Abstract
Pancreatic islet dysfunction and beta cell failure are hallmarks of type 2 diabetes (T2D) pathogenesis. In this review, we discuss how genome-wide association studies (GWASs) and recent developments in islet (epi)genome and transcriptome profiling (particularly single cell analyses) are providing novel insights into the genetic, environmental, and cellular contributions to islet (dys)function and T2D pathogenesis. Moving forward, study designs that interrogate and model genetic variation (e.g., allelic profiling and (epi)genome editing) will be critical to dissect the molecular genetics of T2D pathogenesis, to build next-generation cellular and animal models, and to develop precision medicine approaches to detect, treat, and prevent islet (dys)function and T2D.
Keywords: Type 2 diabetes (T2D), pancreatic islets, epigenomics, transcriptomics, single cell, genomics
Lay of the Land: (Functional) Genomic Landscape of Islets and Type 2 Diabetes
Type 2 diabetes (T2D) is a complex metabolic disorder with both genetic and environmental components. It results from dysfunction and loss of insulin-secreting beta cells in the endocrine pancreas (Islets of Langerhans) as they work to secrete more insulin to counteract insulin resistance in peripheral tissues (adipose, skeletal muscle, liver). Ultimately, T2D manifests as uncontrolled elevations in blood glucose levels. Genome-wide association studies (GWAS) have systematically identified hundreds of single nucleotide variants (SNVs) (see Glossary) representing >150 regions of the genome (loci) [1], that are associated with T2D risk and differences in T2D-related quantitative metabolic traits such as insulin, proinsulin, and glucose levels. The large majority (>90%) of these SNVs reside in non-coding regions of the genome. In parallel, functional (epi)genomics approaches to map open chromatin using DNase I hypersensitive site sequencing (DNase-seq), Assay for transposase-accessible chromatin sequencing (ATAC-seq) and histone modification and transcription factor (TF) binding patterns using Chromatin immunoprecipitation sequencing (ChIP-seq) (see Glossary) have identified genome-wide location of regulatory elements (REs) such as promoters, enhancers, and insulators in >150 human cell types and tissues. T2D SNVs are significantly and specifically enriched in islet-specific REs [2–7], suggesting that changes in islet RE activity and target gene expression is a common mechanism underlying the molecular genetics of islet dysfunction and T2D [8] (Figure 1, left). Indeed, recent studies have identified putative factors binding these REs and detected allelic effects on their binding and target gene expression [9–11].
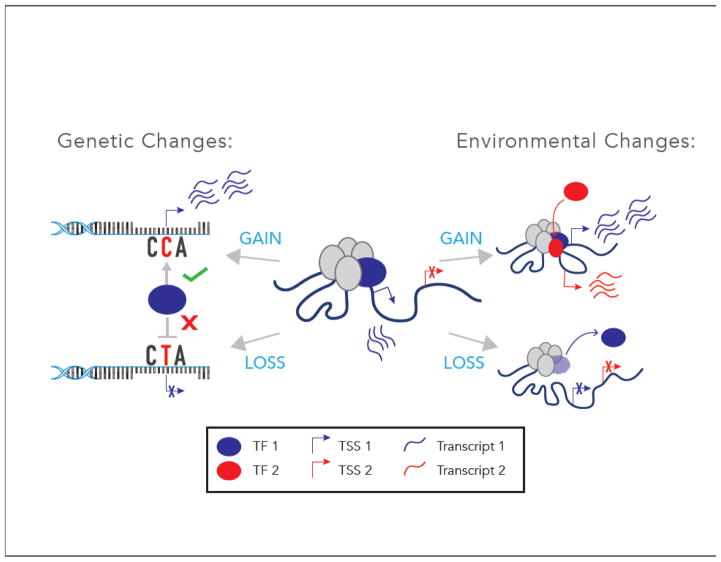
Figure 1. Genomic Effects of Genetic and Environmental Perturbations Contributing to Pancreatic Islet Dysfunction and Type 2 Diabetes (T2D).
(Left) DNA single nucleotide variants (SNVs) may enhance (gain-of-function) or diminish (loss-of-function) transcription element (e.g., enhancer) activity and islet gene expression. The majority of T2D-associated SNVs reside in non-coding regions of the genome and overlap islet REs [2,3,12,14,15,32,47] implicating disruptions in gene regulatory network components as a central molecular feature in disease pathogenesis. A subset of SNVs have been linked to changes in basal islet gene expression [11,31]. (Right) Environmental factors such as inflammation, diet, aging, circadian rhythms, and stress may also influence RE activity, resulting in altered and/or novel transcription of genes essential for islet function [20–25,48–50,57,58]. TF = transcription factor, TSS = transcription start site.
In this review, we discuss how recent studies are improving our understanding of how islet REs are perturbed by SNVs contributing to T2D risk [1,12–19] and are elucidating the transcriptional underpinnings of islet responses to (patho)physiologic environmental changes such as aging, circadian rhythms, Western diet and lifestyle, as well as oxidative, endoplasmic reticulum (ER), and inflammatory stress responses [20–25]. We explore how studies applying next generation sequencing (NGS) to profile individual cells are improving our comprehension of islet biology and reshaping our view of T2D pathogenesis. Finally, we examine similarities and differences between mice and humans in the ‘omics of islet function and T2D (summarized in Figure 2, Key Figure). Throughout, we attempt to highlight future challenges and opportunities and to offer perspectives on how these recent developments set the stage for precision medicine approaches to understand, treat, and prevent T2D.
