3 h of ethanol adaptation, indicating that an itinerary of PpAtg8 during macropexophagy is similar to that during macroautophagy. However, although YFP-PpAtg8 fluorescence was observed at the vacuolar membrane, it was absent from the vacuolar lumen in Ppatg24Δ cells after ethanol adaptation.
These results strongly support the notion that PpAtg24 is not necessary for pexophagosome formation and that PpAtg24 plays a role in membrane fusion between the pexophagosome and vacuole.
Localization of PpAtg24-CFP during Macropexophagy
To further understand the role of PpAtg24, particularly in fusion between the pexophagosome and vacuolar membrane, we followed the localization of functional PpAtg24-CFP, together with FM 4-64-labeled vacuolar membrane and YFP-labeled fusion proteins in macropexophagic cells.
Figure 7A shows representative images from cells coexpressing the peroxisomal marker YFP-PTS1. Before induction of pexophagy, at least one perivacuolar spot of PpAtg24-CFP fluorescence was observed proximal to the peroxisomal cluster (YFP-PTS1) in methanol-grown cells. PpAtg24-CFP also stained some parts of the vacuolar membrane. These results corroborated the biochemical observation that PpAtg24-CFP was recovered as a pellet in the P13 fraction (Supplementary Figure 1A). Similar punctate fluorescence was also observed in glucose- and ethanol-grown cells, suggesting that PpAtg24 is expressed constitutively.
Figure 7.
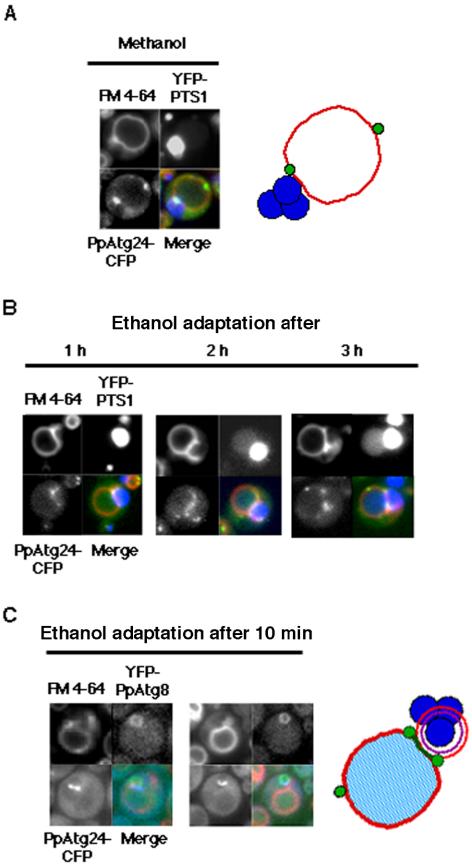
Intracellular behavior of PpAtg24 during macropexophagy. Pichia cells expressing PpAtg24-CFP and YFP-PTS1 in Ppatg24Δ (YAP2405) were transferred from methanol medium to ethanol medium for induction of macropexophagy. (A and B) Superimposed images are shown with (red) FM4-64, (blue) YFP-PTS1, and (green) PpAtg24-CFP signal. (A) Methanol-grown cell. Left, fluorescent images; right, representative localization of PpAtg24-CFP. (B) Fluorescent images after ethanol-adaptation. A major portion of the PpAtg24-CFP spot fluorescence was juxtaposed to the peroxisome cluster and pexophagosome, at the vertex and boundary region. Fusion could be detected by the flow of YFP-PTS1 into the vacuolar lumen after 2 h. (C) PpAtg24-CFP-expressing cells were costained with YFP-PpAtg8 (strain YAP2411) and FM 4-64 (left). In these cells, the fusion was not detected because we could observe a ring-shaped PpAtg8 fluorescence (see Figure 2A). Representative localization of PpAtg24-CFP during macropexophagy (right). Green, PpAtg24-CFP; red, vacuole membrane; blue, peroxisomes.
In the YFP-PTS1 coexpressing strain, macropexophagy could be detected by the flow of YFP-PTS1 into the vacuolar lumen, which can be often seen after ethanol-adaptation for 1 to 2 h. The flow of GFP-PTS1 into the vacuolar lumen was never observed in the Ppatg24Δ strain (Figure 3C).
