Abstract
The gut microbiome plays a key role in energy production, immune system development, and host resistance against invading pathogens, etc. Disruption of gut bacterial homeostasis is associated with a number of human diseases. Several environmental chemicals have been reported to induce alterations of the gut microbiome. Diazinon, one of important organophosphate insecticides, has been widely used in agriculture. Diazinon and its metabolites are readily detected in different environmental settings and human urine. The toxicity of organophosphates has been a long-standing public health concern. We recently demonstrated that organophosphate insecticide diazinon perturbed the gut microbiome composition of mice. However, the functional impact of exposure on the gut microbiome has not been adequately assessed yet. In particular, the molecular mechanism responsible for exposure-induced microbial profile and community structure changes has not been identified. Therefore, in this study, we used metatranscriptomics to examine the effects of diazinon exposure on the gut metatranscriptome in C57BL/6 mice. Herein, we demonstrated for the first time that organophosphate diazinon modulated quorum sensing, which may serve as a key mechanism to regulate bacterial population, composition, and more importantly, their functional genes. In addition, we also found that diazinon exposure activated diverse stress response pathways and profoundly impaired energy metabolism of gut bacteria. These findings provide new understandings of the functional interplay between the gut microbiome and environmental chemicals, such as organophosphates.
Keywords: organophosphate, gut microbiome, metatranscriptomics, quorum sensing
Accumulating evidence has demonstrated that the gut microbiome is related to a wide variety of diseases such as metabolic disorders (Portune et al., 2016) and neurodevelopmental diseases (Santocchi et al., 2016). Microbiome-based therapies such as fecal microbiota transplantation have been developed since a compelling set of links between the gut microbiome and disease exist (Gweon et al., 2016), which points to the critical role of the gut microbiome in human health. Diversity and stability of gut microbiome could be affected by various factors and, as a counteracting mechanism, gut microbiome forms a biotic shield between host and the outside environment to mitigate against untoward effects from the environmental challenges and stressors (Moos et al., 2016). Several environmental chemicals have been reported to induce changes in the gut microbiome such as arsenic (Lu et al., 2014) and cadmium (Zhang et al., 2015). Recently, we demonstrated that organophosphate insecticide diazinon also perturbed the gut microbiome compositions of mice (Gao et al., 2016).
Diazinon is still an active product approved for many agricultural applications, although its residential uses were banned in the U.S in 2004. Diazinon residue has been detected in watersheds and drinking water wells (Aggarwal et al., 2013). According to the CDC Fourth Report on Human Exposure to Environmental Chemicals, diazinon metabolites in urine have been detected in the U.S. population, which raises the public health concern. Diazinon exposure leads to various deleterious outcomes, not only limited to acetylcholinesterase inhibition (Yen et al., 2011), but also oxidative stress (Boussabbeh et al., 2016), DNA damage (Kashanian et al., 2008), and genotoxic effects (Jones et al., 2015). Given the significance of gut microbiome to human health and widespread use and toxicity of diazinon, it is of great interest to understand the interplay between gut microbiome and diazinon. In our previous study, we demonstrated the impact of diazinon on gut microbial community structure, metagenome and associated metabolic profiles (Gao et al., 2016). However, how diazinon regulates microbial gene expression and how microbial gene regulation contributes to host physiology were not characterized. Metatranscriptomics emerges as a highly informative approach to analyze the regulation and dynamics of active microbial community in microbiome research (Gosalbes et al., 2011). The alterations of microbial community gene-expression profiles by diet (McNulty et al., 2011) and antibiotics (Maurice et al., 2013) have been reported; however, no study has been conducted to examine the effects of environmental toxicants on the metatranscriptome of gut microbiome.
We and others have demonstrated that environmental toxicants could alter the gut microbiome compositions or gut microbiome community structures (Chi et al., 2016; Gao et al., 2016; Lu et al., 2014; Zhang et al., 2015). In fact, assessment of exposure-induced gut bacterial profile changes was performed for almost every single published microbiome-exposure study, from which we could elucidate the disrupting effects of diverse toxicants on gut bacterial compositions. However, it is largely unknown how mechanistically these toxicants lead to gut microbiome composition/structure changes, which clearly is a gap in current microbiome research. The population and many behaviors of bacteria are regulated by the cell-cell signaling process called quorum sensing (Nasser and Reverchon, 2007), which utilizes autoinducers, a type of signaling molecules, to control bacterial density, with acylated homoserine lactones (AHL) and autoinducer prepeptides being used in Gram-negative and Gram-positive bacteria, respectively (Waters and Bassler, 2005). Direct measurement of these autoinducer molecules has been challenging and the procedure is complicated due to their structural complexity and extremely low abundance (Thiel et al., 2009). However, alterations of critical genes of quorum sensing, which can be revealed by metatranscriptomics, may offer an alternative approach to examine the effects of exposure on quorum sensing systems. Since quorum sensing plays a key role in bacterial behaviors such as motility, sporulation, and gene regulation, it could provide important information to understand the functional impact of exposure on the gut microbiome. Thus, the major goal of this study was to examine whether essential genes involved in quorum sensing was altered by diazinon exposure. Similarly, emphasis was also placed to determine the effects of diazinon on other quorum sensing-regulated key bacterial events or pathways, such as motility, sporulation, and cell wall components. Finally, we also analyzed the impact of diazinon on important metabolic pathways, such as carbohydrate metabolism, due to their indispensable role in energy production and harvest for both gut bacteria and host. Through metatranscriptomics sequencing, we demonstrated for the first time that organophosphate diazinon modulated quorum sensing, which may serve as a key mechanism to regulate bacterial population and their functions. In addition, we also demonstrated that diazinon exposure activated diverse stress response pathways and profoundly impaired energy metabolism of gut bacteria.