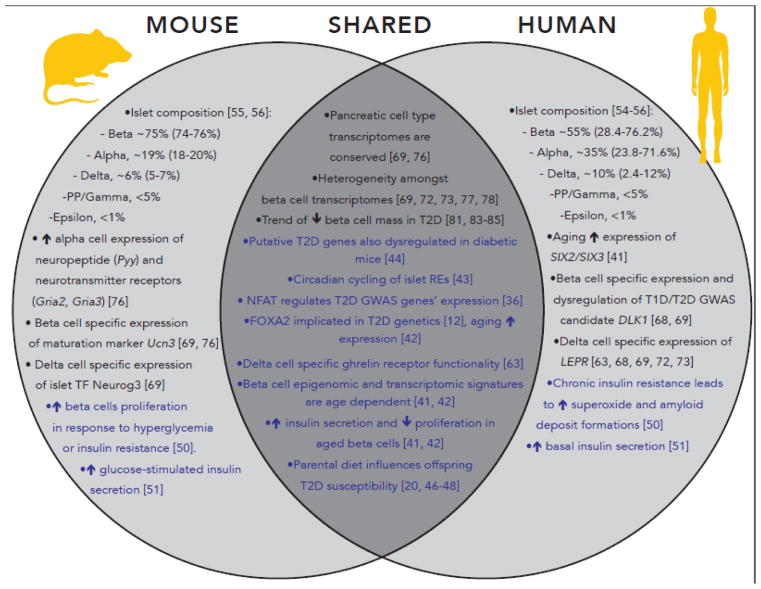
Figure 2 (Key Figure). Converging and Diverging Genetic, Environmental, and Cellular Aspects of Islet (Dys)function and T2D in Mice and Humans.
Parallel analyses of human and mouse islets are revealing important similarities (center) and differences (left, right) between molecular features of islet identity and (dys)function in mice and humans. Text colored in black highlight significant findings regarding islet cellular composition and identity. Text colored in blue highlight longitudinal/comparative analyses of genome-wide molecular datasets and environmental effects on islet (dys)function. These features reaffirm the value of modeling T2D in mice to delineate important species-specific differences in islet biology that may reflect distinct T2D causative mechanisms. RE = regulatory element; TF = transcription factor; ↑ = increase; ↓ = decrease; T2D = type 2 diabetes; T1D = type 1 diabetes; GWAS = genome-wide association study.
Homing in on T2D Genetic Risk and Architecture
Since initial T2D genome-wide association study (GWAS) reports in 2007 [26–29], the list of genomic loci in which sequence variation contributes to T2D risk and variability in quantitative measures of pancreatic islet function has grown to over 150 [1,14,30]. Associated SNVs at each locus contribute modestly to increased T2D risk (odds ratios (OR) 1.05–1.75). Together, these loci only explain a fraction of T2D heritability [13,14]. Genetic consortia continue to dissect the genetic architecture of T2D using larger cohorts with increasing ethnic diversity/representation. Recent efforts have reported [12,14,30] fewer “new” T2D loci (n=10) than previous studies. Importantly, however, they are refining the genetic signals at known (previously associated) T2D loci to define “credible sets” of single nucleotide polymorphisms (SNPs) (see Glossary) that are the most probable causal/functional SNPs driving the association and, consequently, the resulting molecular/phenotypic consequences.
The GOT2D and T2D-GENES consortia sought to identify less common SNVs (0.1%<MAF<5%) with larger effect size that may undergird common variant associations or may account for some of the T2D “missing heritability” using a combined whole genome sequencing (WGS), exome sequencing, and genotype imputation approach [14]. These efforts identified protein-coding variants/mutations that are the most likely causative variant or effector transcripts for 12/78 GWAS loci, confirming five nominated in previous studies (PPARG, KCNJ11-ABCC8, SLC30A8, GCKR, and PAM loci) and identifying seven new ones (FES, TM6SF2, and RREB1 in the PRC1, CILP2, and SSR1 loci, respectively, and TSPAN8, THADA, HNF1A, and HNF4A). For the remaining loci, non-coding SNVs constitute the putative causal SNVs. Comparison of multiple genetic models with the empiric data generated in this study suggest that a long tail of common variants with lower effect sizes may comprise the missing heritability and reaffirm the importance of common, regulatory variation in the genetic architecture of T2D (see Outstanding Questions). Perhaps most importantly, this immense effort has narrowed the list of putative causal SNVs to a handful for 5 loci and by 50% on average for the 78 T2D-associated autosomal loci investigated [14]. Similar themes and reductions in credible sets were reported for fasting glucose- and insulin-associated loci [30].
Outstanding Questions Box.
T2D associated GWAS variants explain only a small portion of T2D heritability with rare variants showing minimal contribution. Do a long tail of common variants with small effect sizes explain this missing heritability? Or are we simply “under-powered” to detect rare variants and their contribution to T2D heritability?
What are the genes targeted by T2D GWAS sequence variant (SV)-containing regulatory elements? Are these links context-specific? Does the risk allele enhance (gain-of-function) or repress (loss-of-function) regulatory element function?
How do the transcriptomes/epigenomes of islets and islet cell types change when subjected to variable environmental stressors (oxidative stress, inflammation, diet, etc.)? How are they changed by intrinsic (aging, circadian rhythms, etc.) environmental factors? Which SVs regulate and alter these islet responses?
What are the precise cellular and molecular pathophysiological changes in each cell type that lead to T2D? Are the major pathologic changes beta cell-specific or do they involve other islet cell types? Or non-islet cell types?
How many islet and single cell samples must be obtained to effectively capture combined cell type heterogeneity while controlling for technical and experimental confounders? How many samples are needed to observe genetic/epigenetic differences between T2D and non-diabetic states? Would stratification of islets by T2D risk genotype improve cell type-specific T2D signatures?