As shown in Figure 7, B and C, the fluorescence images from PpAtg24-CFP-expressing cells bearing either YFP-PTS1 or YFP-PpAtg8 exhibited fluorescence at the boundary region and vertex ring between the pexophagosome and vacuole, suggesting that PpAtg24 is principally localized at the vertex ring and the boundary region. (Note that a ring of PpAtg8 indicates that the pexophagosome has not yet fused with the vacuolar membrane. See Figure 1). Although some other perivacuolar spots existed at the outside edge of vacuolar membrane, these results suggest that PpAtg24 regulates or functions in the fusion between the pexophagosome and vacuole.
Analysis of Macropexophagy using the PtdIns(3)-Binding Probe YFP-2xFYVEHrs
2xFYVEHrs has shown to bind to PtdIns(3)P specifically and has therefore been used as a probe for intercellular PtdIns(3)P (Gillooly et al., 2000; Stenmark et al., 2002). Neither the PX-domain from PpAtg24 nor the FYVEHrs domain could bind to PtdIns(3,5)P2. In cells undergoing macropexophagy, YFP-2xFYVEHrs labeled the whole vacuolar membrane, with brighter labeling at the boundary and vertex regions between the pexophagosome and vacuole (Figure 8).
Figure 8.
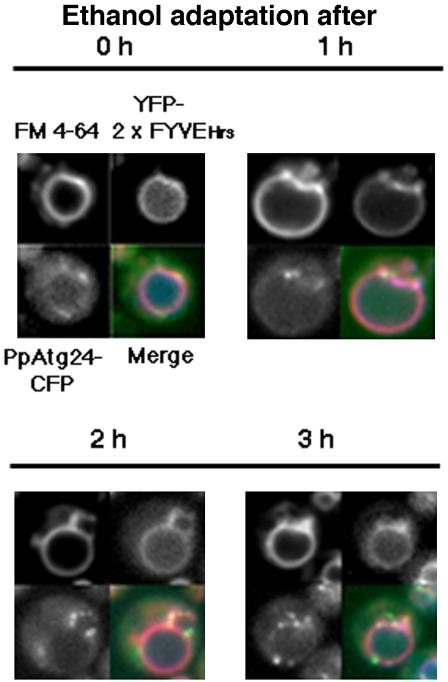
Localization of PtdIns(3)P and PpAtg24 during macropexophagy. Methanol-grown Pichia cells (YAP2406) expressing the YFP-2xFYVE domain from mouse Hrs (PtdIns(3)P marker) and PpAtg24-CFP were transferred from methanol medium to ethanol medium for induction of macropexophagy. Merged images are stained with (red) FM4-64, (green) YFP-2xFYVEHrs domain, and (blue) PpAtg24-CFP.
Localization of PpAtg24-CFP during Micropexophagy
After micropexophagy induction of wild-type cells, the vacuole septated and formed new compartments to engulf a peroxisomal cluster (Mukaiyama et al., 2002). Methanolgrown Ppatg24Δ cells had aberrant vacuoles that retained their abberant morphology after glucose adapation for 2 h and did not exhibit GFP-PTS1 flow into the vacuolar lumen (Figure 3C). This micropexophagy defect in the Ppatg24Δ strain was rescued by introduction of PpAtg24-CFP. As shown in Figure 9A, many spots of PpAtg24-CFP fluorescence were observed proximal to the peroxisomal cluster at a perivacuolar position, especially at the newly formed vacuolar septum in cells undergoing micropexophagy.
Figure 9.
