Abstract
Distinguishing cancer cells from normal cells through surface receptors is vital for cancer diagnosis and targeted therapy. Metabolic glycoengineering of unnatural sugars provides a powerful tool to manually introduce chemical receptors onto the cell surface; however, cancer-selective labeling still remains a great challenge. Herein we report the design of sugars that can selectively label cancer cells both in vitro and in vivo. Specifically, we inhibit the cell-labeling activity of tetraacetyl-N-azidoacetylmannosamine (Ac4ManAz) by converting its anomeric acetyl group to a caged ether bond that can be selectively cleaved by cancer-overexpressed enzymes and thus enables the overexpression of azido groups on the surface of cancer cells. Histone deacetylase and cathepsin L-responsive acetylated azidomannosamine, one such enzymatically activatable Ac4ManAz analog developed, mediated cancer-selective labeling in vivo, which enhanced tumor accumulation of a dibenzocyclooctyne–doxorubicin conjugate via click chemistry and enabled targeted therapy against LS174T colon cancer, MDA-MB-231 triple-negative breast cancer and 4T1 metastatic breast cancer in mice.
Cell-surface receptors play a vital role in regulating the interactions between cells and the extracellular microenvironment1–3. There has been consensus that the recognition of disease-specific, cell-surface receptors enables the targeted delivery of therapeutic agents to diseased tissues and minimizes undesired side effects. Based on lineage-specific genome sequences, different cell types possess varying populations of cell-surface receptors4–6. Additionally, diseased cells harboring genetic mutations have the capacity to express altered receptor populations7–9. For instance, as a consequence of gene amplification, some breast cancer cells overexpress human epidermal growth factor receptor 2 (HER2/neu), which fosters the development of various cancer-targeting drugs10–12. However, the differences in endogenous receptor populations between diseased and normal cells cannot always be exploited as therapeutic targets, as they might be too small to impart high selectivity, or specific receptors may simply not be expressed by the target cells of interest13–15. For example, overexpression of HER2 is only limited to 15–20% of breast cancers, and hence the majority of breast cancer patients cannot benefit from HER2-targeted therapies16,17. Given the limitations of exploiting endogenous receptors as druggable targets, alternative strategies to manually manipulate cell-surface receptors to distinguish cell types are actively being pursued.
Two major strategies have been exploited to manually manipulate cell-surface receptors. The first strategy is to directly amplify the expression of existing cell-surface protein receptors using a gene transfection method in which the overexpression of HER2 in mouse NIH 3T3 cells was achieved by using a HER2-encoding plasmid18. However, this strategy is of limited translational applicability given the stringent technical requirements, nonselectivity over cell types in vitro and in vivo, and potential detrimental effects on the treated cells18,19. A different strategy, instead of directly increasing the amount of cell-surface protein receptors, is to introduce unique chemical functional groups onto the cell surface as an alternative to protein receptors20. These externally introduced chemical functional groups can mimic the receptor function of cell-surface proteins, which can be taken advantage of for cell binding and uptake of extracellular materials via efficient chemical reactions21–26. Previous attempts have been made to site-specifically incorporate unnatural amino acids into transmembrane proteins by using mutant aminoacyl-tRNA synthetases and suppressor tRNAs27–29. However, the necessity for gene insertion and introduction of tRNA synthetase makes this methodology very complicated to use, particularly in vivo30. An alternative method to introduce chemical functional groups onto the cell surface is to use metabolic glycoengineering of unnatural sugars20,31,32. Although this latter methodology can easily incorporate functional groups into cell-surface glycoproteins with desired density, selective labeling of cell types of interest in vitro and in vivo remains a great challenge33,34.
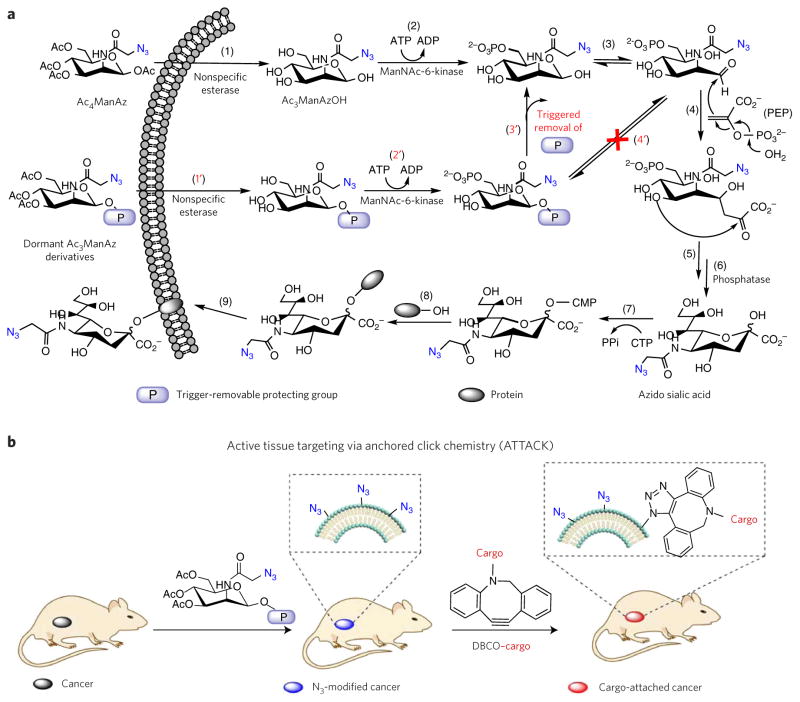
Tetraacetyl-N-azidoacetylmannosamine (Ac4ManAz (1)) is a well-known cell-labeling agent, with the metabolic labeling process illustrated in Figure 1a. Ac4ManAz is hydrolyzed by nonspecific esterase upon entering the cells (step 1); this is followed by the phosphorylation of C6–OH (step 2) and isomerization from the cyclic to the open-chain structure (step 3). The open-chain acetyl azidomannosamine then undergoes an aldol reaction with intracellular phosphoenolpyruvic acid (PEP) (step 4) followed by rearrangement and dephosphorylation reactions to form azido sialic acid (steps 5–6), which is conjugated to glycoprotein (steps 7–8) and expressed on the cell surface (step 9)33,35,36. As steps 4–8 are downstream reactions largely regulated by cellular processes, we reason that controlling the upstream reactions, particularly the site-specific step-2 and step-3 reactions, may allow manipulation of sugar metabolic activity. The C6 position of the sugar has once been modified with a prostate-specific antigen using 4-aminobenzylcarbonate as the self-immolative linker. However, the observed cell-selective labeling was attributed to PSA-protease-dependent cellular uptake of the sugar moiety34. Furthermore, the carbonate linker can be easily cleaved upon entering cells to release the metabolically active sugar moiety; thus, this modification strategy has limited applicability for extracellular triggering and lacks selectivity. Compared to the phosphorylation reaction at the C6 position (step 2), the formation of the open-chain aldehyde form of azidomannosamine at the C1 position (step 3) is more critical to the downstream reactions (steps 4–9), as the step-4 aldol reaction occurs directly at the C1 position. We reason that replacing the anomeric acetyl group (C1–OAc) of Ac4ManAz with a protective ether bond (glycosidic bond; the glycosidic protecting group denoted ‘P’, Fig. 1a) that can efficiently prohibit the deacetylation at the C1 position (Step 1′) and subsequent isomerization (Step 4′) may block the labeling process. Controlled labeling of cell types of interest may therefore be realized by designing target-specific, trigger-responsive ether bond caged Ac4ManAz analogs (steps 1′–3′ followed by steps 3–9). Here, we report the design of unnatural sugars that can selectively label target cells with azido groups both in vitro and in vivo and the development of active tissue targeting via anchored click chemistry (ATTACK; Fig. 1b), an unprecedented in vitro and in vivo targeting technology facilitated by unnatural sugar-mediated tissue-selective labeling followed by reagent-free click chemistry. 1-((4-(2,6-Diacetamidohexanamido)phenyl)(phenyl) methoxy)-3,4,6-triacetyl-N-azidoacetylmannosamine, one such enzymatically activatable Ac4ManAz analog developed, mediated cancer-selective labeling of azide groups in vivo, which subsequently allowed enhanced tumor accumulation of dibenzocyclooctyne (DBCO)–doxorubicin conjugate via click chemistry to provide targeted cancer therapy against several selected tumor types in mice.
Figure 1. Design of sugars for controlled metabolic labeling.
(a) Metabolic labeling processes of Ac4ManAz and trigger-activated labeling processes of dormant Ac3ManAz derivatives. P, trigger-removable protecting group. PEP, phosphoenolpyruvic acid. (b) Schematic illustration of ATTACK (active tissue targeting via anchored click chemistry) technology: in vivo selective labeling of cancer cells with azido groups and subsequent cancer targeting via efficient click chemistry.
RESULTS
Trigger-activatable Ac3ManAz derivatives
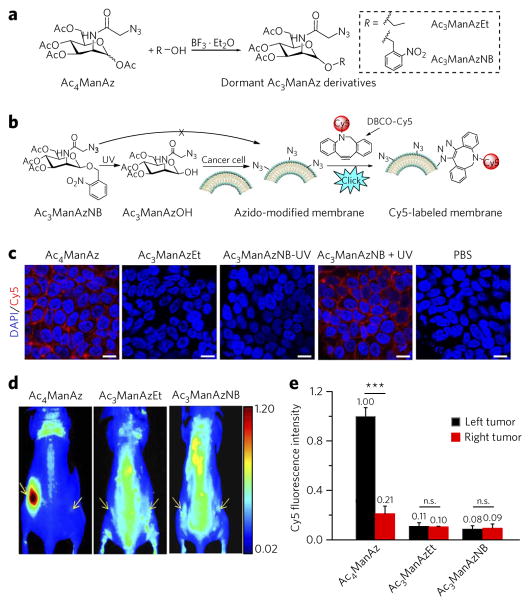
To demonstrate whether replacing the C1–OAc of Ac4ManAz with an ether bond could inhibit its metabolic labeling activity, we first synthesized 1-ethoxy-3,4,6-triacetyl-N-azidoacetylmannosamine (Ac3ManAzEt (2)) with an ethyl ether bond at the C1 position (Fig. 2a) and investigated its cell-labeling capability in vitro. LS174T colon cancer cells were incubated with Ac4ManAz, Ac3ManAzEt, or PBS for 3 d, and the potentially expressed azido groups were detected by DBCO–Cy5 via click chemistry. As shown in Figure 2c, LS174T cells treated with Ac4ManAz displayed strong Cy5 fluorescence intensity on the cell surface, indicating the successful expression of azido groups. Comparatively, LS174T cells treated with Ac3ManAzEt or PBS showed negligible Cy5 fluorescence intensity on the cell surface, suggesting that Ac3ManAzEt failed to label cells with azido groups. Flow cytometry analysis of LS174T cells treated with Ac4ManAz also showed significantly enhanced Cy5 fluorescence intensity compared to cells treated with Ac3ManAzEt or PBS (Supplementary Results, Supplementary Fig. 1a). Notably, passively internalized DBCO–Cy5 was negligible compared to covalently attached DBCO–Cy5 in azido-modified LS174T cells (Supplementary Fig. 2). Western blotting analyses of LS174T cells treated with Ac4ManAz confirmed that azido groups were incorporated into multiple proteins, while LS174T cells treated with Ac3ManAzEt showed no azido-modified proteins (Supplementary Fig. 1b). These experiments demonstrated that Ac3ManAzEt with an ethyl ether bond at the C1 position failed to metabolically label LS174T cells with azido groups.