Ongoing islet epigenomic and transcriptomic analyses are progressively defining the regulatory potential of variant loci, identifying SNV-RE overlaps, and nominating potential target genes, whose dysfunction are likely to contribute to T2D [2,3,11,12,14,15,30–32]. Open chromatin (DNase-seq, ATAC-seq) and histone modification/TF binding profiling (ChIP-seq) indicate that T2D and related trait-associated SNVs are especially prominent in islet distal REs and stretch/super enhancers (see Glossary) [2,3,5,33,34]. Due to the long distances over which REs might act, additional work to elucidate the target genes of T2D SNV-containing REs is needed. Chromosome conformation capture techniques such as 3C, 4C, 5C [35], Hi-C [36], Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) [37], and HiCHIP [38]) (see Glossary) will be important components to effectively map interactions between REs and their target genes (see Outstanding Questions). In two separate studies, RNA-sequencing (RNA-seq) of 89 [31] and 118 [11] human islet samples identified 616 and 2,341 expression quantitative trait loci (eQTLs) (see Glossary), respectively. These analyses are the first studies linking SNVs to gene expression changes in islets to define putative genetic control of islet function and failure. However, of the 216 eQTLs common to both studies, only 14 overlapped T2D associated loci [11]. This may be due to power limitations and inability to detect eQTLs beyond their primary signal. Alternatively, this relatively low overlap could suggest that T2D SNVs may affect islet physiologic or pathophysiologic responses, not just basal expression as has been measured to date. Indeed, a very recent study suggests that several putative T2D GWAS genes may be regulated by NFAT, a TF involved in calcineurin signaling responses [39]. Alternatively, the detection of eQTLs overlapping T2D-associated SNVs in peripheral tissues, such as skeletal muscle [40] and adipose [41] tissues reminds us that these other metabolic tissues should not be ignored in the T2D molecular genetics and pathogenesis and warrants further investigation of genomic variation in these tissues.
Recent islet studies suggest that regulatory non-coding RNAs (ncRNAs) may contribute to diabetes progression and beta cell (dys)function [31,42,43]. Aberrant expression of 17 long non-coding (lncRNAs) has been associated with glycated hemoglobin (HbA1c) (see Glossary) levels [31]. This study identified eQTLs for two of these transcripts (LOC283177 and SNHG5), but the eQTL SNVs did not overlap T2D SNVs [31]. Similarly, a study by Morán and colleagues identified 9/55 T2D-associated loci that contained lncRNAs located within 150 kb of, but do not directly overlap, the reported lead SNVs [42]. In the KCNQ1 locus, T2D risk SNVs overlap both KCNQ1 and KCNQ1OT1 [43,44], a long intergenic non-coding RNA (lincRNA) also found to be significantly induced in T2D islets [42]. We anticipate that additional links will emerge in the coming years. Other studies suggest that islet lncRNA alterations could also contribute to type 1 diabetes (T1D), as a T1D GWAS SNV (rs941576) was identified in the MEG3 lincRNA locus [43,45]. Functional analyses in human islets and rodent models will clarify roles of these ncRNAs in islet development, (dys)function, and diabetes.
DNA methylation studies (see Glossary) of non-diabetic (ND) and T2D islets have suggested that epigenetic dysregulation may promote T2D development [46,47]. DNA methylation profiling of 15 T2D and 34 ND islets using the Illumina 450BeadChip identified 1649 differentially methylated CpG sites (see Glossary) for 853 genes, 17 of which reside in T2D-associated loci [46]. Surprisingly, the majority (97%) of these CpG sites were hypomethylated in T2D islets, suggesting that they may suffer from decreased methyl donor levels or decreased activity of DNA methyltransferases.
Genomics of Islet Responses to Environmental Changes and T2D Pathogenesis
Intrinsic and extrinsic environmental changes, such as aging and Western diet/lifestyle, respectively, are linked to islet dysfunction and T2D risk [23,48–50] (Figure 1, right). Multiple groups have begun to characterize genomic effects of these environmental inputs and insults on islets. Transcriptome profiling of adult and juvenile islet beta cells identified 565 (209 up, 356 down) and 6123 (2083 up, 4040 down) differentially expressed genes in humans and mice, respectively [48,49]. Signatures of decreased proliferative capacity in aged islets/beta cells were apparent in both species, perhaps best illustrated by increased CDKN2A/B expression, a gene cluster with established cellular senescence functions and implicated as “Type 2 Diabetogenes” for a T2D GWAS signal on 9p21 [48,49,51]. Unexpectedly, transcriptome and epigenome signatures suggested superior insulin secretory capacity of adult islets, which was confirmed functionally by glucose-stimulated insulin secretion (GSIS) assays [48,49]. DNA methylation and histone profiling indicated that these expression differences were largely mediated by chromatin remodeling and epigenetic modification of distal REs such as enhancers. Using whole genome bisulfite sequencing (WGBS), Avrahami and colleagues identified ~14,368 aging-related differentially methylated regions (DMRs) between the beta cells of juvenile and adult mice. DMRs overlapping distal REs outnumbered those overlapping promoters 3:1 and exhibited larger changes in magnitude of methylation. Distal DMRs that lost methylation with aging were enriched for binding sites of important islet TFs such as Foxa2, Neurod1, and Pdx1, suggesting these factors mediate the expression differences and improved functionality in adult islets. Finally, genes showing differential expression in adult islets were accompanied by differential methylation at nearby distal REs more often than at their promoters. These data suggest that, in addition to their importance in T2D genetic risk, enhancers also govern important transcriptional regulatory changes accompanying or mediated by aging.
Circadian rhythm links behavior and metabolism to day-night cycles. Notably, insulin secretion oscillates with a circadian periodicity. Analysis of mouse islet transcriptomes revealed that approximately 27% of the beta cell transcriptome (n=3905 genes) demonstrated circadian oscillation, including genes responsible for insulin synthesis, transport, and stimulated exocytosis [50]. The human orthologues of 481 of these genes exhibited circadian oscillations in human islets. ChIP-seq identified 742/3905 of these oscillatory genes as direct targets of the circadian clock TFs CLOCK and BMAL1. As with aging, the majority of differential sites were at distal REs. Beta cell-specific deletion of Bmal1 resulted in islet failure and diabetes in mice. This study demonstrates the importance of circadian rhythms in islet function and suggests that genetic or environmental perturbation of this program could contribute to T2D risk and pathophysiology. GWAS results suggest this could be the case, as SNVs in the CRY2 locus, a component of the circadian machinery, and MTNR1B, a gene encoding a melatonin receptor, are associated with altered islet function and T2D [1,52]. It will be interesting to see if genetic perturbations in circadian clock TFs or their binding sites emerge as one of the molecular mechanisms underlying T2D GWAS.