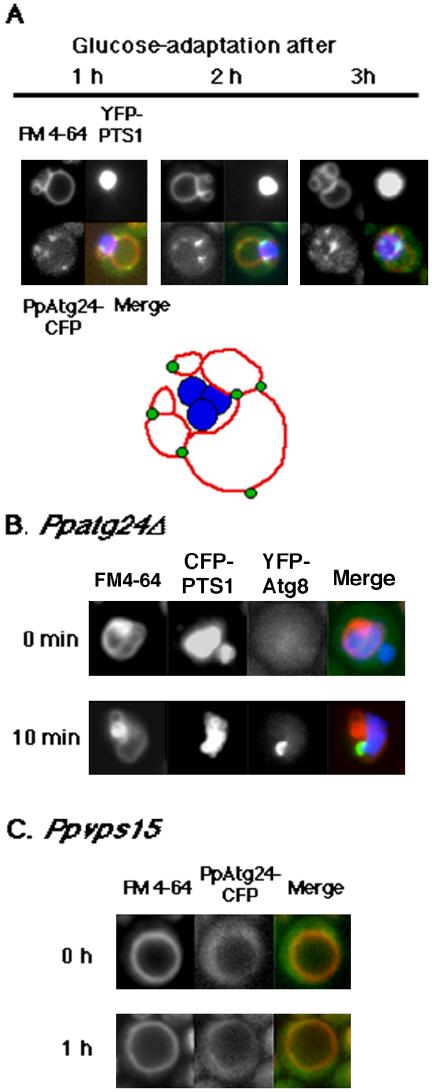
Intracellular behavior of PpAtg24 during micropexophagy. (A) Top: Ppatg24Δ (YAP2405) Pichia cells expressing PpAtg24-CFP and YFP-PTS1, YFP-tagged peroxisomal targeting signal were transferred from methanol medium to glucose medium for induction of micropexophagy. Superimposed images showing (red) FM4-64, (blue) YFP-PTS1, and (green) PpAtg24-CFP signal. A major part of the PpAtg24-CFP spot fluorescence was localized to the vertex region or tips of the septating vacuole. Bottom: representative localization of PpAtg24-CFP during micropexophagy. (B) The extended cup-like YFP-PpAtg8 fluorescence appeared after 10 min of micropexphagy induction, indicating that the MIPA-like structure was formed in Ppatg24Δ cells. (C) Localization of PpAtg24-CFP in the Ppvps15 mutant strain. Wild-type (YAP2404) and PpAtg24-CFP-expressing Ppvps15 (YAP2410) cells were shifted to glucose medium for 60 min. The vacuole failed to invaginate or septate in the vps15 mutant.
Ppatg24Δ cells exhibited a MIPA-like extended structure of YFP-PpAtg8-fluorescence that appeared after glucose-adaptation for 10 min (Figure 9B). Therefore, just as these mutants remain competent in pexophagosome formation, MIPA formation seems to be intact. These results suggest that PpAtg24 regulates septation of the vacuolar compartment during micropexophagy, including the fusion event at the vacuolar membrane, but is not responsible for de novo membrane formation.
By contrast, the Ppvps15 mutant could not septate and remained spherical even after micropexophagy induction (Figure 9C). In the Ppvps15 mutant, the YFP-2xFYVEHrs probe showed neither perivacuolar spot staining nor vacuolar membrane staining in any of the tested conditions, including growth in methanol, ethanol, and glucose. During micropexophagy, YFP-2xFYVEHrs was localized to the vacuolar membrane fluorescence, with intense spots at the points of vacuolar septation (unpublished data).
PpAtg24 Colocalizes with PpAtg17 and PpVac8 during Pexophagy
Perivacuolar spots of ScAtg24 were previously assumed to be part of a preautophaogosomal structure (PAS) necessary for autophagosome formation, because ScAtg24 colocalized with a known preautophagosomal component, ScAtg17 (Nice et al., 2002). However, the formation of membrane structures such as the pexophagosome and MIPA was not inhibited in the Ppatg24Δ strain. In P. pastoris, PpAtg17 exhibited a single (or two) perivacuolar spot(s) in ethanol- and glucose-adapted cells (Figure 10A). During the micropexophagic process, PpAtg24-perivacuolar spots localized mainly to the septation points on the vacuolar membrane, whereas only one intense YFP-PpAtg17 spot colocalized with PpAtg24-CFP. Theses results suggest that PpAtg24-CFP localized not only to the PAS, but also to vertices of the septated vacuole.
Figure 10.