Figure 2. Replacing the C1–OAc of Ac4ManAz with a cleavable ether bond–enabled controlled metabolic cell labeling in vitro and in vivo.
(a) Synthetic route of dormant Ac3ManAz derivatives including Ac3ManAzEt and UV-responsive Ac3ManAzNB. (b) Schematic illustration of UV-irradiation-activated metabolic labeling of Ac3ManAzNB and subsequent detection of the expressed azido groups by DBCO–Cy5 via click chemistry. (c) Confocal laser scanning microscopy (CLSM) images of LS174T colon cancer cells after incubation with Ac4ManAz (50 μM), Ac3ManAzEt (50 μM), Ac3ManAzNB (50 μM) − UV, Ac3ManAzNB (50 μM) + UV or PBS for 72 h and labeling with DBCO–Cy5 (50 μM, red) for 1 h. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Scale bars, 10 μm. (d, e) Ac4ManAz, Ac3ManAzEt or Ac3ManAzNB (25 mM, 20 μl) was injected intratumorally (i.t.) to the left LS174T tumors of athymic nude mice once daily for three consecutive days (days 1–3), and the right LS174T tumors were injected i.t. with PBS as controls. DBCO–Cy5 (5 mg/kg) was i.v. injected on day 4. (d) In vivo fluorescence imaging of mice from different groups at 48 h p.i. of DBCO–Cy5. Tumors are shown by yellow arrows. (e) Ex vivo Cy5 fluorescence intensity of the tumor tissues harvested at 48 h p.i. of DBCO–Cy5. Data are presented as mean ± s.e.m. (n = 3) and analyzed by one-way ANOV A (Fisher; 0.01 < *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). n.s., not significant. Data represent results from at least three experiments.
To further demonstrate that the etherification of C1–OAc of Ac4ManAz was attributable to the blocking effect and that the cleavage of this ether bond to expose C1–OH would reactivate the labeling process, we synthesized 1-((2-nitrobenzyl)oxy)-3,4,6-triacetyl-N-azidoacetylmannosamine (Ac3ManAzNB (3)) with an ultraviolet (UV)-cleavable 2-nitrobenzyl ether bond at the C1 position (Fig. 2b) and investigated its UV-controlled labeling capability in vitro. Ac3ManAzNB can be readily degraded into 3,4,6-triacetyl-N-azidoacetylmannosamine (Ac3ManAzOH) with the C1–OH exposed upon UV irradiation37. LS174T cells treated with Ac3ManAzNB (without UV irradiation) followed by reaction with DBCO–Cy5 showed negligible Cy5 attachment to the cell surface, further demonstrating the blocking effect of etherification of C1–OAc (Fig. 2c). Upon UV treatment, LS174T cells treated with Ac3ManAzNB followed by reaction with DBCO–Cy5 showed strong Cy5 fluorescence on the cell surface (Fig. 2c) as a result of the conversion of Ac3ManAzNB to Ac3ManAzOH, which activated the labeling process (Fig. 2b). Flow cytometry and western blotting analyses also verified successful expression of azido groups on LS174T cells treated with Ac3ManAzNB + UV instead of Ac3ManAzNB − UV (without UV irradiation) (Supplementary Fig. 1). These experiments further demonstrated that etherification of C1–OAc of Ac4ManAz can block its metabolic labeling activity, which can be reactivated in the presence of a specific trigger that cleaves the ether bond to expose C1–OH.
We next studied whether Ac3ManAzEt and Ac3ManAzNB show blocked metabolic labeling activity in vivo. Ac4ManAz, Ac3ManAzEt, or Ac3ManAzNB was injected into an implanted LS174T tumor (left flank) in athymic nude mice once daily for three consecutive days, while PBS was injected into a contralaterally implanted LS174T tumor (right flank) as a control. Western blotting analysis of tumor tissues at 24 h post injection (p.i.) of azido sugars showed a series of azido-modified proteins in tumors treated with Ac4ManAz, while those treated with Ac3ManAzEt and Ac3ManAzNB showed the same endogenous proteins in tumors as those treated with PBS (Supplementary Fig. 3b), suggesting that Ac3ManAzEt and Ac3ManAzNB were unable to label LS174T tumors with azido groups in vivo. To understand how the expressed azido groups would improve the tumor accumulation of DBCO–cargo via in vivo click chemistry, in a separate study, we injected DBCO–Cy5 intravenously (i.v.) at 24 h p.i. of azido sugars and monitored its biodistribution. At 48 h p.i. of DBCO–Cy5, Ac4ManAz-treated tumors showed approximately five-fold Cy5 fluorescence intensity compared to PBS-treated control tumors (Fig. 2d, e; Supplementary Fig. 4). For Ac3ManAzEt and Ac3ManAzNB groups, no significant difference in Cy5 fluorescence intensity between the treated and control tumors was observed (Fig. 2d, e; Supplementary Fig. 4). These experiments not only demonstrated the blocked metabolic activity of Ac3ManAzEt and Ac3ManAzNB in vivo but also indicated the excellent in vivo cancer-targeting effect mediated by sugar labeling and click chemistry.
DCL-AAM for cancer-selective labeling in vitro
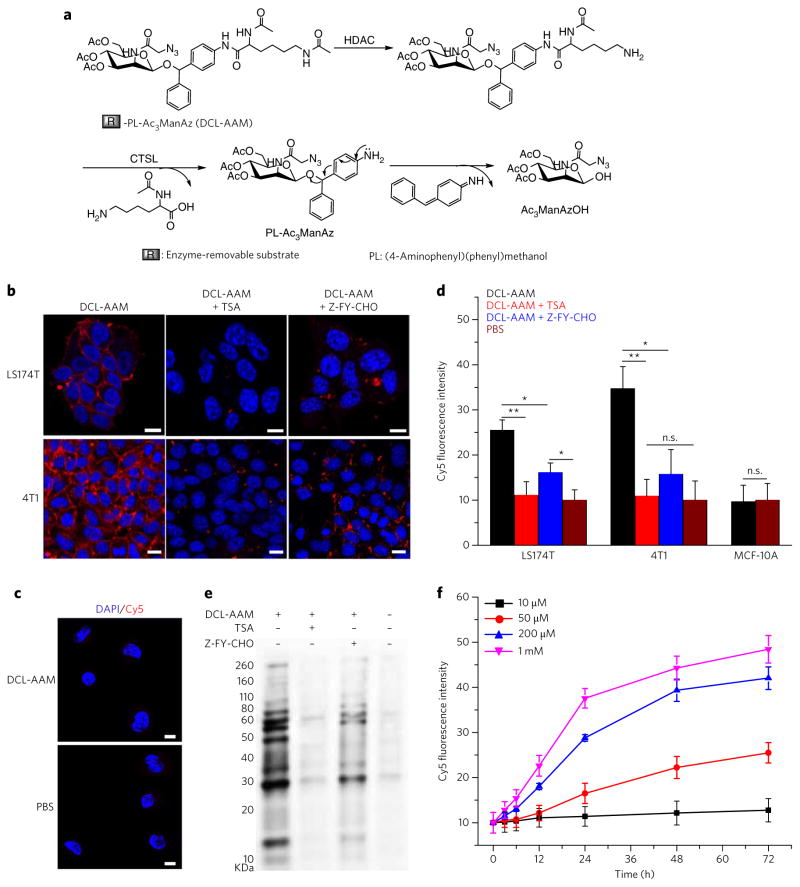
After demonstrating the controlled labeling strategy using UV-responsive Ac3ManAzNB, we next investigated whether endogenous cancer-specific triggers such as overexpressed enzymes can be used to achieve selective labeling of cancer cells over normal cells. In contrast to UV irradiation, which can cleave a 2-nitrobenzyl ether bond to a hydroxyl group, these triggers cannot directly cleave an ether bond, necessitating the incorporation of a self-immolative linker. We designed (4-aminophenyl)(phenyl)methanol (abbreviated as PL) as the self-immolative moiety to link Ac3ManAz via an ether bond and an enzyme-removable substrate via an amide bond (Fig. 3a). To build an activatable Ac3ManAz derivative with high cancer-labeling specificity, 1-((4-(2,6-diacetamidohexanamido)phenyl) (phenyl)methoxy)-3,4,6-triacetyl-N-azidoacetylmannosamine (histone deacetylase (HDAC)/cathepsin L (CTSL)-responsive acetylated azidomannosamine, abbreviated as DCL-AAM (4)) was designed by using N,N′-L-diacetyllysine as the enzyme-removable substrate, which can be serially cleaved by two separate cancer-overexpressed enzymes, HDAC and CTSL38,39. HDAC removes the N6-acetyl group of the lysine residue first, and this is followed by the cleavage of the amide bond by CTSL40–42. Such dual-enzymatically triggering design ensures improved cancer selectivity of DCL-AAM labeling. Upon removal of the HDAC/CTSL-responsive substrate, PL undergoes structural rearrangement to cleave off the conjugated 4-benzylidenecyclohexa-2,5-dien-1-imine and release the metabolically active Ac3ManAzOH (Fig. 3a).
Figure 3. DCL-AAM-mediated cancer-selective labeling in vitro.
(a) Schematic illustration of HDAC/CT SL-induced degradation of DCL-AAM. HDAC removes the N6-acetyl group of the lysine residue first, which is followed by cleavage of the C1-amide bond by CT SL and self-cleavage of (4-aminophenyl) (phenyl)methanol (PL) to release Ac3ManAzOH. (b, c) CLSM images of LS174T and 4T1 cells (b) and MCF-10A breast basal epithelial cells (c) after incubation with DCL-AAM (50 μM), DCL-AAM (50 μM) + TSA (1 μM), DCL-AAM (50 μM) + Z-FY-CHO (50 μM), or PBS for 72 h and treatment with DBCO–Cy5 (50 μM, red) for 1 h. The cell nuclei were stained with DAPI (blue). Scale bars, 10 μm. (d) Average Cy5 fluorescence intensity of LS174T, 4T1 and MCF-10A cells following the same treatment as described in b and c. Data are presented as mean ± s.e.m. (n = 6) and analyzed by one-way ANOV A (Fisher; 0.01 < *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). The PBS group as the negative control was normalized to 10. (e) Western blotting analysis of LS174T cells after treated with DCL-AAM, DCL-AAM + TSA or DCL-AAM + Z-FY-CHO for 72 h. Cell lysates were incubated with DBCO–Cy3 for 1 h at 37 °C before gel running. Protein bands were visualized using ImageQuant LAS 4010 system. (f) Concentration- and time-dependent DCL-AAM-mediated labeling in LS174T cells (n = 4). LS174T cells were treated with various concentrations of DCL-AAM (10, 50, 200 and 1 mM) for different time (0, 3, 6, 12, 24, 48 and 72 h), and labeled with DBCO–Cy5 (50 μM) for 1 h. Each experiment was repeated 2–3 times in triplicate for each group; the data from the representative experiment are used to prepare the figure and are presented as mean ± s.e.m.