MATERIALS AND METHODS
Animals and exposure
Animal exposure was performed as described previously (Gao et al., 2016). In brief, specific pathogen free C57BL/6 mice (∼7-week-old) were purchased from Jackson Laboratory (Bar Harbor, ME) and housed in the University of Georgia animal facility for a week before the start of experimentation, where they were allowed to consume tap water ad libitum. Before and throughout the experimental period, mice were housed under environmental conditions of 22°C, 40%–70% humidity, and a 12:12 h light:dark cycle and provided with standard pelleted rodent diet. At the start of experimentation, 10 male mice were randomly assigned to either a control or diazinon treated group. Diazinon (Sigma Aldrich, Oceanside, CA) was administered to individual mouse in drinking water at concentrations of 4 ppm for a period of 13 weeks like the previous study (Gao et al., 2016). Fresh diazinon solution was prepared weekly. Controls received water alone.
Metatranscriptomics library preparation
Individual mouse was transferred to a clean cage until defecation. Fresh fecal pellets collected at 13 weeks were flash frozen in liquid nitrogen and preserved in a −80°C freezer until RNA isolation. Total RNA from individual mouse was extracted using PowerSoil Total RNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA) followed by genomic DNA removal. Total RNA quality was assessed by Agilent 2100 Bioanalyzer. Contamination of mouse RNA (both mRNA and rRNA) was removed using MICROBEnrich Kit (Thermo-Scientific, Waltham, MA). Bacterial mRNA was purified from total RNA by rRNA depletion using MICROBExpress Bacterial mRNA Enrichment kit (Thermo-Scientific, Waltham, MA). Quality of enriched bacterial mRNA was assessed by Agilent 2100 Bioanalyzer again. The RNAseq libraries were made using a KAPA Stranded RNA-Seq Library Preparation Kit (KAPA Biosystems, Wilmington, MA). In brief, bacterial mRNA was fragmented at 85°C for 2 min, followed by first strand cDNA synthesis with random primers and second strand cDNA synthesis which converted cDNA:RNA hybrid to dscDNA. dAMP was added to 3′end of dscDNA fragments as A-tailing, followed by adaptor ligation and library amplification. The PCR conditions were as follows: initial denaturation at 98°C for 45 s, 10 cycles of 98°C for 15 s, 60°C for 30 s, 72°C for 30 s, and final extension at 72°C for 5 min. Each 25 μL reaction solution consisted of: 5 μL 5× KAPA HiFi buffer, 0.75 μL 10 mM dNTPs, 0.5 μL KAPA HiFi Hotstart, 2.5 μL of 5 μM forward primer, 2.5 μL of 5 μM reverse primer, and 5 μL microbial dscDNA. PCR products were purified by Sera-Mag Speedbeads (Thermo-Scientific, Waltham, MA) at a ratio of 1:1 and quantified by Qubit 2.0 Fluorometer. The resultant libraries were sequenced at the Georgia Genomics Facility using Illumina Nextseq PE150 reads.
Bioinformatics analysis of metatranscriptomics data
The raw fastq files of each animal were analyzed with our metatranscriptomic analysis workflow, built on several previous studies (Gaur et al., 2016; Haas et al., 2013; Livernois et al., 2016; Mora-Ortiz et al., 2016; Schulze et al., 2016). The process is briefly described as follows. First, quality assessment by FastQC (Version 0.11.4) and trimming by Trimmomatic (Version 0.32) were performed on raw data, followed by de novo transcriptome assembly using Trinity (Version 2.0.6) de novo assembler (Grabherr et al., 2011). After assembly, original sample reads were aligned to transcriptome assembly on a per sample basis using Bowtie 2 (Version 2.2.9). Sample-specific expression abundance estimation was determined by RSEM (Li and Dewey, 2011). Bioconductor software edgeR (Version 3.4) was applied to the matrix containing RSEM abundant estimates for differential expression analysis (Robinson et al., 2010). Fragments Per Kilobase of transcript per Million mapped reads were used for the normalization of sequencing depth and transcripts length (Trapnell et al., 2010). Differentially expressed contigs and transcripts were annotated by BlastN and BlastX. Only transcripts with false discovery rates (FDR, based on the Benjamini–Hochberg method) <0.05 were considered significant hits and subject to further analysis. Functional categories of annotated contigs and transcripts were assigned by cluster of orthologous genes (COG) functional annotations (Galperin et al., 2015). The entire set of raw fastq data have been deposited to MG-RAST server (http://metagenomics.anl.gov; last accessed March 22, 2016) with job IDs 292137, 292138, 292139, 292140, 292141, 292142, 292143, 292144, 292145, 292146, 292147, 292148, 292149, 292150, 292151, 292152, 292153, 292154, 292195, and 292156. In addition, processed data have been uploaded into the MG-RAST server with the submission job IDs 302500 and 302501.
RESULTS
Diazinon Exposure Altered the Expression Profile of Gut Metatranscriptome
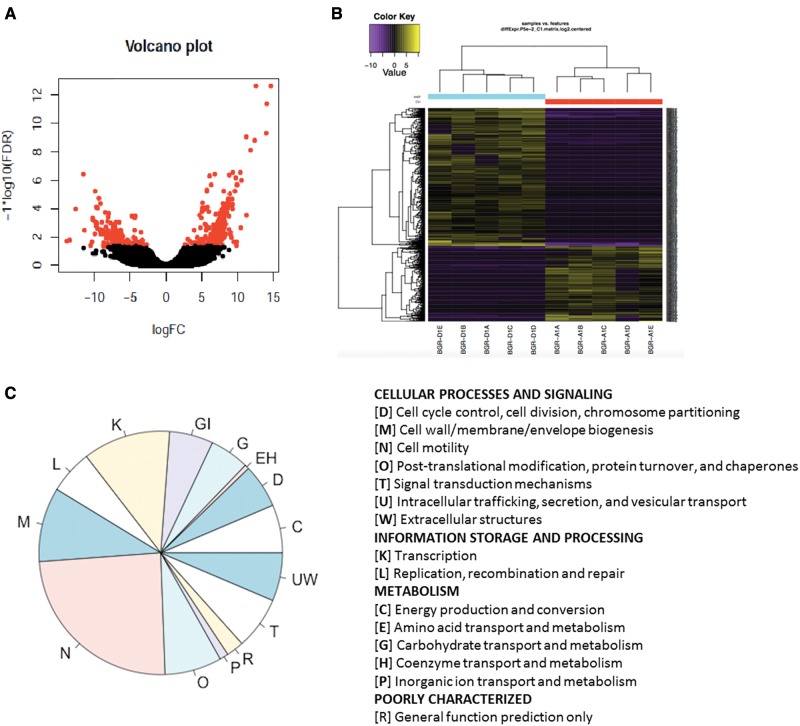
We found that diazinon exposure altered the expression profile of gut metatranscriptome, with 278 and 399 microbial transcripts being down- and up-regulated, respectively (Figs. 1A and B). Annotation of these differentially expressed transcripts by Blast shows that they have a wide range of functions, which belong to different COG functional categories in cellular processes and signaling, information storage and processing and metabolism (Figure 1C). These significant perturbed functional categories include cell motility, and cell wall/membrane/envelope biogenesis, and metabolism pathways of diverse compounds.