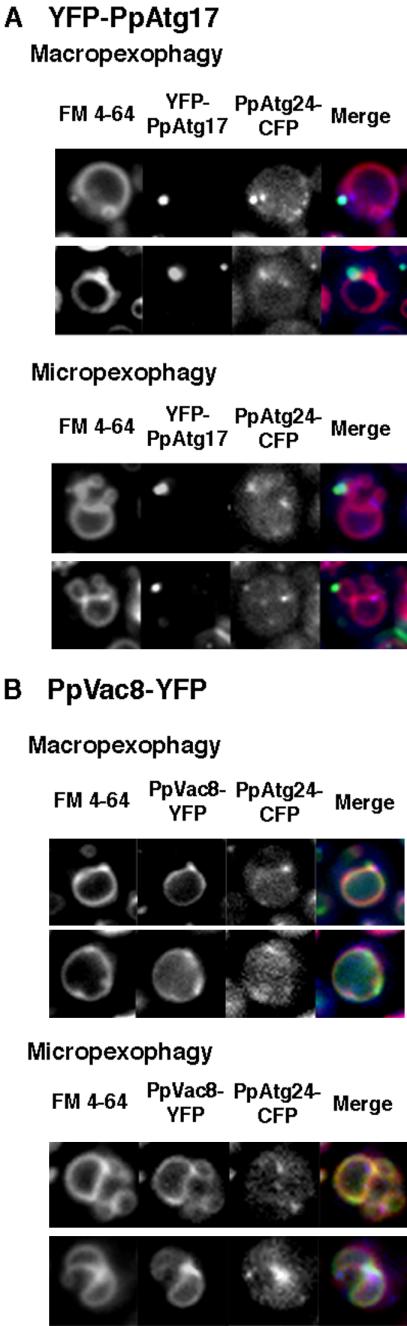
A portion of PpAtg24-CFP spot fluorescence colocalized with YFP-Atg17 and PpVac8-YFP during pexophagy. (A) YFP-PpAtg17 and (B) PpVac8-YFP were introduced into the PpAtg24-CFP-expressing strain, yielding the YAP2413 strain and the YAP2414 strain, respectively. Macropexophagy or micropexophagy was induced in these cells.
ScVac8 was previously shown to be concentrated at the vertex of the homotypic vacuolar fusion complex (Wang et al., 2002, 2003). After macropexophagy induction, PpVac8 is located at both the vacuolar membrane and the perivacuolar spots. As shown in Figure 10B, PpAtg24-CFP colocalized with regions of intense PpVac8 fluorescence, which may mark vertices of the pexophagosome-vacuole fusion complex. Punctate perivacuolar fluorescence of PpAtg24-CFP was observed at the point between the peroxisomal cluster and vacuole even before pexophagy induction, and the depletion of PpAtg24 resulted in aberrantly septated vacuoles. These results indicate that PpAtg24 regulates vacuolar dynamics not only in pexophagy, but also in other phases of the yeast cell cycle.
DISCUSSION
This study was conducted to reveal the function of a Snx family protein PpAtg24 during pexophagy. First, our fluorescence analysis revealed that heterotypic fusion between the pexophagosome and vacuole (PV fusion) occurs at the vertex domain, resulting in internalization of the boundary membranes. A major portion of PpAtg24 was localized to the vertex and boundary regions in this PV fusion complex during macropexophagy. Because the pexophagosome was formed and macropexophagy was blocked before the PV fusion step in the Ppatg24Δ strain, PpAtg24 is likely to be directly involved in PV fusion. On the other hand, during micropexophagy, yeast vacuoles septated toward appropriate directions to engulf peroxisomes and PpAtg24 was present at tips or vertexes of septating vacuoles. The aberrant vacuolar morphology of the Ppatg24Δ strain was also reminiscent of the phenotype of the mutants defective in vacuolar fusion, such as erg6 (Kato and Wickner, 2001). Under normal (nonpexophagic) growth conditions, vacuoles are known to fuse after segregation of vacuolar compartments into daughter cells. These observations are consistent with PpAtg24 being responsible for vacuolar fusion or the spatiotemporal regulation of fusion at the vacuolar membrane surface.
Snx is recently identified as a family of conserved hydrophilic cytoplasmic proteins that have been found to be associated with membranes of the endocytic system and have been implicated in the trafficking of many endosomal membrane proteins, including the epidermal growth factor (EGF) receptor and transferrin receptor (Kurten et al., 1996; Phillips et al., 2001; Teasdale et al., 2001; Xu et al., 2001; Hettema et al., 2003). Most Snx proteins contain a PX domain, which functions as a phosphoinoside-binding motif. They have a predicted coiled-coil structure and have been shown to form heterodimers with other members of the Snx family. ScAtg24 was previously shown to form a dimer with Sc-Snx41 and ScAtg20 (ScSnx42; Hettema et al., 2003).