HDAC/CTSL activity in different cell lines was analyzed using Naph-Lys (5), a fluorescence turn-on reporter with the same HDAC/CTSL-responsive moiety as DCL-AAM (Supplementary Fig. 5a, b). All selected cancer cells of investigation including HeLa cells, LS174T colon cancer cells, MCF-7 breast cancer cells, HepG2 liver cancer cells, and 4T1 and MDA-MB-231 triple-negative breast cancer (TNBC) cells showed much higher HDAC/CTSL activity than MCF-10A breast basal epithelial cells, HBEC-5i cerebral microvascular endothelium cells and IMR-90 human fibroblast cells, the three selected noncancerous cells (Supplementary Fig. 5c, d). In the presence of the inhibitor for either HDAC (trichostatin A (TSA)) or CTSL (Z-FY-CHO), turn-on fluorescence intensity of Naph-Lys greatly decreased as a result of the reduced enzymatic activity (Supplementary Fig. 5d). The controlled labeling capability of DCL-AAM in vitro was studied by incubating different cell lines with DCL-AAM for 3 d, and the surface-expressed azido groups were analyzed by click reaction with DBCO–Cy5. Confocal laser scanning microscopy (CLSM) images showed strong Cy5 fluorescence intensity in LS174T, 4T1, MCF-7 and HepG2 cells (Fig. 3b; Supplementary Fig. 6), in sharp contrast to the very low Cy5 fluorescence intensity observed in MCF-10A, HBEC-5i and IMR-90 cells (Fig. 3c; Supplementary Fig. 6), indicating the selective labeling capability of DCL-AAM in cancer cells with high HDAC and CTSL activity over normal cells with low HDAC and CTSL activity. The labeling efficiency of DCL-AAM in cancer cells was significantly reduced in the presence of TSA or Z-FY-CHO (Fig. 3b, d), substantiating HDAC–CTSL induced activation of DCL-AAM for metabolic labeling. Western blotting analysis of LS174T cells treated with DCL-AAM, DCL-AAM + TSA, and DCL-AAM + Z-FY-CHO also substantiated the inhibitory effect of TSA and Z-FY-CHO against the metabolic activity of DCL-AAM, which otherwise would be enabled by HDAL and CTSL (Fig. 3e). It is noted that neither DCL-AAM nor its degradation products showed significant cytotoxicity after 3-d incubation with cells (Supplementary Fig. 7a). In vitro labeling kinetics study in LS174T cells showed that DCL-AAM-mediated metabolic labeling was time and concentration dependent, with the number of cell-surface azido groups approaching a plateau value in the order of 106–107 per cell after 48-h incubation with 200 μM DCL-AAM (Fig. 3f; Supplementary Fig. 8). Compared to the reported number density of endogenous protein receptors (103–105 per cell)43,44, the number of azido groups that can be placed on the cell surface is 2–4 orders of magnitude higher, which makes for potentially more efficient cancer targeting.
DCL-AAM for cancer-selective labeling in vivo
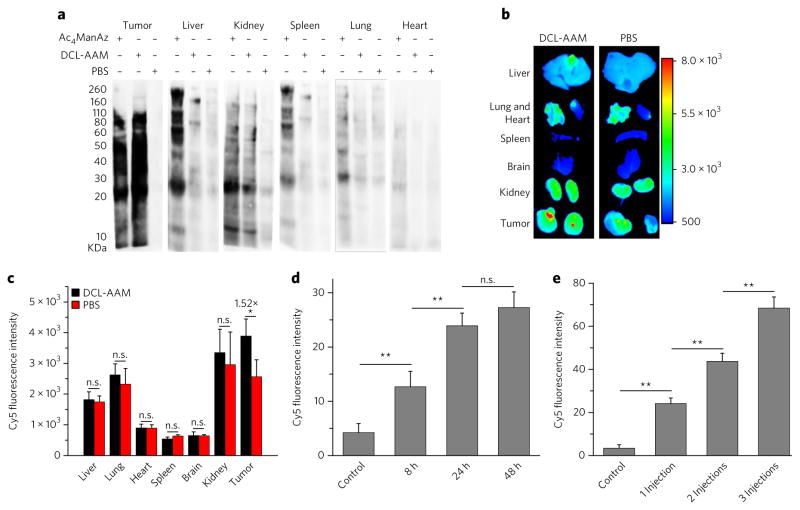
We next studied the cancer-selective labeling capability of DCL-AAM in vivo. DCL-AAM was i.v. injected into athymic nude mice bearing LS174T tumors once daily for three consecutive days. Mice treated with i.v. Ac4ManAz or PBS were used as controls. Western blotting analyses of tumor tissues at 24 h p.i. of azido sugars showed an increase in the amount of azido-modified proteins in the DCL-AAM group compared to the PBS group, while there was a negligible difference observed in the amount of azido-modified proteins in liver, spleen, heart and lung between DCL-AAM and PBS groups (Fig. 4a). In contrast to those results, Ac4ManAz with no labeling selectivity showed nonspecific labeling in normal tissues, with a considerable amount of azido groups expressed in liver, spleen, lung and kidney tissues (Fig. 4a). These experiments demonstrated the superior cancer-selective labeling capability of DCL-AAM in vivo in comparison to Ac4ManAz. In a separate study, DBCO–Cy5 was i.v. injected at 24 h post azido-sugar injections and its biodistribution was monitored. At 48 h p.i. of DBCO–Cy5, the tumors in the DCL-AAM group showed significantly enhanced Cy5 retention compared to the tumors in the PBS group (Fig. 4b). Imaging of the harvested tumors in the DCL-AAM group showed a Cy5 fluorescence intensity of 1.52-fold that of the PBS group (Fig. 4b, c), while there were no significant changes in Cy5 retention in the liver, spleen, heart, lung and kidney tissues (Fig. 4b, c). Compared to the Ac4ManAz group, the DBCO–Cy5 in the DCL-AAM group showed improved accumulation in tumors (Supplementary Fig. 9). Together, these experiments demonstrated that DCL-AAM can selectively label LS174T tumors in vivo and that the expressed azido groups can significantly enhance the tumor accumulation of DBCO–Cy5 via click chemistry.
Figure 4. DCL-AAM mediated cancer-selective labeling in vivo.
(a–c) DCL-AAM (60 mg/kg), Ac4ManAz (40 mg/kg) or PB S was i.v. injected into athymic nude mice bearing subcutaneous LS174T tumors once daily for 3 d. At 24 h after the last injection, the metabolic expression of azido groups in tumor tissues was analyzed by western blotting (a) or by monitoring the biodistribution of i.v.-injected DBCO–Cy5 (5 mg/kg) (b, c). (a) Western blotting analysis of tissues harvested from mice treated with Ac4ManAz, DCL-AAM or PBS. Tissue lysates were incubated with DBCO–Cy3 for 1 h at 37 °C before the gel was run. Protein bands were visualized using ImageQuant LAS 4010 system. (b) Ex vivo fluorescence imaging of tissues harvested from mice treated with DCL-AAM or PBS at 48 h p.i. of DBCO–Cy5. (c) Average Cy5 fluorescence intensity of tissues (n = 3) from (b). (d) Normalized Cy5 fluorescence intensity of LS174T tumor tissues (20 sections per tumor, n = 3) harvested and sectioned from mice at 8, 24 and 48 h p.i. of DCL-AAM (60 mg/kg), respectively. Tumor tissue sections were labeled with DBCO–Cy5 for 30 min. (e) Normalized Cy5 fluorescence intensity of LS174T tumor tissues (20 sections per tumor, n = 3) harvested from mice with 0, 1, 2 and 3 DCL-AAM injections (60 mg/kg, 24 h interval). Tumor tissue sections were labeled with DBCO–Cy5 for 30 min. All the numerical data are presented as mean ± s.e.m. and analyzed by one-way ANOV A (Fisher; 0.01 < *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
The tumor-labeling kinetics of DCL-AAM in vivo was studied to better understand DCL-AAM-mediated cancer labeling. DCL-AAM was i.v. injected into LS174T tumor-bearing nude mice, and the tumors were harvested and sectioned at 8, 24 and 48 h p.i. Metabolically expressed azido groups in tumor tissues were detected by staining with DBCO–Cy5. Tumor tissues harvested at 8 h p.i. of DCL-AAM showed significantly enhanced Cy5 fluorescence intensity on the cell surface compared to the control group without DCL-AAM treatment (Fig. 4d; Supplementary Fig. 10b), indicating that DCL-AAM successfully labeled tumor cells with azido groups. The amount of expressed azido groups in tumors significantly increased over the course of 24 h treatment with DCL-AAM. No significant difference in the amount of expressed azido groups between 24 h and 48 h groups was observed, indicating that the metabolic labeling process of DCL-AAM was largely complete within 24 h (Fig. 4d; Supplementary Fig. 10b). Dose escalation through multiple injections of DCL-AAM (24 h interval between each injection) resulted in a nearly linear increase of the amount of expressed azido groups in tumor tissues (Fig. 4e; Supplementary Fig. 10d).