Maternal nutrition and in utero stresses have been linked to T2D risk for offspring in humans and rodents [23,53–55]. Although changes in fetal nutrition are suggested to influence offspring metabolism via epigenetic modifications [23,56], the genome-wide effects on the islet (epi)genome have not been determined. Similarly, stress responses to elevated oxidative/ER stress lead to islet failure, impaired insulin secretion, and T2D susceptibility [57–59]. Ultimately, these responses converge on the nucleus and involve re-distribution or covalent modifications of master TFs (MAFB, NKX6-1, PDX1) or stress response factors (FOXO1, ATF4, HIF1alpha) [20,22,53,57,58]. (Epi)genomic analyses of these stress responses are warranted and may reveal important connections between T2D SNVs and altered islet stress responses. Moving forward, it will be crucial to understand the extent to which genetic and epigenetic changes interact in T2D pathogenesis (see Outstanding Questions). Response QTL (reQTL) and epigenome-wide association studies (EWAS) [56]should provide these important links (see Outstanding Questions). Indeed, studies of SNV effects on immune cell responses identified 121 reQTLs, 38 of which overlapped autoimmune disease-associated SNVs [60]. Specific factor(s) and pathway(s) activated by insulin resistance appear to differ between mouse and human islets [57,58] (Figure 2); thus, we emphasize that caution must be taken in study design and interpretation to interrogate this and possibly other islet responses.
Deconstructing Pancreatic Islet Cellular/Functional Heterogeneity
Islets comprise 1–5% of the pancreas and consist of at least five endocrine cell types performing coordinated but distinct functions and each producing a unique hormone in the islet: beta (insulin), alpha (glucagon), delta (somatostatin), gamma (pancreatic polypeptide), and epsilon (ghrelin) cells [61–64]. Precise understanding of islet molecular changes during T2D development is likely complicated by variability in islet cell type composition. On average, islets are composed of 55% beta cells, 35% alpha, 10% delta, and less than 5% and 1% gamma/PP and epsilon cells, respectively [61–63]. However, this can vary considerably between donors, with ranges of 28.4%–76.2%, 23.8%–71.6%, and 2.4%–12% for beta, alpha, and delta cell compositions, respectively [61] (Figure 2). This cellular heterogeneity, combined with donor-to-donor variability, masks the molecular repertoire of each cell type and impedes clear understanding of the molecular programs perturbed in each cell type by T2D pathogenesis.
Until recently, most studies have focused on epigenetic and transcriptional analyses of whole islets or, at the expense of other cell types, beta cells. However, recent studies demonstrating roles for alpha [65–67] and delta cells [68–71] in modulating beta cell function/resilience and in T2D pathogenesis are fueling a renewed interest in these cell types. First attempts to overcome these obstacles and understand the molecular repertoire of each islet cell type, focused on transcriptomic analyses of sorted and enriched cell type populations [61,72–74]. However, such methods were unable to effectively isolate and enrich the less abundant non-beta cells [75], leaving much of the functional genomic landscape of islets imprecisely assigned/classified or, in the case of rarer islet cell types, undefined.
Within the past year, multiple groups have applied single cell transcriptome profiling to islets to begin to address questions about islet physiology [75–83] (see Outstanding Questions) with single cell resolution, such as: (1) What is the gene repertoire of each islet cell type? (2) Does the gene repertoire reveal any new/unexpected roles for each cell type in islet (patho)physiology? (3) Are there novel cell types or unappreciated subpopulations in islets? These studies are providing new appreciation of the repertoire of both islet beta and non-beta cells. As much of the beta cell transcriptional repertoire has been extensively studied [61,72–74], several features were validated including genes involved in cell survival/maturation (PDX1), regulation of insulin secretion (RGS16, SYT13, ENTPD3), and diabetes associated genes (DLK1, MEG3, SLC2A2) [75,76,78–81,83]. Unique expression of genes encoding TFs (IRX2), membrane glycoproteins (DPP4), and hormone transporters (TTR) were also validated in alpha cells. Analysis of single alpha cell transcriptomes uncovered signatures involved in wound healing (FAP), blood clotting (F10) and tissue biogenesis (LOXL4) [75,76,78–81,83] suggesting they share functions akin to pancreatic fibroblast/mesenchymal cells.
Single cell profiling has provided new views of delta and PP/gamma cell roles in islet physiology and the molecular genetics of islet failure and diabetes. For example, these studies revealed that delta cells uniquely express appetite-suppressing leptin (LEPR) and appetite-stimulating ghrelin (GHSR) hormone receptors [75,79,80], implicating them as the integrators and regulators of these pathways in the islet. GHSR functionality has been demonstrated in both human and mouse delta cells [70]. LEPR expression is unique to human delta cells, suggesting that these cells may uniquely mediate the leptin response in human islets [70,75,76,79,80] (Figure 2). Expression of genes associated with congenital hyperinsulinemia (CHI) (UCP2, HADH) in delta cells further implicates this cell type in the molecular genetics of CHI [75]. PP/gamma cell transcriptomes exhibited enrichment of genes involved in neuronal development (MEIS2, FEV) [75,78–80] and serotonin catalysis and reuptake (TPH1, SLC6A4) [75,79,80,83]. Together these findings suggest that delta and PP/gamma cells act as the “brains” of the pancreatic islets, capable of receiving and integrating various neuronal signals to coordinate islet function. Due to their scarcity in human pancreatic islets (<1% of islet volume), our knowledge of the epsilon cell repertoire and its putative function(s) remain speculative. Nonetheless, the insights gleaned from these initial studies undoubtedly motivate follow-up studies that continue transitioning from whole islet to functional constituent cell studies. Identification of genes encoding cell type-specific surface markers (beta – LRRTM3, CASR; alpha – DPP4, PLCE1; delta – LEPR, GHSR, ERBB4; PP/gamma – SLC6A4, PTGFR; epsilon – ANXA13) [75,79], provide new targets that may be exploited for more accurate purification of each islet cell type and analysis of its specific responses to genetic and environmental stressors.