Nice et al. (2002) also reported that ScAtg24 and ScAtg20 formed a complex necessary for both Cvt and macropexophagy. They suggested that perivacuolar punctate structures represent a preautophagosomal structure (PAS), which is considered to be a precursor for autophagosome formation, and discussed the role of Snx proteins in the formation of Cvt vesicles. However, it is hard to discriminate between the PAS and other organelles among all the perivacuolar spots in S. cerevisiae. A minor subgroup of PpAtg24 indeed colocalized with the PAS component PpAtg17 in P. pastoris (Figure 10A), although Atg17 is not necessary for the Cvt and pexophagy pathways. Our present results demonstrate clearly that PpAtg24 is not necessary for either pexophagosome or MIPA formation. Although PpAtg24-CFP was localized to multiple perivacuolar spots, a minor portion of PpAtg24-CFP could colocalize with PpAtg17. Multiple PpAtg24-CFP spots that did not overlap with PpAtg17-YFP appeared to localize to the PV fusion complex or vacuolar septum (Figure 10A). We suggest that localization of PpAtg24 to multiple organelles reflects a regulatory function for PpAtg24 at various targeted organelles. Because PpAtg24 was not necessary for pexophagosome formation, the role of PpAtg24 at the PAS still remains unclear.
Previous studies on the PtdIns(3)P-kinase ScVps34 in macroautophagy revealed that ScVps34 functions in a complex with Vps15, Vps30/Atg6, and Atg14 (Kihara et al., 2001) and that the PtdIns(3) kinase complex colocalized with the Atg1 kinase complex at PAS. These data suggested that PtdIns(3)P regulates membrane formation at the PAS. However, because the PtdIns(3)P-binding protein PpAtg24 was not involved in formation of the pexophagosome, PpAtg24 must have independent function downstream of the PtdIns(3)P-kinase complex. Indeed, it appears to be mainly involved in PV fusion during macropexophagy. Therefore, PtdIns(3)P signaling has two distinct functions during macropexophagy: de novo membrane formation from the PAS and PV fusion.
The requirement for PtdIns(3)P-signaling in micropexophagy was demonstrated by the following experiments: i) The Ppvps15 mutant did not show any septation of the vacuole (Stasyk et al., 1999; Mukaiyama et al., 2002). ii) Overexpression of FYVE-domain blocked micropexophagy at a very early stage (unpublished data), similar to the Ppvps15 mutant (Figure 9C). On the basis of the following observations, we postulate that PpAtg24 regulates vacuolar membrane dynamics in micropexophagy, which includes three fusion events (see Figure 1): Deletion of PpAtg24 and the PX-domain of PpAtg24 induced uncontrolled septation of the vacuole together with the loss of perivacuolar spots, resulting in inhibition of proper vacuolar maintenance (unpublished data). Furthermore, addition of PX domain proteins at the vertex negatively regulates vacuolar homotypic fusion in in vitro assays (Wang et al., 2003). Alternatively, the aberrant vacuolar morphology of the Ppatg24Δ strain may represent the impairment of membrane scission into the vacuolar lumen. Such an event should occur at the last stage of micropexophagy (Figure 1) or after fusion of vacuoles after segregation to daughter cells (Wang et al., 2002).
This report shows the first evidence that heterotypic fusion at the vacuolar membrane occurs at the vertex region, concomitant with internalization of the boundary membrane (Figure 2). However, whether fusion occurs at the vertex region at the surface of other organelles remains to be solved. Because vacuoles contain many hydrolytic enzymes, the internalized membrane could be degraded more easily than in other organelles.