DCL-AAM coupled with DBCO–drug for cancer treatment
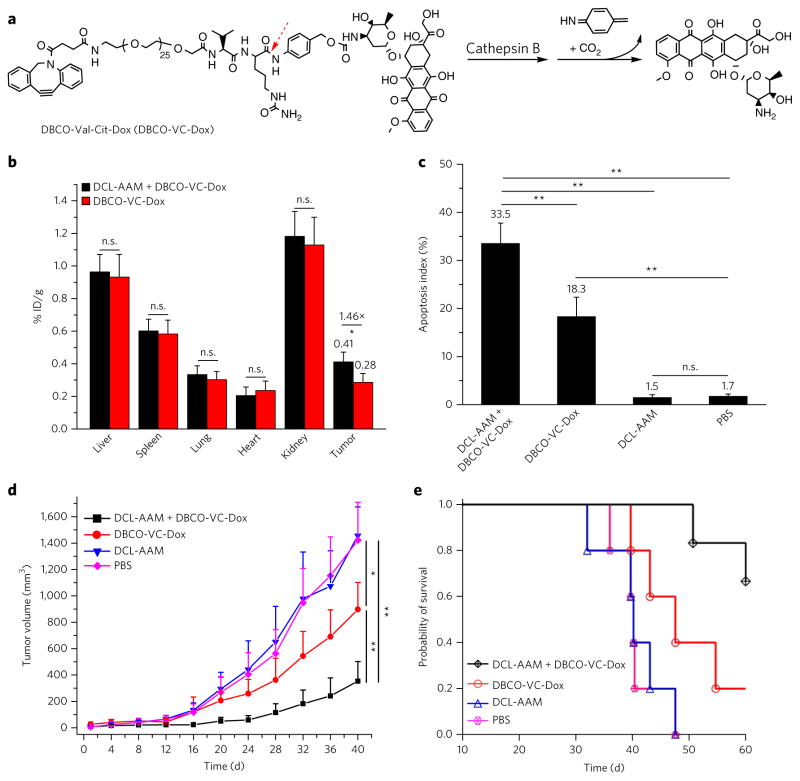
After demonstrating the superior cancer-selective labeling capability of DCL-AAM and the resulting excellent targeting effect of DBCO–Cy5 via click chemistry, we next explored cancer-targeted therapy using DCL-AAM and a DBCO–doxorubicin conjugate (DBCO-VC-Dox (6), Fig. 5a). DBCO-VC-Dox underwent rapid release of Dox in the presence of cathepsin B (Supplementary Fig. 11a) and showed enhanced cellular uptake in DCL-AAM-treated LS174T cells in vitro (Supplementary Fig. 11d). CLSM imaging of LS174T cells showed the overlay of DBCO-VC-Dox with membrane stain (Supplementary Fig. 12), and these covalently attached DBCO-VC-Dox by DCL-AAM-treated LS174T cells demonstrated its capacity to enter late endosomes and lysosomes, as exemplified by DBCO–Cy5 with low passive cell uptake (Supplementary Fig. 13a). The highest Cy5 fluorescence intensity clearly migrated from the edges of the cells toward the center over time (Supplementary Fig. 13b). In a short-term antitumor efficacy study, athymic nude mice bearing LS174T tumors were administered i.v. DCL-AAM or PBS once daily for 3 d (day 0, 1 and 2) and then i.v. DBCO-VC-Dox on day 3. At 48 h p.i. of DBCO-VC-Dox, quantification of the retained drugs in the tissues showed a 1.46-fold tumor accumulation of DBCO-VC-Dox in mice treated with DCL-AAM compared to the control mice treated with PBS, while nonsignificant changes of DBCO-VC-Dox accumulation were observed in the liver, spleen, lung, heart and kidney (Fig. 5b). Tumors treated with DCL-AAM + DBCO-VC-Dox showed an apoptosis index of 33.5 ± 4.2%, which was significantly greater than that of tumors treated with DBCO-VC-Dox only (18.3 ± 4.0%) (Fig. 5c). As negative controls, tumors treated with DCL-AAM alone or PBS showed a much lower apoptosis index, 1.5 ± 0.6% or 1.7 ± 0.5%, respectively (Fig. 5c). These experiments confirmed that DCL-AAM-mediated cancer-selective labeling could significantly improve the tumor accumulation and acute antitumor efficacy of DBCO-VC-Dox.
Figure 5. DCL-AAM-mediated tumor labeling improved antitumor efficacy of DBCO–drug conjugate against primary LS174T colon tumor and MDA-MB-231 triple-negative breast tumor models.
(a) Structure of DBCO-VC-Dox with a cathepsin-B-cleavable linker that consists of a dipeptide (Val–Cit), a self-immolative p-aminobenzylcarbamate and a short polyethylene glycol segment (1 kDa). (b–d) Short-term efficacy study in athymic nude mice bearing subcutaneous LS174T tumors. DCL-AAM (60 mg/kg) was injected i.v. once daily for 3 d (days 0, 1 and 2). Subsequently, DBCO-VC-Dox (10 mg/kg in Dox equivalent) was injected i.v. on day 3. Tumors were harvested for analysis at 48 h p.i. of DBCO-VC-Dox (on day 5). (b) Retained drugs in tissues of drug-treated mice (n = 3). % ID/g, percent of injected dose normalized by gram of tissue. (c) Quantification of TUNEL stains of LS174T tumors from mice treated with DCL-AAM + DBCO-VC-Dox, DBCO-VC-Dox, DCL-AAM and PBS, respectively (20 sections per tumor, n = 6). (d, e) Long-term antitumor efficacy study in nude mice bearing subcutaneous MDA-MB-231 tumors. DCL-AAM (60 mg/kg) was injected i.v. on days 0, 1 and 2. Subsequently DBCO-VC-Dox (12 mg/kg in Dox equivalent) was injected i.v. on days 3, 7 and 11. (d) Average MDA-MB-231 tumor volume of each group of mice (DCL-AAM + DBCO-VC-Dox and DBCO-VC-Dox group, n = 6; DCL-AAM and PBS group, n = 5) over the course of the efficacy study. (e) Kaplan–Meier plots for all groups. Loss of mice was a result of treatment-related death or euthanasia after the predetermined end point was reached. All the numerical data are presented as mean ± s.e.m. and analyzed by one-way ANOV A (Fisher; 0.01 < *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
To further understand how the enhanced tumor accumulation of DBCO-VC-Dox mediated by DCL-AAM labeling would impart improved antitumor activity, a separate efficacy study was conducted by monitoring the tumor volume over a prolonged period. Athymic nude mice bearing LS174T tumors were divided into four groups: DCL-AAM + DBCO-VC-Dox, DBCO-VC-Dox, DCL-AAM and PBS. Compared to the PBS and DCL-AAM groups, the DCL-AAM + DBCO-VC-Dox group exerted greater tumor growth inhibition (Supplementary Fig. 14a). In comparison to DBCO-VC-Dox, DCL-AAM + DBCO-VC-Dox also significantly reduced tumor growth rate (Supplementary Fig. 14a). DCL-AAM + DBCO-VC-Dox improved the survival time of mice by 86.0% compared to that of the PBS group, which was substantially greater than that of the DBCO-VC-Dox group (17.1%) (Supplementary Fig. 14b, c). Together, DCL-AAM + DBCO-VC-Dox exerted greatly improved anticancer efficacy over DBCO-VC-Dox alone, as a result of the enhanced tumor accumulation of drugs via click chemistry.
To extend the applicability of ATTACK to address unmet clinical needs, we evaluated the activity of DCL-AAM + DBCO-VC-Dox in additional preclinical models such as TNBC, which is characterized by the limited expression of estrogen receptors, progesterone receptors and HER2 (refs. 45,46). DCL-AAM was able to efficiently label MDA-MB-231 TNBC cells with azido groups, which improved the cellular uptake of DBCO-VC-Dox in vitro (Supplementary Fig. 15a–c). In a long-term efficacy study, athymic nude mice bearing MDA-MB-231 tumors were divided into four groups: DCL-AAM + DBCO-VC-Dox, DBCO-VC-Dox, DCL-AAM and PBS. DCL-AAM was administered i.v. on days 0, 1 and 2 for groups 1 and 3. Subsequently, DBCO-VC-Dox was administered i.v. on days 3, 7 and 11 for groups 1 and 2. Compared to the PBS group, the DCL-AAM group showed a negligible difference in the tumor growth rate (Fig. 5d), excluding the influence of DCL-AAM alone on any observed antitumor effect. DCL-AAM + DBCO-VC-Dox and DBCO-VC-Dox increased the survival time of mice by 62.6% and 29.0%, respectively, compared to PBS group (Fig. 5e; Supplementary Fig. 16a). DCL-AAM + DBCO-VC-Dox exerted greater antitumor efficacy than DBCO-VC-Dox alone, with significantly reduced tumor volume from as early as day 24 (Fig. 5d; Supplementary Fig. 16b). These experiments demonstrated that DCL-AAM-mediated labeling of MDA-MB-231 tumors could improve the antitumor efficacy of DBCO-VC-Dox.
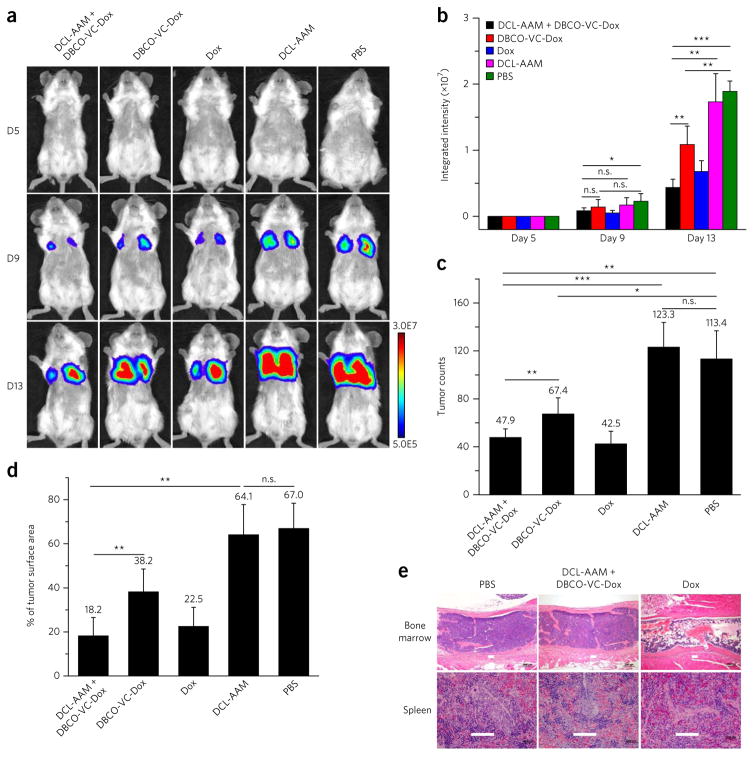
Metastatic cancers are the leading cause of cancer-related deaths, as metastases can evade conventional cancer therapies such as surgery, radiotherapy and chemotherapy47. After demonstrating the effectiveness of DCL-AAM + DBCO-VC-Dox for the treatment of primary LS174T colon tumor and MDA-MB-231 breast tumor models, we investigated whether ATTACK-mediated targeted therapy would be effective in inhibiting the growth of 4T1 metastatic breast cancers. The efficacy study was designed similarly to the previous studies except that Dox was added as a control group to evaluate the therapeutic benefits of DCL-AAM + DBCO-VC-Dox. 4T1 metastatic cancers were established in BALB/c mice by i.v. injection of luciferase-engineered 4T1 cells on day 0. After treatment with DCL-AAM once daily for 3 d (days 1–3), mice were i.v. administered with DBCO-VC-Dox or Dox starting from day 4 to examine their efficacy in metastasis inhibition. The intensifying bioluminescence signals of PBS group mice over time confirmed the proliferation of 4T1 metastases in the lung parenchyma (Fig. 6a). All drug treatment groups showed significantly reduced bioluminescence signals and tumor nodule counts in comparison to PBS and DCL-AAM groups (Fig. 6a–d). Compared to the DBCO-VC-Dox group, the mice in the DCL-AAM + DBCO-VC-Dox group showed significantly reduced lung metastases, evidenced by the greatly reduced bioluminescence signals (Fig. 6a, b), lower counts of tumor nodules (47.9 ± 7.1% compared to 67.4 ± 13.5%) (Fig. 6c) and decreased percentage of tumor surface area (18.2 ± 8.3% compared to 38.2 ± 10.4%) (Fig. 6d). Despite yielding similar lung metastasis severity to DCL-AAM + DBCO-VC-Dox, Dox exerted much greater toxicity in mouse tissues, especially in the bone marrow and spleen (Fig. 6e; Supplementary Fig. 17d). Together, DCL-AAM + DBCO-VC-Dox exerted the best anticancer efficacy, with greatly reduced toxicity, as a collective result of enhanced tumor accumulation of DBCO-VC-Dox via click chemistry and its cancer-preferential drug release imparted by differences in cathepsin B activity between cancerous and normal tissues48.