FIG. 1.
Diazinon exposure altered the expression profile of gut metatranscriptome (FDR <0.05 for the points shown on the top left and right of the volcano plot). (A); 677 microbial transcripts were significantly regulated, with 278 and 399 being down- and up-regulated, respectively (B); Classification of the COGs by functional categories for differentially expressed transcripts (C).
Diazinon Exposure Disturbed Quorum Sensing System
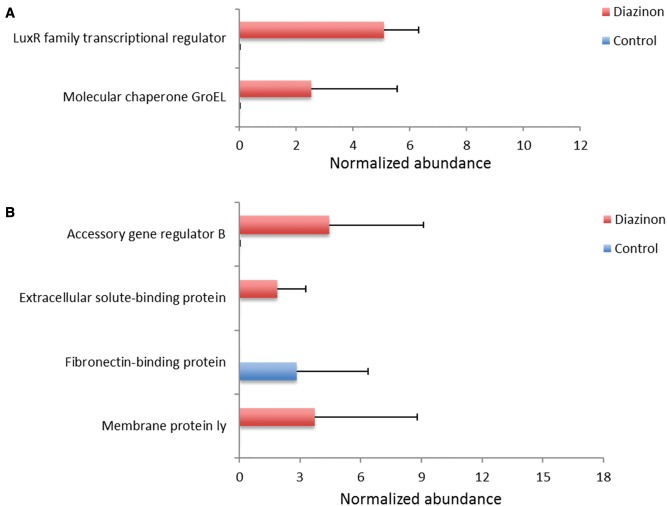
Previously, we and others reported that toxicant exposure perturbed the gut microbiome community structures or compositions (Chi et al., 2016; Gao et al., 2016; Lu et al., 2014; Zhang et al., 2015). However, specific mechanism responsible for such changes remains unknown. Bacteria control behaviors including sporulation, motility, and virulence according to the population density fluctuation by the cell-cell signaling process called quorum sensing (Nasser and Reverchon, 2007). The detection of an autoinducer at the threshold concentration leads to the alteration of gene expression. In general, AHLs are used as autoinducers in Gram-negative bacteria and quorum sensing is mediated by AHL-dependent LuxR family transcriptional regulators (Fuqua et al., 1994). The expression of LuxR family transcriptional regulator was up-regulated by diazinon exposure (Figure 2A). The folding of LuxR polypeptide requires GroEL chaperonin (Manukhov et al., 2010), which was also up-regulated by diazinon exposure (Figure 2A). In Gram-positive bacteria, accessory gene regulator (Agr) is a conventional quorum sensing system with a four-gene operon carrying agrA, agrB, agrC, and agrD, which influences the expression of many virulence genes (Darkoh et al., 2015). Autoinducer prepeptide is produced by agrD gene, processed by transmembrane protein AgrB, and then released into extracellular milieu (Chitra et al., 2015). AgrB was up-regulated by diazinon exposure (Figure 2B), so was the associated extracellular solute-binding protein. In addition, fibronectin-binding protein, a cell surface protein, was down-regulated by diazinon exposure (Figure 2B), which is typically negatively regulated by agr system (Saravia-Otten et al., 1997). Besides, membrane protein ly involved in post-translational modification of the autoinducing quorum-sensing peptide was also up-regulated (Figure 2B). Taken together, the alteration of these key transcripts clearly indicated that quorum sensing system has been significantly altered by diazinon exposure, which could be used by bacteria to control community structures, functional systems, and bacterial signaling.
FIG. 2.
Diazinon exposure altered quorum sensing systems of the gut bacteria in mice (A: Gram-negative; B: Gram-positive bacteria), with key regulators of quorum sensing being significantly changed. (FDR <0.05, n = 5, Mean ± SD).
Diazinon Exposure Increased Motility- and Sporulation-Related Genes
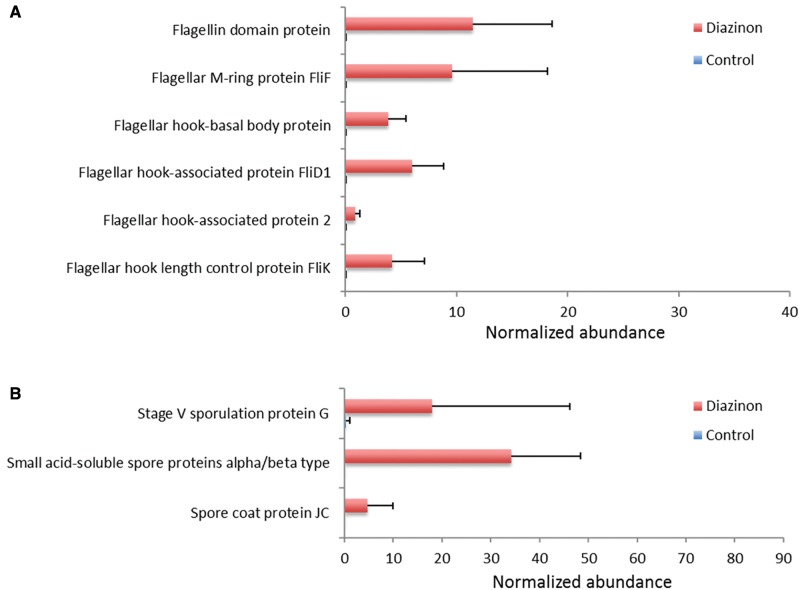
Quorum sensing controls and regulates many important bacterial behaviors and functional genes. Along with the up-regulation of LuxR family transcriptional regulator and AgrB, bacterial sporulation and motility activity were also increased in diazinon-treated animals (Figure 3). Bacteria use flagella to drive cell locomotion and move toward favorable environment. Bacterial flagella can function as a sensory organelle, being sensitive to toxicants and environmental changes surrounding the cells. As shown in Figure 3A, a number of flagellar associated transcripts were significantly up-regulated in the gut microbiome of diazinon-treated animals. Moreover, several sporulation-related genes were also up-regulated by diazinon exposure (Figure 3B). Sporulation is one of the protection mechanisms for some bacteria to respond to environmental changes, such as nutrition starvation and extreme growth conditions (Gaidenko and Price, 1998). In summary, these results indicated that diazinon exposure may have deteriorated the gut ecology and caused stress to the gut microbiome.