Yeast Snx proteins (ScAtg24, ScAtg20, and ScSnx41) are also known to function in post-Golgi endosomes to recruit Snc1 to the late Golgi compartment (Hettema et al., 2003). Therefore, the function of Snx proteins is not limited to the Cvt or pexophagic pathways. One important finding uncovered in this study is that the Ppatg24Δ strain was impaired in the clustering of peroxisomes and showed retarded growth on methanol compared with the wild-type strain (unpublished data). So far, several PEX gene products, including Pex11, Pex 25, Pex27, Pex30, Pex31, and Pex32, are known to be involved in the division or clustering of peroxisomes (Rottensteiner et al., 2003; Tam et al., 2003; Vizeacoumar et al., 2004). Deletion of PpATG24 appears to result in a phenotype that closely resembles that of several pex mutants. The fluorescent perivacuolar spots of PpAtg24-CFP, which is necessary for both peroxisome proliferation and degradation, may represent the formation of complexes with other proteins, such as Snx proteins, PpAtg17, and PpVac8 (Nice et al., 2002; Hettema et al., 2003). Therefore, future studies examining the Atg24 complex in greater detail from the viewpoint of peroxisome homeostasis may be interesting.
Supplementary Material
Acknowledgments
We thank Dr. H. Yurimoto and other members of the Laboratory of Microbial Biotechnology at Kyoto University for helpful advice and technical assistance. This research was supported in part by a Grant-in-Aid for Scientific Research (S) 13854008 and a Grant-in-Aid for Scientific Research on Priority Areas 12146202 and 504 to Y.S., the COE program from the Ministry of Education, Science, Sports, and Culture of Japan, and the National Institute for Basic Biology Cooperative Research Program.
Article published online ahead of print in MBC in Press on November 24, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0842).
Abbreviations used: Snx(s), sorting nexin(s); PX, Phox homology; FYVE domain, Fab1-YOTB-Vac1-EEA1 homology domain; PtdIns, phosphoinoside; PtdIns(3)P, phosphatidylinositol 3-phosphate; PV-fusion, pexophagosome-vacuole fusion; MIPA, micropexophagy-specific membrane apparatus; PAS, preautophagosomal structure; GFP, green fluorescent protein; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; PTS1, peroxisome targeting signal type 1; GFP-PTS1, GFP fused to PTS1; FM4-64, N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide; GST, glutathione S-transferase; Aox, alcohol oxidase.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Ago, T., Takeya, R., Hiroaki, H., Kuribayashi, F., Ito, T., Kohda, D., and Sumimoto, H. (2001). The PX domain as a novel phosphoinositide-binding module. Biochem. Biophys. Res. Commun. 287, 733-738. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (eds.) (1987). Current Protocols in Molecular Biology, New York: Greene Publishing Associates and Wiley-Interscience.
- Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J., and Ohsumi, Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139, 1687-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever, M. L., Sato, T. K., de Beer, T., Kutateladze, T. G., Emr, S. D., and Overduin, M. (2001). Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3, 613-618. [DOI] [PubMed] [Google Scholar]
- Gillooly, D. J., Morrow, I. C., Lindsay, M., Gould, R., Bryant, N. J., Gaullier, J. M., Parton, R. G., and Stenmark, H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema, E. H., Lewis, M. J., Black, M. W., and Pelham, H. R. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22, 548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, F., Liu, H., Field, S. J., Akbary, H., Matsuo, T., Brown, G. E., Cantley, L. C., and Yaffe, M. B. (2001). The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3, 675-678. [DOI] [PubMed] [Google Scholar]
- Kato, M., and Wickner, W. (2001). Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO J. 20, 4035-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel, J. A., Rechinger, K. B., van der Klei, I. J., Salomons, F. A., Titorenko, V. I., and Veenhuis, M. (1999). The Hansenula polymorpha PDD1 gene product, essential for the selective degradation of peroxisomes, is a homologue of Saccharomyces cerevisiae Vps34p. Yeast 15, 741-754. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Takao, T., Noda, T., and Ohsumi, Y. (2000). The reversible modification regulates the membrane-binding stage of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J. et al. (2003). A unified nomenclature for yeast autophagyrelated genes. Dev. Cell 5, 539-545. [DOI] [PubMed] [Google Scholar]