Figure 6. DCL-AAM-mediated tumor labeling improved anticancer efficacy of DBCO–drug conjugate against 4T1 lung metastases.
4T1 lung metastases were established in BALB/c mice by i.v. injection of luciferase-engineered 4T1 cells on day 0, and mice were randomly divided into five groups (group 1: DCL-AAM + DBCO-VC-Dox; group 2: DBCO-VC-Dox; group 3: Dox; group 4: DCL-AAM; group 5: PB S; DCL-AAM + DBCO-VC-Dox, DBCO-VC-Dox and Dox group, n = 8; DCL-AAM and PBS group, n = 7). DCL-AAM (60 mg/kg) was injected i.v. once daily on days 1, 2 and 3. Subsequently, DBCO-VC-Dox (12 mg/kg) or Dox (7.5 mg/kg) was injected i.v. on days 4, 8 and 12. Tumor growth was monitored by bioluminescence imaging on days 5, 9 and 13. (a) Representative bioluminescence images of BALB/c mice on days 5, 9 and 13 (D5, D9, D13, respectively). (b) Average integrated bioluminescence intensity of mice over the course of the efficacy study (n = 7 or 8). (c) Average tumor nodule counts on the lung tissues of mice (n = 7 or 8). (d) Percentage of tumor surface area over total lung tissue area for mice from different groups (n = 7 or 8). (e) Histopathology of BALB/c mice tissues harvested at the end of 4T1 lung metastases efficacy study showed severe toxicity of Dox (22.5 mg/kg in total) in bone marrow (upper row, marked bone marrow lysis) and spleen (lower row, lymphoid depletion), while DCL-AAM + DBCO-VC-Dox (36.0 mg/kg Dox in total) showed minimal toxicity. Scale bar, 100 μm. All the numerical data are presented as mean ± s.e.m. and analyzed by one-way ANOV A (Fisher; 0.01 < *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
DISCUSSION
Cell-surface receptors provide the molecular recognition moieties for the design of cancer-targeting technologies49. Nearly all current targeting strategies rely on specific recognition of the endogenous protein receptors; one such technology that has achieved great success is the antibody and antigen technology. Antibody–antigen technology for cancer targeting has several limitations, however, such as the large size of both receptor and targeting ligand, the limited number of endogenous receptors, immunogenicity, high cost, the difficulty of handling antibody, and the synthetic challenges of structure-specific drug–antibody conjugates. In this paper, we report the development of ATTACK (Fig. 1b), an unprecedented in vitro and in vivo targeting technology that is based on selective exogenous introduction of an azido group—an artificial antigen—onto the surface of the target cancer cells followed by efficient in vivo click chemistry targeting the introduced azido group. ATTACK represents a completely small-molecule-based targeting technology complementary to antibody–antigen targeting technology with several obvious advantages, such as greatly reduced size of the receptor or targeting moieties, substantially increased cell-surface receptor number, ease of handling and synthesis of both azido sugars and targeting small-molecule substrates, low cost and nonimmunogenicity. One great advantage of ATTACK over antibody–antigen technology is that ATTACK-mediated targeting hinges upon cancer-specific triggers for intracellular activation of metabolic precursors instead of endogenously expression of tumor antigen. Thus, ATTACK may provide a clinical solution for targeted therapy to some tough diseases lacking targetable antigens, such as TNBC50, which still remains a clinical challenge for any type of targeted therapy.
In conclusion, we report the design of ATTACK, an in vitro and in vivo cancer targeting technology facilitated by nonproteinic small molecules. ATTACK-mediated enhanced antitumor efficacies with the use of DCL-AAM and DBCO-VC-Dox were demonstrated in several tumor models, including primary LS174T colon cancer, MDA-MB-231 TNBC and 4T1 lung metastases. Substantially reduced toxicity was also observed with the use of ATTACK therapy (Fig. 6e and Supplementary Fig. 17d). The strategy for the design of DCL-AAM should be expandable to the design of other sugar derivatives that can undergo selective metabolism and azide cell labeling in various other types of diseases, as long as endogenous disease-specific triggers (such as oxidants, reductases, matrix metalloproteinases and cathepsin subsets) can be identified and used to trigger the activation of azido-sugar derivatives. Further improved in vivo targeting efficiency can potentially be achieved through the use of drugs or agents with low passive accumulation in cancer cells, such as protein- and nucleic-acid-based therapeutics. We believe that ATTACK, as a powerful small molecule based targeting technology, has tremendous potential for eventual clinical applications.
ONLINE METHODS
Materials
D-Mannosamine hydrochloride and other materials were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted. N-hydroxysuccinimide (NHS) was purchased from Acros Organics (Renningen, Germany). DBCO–Cy3 and DBCO–Cy5 were purchased from KeraFAST, Inc (Boston, MA, USA). EZ-Link phosphine–PEG3–biotin, streptavidin–horseradish peroxidase (HRP), and Pierce ECL western blotting substrate were purchased from Thermo Fisher Scientific (Waltham, MA, USA). DBCO–NHS was purchased from Conju-Probe (San Diego, CA, USA). Doxorubicin hydrochloride was purchased from Cayman Chemical Company (Ann Arbor, MI, USA). Fmoc–Val–Cit–PABC–PNP was purchased from Concortis (San Diego, CA, USA). HOOC–PEG–NH2 (1 k molecular weight) was purchased from Nanocs Inc. (New York, NY, USA). Ac-Lys(Ac)-OH was purchased from Neobits Inc. (Sunnyvale, CA, USA). Cathepsin L Inhibitor II (Z-FY-CHO) was purchased from EMD Millipore (Billerica, MA, USA). Trichostatin A was purchased from Abcam (Cambridge, UK). 64Cu chloride was obtained from the Washington University (St. Louis, MO, USA). Anhydrous dichloromethane (DCM), hexane, ethyl acetate, diethyl ether, tetrahydrofuran (THF) and dimethylformamide (DMF) were purified by passing them through alumina columns and kept anhydrous by storing them in the presence of molecular sieves. Phosphate-buffered saline (PBS) and Dulbecco’s Modified Eagle Medium (DMEM) were obtained from Invitrogen (Carlsbad, CA, USA). MEGM BulletKit was purchased from Lonza Inc. (Allendale, NJ, USA). Eagle’s minimum essential medium (EMEM) was purchased from American Type Culture Collection (Manassas, VA, USA). Fetal Bovine Serum (FBS) was obtained from Lonza Walkersville Inc (Walkersville, MD, USA). BD Falcon culture plates were purchased from Fisher Scientific (Hampton, NH, USA). ProLong Gold antifade reagent was purchased from Life Technologies (Carlsbad, CA, USA).
Instrumentation
Nuclear magnetic resonance (NMR) analyses were conducted on a Varian U500 (500 MHz) or a VXR500 (500 MHz) spectrometer. Infrared spectra were recorded on a PerkinElmer 100 serial FTIR spectrophotometer (PerkinElmer, Waltham, MA, USA). Electrospray ionization (ESI) mass spectra were obtained from Waters ZMD Quadrupole Instrument (Waters, Milford, MA, USA). MALDI spectra were collected on a Bruker Daltonics UltrafleXtreme MALDI TOFTOF (Bruker, Billerica, MA, USA). HPLC analyses were performed either on a Beckman Gold system (Beckman Coulter, Fullerton, CA, USA) equipped with a 126P solvent module, a System Gold 128 UV detector and an analytical C18 column (Luna C18, 250*4.6 mm, 5 μm, Phenomenex, Torrance, CA, USA) or on a Shimadzu CBM-20A system (Shimadzu, Kyoto, Japan) equipped with a SPD20A PDA detector (190–800 nm), a RF10Axl fluorescence detector, and an analytical C18 column (Shimadzu, 3 μm, 50*4.6 mm, Kyoto, Japan). Lyophilization was conducted in a Labconco FreeZone lyophilizer (Kansas City, MO, USA). UV light (365 nm) was generated from an OmiCure S1000 UV lamp (EXFO, Mississauga, Canada). Confocal laser scanning microscopy (CLSM) images were taken on a Zeiss LSM 700 Confocal Microscope (Carl Zeiss, Thornwood, NY, USA). Flow cytometry analyses of cells were conducted with a BD FACS Canto 6-color flow cytometry analyzer (BD, Franklin Lakes, NJ, USA). Fluorescence intensity of cells was measured on an IN Cell Analyzer 2200 system (GE Healthcare Life Sciences, Pittsburgh, PA, USA). In vivo and ex vivo images of animals were taken either on the Maestro In Vivo Fluorescence Imaging System (CRI, Woburn, MA, USA) or on the Bruker In Vivo Imaging System (Bruker, Billerica, MA, USA). Bioluminescence images of animals were taken on the Bruker In Vivo Imaging System (Bruker, Billerica, MA, USA). Ex vivo measurement of the radioactivity was conducted on a 2480 Wizard2 Automatic Gamma Counter (PerkinElmer, Waltham, MA, USA). Protein bands were visualized by using either X-ray imaging or Image Quant LAS 4010 (GE Healthcare, Little Chalfont, UK). Frozen tissues were embedded with optimum cutting temperature (O.C.T.) compound (Sakura Finetek USA, Torrance, CA, USA), sectioned by a Leica CM3050S Cryostat, and mounted onto glass slides. Histological images were taken on a Nanozoomer slide scanning system (Hamamatsu Photonics, Japan).