Islet Subpopulations and Cell Type Heterogeneity
Detection of heterogeneous beta cell subpopulations were reported for enriched cell and single cell studies. These include four subpopulations with differing expression of ST8SIA1 and CD9 [84], five subpopulations defined by RBP4, FFAR4/GPR120, ID1, ID2, and ID3 expression [80], and subpopulations characterized by ER stress-associated [76] and oxidative stress-associated genes [79]. Fltp/CFAP126 expression has been reported to distinguish proliferating and mature beta cell subpopulations in mice [85], but single cell transcriptome analyses failed to detect this distinction in human beta cells [75,83]. However, proliferative and mature human beta cells were identified by single cell mass cytometry analysis [86], suggesting mice and humans may make use of distinct cell growth pathways. As each study detected distinct beta cell subpopulations with different gene signatures, it remains difficult to distinguish whether these subpopulations are functionally distinct cells or the result of technical confounders such as the time to sort and enrich in a harsh cell sorting environment, time for cell capture, or cell and transcript capture efficiency [87].
Single Cell Dissection of Islet Dysfunction and T2D
Single cell transcriptome analyses provide a fresh and agnostic opportunity to investigate the putative mechanisms underlying islet dysfunction in T2D. To date, single cell transcriptome profiling has been completed for a total of 1831 and 1970 islet cells from 26 ND and 15 T2D donors respectively [75,80,81,83]. Comparison of T2D and ND single cell transcriptomes suggest that specific alterations in islet cell type transcriptomes may underlie T2D pathogenesis (Figure 3A). However, changes in cell proportions (Figure 3B), identity, and plasticity (Figure 3C, 3D) have also been regarded as potential contributors to T2D [72,88–92]. Specifically, decreases in diabetic beta cell mass were suggested to be caused by reversion to endocrine progenitor (hormone-negative) cells (Figure 3C) or different islet cell types (Figure 3D) rather than apoptosis. The model of transformed beta cell identity remains controversial. A recent study concluded that the observed magnitude of decline in beta cell numbers in T2D islets are not accompanied by proportionate increases in cells exhibiting trans-differentiation (see Glossary) markers or increases in other islet cell types [93]. Rather, the presence of endocrine progenitor-like cells in T2D islets may represent newly forming endocrine cells [93]. Single cell profiling also did not identify transcriptomic evidence of de-differentiated (see Glossary) or trans-differentiated cells in T2D islets (Figure 3C, D) [75,80,83]. Similar trends were observed in whole islet RNA-seq data upon deconvolution (see Glossary) where cell type proportions did not significantly vary between hypoglycemic and hyperglycemic islets [76]. Thus, the transcriptome data to date do not provide supporting evidence of de-differentiation in T2D islets.
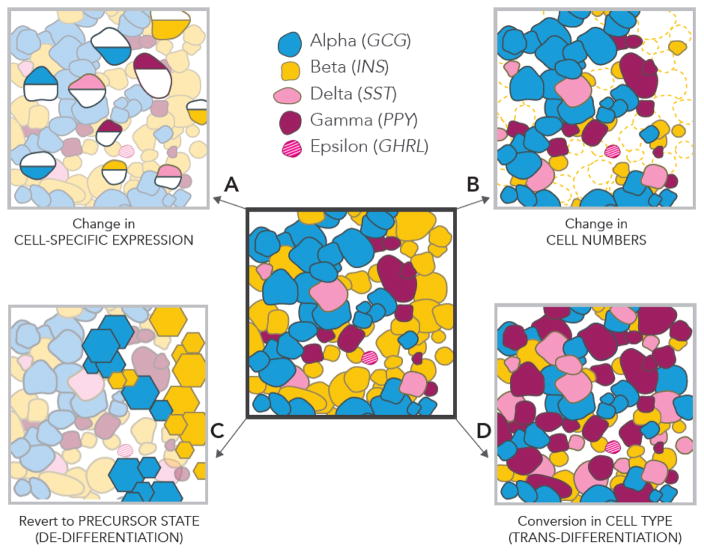
Figure 3. Proposed Cellular Mechanisms Contributing to T2D Development.
(Center) Cartoon representation of human islet cellular composition. Studies have described the following phenomena: (A) Islet single cell transcriptomic studies [75,80,83] suggest that cell type-specific changes in gene expression (depicted as half-shaded cells) contribute to T2D pathogenesis. These studies suggest that potential pathogenic expression changes may occur in each islet cell type, not just beta cells. (B) Decreases in beta cell (depicted in yellow) numbers [25,92,100,101], thought to precede islet dysfunction and development of insulin resistance. (C) Alterations in islet cellular identity may also account for islet failure. De-differentiation of islet cell types to precursor cell types/states (depicted via hexagons) has been proposed to underlie loss of beta cell mass and function in T2D [88,90–92]. (D) Similarly, trans-differentiation of islet cell types may lead to imbalances in islet cell proportions and improper function [72,88,89].