- Kurten, R. C., Cadena, D. L., and Gill, G. N. (1996). Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science 272, 1008-1010. [DOI] [PubMed] [Google Scholar]
- Mukaiyama, H., Baba, M., Osumi, M., Aoyagi, S., Kato, N., Ohsumi, Y., and Sakai, Y. (2004). Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama, H., Oku, M., Baba, M., Samizo, T., Hammond, A. T., Glick, B. S., Kato, N., and Sakai, Y. (2002). Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells 7, 75-90. [DOI] [PubMed] [Google Scholar]
- Nice, D. C., Sato, T. K., Stromhaug, P. E., Emr, S. D., and Klionsky, D. J. (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to PtdIns(3)P at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30198-30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku, M., Warnecke, D., Noda, T., Muller, F., Heinz, E., Mukaiyama, H., Kato, N., and Sakai, Y. (2003). Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 22, 3231-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, S. A., Barr, V. A., Haft, D. H., Taylor, S. I., and Haft, C. R. (2001). Identification and characterization of SNX15, a novel sorting nexin involved in protein trafficking. J. Biol. Chem. 276, 5074-5084. [DOI] [PubMed] [Google Scholar]
- Rottensteiner, H., Stein, K., Sonnenhol, E., and Erdmann, R. (2003). Conserved function of Pex11p and the novel Pex25p and Pex27p in peroxisome biogenesis. Mol. Biol. Cell 14, 4316-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, Y., Koller, A., Rangell, L. K., Keller, G. A., and Subramani, S. (1998). Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 141, 625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu, P. V., Takegawa, K., Fry, M. J., Stack, J. H., Waterfield, M. D., and Emr, S. D. (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88-91. [DOI] [PubMed] [Google Scholar]
- Song, X., Xu, W., Zhang, A., Huang, G., Liang, X., Virbasius, J. V., Czech, M. P., and Zhou, G. W. (2001). Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry 40, 8940-8944. [DOI] [PubMed] [Google Scholar]
- Stack, J. H., DeWald, D. B., Takegawa, K., and Emr, S. D. (1995). Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack, J. H., Herman, P. K., Schu, P. V., and Emr, S. D. (1993). A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasyk, O. V., van der Klei, I. J., Bellu, A. R., Shen, S., Kiel, J.A.K.W., Cregg, J. M., and Veenhuis, M. (1999). A Pichia pastoris VPS15 homolog is required in selective peroxisome autophagy. Curr. Genet. 36, 262-269. [DOI] [PubMed] [Google Scholar]
- Stenmark, H., Aasland, R., and Driscoll, P. C. (2002). The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 513, 77-84. [DOI] [PubMed] [Google Scholar]
- Tam, Y. Y., Torres-Guzman, J. C., Vizeacoumar, F. J., Smith, J. J., Marelli, M., Aitchison, J. D., and Rachubinski, R. A. (2003). Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 4089-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale, R. D., Loci, D., Houghton, F., Karlsson, L., and Gleeson, P. A. (2001). A large family of endosome-localized proteins related to sorting nexin 1. Biochem. J. 358, 7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169-174. [DOI] [PubMed] [Google Scholar]
- Tuttle, D. L., and Dunn, W. A. (1995). Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108, 25-35. [DOI] [PubMed] [Google Scholar]
- Vizeacoumar, F. J., Torres-Guzman, J. C., Bouard, D., Aitchison, J. D., and Rachubinski, R. A. (2004). Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Merz, A. J., Collins, K. M., and Wickner, W. (2003). Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J. Cell Biol. 160, 365-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Seeley, E. S., Wickner, W., and Merz, A. J. (2002). Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108, 357-369. [DOI] [PubMed] [Google Scholar]
- Weisman, L. S. (2003). Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37, 435-460. [DOI] [PubMed] [Google Scholar]
- Wickner, W. (2002). Yeast vacuoles and membrane fusion pathways. EMBO J. 21, 1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, W., and Haas, A. (2000). Yeast homotypic vacuole fusion: a window on organelle trafficking mechanisms. Annu. Rev. Biochem. 69, 247-275. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Hortsman, H., Seet, L., Wong, S. H., and Hong, W. (2001). SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 3, 658-666. [DOI] [PubMed] [Google Scholar]
- Yu, J. W., and Lemmon, M. A. (2001). All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276, 44179-44184. [DOI] [PubMed] [Google Scholar]
- Zhou, C. Z. et al. (2003). Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J. Biol. Chem. 278, 50371-50376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