FIG. 3.
Bacterial motility- and sporulation-related genes were significantly increased in gut bacteria of mice exposed to diazinon for 3 months (A: motility-related genes; B: genes involved in sporulation). (FDR <0.05, n = 5, Mean ± SD).
Diazinon Exposure Altered Genes in Cell Wall Components
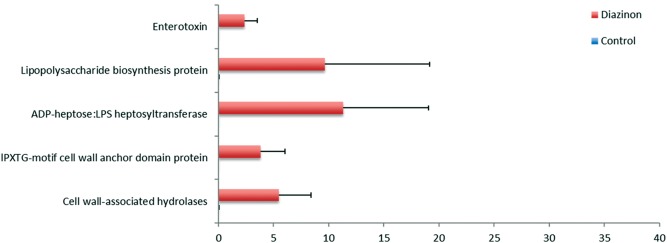
Cell wall components, such as lipopolysaccharide (LPS), largely contribute to the toxic effects of bacteria, which can translocate into host circulation and tissues, leading to inflammatory responses. LPS consists of lipid A, core polysaccharide and O polysaccharide, while lipid A is the main toxic component (Schromm et al., 2000). As shown in Figure 4, the genes for enterotoxin production, LPS biosynthesis, ADP-heptose:LPS heptosyltransferase I, lPXTG-motif cell wall anchor domain protein, cell wall-associated hydrolases, were all significantly up-regulated in the gut microbiome of mice exposed to diazinon. These increased genes of LPS pathways may contribute to elevated serum levels of LPS in diazinon-treated mice (Supplementary Figure 1).
FIG. 4.
Diazinon exposure stimulated the enterotoxin production, LPS biosynthesis, and expression of other genes involved in bacterial cell wall components. (FDR <0.05, n = 5, Mean ± SD).
Diazinon Exposure Activated Stress Response Pathways
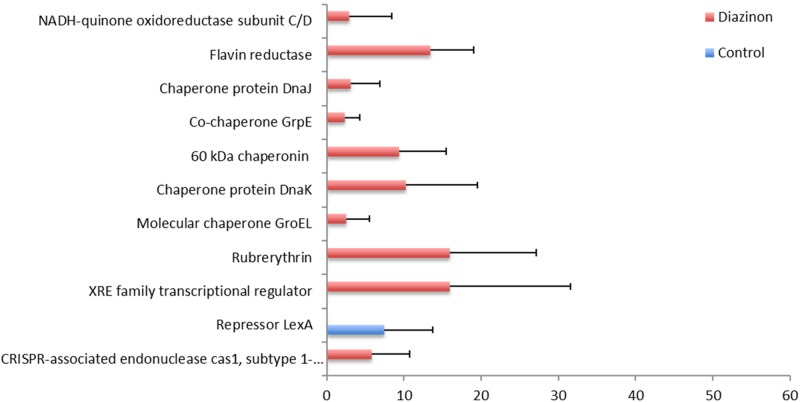
As mentioned above, diazinon exposure led to adverse gut ecological environmental and challenges to the gut microbiome. Bacteria have developed diverse pathways to respond to environmental changes and stress. As shown in Figure 5, a number of genes related to stress response pathways were activated by diazinon exposure, including NADH-quinone oxidoreductase, flavin reductase, heat shock proteins, chaperones, rubrerythin, and xenobiotic response element (XRE) family transcriptional regulator, etc. Diazinon-induced DNA damage has been widely studied (Kashanian et al., 2008) and SOS is a global response to DNA damage. As shown in Figure 5, LexA was down-regulated by diazinon exposure, which cleaves itself and initiates the SOS response upon DNA damage (Qin et al., 2015), suggesting that diazinon may have induced bacterial DNA damage and bacteria activated defense pathways to maintain genome stability.
FIG. 5.
Diazinon exposure activated the expression of multiple stress response genes in oxidative stress and DNA damage (FDR <0.05, n = 5, Mean ± SD).