Cell culture
LS174T colon cancer, MDA-MB-231 triple-negative breast cancer, HepG2 liver cancer, MCF-7 breast cancer, HeLa, MCF-10A breast basal epithelial cell, HBEC-5i cerebral microvascular endothelium cell, and IMR-90 human fibroblast cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). 4T1 cells engineered with firefly luciferase were provided by D. Piwnica-Worms from Washington University (St. Louis, MO, USA). Cells were cultured in DMEM containing 10% FBS, 100 units/ml Penicillin G and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C in 5% CO2 humidified air unless otherwise noted. MCF-10A cells were cultured in MEGM BulletKit. IMR-90 cells were cultured in EMEM containing 10% FBS. HBEC-5i were cultured in EMEM with 10% FBS and 40 μg/ml endothelial growth supplement (ECGS).
Animals
Female athymic nude mice and BALB/c mice were purchased from Charles River (Wilmington, MA, USA) or Shanghai Slaccas Experimental Animal Co. Ltd. (Shanghai, China). Feed and water were available ad libitum. Artificial light was provided in a 12 h/12 h cycle. The animal protocol was reviewed and approved by the Illinois Institutional Animal Care and Use Committee (IACUC) of University of Illinois at Urbana–Champaign and IACUC of Soochow University. For all the animal studies, mice were randomly allocated to each group. The investigator was aware of the group allocation during the animal studies, as demanded by the experimental designs.
In vitro and in vivo cancer labeling of Ac3ManAzEt and Ac3ManAzNB
UV induced degradation of Ac3ManAzNB
Ac3ManAzNB (1 mg) was dissolved in methanol/H2O (1.0 ml) and stirred at room temperature under UV irradiation. The intensity of UV irradiation was set at 10 mW/cm2. At selected time points (0, 5, 15, and 30 min), HPLC and ESI MS measurements were conducted.
General procedures for confocal imaging of azido-sugar labeled cells
Cells were seeded onto coverslips in a 6-well plate at a density of 4 × 104 cells per well and allowed to attach for 12 h. Azido sugar (50 μM) was added and the cells were incubated at 37 °C for 72 h. After washing with PBS, cells were incubated with DBCO–Cy5 (50 μM) for 1 h and fixed with 4% paraformaldehyde (PFA) solution, followed by staining of cell nuclei and membrane with DAPI (2 μg/ml) and CellMask orange plasma membrane stain (1 μg/ml), respectively. The coverslips were mounted onto microscope slides and imaged under a confocal laser scanning microscope.
General procedures for flow cytometry analysis of azido-sugar labeled cells
Cells were seeded in a 24-well plate at a density of 1 × 104 cells per well and allowed to attach for 12 h. Ac4ManAz or Ac3ManAz derivative (50 μM) was added and incubated with cells for 72 h. After washing with PBS, cells were incubated with DBCO–Cy5 (50 μM) for 1 h. Cells were lifted by incubating with trypsin solution and analyzed by flow cytometry.
Comparison of passive and active cellular uptake of DBCO-Cy5
LS174T cells were seeded in a 24-well plate at a density of 1 × 104 cells per well and allowed to attach for 12 h. Ac4ManAz (50 μM) was added to designated wells. After 72 h, nontreated cells were incubated with DBCO–Cy5 (50 μM) for 30 min, 1 h, or 2 h. Cells treated with Ac4ManAz were incubated with DBCO–Cy5 (50 μM) for 1 h. After washing with PBS, cells were lifted by incubating with trypsin solution and analyzed by flow cytometry.
Ac3ManAzNB-mediated controlled cell labeling
LS174T cells were seeded onto coverslips in a 6-well plate at a density of 4 × 104 cells per well and allowed to attach for 12 h. Ac3ManAzNB (50 μM) was added. After 4 h, UV light (10 mW/m2) was applied for 10 min, and the cells were further incubated for 68 h. Cells without UV irradiation were continuously incubated for 72 h. Cell samples for confocal imaging and flow cytometry were prepared following the above-mentioned general procedures.
Western blotting analysis of cells treated with azido sugars
(1) Chemiluminescence method: azido-modified proteins in cell lysates were biotinylated by incubation with phosphine–PEG3–biotin and then detection by streptavidin–HRP and ECL Western Blotting Substrate. The experimental procedures were reported elsewhere22. (2) Fluorescence method: 20 μl of cell lysate was incubated with DBCO–Cy3 (10 μM) in 5% bovine serum albumin (BSA) for 1 h. After gel running and membrane transfer, protein bands were visualized using an Image Quant LAS 4010 system with a Cy3/Cy3 (excitation/emission) channel.
In vivo tumor labeling of Ac4ManAz, Ac3ManAzEt, and Ac3ManAzNB. LS174T colon tumors were established in 6-week-old female athymic nude mice by subcutaneous injection of LS174T cells (1.5 × 106 cells) in Hank’s balanced salt solution (HBSS)/Matrigel (1/1, v/v) into both flanks. When the tumors reached ~50 mm3, Ac4ManAz or Ac3ManAzEt or Ac3ManAzNB (25 mM, 20 μl) was injected into the left tumors once daily for three consecutive days while the same amount of PBS was injected to the right tumors as control (n = 4 per group): (1) Tumors were harvested from mice (n = 1 per group) at 24 h p.i. of azido sugars and bisected. Half of the tumor was homogenized and lysed. The concentration of soluble proteins in cell lysates was determined by bicinchoninic acid (BCA) assay and adjusted to 5 mg/ml for each group. The rest of procedures were the same as the abovementioned western blotting analysis of cells. The other half of tumor was directly frozen in optimal cutting temperature (O.C.T.) compound and sectioned with a thickness of 8 μm. Tumor tissue sections were incubated with 5% BSA for 2 h and then labeled with DBCO–Cy5 (20 μM) for 30 min. After washing with PBS, DAPI (2 μg/ml) and CellMask orange plasma membrane stain (1 μg/ml) were added to stain cell nuclei and membrane, respectively. Tumor sections were imaged under a confocal laser scanning microscope. (2) In a separate study, at 24 h p.i. of azido sugars, DBCO–Cy5 (5 mg/kg) was i.v. injected and its biodistribution was monitored using the Maestro In Vivo Fluorescence Imaging System (n = 3 per group). The excitation filter of 575–605 nm was used. Tumors and organs were harvested from mice at 24 h p.i. of DBCO–Cy5 and imaged ex vivo. Cy5 fluorescence intensity at selected regions of interest (ROIs) was quantified using the Maestro imaging software. After ex vivo imaging, tumors were bisected. Half of the tumor was directly frozen in O.C.T. compound, sectioned with a thickness of 8 μm, stained with DAPI (2 μg/ml) and CellMask orange plasma membrane stain (1 μg/ml), and imaged under a confocal laser scanning microscope. Organs and the other half of tumors were homogenized and lysed. The lysates were measured on a fluorescence spectrometer to determine the amount of Cy5 retained in the tissues based on the standard curve of Cy5 fluorescence intensity. Data were presented as the percentage of injected dose per gram of tissue (% I.D./g).
DCL-AAM mediated cancer-selective labeling in vitro and in vivo
Detection of cellular HDAC/CTSL activity
Cells were seeded in a 24-well plate at a density of 1 × 104 cells per well and allowed to attach overnight. Naph-Lys (50 μM, see Supplementary Section) was added and incubated with cells for different time (30 min, 1 h, 2 h, and 4 h). After washing with PBS, cells were fixed with 4% PFA and stained with DAPI (2 μg/ml). Average fluorescence intensity of cells was measured on the IN Cell Analyzer 2200 using DAPI and FITC channels. The DAPI and FITC channels were used for determining the number of cells per well and the total fluorescence intensity of released Naph-NH2 per well, respectively. Data were presented as the mean fluorescence intensity ± s.e.m. per cell (n = 6).
Inhibitory effect of TSA and Z-FY-CHO on cellular HDAC/CTSL activity
Cells were seeded in a 24-well plate at a density of 1 × 104 cells per well and allowed to attach overnight. The cells were incubated with Naph-Lys (50 μM), Naph-Lys (50 μM) + TSA (1 μM), Naph-Lys (50 μM) + Z-FY-CHO (50 μM), or PBS for 4 h, fixed with 4% PFA, and stained with DAPI (2 μg/ml). Average fluorescence intensity of cells was measured on the IN Cell Analyzer 2200 using DAPI and FITC channels.
DCL-AAM mediated controlled cell labeling in vitro
Cells were seeded onto coverslips in a 6-well plate at a density of 4 × 104 cells per well and allowed to attach overnight. The cells were incubated with DCL-AAM (50 μM), DCL-AAM (50 μM) + TSA (1 μM), DCL-AAM (50 μM) + Z-FY-CHO (50 μM), and PBS, respectively, for 72 h and subsequently labeled with DBCO–Cy5 (50 μM) for 1 h. Cell samples for confocal imaging and flow cytometry measurements were prepared following the above-mentioned procedures.
Labeling kinetics of DCL-AAM in vitro
LS174T cells were seeded in black 96-well plates and incubated with various concentrations of DCL-AAM (2 μM, 10 μM, 50 μM, 200 μM, and 1 mM) for different time (3 h, 6 h, 12 h, 24 h, 48 h, and 72 h). After washing with PBS, cells were incubated with DBCO–Cy5 (50 μM) for 1 h, washed, and analyzed by flow cytometry.
Western blotting analysis of DCL-AAM treated cells
Cells were seeded onto cell culture flasks (25 cm2 growth area) at a density of 5 × 105 cells per flask and allowed to attach overnight. The cells were then treated with DCL-AAM (50 μM), DCL-AAM (50 μM) + TSA (1 μM), DCL-AAM (50 μM) + Z-FY-CHO (50 μM), and PBS, respectively, for 72 h. The rest of procedures were the same as the abovementioned general procedures for western blotting analysis of cells. Protein bands were visualized using the fluorescence method.
Pharmacokinetics study of 64Cu-labeled DCL-AAM
64Cu-labeled DCL-AAM (~100 μCi) was i.v. injected into female athymic nude mice bearing subcutaneous LS174T tumors (n = 3). At selected time points (10 min, 30 min, 1 h, 3 h, 6 h, 12 h, and 24 h p.i.), blood was collected from orbital sinus using capillary tubes. The collected blood samples were weighed and measured for 64Cu radioactivity on a Wizard2 automatic γ-counter. Raw counts were corrected for background, decay, and weight. Corrected counts were converted to μCi per gram blood via a previously determined calibration curve and presented as % I.D./g.