Transcriptomes of each cell type from ND and T2D donors exhibited remarkable correlation overall. However, specific changes in gene expression were reported in T2D beta cells including reduced expression of INS [75,80], genes important for insulin secretion (STX1A) [75] and beta cell proliferation (FXYD2) [80,83], as well as elevated expression of genes implicated in T2D GWAS (DLK1, DGKB) [75]. Transcriptional differences were also identified in T2D alpha cells including expression of CD36 [75,80], a crucial activator of the NLRP3 inflammasome [94] and RGS4, a negative regulator of GSIS [80]. Several genes were dysregulated in T2D delta cell transcriptomes [75,83]. However, the underlying biology of these candidates remains undefined with no association with islet growth or function [83]. Aside from these encouraging examples, these single cell studies have not reached consensus regarding differentially expressed genes between T2D and ND cell types. Differences in islet donor variability, islet isolation/transport, and single cell dissociation/sequencing protocols may explain these inconsistencies across studies. We expect that sampling thousands of single cells each from hundreds of individuals for large-scale meta-analyses will provide a more convergent list of cell type-specific genes and pathways disrupted in T2D islets. It will also be important for future studies to profile cells from individuals at different points along the T2D pathogenesis spectrum, such as pre-diabetic individuals (5.5<HbA1C<6.0) (see Glossary) to identify and distinguish primary from secondary genomic changes that may be cause or consequence of progression to T2D.
Concluding Remarks and Future Directions
The past few years have marked exciting developments in our understanding of the underlying genomic, environmental, and cellular components driving T2D pathogenesis. Numerous common (and only few rare) genetic SNVs have been implicated in T2D progression [13,14]. It is unclear if the “missing T2D heritability” is explained by a larger distribution of common SNVs with minimal effect sizes, if current methods have missed critical rare SNVs, or if it will be captured by gene-gene and gene-environment interactions (such as detected by reQTL). Thus far, most catalogued T2D-SNVs occur in and disrupt islet RE function, however, the causal connections between the two remain challenging to decipher. eQTL and chromatin accessibility QTL (caQTL) [95,96] studies have and will continue to be essential for linking genetic variants to molecular phenotypes. A subsequent challenge will be to link these molecular effects to pathways [39] and (patho)physiologic phenotypes [97].
Functional genomic studies have identified minimal overlap between islet eQTLs and T2D-SNVs [11,31] suggesting that responses to environmental stress factors may be key mediators of T2D pathogenesis. Mouse models have been instrumental to elucidate the genetic and molecular regulation of these responses and how environmental stressors influence islet (dys)function. However, observed differences between mice and humans in islet morphology, composition, expression, and function remind us to exercise caution when extrapolating findings in mice to human T2D. Studies comparing the genomic features of human islets and models are essential to define conserved features and those that require modification to determine what aspects of islet dysfunction and T2D we can model effectively and to decide how/where we should manipulate or humanize the mouse (epi)genome to better model human T2D. (Epi)genome editing technologies such as CRISPR/Cas9 can then be applied to develop new cellular and animal models to more effectively study islet phenotypic changes resulting from genetic and environmental variation. We anticipate that these integrative genomic studies and techniques will also serve as valuable resources to determine the underlying genetic changes and mechanisms of beta cell dysfunction that lead to T1D [98].
Rapid developments in single cell NGS technologies have renewed interest in the less-studied islet cell types. Deconstructing the major molecular changes that occur in each cell type during T2D progression has proven challenging, yielding inconsistent results between studies due to patient donor variability and technical sequencing artifacts. This is also likely the result of limited statistical power. In the future, it will be interesting to perform meta-analyses of available transcriptomic data to maximize our confidence of changes in cell specific expression programs. Together, the innovative new genomic technologies of the past few years will allow us to more precisely define, model, and manipulate the genes and pathways gone awry in T2D with the ultimate goal of designing novel therapeutic approaches.
Trends.
Type 2 diabetes (T2D) is a multi-tissue metabolic disorder that results when pancreatic islets fail to compensate for insulin resistance in peripheral tissues.
Recent studies reaffirm the common variant origins of T2D genetic risk. Variants overlap noncoding genomic regions, implicating regulatory defects in T2D etiology.
Environmental stressors are associated with changes in gene expression programs leading to T2D progression.
Single cell sequencing technologies permit investigation of islet cell type transcriptomes and epigenomes with single cell resolution/precision. Such methods provide greater insight into cell type-specific perturbations and their roles in T2D
Recent studies suggest that other cells (alpha, delta, PP/gamma) in the islet may play important roles in islet/beta cell function, resilience, and T2D pathogenesis.
Acknowledgments
Work in the Stitzel Lab is supported by the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program under Award No. W81XWH-16-1-0130 and by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) under award number R00DK092251. Opinions, interpretations, conclusions and recommendations are solely the responsibility of the authors, are not necessarily endorsed by the Department of Defense, and do not necessarily represent the official views of the National Institutes of Health. We thank Jane Cha for her expertise and help in figure design and artwork. We gratefully acknowledge members of the Stitzel and Ucar labs for helpful discussion and feedback on this work, and we thank the anonymous reviewers for their helpful suggestions to improve the perspective and content of this manuscript.
Glossary
- Assay for transposase-accessible chromatin sequencing (ATAC-seq)
a technique used to profile regions of open chromatin from small cell numbers.
- Chromatin immunoprecipitation sequencing (ChIP-seq)
a method used to study DNA-protein interactions.
- Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET)
a method used to study three-dimensional chromatin interactions genome-wide.
- CpG sites
areas of DNA containing a cytosine nucleotide directly linked to a single phosphate group and guanine nucleotide. These sites are often methylated and influence transcription.
- Credible set SNPs
list of sequence variants with 95% posterior probability of containing a/the causal disease-associated SNP(s) [99].
- Deconvolution
a statistical framework to resolve a heterogeneous mixture into its constituent elements.
- De-differentiation
the process in which a mature differentiated cell type reverts to an earlier developmental/precursor state.
- DNA methylation
molecular process wherein a methyl group is covalently attached to a DNA base without altering the DNA sequence.
- DNase I hypersensitive site sequencing (DNase-seq)
a method used to characterize regulatory and open chromatin regions of the genome.