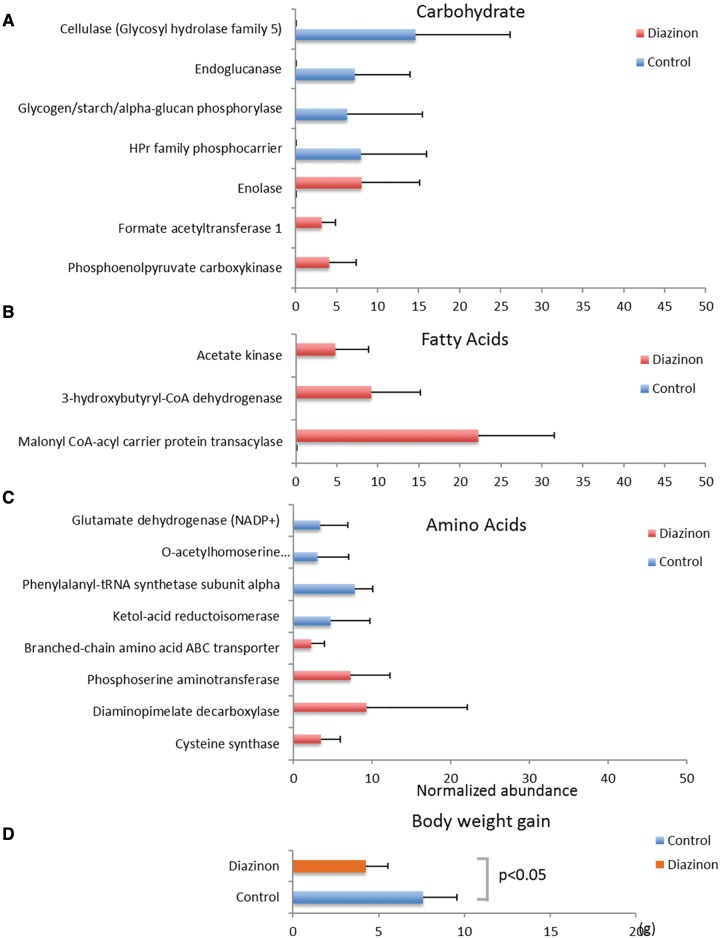
Diazinon Disturbed Carbohydrate, Fatty Acid, Amino Acid Metabolic Homeostasis
A widespread regulation has been observed for carbohydrate, fatty acid, and amino acid pathways (Figure 6). For example, several enzymes involved in carbohydrate metabolism and transport were significantly regulated by diazinon exposure (Figure 6A). Down-regulated enzymes are involved in cellulose decomposition (cellulose and endoglucanase), polysaccharides biosynthesis (glycogen/starch/alpha-glucan phosphorylase) and phosphoenolpyruvate-dependent sugar phosphotransferase system (phosphocarrier HPr protein). Other enzymes were up-regulated, including those in glycolysis (enolase) and pyruvate metabolism (formate acetyltransferase 1 and phosphoenolpyruvate carboxykinase). Fatty acid metabolism was also altered by diazinon exposure (Figure 6B). Malonyl CoA-acyl carrier protein transacylase (MCAT) (type II fatty acid synthesis) was up-regulated. 3-hydroxybutyryl-CoA dehydrogenase and acetate kinase were up-regulated in diazinon-treated animals, which are involved in butyric acid metabolism and production of acetyl-CoA, respectively. Several enzymes involved in amino acid metabolism and transport were also significantly regulated by diazinon exposure (Figure 6C). Besides the changes of gene expression, perturbation of homeostasis of fatty acids and amino acids was confirmed at the metabolite level (Supplementary Figure 2). Regulation of these key genes in carbohydrate, fatty acid, amino acid pathway suggests impaired energy metabolism and harvest in the gut microbiome, which is consistent with the observation that diazinon-treated mice had a significantly reduced body weight gain over a 3-month exposure period when compared with controls (4.3 vs 7.6 g, P < .05) (Figure 6D).
FIG. 6.
Diazinon exposure disturbed metabolic homeostasis of carbohydrate (A), fatty acid (B), amino acid metabolism (C), which is associated with a significantly reduced body weight gain in diazinon-treated mice (P < .05) (D). (FDR <0.05, n = 5, Mean ± SD).
DISCUSSION
We used metatranscriptomic sequencing to study the effect of diazinon exposure on gut metatranscriptome, with the goal of understanding the functional impact of organophosphate on the gut microbiome. We demonstrated that diazinon exposure changed the gut metatranscriptome expression profile and a number of key pathways. In particular, organophosphate diazinon disturbed quorum sensing systems, increased motility and sporulation-related genes, activated stress response pathways, and perturbed carbohydrate, fatty acid, and amino acid metabolic homeostasis. These findings may provide important implications in understanding and defining gut microbiome toxicity induced by organophosphate and other toxicants.
In our previous study, we found that 19 bacterial genera were altered in diazinon-exposed mice (Gao et al., 2016). Similarly, we also demonstrated that other environmental toxicants, like arsenic, could also largely alter the bacterial compositions, abundance and community structures of gut bacteria (Chi et al., 2016; Lu et al., 2014). However, it remains unknown what mechanism is responsible for regulating the bacterial community. This is an important question to be addressed in microbiome-exposure research. Bacteria control behaviors such as sporulation, motility and virulence according to the population density fluctuation by quorum sensing (Nasser and Reverchon, 2007). Autoinducers play a key role in quorum sensing, however, the detection of autoinducers is challenging due to their low abundance and structural complexity, so the emphasis is being placed to examine key genes of quorum sensing system in this study. As illustrated in Figure 2, key gene regulators of quorum sensing system were significantly modulated by diazinon exposure. Bacteria use autoinducers to coordinately modulate gene expression as a response to external environment stimuli (Miller and Bassler, 2001). Diverse bacterial species use quorum sensing to boost their competitive advantages and quorum sensing controls important functions including virulence factors (Knecht et al., 2016). Therefore, quorum sensing may serve as a key mechanism to modulate gut microbiome community structure under external chemical stress. To our knowledge, this is the first study to demonstrate that environmental toxicants can modulate quorum sensing-related key genes to influence bacterial community structures.
Along with the up-regulation of quorum sensing-related key genes, diazinon exposure significantly stimulated the expression of genes in bacterial motility and sporulation-related genes in the gut microbiome of mice (Figure 3). The increase of bacterial motility and sporulation indicated that diazinon exposure has deteriorated the gut ecology and caused stress to the gut bacteria. On the other hand, activation of flagellar motility and cell wall components is often related to bacterial pathogenicity and regulated by quorum sensing (Nakamura et al., 2008; Yang and Defoirdt, 2015). As reported in our previous study (Gao et al., 2016), gut microbiome community structures were significantly disrupted by diazinon. In particular, the prevalence of several potentially pathogenic bacteria was only observed after diazinon treatment, such as bacteria belonging to Burkholderiales order, which have been associated with respiratory infection, chronic granulomatous and inflammatory bowel disease. A sub-group of LuxR transcriptional regulators with N-termianl autoinducer binding domains and C-terminal Helix-Turn-Helix DNA binding domains were found in Burkholderiales order (Subramoni et al., 2015). Diazinon exposure increased the opportunity for the invasion of bacteria belonging to Burkholderiales, which could utilize the quorum sensing master regulator, LuxR family transcriptional regulator, to boost their competitive strength. Similarly, genes of enterotoxin production, LPS synthesis protein and ADP-heptose:LPS heptosyltransferase I were all increased in diazinon-exposed animals (Figure 4), which could partially contribute to well-documented organophosphate-induced systemic inflammation in host tissues (Banks and Lein, 2012).