DCL-AAM mediated cancer-selective labeling in vivo
(1) Western blotting method: LS174T tumor models were established in 6-week-old female athymic nude mice by subcutaneous injection of LS174T colon cancer cells (1.5 × 106 cells) in HBSS/Matrigel (1/1, v/v). When the tumors reached ~50 mm3, DCL-AAM (60 mg/kg, 1.0 molar equivalent of azido groups) was i.v. injected once daily for three consecutive days. Mice i.v. administered with Ac4ManAz (40 mg/kg, 1.2 molar equivalent of azido groups) or PBS were used as controls. At 24 h post the last injection, tumors and organs were harvested for western blotting analyses following the above-mentioned procedures. Azido-labeled proteins were visualized using the fluorescence method. (2) Fluorescence imaging method: in a separate study, at 48 h post the last injection of DCL-AAM or Ac4ManAz or PBS, DBCO–Cy5 (5 mg/kg) was i.v. injected and its biodistribution was monitored using the Bruker In Vivo Xtreme Imaging System. The excitation/emission filter of 630 nm/700 nm was used. Tumors and organs were harvested from mice at 48 h p.i. of DBCO–Cy5 and imaged ex vivo. Fluorescence intensity at selected ROIs was quantified using the Bruker imaging software. After ex vivo imaging, tumors were bisected. Half of the tumor was directly frozen in O.C.T. compound, sectioned with a thickness of 8 μm, stained with DAPI (2 μg/ml) and CellMask orange plasma membrane stain (1 μg/ml), and imaged under a confocal laser scanning microscope. Organs and the other half of tumors were homogenized and lysed. The lysates were measured on a fluorescence spectrometer to determine the amount of Cy5 retained in the tissues. Data were presented as % I.D./g.
Tumor labeling kinetics of DCL-AAM in vivo
Athymic nude mice bearing subcutaneous LS174T tumors were i.v. injected with DCL-AAM (60 mg/kg). Mice without DCL-AAM injection were used as controls. At different time (8, 24, and 48 h) p.i. of DCL-AAM, tumors were harvested, frozen in O.C.T. compound, and sectioned with a thickness of 8 μm. Tumor tissue sections were incubated with 5% BSA for 2 h and then labeled with DBCO–Cy5 (20 μM) for 30 min. After washing with PBS, DAPI (2 μg/ml) and CellMask orange plasma membrane stain (1 μg/ml) were added to stain cell nuclei and membrane, respectively. Tumor tissue sections were imaged under a confocal laser scanning microscope. The imaging parameters were kept the same for all samples imaged. Data analyses were performed with the ZEN 2011 software. The mean Cy5 fluorescence intensity of each image was extracted and averaged over 20 images to obtain the mean Cy5 fluorescence intensity of each tissue section, which was then averaged over 20 tissue sections to obtain the mean Cy5 fluorescence intensity for each tumor.
Dose-frequency-dependent DCL-AAM-mediated tumor labeling in vivo
Athymic nude mice (n = 3 per group) bearing subcutaneous LS174T tumors were i.v. administered with DCL-AAM (60 mg/kg) for different times (one, two, or three injections) with a 24 h interval. Mice without DCL-AAM injection were used as controls. At 24 h post the last injection of DCL-AAM, tumors were harvested, frozen in O.C.T. compound, and sectioned with a thickness of 8 μm. Tumor tissue sections were stained, imaged, and analyzed following the procedures described above.
Characterizations of DBCO-VC-Dox
Cathepsin-B-induced degradation of DBCO-VC-Dox
Bovine spleen cathepsin B was activated following the reported procedures51. DBCO-VC-Dox was added to the activated cathepsin B solution or PBS or 10% FBS, and incubated at 37 °C. Aliquots of the mixture were taken out for HPLC measurements at selected time points.
Uptake of DBCO-VC-Dox in DCL-AAM treated cancer cells
Cells were seeded in a 24-well plate at a density of 1 × 104 cells per well and incubated with DCL-AAM (50 μM) or PBS for three days. After washing with PBS, the cells were incubated with DBCO-VC-Dox (20 μM) for different time (30 min, 1 h, 2 h, and 4 h), lysed, and measured on a plate reader for fluorescence intensity of DBCO-VC-Dox. After fluorescence measurements, protein concentration in each well was determined via BCA assay. The final data were presented as fluorescence intensity of DBCO-VC-Dox per milligram protein.
Uptake mechanism of DBCO–Cy5 or DBCO-VC-Dox in DCL-AAM-treated cells
LS174T cells were seeded onto cell culture dishes with cover glass bottom and incubated with DCL-AAM (50 μM) for 3 d. After washing with PBS, the cells were incubated with DBCO–Cy5 (20 μM) and LysoTracker Green (2 μg/ml) for 1 h. Hoechst 33342 (10 μg/ml) and CellMask orange plasma membrane stain (1 μg/ml) were then added to stain cell nuclei and membrane, respectively, for 10 min. After washing with PBS, the cells were further incubated in fresh medium at 37 °C for different time (1, 3, 6, 12, and 24 h) before being imaged under a fluorescence microscope to monitor the cellular internalization of DBCO–Cy5. Uptake of DBCO-VC-Dox was monitored similarly except that CellMask deep-red plasma membrane stain was used to stain cell membrane and that LysoTracker Green was not added, because the fluorescence signals of DBCO-VC-Dox overlapped with those of LysoTracker Green and CellMask orange plasma membrane stain.
Pharmacokinetics study of DBCO-VC-Dox
DBCO-VC-Dox (5 mg/kg in Dox equivalent) was i.v. injected into female athymic nude mice (n = 3). At selected time points (10 min, 30 min, 1 h, 2 h, 4 h, 6 h, 12 h, and 24 h p.i.), blood was collected from orbital sinus using capillary tubes and diluted with methanol/H2O. After centrifugation, the supernatant (20 μl) was injected into HPLC for the quantification of DBCO-VC-Dox or released Dox. The plasma concentration of DBCO-VC-Dox was calculated in μg/ml.
Anticancer efficacy studies of DCL-AAM + DBCO-VC-Dox
Short-term efficacy study of DCL-AAM + DBCO-VC-Dox against subcutaneous LS174T tumors
LS174T tumors were established in 6 week-old female athymic nude mice by subcutaneous injection of LS174T colon cancer cells (1.5 × 106 cells) in HBSS/Matrigel (1/1, v/v, 50 μl) into both flanks. When the tumors reached ~50 mm3, mice were randomly divided into four groups (group 1: DCL-AAM+ DBCO-VC-Dox; group 2: DBCO-VC-Dox; group 3: DCL-AAM; group 4: PBS; n = 3). For group 1 and group 3, DCL-AAM (60 mg/kg) was i.v. injected once daily for three days (days 0, 1, and 2). Mice in the other two groups were i.v. injected with PBS as control. At 24 h post the last injection of DCL-AAM, DBCO-VC-Dox (10 mg/kg in Dox equivalent) or PBS was i.v. injected. At 48 h p.i., tumors were harvested and bisected. Half of the tumors were frozen with O.C.T. compound, sectioned with a thickness of 8 μm, and analyzed for apoptosis via terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. Organs and the other half of tumors were weighed, homogenized, and lysed. After the addition of acidified isopropanol, the mixture was vortexed and frozen overnight at −20 °C. After centrifugation, the supernatant was injected into HPLC for the quantification of retained Dox in tissues. Data were presented as %I.D./g.
Long-term efficacy study of DCL-AAM + DBCO-VC-Dox against subcutaneous LS174T tumors
LS174T tumors were established in 6-week-old female athymic nude mice by subcutaneous injection of LS174T cells (1.5 × 106 cells in HBSS/Matrigel (1/1, v/v, 50 μl)) into both flanks. When the tumors reached ~50 mm3, mice were randomly divided into four groups (group 1: DCL-AAM+DBCO-VC-Dox; group 2: DBCO-VC-Dox; group 3: DCL-AAM; group 4: PBS; n = 5–6). For group 1 and group 3 mice, DCL-AAM (60 mg/kg) was i.v. injected on days 0, 1, and 2. Mice in the other two groups were i.v. injected with PBS as control. DBCO-VC-Dox (12 mg/kg in Dox equivalent) or PBS was i.v. injected on days 3, 7, and 11. Tumor volume and body weight of mice were measured every other day. The tumor volume was calculated using the formula (length) × (width)2/2, where the long axis diameter was regarded as the length and the short axis diameter was regarded as the width. When the tumor volume reached 2,000 mm3 (as predetermined endpoint) or the animal had become moribund or the body weight loss was beyond 20% of original weight, the animal was sacrificed. When an animal exited the study due to tumor volume or treatment-related death, the final tumor volume recorded for the animal was used to calculate the mean tumor volume at subsequent time points. The time to endpoint (TTE) of each animal was defined as the day when its tumor volume had reached the predetermined endpoint. Animals classified as treatment-related deaths were assigned a TTE value equal to the day of death. Treatment efficacy was determined by tumor growth delay (TGD), which was defined as the increase in the median TTE in a treatment group compared to the control (PBS) group: TGD = TTE(T)-TTE(C) which was expressed in days, or as a percentage of the median TTE of the control group: %TGD = 100% × (TTE(T)-TTE(C))/TTE(C)52.
Long-term efficacy study of DCL-AAM + DBCO-VC-Dox against subcutaneous MDA-MB-231 tumors
MDA-MB-231 tumors were established in 6 week-old female athymic nude mice by subcutaneous injection of MDA-MB-231 cells (1.5 × 106 cells in HBSS/Matrigel (50 μL, 1/1, v/v)) into both flanks. When the tumors reached ~50 mm3, mice were randomly divided into four groups (group 1: DCL-AAM + DBCO-VC-Dox; group 2: DBCO-VC-Dox; group 3: DCL-AAM; group 4: PBS; n = 5). DCL-AAM (60 mg/kg) was i.v. injected to group 1 and group 3 mice once daily for three consecutive days (days 0, 1, and 2). DBCO-VC-Dox (12 mg/kg in Dox equivalent) was i.v. injected on days 3, 7, and 11. Tumor volume and body weight of each mouse were measured every 4 d. The endpoint of tumor volume was set at 1,500 mm3. Data analyses were the same as the above-mentioned long-term efficacy study against LS174T tumors.
Anticancer efficacy study against 4T1 lung metastases in BALB/c mice
4T1 metastatic cancer model was established by tail vein injection of luciferase-engineered 4T1 cells (1 × 105 cells in 200 μl HBSS) into 6-week old BALB/c mice on day 0. Mice were then randomly divided into five groups (group 1: DCL-AAM + DBCO-VC-Dox; group 2: DBCO-VC-Dox; group 3: Dox; group 4: DCL-AAM; group 5: PBS; n = 7–8). DCL-AAM (60 mg/kg) was i.v. injected once daily for 3 d (days 1, 2, and 3). DBCO-VC-Dox (12 mg/kg in Dox equivalent) or Dox (7.5 mg/kg, maximum tolerated dose) was i.v. injected on days 4, 8, and 12. Body weight and food intake of each mouse were measured every other day. Lung metastases were monitored via bioluminescence imaging of BALB/c mice using the Bruker In Vivo Xtreme imaging system every 4 d starting from day 5. D-luciferin potassium salt (150 mg/kg) was intraperitoneally injected 3 min before imaging. Bioluminescence imaging data were processed using the Bruker imaging software. After the last imaging on day 13, all mice were sacrificed under anesthesia. The lung and heart of each animal were resected as a whole, weighed, and injected with 10% formalin into trachea until the lungs inflated. Tumor nodules on lungs (n = 7–8 per group) were counted under a dissecting microscope. Each lung lobe was separated after fixation in formalin. All lung tissues were paraffin embedded, sectioned with a thickness of 4 μm, and stained with hematoxylin and eosin (H&E). All the lung sections were then scanned and analyzed. The surface areas of tumors and lungs were measured to calculate the percentage of tumor surface area over the total lung surface area (Atumor/Atotal).