- Expression quantitative trait loci (eQTL)
approach to link sequence variation at a position in the genome to expression of target gene(s).
- Genome-wide association study (GWAS)
statistical association of sequence variation with disease risk or variability in a measurable phenotypic trait/feature.
- Glycated hemoglobin (HbA1C)
a type of hemoglobin modification that is measured to determine plasma glucose concentration.
- RNA-sequencing (RNA-seq)
measures the amount of RNA in a sample at a given time.
- Single nucleotide polymorphism (SNP)
nucleotide variation at a specific location in the genome that exists with >5% frequency in the population.
- Single nucleotide variant (SNV)
changes in a given nucleotide sequence in the genome.
- Stretch/Super enhancers
extended (>3 kilobase) regions of the genome marked by enhancer chromatin states; enriched near genes important for cell type identity and cell type-specific functions.
- Subpopulation
a subset of cells within a tissue distinguished by expression of specific marker genes/proteins.
- Trans-differentiation
the process in which a mature cell type converts into another mature cell type.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genet. 2015;24:R85–R92. doi: 10.1093/hmg/ddv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker SCJ, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quang DX, et al. Motif signatures in stretch enhancers are enriched for disease-associated genetic variants. Epigenetics Chromatin. 2015;8:23. doi: 10.1186/s13072-015-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stitzel ML, et al. Transcriptional Regulation of the Pancreatic Islet: Implications for Islet Function. Curr Diab Rep. 2015;15:66. doi: 10.1007/s11892-015-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulzer JR, et al. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am J Hum Genet. 2014;94:186–197. doi: 10.1016/j.ajhg.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogarty MP, et al. Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the CDC123/CAMK1D Type 2 Diabetes GWAS Locus. PLOS Genet. 2014;10:e1004633. doi: 10.1371/journal.pgen.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Bunt M, et al. Transcript Expression Data from Human Islets Links Regulatory Signals from Genome-Wide Association Studies for Type 2 Diabetes and Glycemic Traits to Their Downstream Effectors. PLoS Genet. 2015:11. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016 doi: 10.1038/nature18642. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwala V, et al. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet. 2013;45:1418–1427. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strawbridge RJ, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinthorsdottir V, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 19.Hara K, et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet. 2014;23:239–246. doi: 10.1093/hmg/ddt399. [DOI] [PubMed] [Google Scholar]
- 20.Aouacheri O, et al. The Investigation of the Oxidative Stress-Related Parameters in Type 2 Diabetes Mellitus. Can J Diabetes. 2015;39:44–49. doi: 10.1016/j.jcjd.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhari N, et al. A Molecular Web: Endoplasmic Reticulum Stress, Inflammation, and Oxidative Stress. Front Cell Neurosci. 2014:8. doi: 10.3389/fncel.2014.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorasia DG, et al. Pancreatic Beta Cells Are Highly Susceptible to Oxidative and ER Stresses during the Development of Diabetes. J Proteome Res. 2015;14:688–699. doi: 10.1021/pr500643h. [DOI] [PubMed] [Google Scholar]
- 23.Rando OJ, Simmons RA. I’m eating for two: parental dietary effects on offspring metabolism. Cell. 2015;161:93–105. doi: 10.1016/j.cell.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zephy D, Ahmad J. Type 2 diabetes mellitus: Role of melatonin and oxidative stress. Diabetes Metab Syndr Clin Res Rev. 2015;9:127–131. doi: 10.1016/j.dsx.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Laybutt DR, et al. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 2007;50:752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 26.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 28.Burton PR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diabetes Genetics Initiative of Broad Institute of Harvard MIT Lund University and Novartis Institutes of BioMedical Research et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 30.Liu CT, et al. Trans-ethnic Meta-analysis and Functional Annotation Illuminates the Genetic Architecture of Fasting Glucose and Insulin. Am J Hum Genet. 2016;99:56–75. doi: 10.1016/j.ajhg.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stitzel ML, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ackermann AM, et al. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol Metab. 2016;5:233–244. doi: 10.1016/j.molmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mumbach MR, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller MP, et al. The Transcription Factor Nfatc2 Regulates β-Cell Proliferation and Genes Associated with Type 2 Diabetes in Mouse and Human Islets. PLOS Genet. 2016;12:e1006466. doi: 10.1371/journal.pgen.1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott LJ, et al. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium the M. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morán I, et al. Human β Cell Transcriptome Analysis Uncovers lncRNAs That Are Tissue-Specific, Dynamically Regulated, and Abnormally Expressed in Type 2 Diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnes L, Sussel L. Epigenetic modifications and long noncoding RNAs influence pancreas development and function. Trends Genet TIG. 2015;31:290–299. doi: 10.1016/j.tig.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace C, et al. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dayeh T, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkmar M, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arda HE, et al. Age-Dependent Pancreatic Gene Regulation Reveals Mechanisms Governing Human β Cell Function. Cell Metab. 2016;23:909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avrahami D, et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved β Cell Function. Cell Metab. 2015;22:619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perelis M, et al. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350:aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kluth O, et al. Differential Transcriptome Analysis of Diabetes-Resistant and -Sensitive Mouse Islets Reveals Significant Overlap With Human Diabetes Susceptibility Genes. Diabetes. 2014;63:4230–4238. doi: 10.2337/db14-0425. [DOI] [PubMed] [Google Scholar]
- 52.Persaud SJ, Jones PM. A Wake-up Call for Type 2 Diabetes? N Engl J Med. 2016;375:1090–1092. doi: 10.1056/NEJMcibr1607950. [DOI] [PubMed] [Google Scholar]
- 53.Halban PA, et al. β-Cell Failure in Type 2 Diabetes: Postulated Mechanisms and Prospects for Prevention and Treatment. Diabetes Care. 2014;37:1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulz LC. The Dutch Hunger Winter and the developmental origins of health and disease. Proc Natl Acad Sci. 2010;107:16757–16758. doi: 10.1073/pnas.1012911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, et al. Prenatal exposure to famine and the development of hyperglycemia and type 2 diabetes in adulthood across consecutive generations: a population-based cohort study of families in Suihua, China. Am J Clin Nutr. 2016 doi: 10.3945/ajcn.116.138792. [DOI] [PubMed] [Google Scholar]