Diazinon induces oxidative stress, leading to excessive production of reactive oxygen species, DNA oxidation and aggregation of proteins, which has been demonstrated both in vivo and in vitro (Giordano et al., 2007; Jafari et al., 2012). However, it remains unknown whether diazinon causes oxidative stress in gut bacteria and how gut microbiome responds to the stress. In the present study, several key enzymes/proteins were up-regulated, which could be used by the gut microbiome to counteract the oxidative stress induced by diazinon. For example, NADH-quinone oxidoreductase and flavin reductase are crucial mediators of cellular defense against oxidative stress (Martinez-Hernandez et al., 2015). The expression of NADH-quinone oxidoreductase subunit C/D and flavin reductase was up-regulated in diazinon-treated animals (Figure 5). In addition, heat shock proteins, which are ubiquitously expressed stress response proteins, also play a critical role in oxidative stress (Kalmar and Greensmith, 2009). Among heat shock proteins, GroEL and DnaK chaperones are representative and both of them were up-regulated in diazinon-treated animals. DnaK has been demonstrated with the protection role against oxidative stress. First, it plays a key role in protein sorting, quality control, and aiding the repair and clearance of damaged proteins. Second, it could bind and inhibit members of the apoptotic cascade and negatively regulate apoptosis (Kalmar and Greensmith, 2009). The up-regulation of DnaK by diazinon exposure indicated that the gut microbiome responded against the oxidative stress. Besides, rubrerythin was up-regulated by diazinon exposure (Figure 5), which has been demonstrated to play a protection role against oxidative stress in anaerobic bacteria and archaea (Lumppio et al., 2001). Furthermore, XRE family transcriptional regulator was up-regulated (Figure 5). XRE is required for the transcriptional activation of Cu/Zn superoxide dismutase, which catalyzes the dismutation of superoxide radicals produced from biological oxidation (Park and Rho, 2002).
Diazinon-induced DNA damage has been widely studied (Kashanian et al., 2008) and SOS is a global response to DNA damage. SOS response is the transcriptional regulatory network controlled by repressor protein LexA, which represses transcription of SOS genes by binding to SOS promoters (Butala et al., 2009). We observed that LexA was down-regulated by diazinon exposure (Figure 5). It is also interesting to notice that CRISPR-associated endonuclease cas1, was up-regulated by diazinon exposure (Figure 5). CRISPR-Cas system is a sophisticated adaptive immunity system in bacteria and archaea to combat phage infection (Datsenko et al., 2012). CRISPR-Cas system functions as a self and non-self recognition mechanism, which targets and inactivates invading viruses and plasmids (van der Oost et al., 2014). Resistance is acquired by integrating a short invader sequence, spacer, into the CRISPR loci. Upon repeated infection, CRISPRs are processed into short interfering RNAs (siRNAs), which guide the Cas proteins to complementary invading nucleic acid and targets this for destruction. Several Cas proteins were required for spacer acquisition including Cas1 (Fineran and Charpentier, 2012). In addition to the spacer acquisition, Cas1 protein has also been shown to be involved in DNA repair (Serbanescu et al., 2015). A previous study using Escherichiacoli demonstrated that a mutant deficient in Cas1 had a DNA repair-deficient phenotype, highlighting the role of Cas1 in DNA repair beyond antivirus immunity (Babu et al., 2011). Herein, the upregulation of Cas1 may also reflect its participation in repairing bacterial DNA damage arising from diazinon exposure.
The metabolism of gut bacteria is essential for normal host food digestion, energy metabolism, and signal communication (Nicholson et al., 2012). In this study, we found diazinon exposure dramatically changed gut bacteria metabolism. This perturbation could alter the function of gut bacteria and further affect host health. For example, several enzymes involved in carbohydrate metabolism and transport were significantly regulated by diazinon exposure (Figure 6A). Cellulase and endoglucanase were both down-regulated, which are involved in cellulose and polysaccharide decomposition by catalyzing cellulolysis. Mammalian cells have limited ability to digest dietary fiber like cellulose, while gut bacteria could perform this task well. In addition, glycogen/starch/alpha-glucan phosphorylase, the gene involved in polysaccharides biosynthesis, was also significantly down-regulated in the gut bacteria of treated animals. Moreover, phosphocarrier HPr protein was also down-regulated by diazinon, which serves as a major bacterial carbohydrate transport system to catalyze the phosphorylation of sugar substrates when they are being transported across the cell membrane. These results indicated that diazinon exposure affected the expression of key enzymes in cellulolysis, polysaccharides synthesis, and sugar transportation, which may affect the energy metabolism and harvest for both gut bacteria and host. In agreement with this, the body gain of diazinon-treated mice was significantly lower than that of controls (Figure 6D). Meanwhile, enzymes involved in glycolysis and pyruvate metabolism were up-regulated, such as enolase, formate acetyltransferase 1, and phosphoenolpyruvate carboxykinase. Activation of these bacterial enzymes may be used to compensate impaired energy production or harvest. Similarly, MCAT (type II fatty acid synthesis), a critical gene in bacterial fatty acid metabolism, was up-regulated. And, 3-hydroxybutyryl-CoA dehydrogenase and acetate kinase were up-regulated in diazinon-treated animals. 3-hydroxybutyryl-CoA dehydrogenase is involved in butyric acid metabolism and acetate kinase could convert acetate to a key metabolic intermediate acetyl-CoA of energy metabolism. Similarly, genes involved in amino acid metabolism and transport were also regulated by diazinon exposure. Genes related to cysteine, lysine and serine biosynthesis were up-regulated, while genes related to phenylalanine, leucine/isoleucine and glutamate biosynthesis were down-regulated. Meanwhile, branched-chain amino acid transporter was up-regulated. It is difficult to precisely track down what the biological significance of these regulations as amino acids are being utilized by many metabolic pathways. Nevertheless, some regulations may still reflect bacterial response to energy deficiency. For example, up-regulated branched-chain amino acid transporter may be used to boost the utilization of branched-chain amino acids. Taken together, profound effects of diazaion on carbohydrate, fatty acid, and amino acid metabolism will not only influence bacteria but also host.