Toxicity evaluation of DCL-AAM + DBCO-VC-Dox and Dox
Liver, heart, kidneys, spleen, brain, sternum and spinal cord (cervical, thoracic and lumbar) were fixed in formalin, paraffin-embedded, sectioned with a thickness of 4 μm, and stained with H&E. Tissues were analyzed by a board-certified pathologist to investigate treatment-mediated toxicity.
Statistical analyses
The statistical analysis was performed by one-way analysis of variance (ANOVA) with post hoc Fisher’s LSD test (OriginPro 8.5), and P values < 0.05 were considered statistically significant. The results were deemed significant at 0.01 < *P ≤ 0.05, highly significant at 0.001 < **P ≤ 0.01, and extremely significant at ***P ≤ 0.001. Sample size was empirically set at n = 3–6 for in vitro cell experiments, n = 3 or 4 for in vivo biodistribution and imaging studies, n = 5 or 6 for xenograft tumor studies, and n = 7 or 8 for metastatic tumor studies.
Supplementary Material
Acknowledgments
J.C. acknowledges support from the United States National Institute of Health (Director’s New Innovator Award 1DP2OD007246), which partially supported the in vivo part of the research, and National Science Foundation (DMR 1309525), which partially supported the chemical design and synthesis of the work. L.Y. acknowledges the support from the National Natural Science Foundation of China (51403145 and 51573123), the Ministry of Science and Technology of China (2016YFA0201200), the Collaborative Innovation Center of Suzhou Nano Science and Technology, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). X.C. acknowledges the support from the National Natural Science Foundation of China (51528303). H.W. was supported via a Howard Hughes Medical Institute International Student Research Fellowship. K.C. and Q.Y. acknowledge Beckman Institute Graduate Fellowship support at the University of Illinois at Urbana–Champaign. K.C., Q.Y., and L.T. acknowledge support from the NIH National Cancer Institute Alliance for Nanotechnology in Cancer “Midwest Cancer Nanotechnology Training Center” Grant R25 CA154015A. R.W. acknowledges the support of a CSTAR/T32 Fellowship through the NIH T32 Tissue Microenvironment Training Program. We acknowledge R. Tong and V. Mirshafiee for their early work related to the design of this project. We also acknowledge H. Ying and Y. Zhang for their useful discussions.
Footnotes
Author contributions
J.C. conceived the original concept of the two-step targeting strategy and supervised the entire project. J.C., H.W., Q.Y. and L.T. initiated this project. H.W. demonstrated the controlled labeling strategy using ether-caged Ac3ManAz derivatives and designed DCL-AAM under the supervision of J.C. H.W. performed the initial synthesis of DCL-AAM. R.W. and K.C. enabled the synthesis of high-purity DCL-AAM. H.W. performed the experiments of flow cytometry and confocal imaging with the help of Y.L. and J.Y. H.W., M.X., Y.S., X.Z. and E.J.C. designed and performed in vivo imaging studies under the supervision of J.C., I.T.D., L.W.D. and S.A.B. H.W., H.H., R.W., Z.W. and K.C. designed and performed tumor efficacy studies under the supervision of J.C., L.Y., X.C., S.L. and T.M.F. H.W., J.C., L.Y., S.L., T.M.F. and X.C. analyzed data. L.Y., X.C., T.M.F. and S.L. provided other expertise and critical feedback. H.W., J.C., L.Y., S.L. and T.M.F. wrote the paper with input from other authors.
Competing financial interests
The authors declare no competing financial interests.
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
References
- 1.Hynes RO. Integrins: a family of cell surface receptors. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 2.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 3.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 4.Nusser Z, et al. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998;21:545–559. doi: 10.1016/s0896-6273(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 5.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 6.Raff MC, et al. Cell-type-specific markers for distinguishing and studying neurons and the major classes of glial cells in culture. Brain Res. 1979;174:283–308. doi: 10.1016/0006-8993(79)90851-5. [DOI] [PubMed] [Google Scholar]
- 7.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 8.Mombaerts P, et al. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–282. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 9.Puffenberger EG, et al. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung’s disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 10.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 11.Vogel CL, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 12.Romond EH, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 13.Tang L, et al. Aptamer-functionalized, ultra-small, monodisperse silica nanoconjugates for targeted dual-modal imaging of lymph nodes with metastatic tumors. Angew Chem Int Ed Engl. 2012;51:12721–12726. doi: 10.1002/anie.201205271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santra S, Kaittanis C, Santiesteban OJ, Perez JM. Cell-specific, activatable, and theranostic prodrug for dual-targeted cancer imaging and therapy. J Am Chem Soc. 2011;133:16680–16688. doi: 10.1021/ja207463b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slamon DJ, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 17.Goldhirsch A, et al. 2 years versus 1 year of adjuvant trastuzumab for HER2-positive breast cancer (HERA): an open-label, randomised controlled trial. Lancet. 2013;382:1021–1028. doi: 10.1016/S0140-6736(13)61094-6. [DOI] [PubMed] [Google Scholar]
- 18.Hudziak RM, Schlessinger J, Ullrich A. Increased expression of the putative growth factor receptor p185HER2 causes transformation and tumorigenesis of NIH 3T3 cells. Proc Natl Acad Sci USA. 1987;84:7159–7163. doi: 10.1073/pnas.84.20.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 21.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, et al. Chemical tumor-targeting of nanoparticles based on metabolic glycoengineering and click chemistry. ACS Nano. 2014;8:2048–2063. doi: 10.1021/nn406584y. [DOI] [PubMed] [Google Scholar]
- 23.Koo H, et al. Bioorthogonal copper-free click chemistry in vivo for tumor-targeted delivery of nanoparticles. Angew Chem Int Ed Engl. 2012;51:11836–11840. doi: 10.1002/anie.201206703. [DOI] [PubMed] [Google Scholar]
- 24.Xie R, Hong S, Feng L, Rong J, Chen X. Cell-selective metabolic glycan labeling based on ligand-targeted liposomes. J Am Chem Soc. 2012;134:9914–9917. doi: 10.1021/ja303853y. [DOI] [PubMed] [Google Scholar]
- 25.Xie R, et al. Targeted imaging and proteomic analysis of tumor-associated glycans in living animals. Angew Chem Int Ed Engl. 2014;53:14082–14086. doi: 10.1002/anie.201408442. [DOI] [PubMed] [Google Scholar]
- 26.Xie R, et al. In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc Natl Acad Sci USA. 2016;113:5173–5178. doi: 10.1073/pnas.1516524113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saks ME, et al. An engineered Tetrahymena tRNAGln for in vivo incorporation of unnatural amino acids into proteins by nonsense suppression. J Biol Chem. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 29.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez EA, Lester HA, Dougherty DA. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc Natl Acad Sci USA. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- 32.Breidenbach MA, et al. Targeted metabolic labeling of yeast N-glycans with unnatural sugars. Proc Natl Acad Sci USA. 2010;107:3988–3993. doi: 10.1073/pnas.0911247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luchansky SJ, et al. Constructing azide-labeled cell surfaces using polysaccharide biosynthetic pathways. Methods Enzymol. 2003;362:249–272. doi: 10.1016/S0076-6879(03)01018-8. [DOI] [PubMed] [Google Scholar]
- 34.Chang PV, Dube DH, Sletten EM, Bertozzi CR. A strategy for the selective imaging of glycans using caged metabolic precursors. J Am Chem Soc. 2010;132:9516–9518. doi: 10.1021/ja101080y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao J, Vann WF, Hinderlich S, Sundaramoorthy M. Elimination of 2-keto-3-deoxy-D-glycero-D-galacto-nonulosonic acid 9-phosphate synthase activity from human N-acetylneuraminic acid 9-phosphate synthase by a single mutation. Biochem J. 2006;397:195–201. doi: 10.1042/BJ20052034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunawan J, et al. Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J Biol Chem. 2005;280:3555–3563. doi: 10.1074/jbc.M411942200. [DOI] [PubMed] [Google Scholar]
- 37.Il’ichev YV, Schwörer MA, Wirz J. Photochemical reaction mechanisms of 2-nitrobenzyl compounds: methyl ethers and caged ATP. J Am Chem Soc. 2004;126:4581–4595. doi: 10.1021/ja039071z. [DOI] [PubMed] [Google Scholar]
- 38.Chauhan SS, Goldstein LJ, Gottesman MM. Expression of cathepsin L in human tumors. Cancer Res. 1991;51:1478–1481. [PubMed] [Google Scholar]
- 39.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Ueki N, Lee S, Sampson NS, Hayman MJ. Selective cancer targeting with prodrugs activated by histone deacetylases and a tumour-associated protease. Nat Commun. 2013;4:2735. doi: 10.1038/ncomms3735. [DOI] [PubMed] [Google Scholar]
- 41.Bonfils C, et al. Evaluation of the pharmacodynamic effects of MGCD0103 from preclinical models to human using a novel HDAC enzyme assay. Clin Cancer Res. 2008;14:3441–3449. doi: 10.1158/1078-0432.CCR-07-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wegener D, Wirsching F, Riester D, Schwienhorst A. A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chem Biol. 2003;10:61–68. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 43.Lodish H, et al. Molecular Cell Biology. 4. Ch. 20.2. W.H. Freeman; 2000. [Google Scholar]
- 44.Malinoff HL, Wicha MS. Isolation of a cell surface receptor protein for laminin from murine fibrosarcoma cells. J Cell Biol. 1983;96:1475–1479. doi: 10.1083/jcb.96.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dent R, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 46.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 47.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 48.Yan S, Sameni M, Sloane BF. Cathepsin B and human tumor progression. Biol Chem. 1998;379:113–123. [PubMed] [Google Scholar]
- 49.Deller MC, Yvonne Jones E. Cell surface receptors. Curr Opin Struct Biol. 2000;10:213–219. doi: 10.1016/s0959-440x(00)00072-5. [DOI] [PubMed] [Google Scholar]
- 50.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubowchik GM, et al. Cathepsin B-labile dipeptide linkers for lysosomal release of doxorubicin from internalizing immunoconjugates: model studies of enzymatic drug release and antigen-specific in vitro anticancer activity. Bioconjug Chem. 2002;13:855–869. doi: 10.1021/bc025536j. [DOI] [PubMed] [Google Scholar]
- 52.Tang L, et al. Investigating the optimal size of anticancer nanomedicine. Proc Natl Acad Sci USA. 2014;111:15344–15349. doi: 10.1073/pnas.1411499111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.