- 56.Rakyan VK, et al. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai C, et al. Stress-impaired transcription factor expression and insulin secretion in transplanted human islets. J Clin Invest. 2016;126:1857–1870. doi: 10.1172/JCI83657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai C, et al. Islet-enriched gene expression and glucose-induced insulin secretion in human and mouse islets. Diabetologia. 2012;55:707–718. doi: 10.1007/s00125-011-2369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo S, et al. Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest. 2013;123:3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee MN, et al. Common Genetic Variants Modulate Pathogen-Sensing Responses in Human Dendritic Cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blodgett DM, et al. Novel Observations From Next-Generation RNA Sequencing of Highly Purified Human Adult and Fetal Islet Cell Subsets. Diabetes. 2015;64:3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brissova M, et al. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem Off J Histochem Soc. 2005;53:1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 63.Cabrera O, et al. The unique cytoarchitecture of human pancreatic islets has implications for islet cell function. Proc Natl Acad Sci U S A. 2006;103:2334–2339. doi: 10.1073/pnas.0510790103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ionescu-Tirgoviste C, et al. A 3D map of the islet routes throughout the healthy human pancreas. Sci Rep. 2015:5. doi: 10.1038/srep14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanojevic V, Habener JF. Evolving function and potential of pancreatic alpha cells. Best Pract Res Clin Endocrinol Metab. 2015;29:859–871. doi: 10.1016/j.beem.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jamison RA, et al. Hyperglucagonemia precedes a decline in insulin secretion and causes hyperglycemia in chronically glucose-infused rats. Am J Physiol Endocrinol Metab. 2011;301:E1174–1183. doi: 10.1152/ajpendo.00175.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez-Diaz R, et al. Alpha cells secrete acetylcholine as a non-neuronal paracrine signal priming beta cell function in humans. Nat Med. 2011;17:888–892. doi: 10.1038/nm.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hauge-Evans AC, et al. Somatostatin secreted by islet delta-cells fulfills multiple roles as a paracrine regulator of islet function. Diabetes. 2009;58:403–411. doi: 10.2337/db08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Meulen T, et al. Urocortin3 mediates somatostatin-dependent negative feedback control of insulin secretion. Nat Med. 2015;21:769–776. doi: 10.1038/nm.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiGruccio MR, et al. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol Metab. 2016;5:449–458. doi: 10.1016/j.molmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molina J, et al. Control of insulin secretion by cholinergic signaling in the human pancreatic islet. Diabetes. 2014;63:2714–2726. doi: 10.2337/db13-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bramswig NC, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123:1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dorrell C, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011:54. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nica AC, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawlor N, et al. Single cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 2016 doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360. e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grün D, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J, et al. Single cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muraro MJ, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016 doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segerstolpe Å, et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang YJ, et al. Single cell transcriptomics of the human endocrine pancreas. Diabetes. 2016 doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xin Y, et al. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc Natl Acad Sci U S A. 2016;113:3293–3298. doi: 10.1073/pnas.1602306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xin Y, et al. RNA Sequencing of Single Human Islet Cells Reveals Type 2 Diabetes Genes. Cell Metab. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 84.Dorrell C, et al. Human islets contain four distinct subtypes of β cells. Nat Commun. 2016;7:11756. doi: 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bader E, et al. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature. 2016;535:430–434. doi: 10.1038/nature18624. [DOI] [PubMed] [Google Scholar]
- 86.Wang YJ, et al. Single-Cell Mass Cytometry Analysis of the Human Endocrine Pancreas. Cell Metab. 2016;24:616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Research. 2016:5. doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cinti F, et al. Evidence of β-Cell Dedifferentiation in Human Type 2 Diabetes. J Clin Endocrinol Metab. 2016;101:1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu J, et al. Transdifferentiation of pancreatic α-cells into insulin-secreting cells: From experimental models to underlying mechanisms. World J Diabetes. 2014;5:847–853. doi: 10.4239/wjd.v5.i6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Talchai C, et al. Pancreatic β-Cell Dedifferentiation As Mechanism Of Diabetic β-Cell Failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z, et al. Pancreatic β-cell Dedifferentiation in Diabetes and Re-differentiation following Insulin Therapy. Cell Metab. 2014;19:872–882. doi: 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butler AE, et al. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 93.Butler AE, et al. β-Cell Deficit in Obese Type 2 Diabetes, a Minor Role of β-Cell Dedifferentiation and Degranulation. J Clin Endocrinol Metab. 2016;101:523–532. doi: 10.1210/jc.2015-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sheedy FJ, et al. CD36 coordinates NLRP3 inflammasome activation by facilitating the intracellular nucleation from soluble to particulate ligands in sterile inflammation. Nat Immunol. 2013;14:812–820. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumasaka N, et al. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thomsen SK, et al. Systematic Functional Characterization of Candidate Causal Genes for Type 2 Diabetes Risk Variants. Diabetes. 2016 doi: 10.2337/db16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soleimanpour SA, Stoffers DA. The pancreatic β cell and type 1 diabetes: innocent bystander or active participant? Trends Endocrinol Metab TEM. 2013;24:324–331. doi: 10.1016/j.tem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cerf ME. Beta Cell Dysfunction and Insulin Resistance. Front Endocrinol. 2013:4. doi: 10.3389/fendo.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Halban PA, et al. β-Cell Failure in Type 2 Diabetes: Postulated Mechanisms and Prospects for Prevention and Treatment. Diabetes Care. 2014;37:1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]