CONCLUSIONS
In summary, we found that diazinon exposure altered the expression profile of gut metatranscriptome. Annotation of these transcripts reveals that they have a wide range of biological functions. Especially, we demonstrated that organophosphate exposure altered quorum sensing systems of gut bacteria, leading to the regulation of many critical downstream genes and pathways. For example, we found that diazinon activated bacterial motility and cell wall components, which could contribute to increased pathogenicity of bacteria and systemic inflammation in the host. Similarly, diazinon exposure also activated diverse genes in multiple stress-response pathways and profoundly altered the metabolic homeostasis of carbohydrate, fatty acid and amino acid metabolism. These results improved our understanding of how environmental toxicants, such as organophosphates, interact with the gut microbiome to affect the community structures and its function.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Travis Glenn for his helpful suggestions on metatranscriptomics library preparation. We also thank Dr. Lorenz W. Walter for the support on bioinformatics analysis of RNAseq data. There is not a conflict of interest for authors.
FUNDING
This study was partially supported by the University of Georgia, University of North Carolina at Chapel Hill and the National Institute of Health/National Institute of Environmental Health Sciences (grant number R01ES024950).
REFERENCES
- Aggarwal V., Deng X., Tuli A., Goh K. S. (2013). Diazinon-chemistry and environmental fate: a California perspective. Rev. Environ. Contam. Toxicol. 223, 107–140. [DOI] [PubMed] [Google Scholar]
- Babu M., Beloglazova N., Flick R., Graham C., Skarina T., Nocek B., Gagarinova A., Pogoutse O., Brown G., Binkowski A., et al. (2011). A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79, 484–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks C. N., Lein P. J. (2012). A review of experimental evidence linking neurotoxic organophosphorus compounds and inflammation. Neurotoxicology 33, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussabbeh M., Ben Salem I., Hamdi M., Ben Fradj S., Abid-Essefi S., Bacha H. (2016). Diazinon, an organophosphate pesticide, induces oxidative stress and genotoxicity in cells deriving from large intestine. Environ. Sci. Pollut. Res. 23, 2882–2889. [DOI] [PubMed] [Google Scholar]
- Butala M., Zgur-Bertok D., Busby S. J. (2009). The bacterial LexA transcriptional repressor. CMLS 66, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L., Bian X., Gao B., Ru H., Tu P., Lu K. (2016). Sex-specific effects of arsenic exposure on the trajectory and function of the gut microbiome. Chem. Res. Toxicol. 29, 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitra M. A., Jayanthy C., Nagarajan B. (2015). Detection and sequence analysis of accessory gene regulator genes of Staphylococcus pseudintermedius isolates. Vet. World 8, 902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkoh C., DuPont H. L., Norris S. J., Kaplan H. B. (2015). Toxin synthesis by Clostridium difficile is regulated through quorum signaling. MBIO 6, e02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Pougach K., Tikhonov A., Wanner B. L., Severinov K., Semenova E. (2012). Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3, 945–951. [DOI] [PubMed] [Google Scholar]
- Fineran P. C., Charpentier E. (2012). Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology 434, 202–209. [DOI] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S. C., Greenberg E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidenko T. A., Price C. W. (1998). General stress transcription factor ςB and sporulation transcription factor ςH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180, 3730–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Bian X., Mahbub R., Lu K. (2016). Gender-specific effects of organophosphate diazinon on the gut microbiome and its metabolic functions. Environ. Health Perspect. 125, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur M., Das A., Sahoo R. K., Kar B., Nayak S., Subudhi E. (2016). De Novo transcriptome assembly of Zingiber officinale cv. Suruchi of Odisha. Genom. Data 9, 87–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano G., Afsharinejad Z., Guizzetti M., Vitalone A., Kavanagh T. J., Costa L. G. (2007). Organophosphorus insecticides chlorpyrifos and diazinon and oxidative stress in neuronal cells in a genetic model of glutathione deficiency. Toxicol. Appl. Pharmacol. 219, 181–189. [DOI] [PubMed] [Google Scholar]
- Gosalbes M. J., Durban A., Pignatelli M., Abellan J. J., Jimenez-Hernandez N., Perez-Cobas A. E., Latorre A., Moya A. (2011). Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6, e17447.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gweon T. G., Kim J., Lim C.-H., Park J. M., Lee D.-G., Lee I. S., Cho Y.-S., Kim S. W., Choi M. G. (2016). Fecal microbiota transplantation using upper gastrointestinal tract for the treatment of refractory or severe complicated Clostridium difficile infection in elderly patients in poor medical condition: the first study in an Asian country. Gastroenterol. Res. Pract. 2016, 2687605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari M., Salehi M., Ahmadi S., Asgari A., Abasnezhad M., Hajigholamali M. (2012). The role of oxidative stress in diazinon-induced tissues toxicity in Wistar and Norway rats. Toxicol. Mech. Methods 22, 638–647. [DOI] [PubMed] [Google Scholar]
- Jones R. R., Barone-Adesi F., Koutros S., Lerro C. C., Blair A., Lubin J., Heltshe S. L., Hoppin J. A., Alavanja M. C. R., Beane Freeman L. E. (2015). Incidence of solid tumours among pesticide applicators exposed to the organophosphate insecticide diazinon in the Agricultural Health Study: an updated analysis. Occup. Environ. Med. 72, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar B., Greensmith L. (2009). Induction of heat shock proteins for protection against oxidative stress. Adv. Drug Deliv. Rev. 61, 310–318. [DOI] [PubMed] [Google Scholar]
- Kashanian S., Gholivand M. B., Ahmadi F., Ravan H. (2008). Interaction of diazinon with DNA and the protective role of selenium in DNA damage. DNA Cell Biol. 27, 325–332. [DOI] [PubMed] [Google Scholar]
- Knecht L. D., O’Connor G., Mittal R., Liu X. Z., Daftarian P., Deo S. K., Daunert S. (2016). Research paper: serotonin activates bacterial quorum sensing and enhances the virulence of Pseudomonas aeruginosa in the host. EbioMedicine 9, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livernois A., Hardy K., Domaschenz R., Papanicolaou A., Georges A., Sarre S. D., Rao S., Ezaz T., Deakin J. E. (2016). Identification of interleukin genes in Pogona vitticeps using a de novo transcriptome assembly from RNA-seq data. Immunogenetics 68, 719–731. [DOI] [PubMed] [Google Scholar]
- Lu K., Abo R. P., Schlieper K. A., Graffam M. E., Levine S., Wishnok J. S., Swenberg J. A., Tannenbaum S. R., Fox J. G. (2014). Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumppio H. L., Shenvi N. V., Summers A. O., Voordouw G., Kurtz D. M. Jr. (2001). Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manukhov I. V., Melkina O. E., Goryanin I. I., Baranova A. V., Zavilgelsky G. B. (2010). The N-terminal domain of Aliivibrio fischeri LuxR is a target of the GroEL chaperonin. J. Bacteriol. 192, 5549–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez A., Cordova E. J., Rosillo-Salazar O., Garcia-Ortiz H., Contreras-Cubas C., Islas-Andrade S., Revilla-Monsalve C., Salas-Labadia C., Orozco L. (2015). Association of HMOX1 and NQO1 polymorphisms with metabolic syndrome components. PLoS One 10, e0123313.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice C. F., Haiser H. J., Turnbaugh P. J. (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty N. P., Yatsunenko T., Hsiao A., Faith J. J., Muegge B. D., Goodman A. L., Henrissat B., Oozeer R., Cools-Portier S., Gobert G., et al. (2011). The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3, 106ra106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. [DOI] [PubMed] [Google Scholar]
- Moos W. H., Faller D. V., Harpp D. N., Kanara I., Pernokas J., Powers W. R., Steliou K. (2016). Microbiota and neurological disorders: a gut feeling. Biores. Open Access 5, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Ortiz M., Swain M. T., Vickers M. J., Hegarty M. J., Kelly R., Smith L. M. J., Skøt L. (2016). De-novo transcriptome assembly for gene identification, analysis, annotation, and molecular marker discovery in Onobrychis viciifolia. BMC Genomics 17, 756.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Higashiyama Y., Izumikawa K., Seki M. (2008). The roles of the quorum-sensing system in the release of extracellular DNA, lipopolysaccharide, and membrane vesicles from Pseudomonas aeruginosa. Jpn. J. Infect. Dis. 61, 375–378. [PubMed] [Google Scholar]
- Nasser W., Reverchon S. (2007). New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem. 387, 381–390. [DOI] [PubMed] [Google Scholar]
- Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- Park E. Y., Rho H. M. (2002). The transcriptional activation of the human copper/zinc superoxide dismutase gene by 2,3,7,8-tetrachlorodibenzo-p-dioxin through two different regulator sites, the antioxidant responsive element and xenobiotic responsive element. Mol. Cell Biochem. 240, 47–55. [DOI] [PubMed] [Google Scholar]
- Portune K. J., Benítez-Páez A., Del Pulgar E. M. G., Cerrudo V., Sanz Y. (2016). Gut microbiota, diet and obesity-related disorders—the good, the bad and the future challenges. Mol. Nutr. Food Res. doi: 10.1002/mnfr.201600252. [DOI] [PubMed] [Google Scholar]
- Qin T. T., Kang H. Q., Ma P., Li P. P., Huang L. Y., Gu B. (2015). SOS response and its regulation on the fluoroquinolone resistance. Ann. Transl. Med. 8, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocchi E., Guiducci L., Fulceri F., Billeci L., Buzzigoli E., Apicella F., Calderoni S., Grossi E., Morales M. A., Muratori F. (2016). Gut to brain interaction in Autism Spectrum Disorders: a randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry 16, 183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravia-Otten P., Müller H. P., Arvidson S. (1997). Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179, 5259–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schromm A. B., Brandenburg K., Loppnow H., Moran A. P., Koch M. H., Rietschel E. T., Seydel U. (2000). Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 267, 2008–2013. [DOI] [PubMed] [Google Scholar]
- Schulze T. T., Ali J. M., Bartlett M. L., McFarland M. M., Clement E. J., Won H. I., Sanford A. G., Monzingo E. B., Martens M. C., Hemsley R. M., et al. (2016). De novo assembly and analysis of the chilean pencil catfishtrichomycterus areolatus transcriptome. J. Genomics 4, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbanescu M. A., Cordova M., Krastel K., Flick R., Beloglazova N., Latos A., Yakunin A. F., Senadheera D. B., Cvitkovitch D. G. (2015). Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology. J. Bacteriol. 197, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S., Florez Salcedo D. V., Suarez-Moreno Z. R. (2015). A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front. Cell Infect. Microbiol. 5, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Vilchez R., Sztajer H., Wagner-Döbler I., Schulz S. (2009). Identification, quantification, and determination of the absolute configuration of the bacterial quorum-sensing signal autoinducer-2 by gas chromatography-mass spectrometry. ChemBioChem 10, 479–485. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B., Pertea G., Mortazavi A., Kwan G., van Baren M., Salzberg S., Wold B., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J., Westra E. R., Jackson R. N., Wiedenheft B. (2014). Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. M., Bassler B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 21, 319–346. [DOI] [PubMed] [Google Scholar]
- Yang Q., Defoirdt T. (2015). Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ. Microbiol. 17, 960–968. [DOI] [PubMed] [Google Scholar]
- Yen J., Donerly S., Levin E. D., Linney E. A. (2011). Differential acetylcholinesterase inhibition of chlorpyrifos, diazinon and parathion in larval zebrafish. Neurotoxicol. Teratol. 33, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Jin Y., Zeng Z., Liu Z., Fu Z. (2015). Subchronic exposure of mice to cadmium perturbs their hepatic energy metabolism and gut microbiome. Chem. Res. Toxicol. 28, 2000–2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.