Abstract
ASPM/Asp/ASPM-1 is a microcephaly-associated protein family that regulates spindle architecture, but the underlying mechanism is poorly understood. Here, we show that ASPM forms a complex with another protein linked to microcephaly, the microtubule severing ATPase katanin. ASPM and katanin localize to spindle poles in a mutually dependent manner and regulate spindle flux. X-ray crystallography revealed that the heterodimer formed by the N- and C-terminal domains of the katanin subunits p60 and p80, respectively, binds conserved motifs in ASPM. Reconstitution experiments demonstrated that ASPM autonomously tracks growing microtubule minus ends and inhibits their growth, while katanin decorates and bends both ends of dynamic microtubules and potentiates the minus-end blocking activity of ASPM. ASPM also binds along microtubules, recruits katanin and promotes katanin-mediated severing of dynamic microtubules. We propose that the ASPM/katanin complex controls microtubule disassembly at spindle poles and that misregulation of this process can lead to microcephaly.
Microcephaly is a neurodevelopmental disorder characterized by small brain and mental retardation. A significant number of proteins implicated in microcephaly, including ASPM (abnormal spindle-like microcephaly associated) participate in organizing mitotic spindle poles1. The function of a spindle pole depends on microtubule nucleation as well as stabilization, disassembly and bundling of microtubule minus ends. Microtubule minus ends can grow and shrink in vitro2, however, it is generally believed that within mitotic spindles, minus ends are either stable or slowly depolymerize, resulting in poleward microtubule flux3. The molecular mechanisms controlling microtubule minus-end behavior in the spindle are poorly understood.
Previous work indicated that ASPM (known as Asp in fly and ASPM-1 in worm) is an excellent candidate for being a mitotic microtubule minus-end regulator4–13. It is encoded by the gene that is most frequently mutated in microcephaly14, but the molecular mechanism underlying ASPM activity is not understood. Katanin, a conserved microtubule-severing protein complex, which consists of the AAA ATPase containing enzymatic subunit p60 and the regulatory subunit p8015–17, is also linked to microcephaly18,19. Formation of the katanin p60/p80 heterodimer is mediated by the N-terminal Microtubule Interacting and Trafficking domain of p60 and a conserved C-terminal domain of p8020. Katanin localizes to spindle poles during mitosis in most animal cells and plays an important role in spindle organization18,19,21–25. Mutations in the katanin-encoding gene in microcephaly patients lead to a dramatic increase of centrosome numbers and multipolar spindles, suggesting a role in preventing centriole overduplication19.
Most of the past in vitro work on katanin focused on the p60 subunit using stable microtubules as a substrate. Importantly, the severing activity of p60 is strongly suppressed by free tubulin26, suggesting that additional factors are needed for katanin-mediated microtubule severing in cells. Here, we show that ASPM forms a physiological complex with katanin p60/80 and promotes its activity. Through a combination of structural analysis, genome editing in cells, in vitro reconstitution assays and live cell imaging, we demonstrate that the ASPM/katanin complex likely acts through a combination of microtubule-severing and minus-end blocking activities and regulates spindle flux. Our data thus identify the ASPM/katanin complex as a microtubule minus-end regulator and suggest that its perturbation might lead to neurodevelopmental disease.
Results
ASPM and katanin form a complex and regulate spindle flux
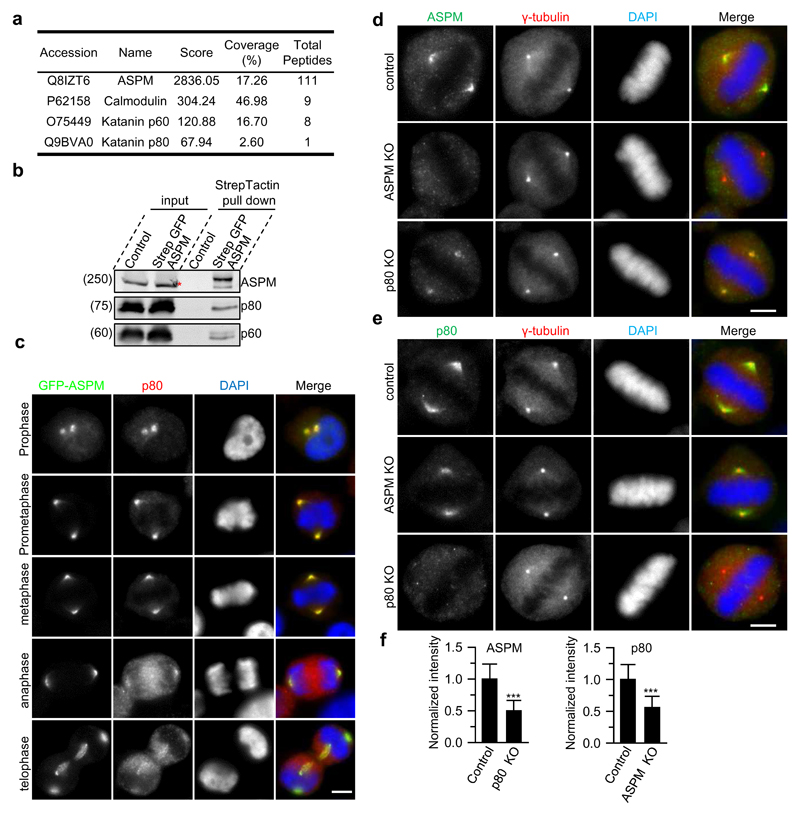
To gain insight into the function of ASPM, we tagged the endogenous protein with a strep tag and GFP by CRISPR/Cas9 technology in cultured human cells (Supplementary Fig. 1a and Table 1). As expected5,8, GFP-ASPM localized to spindle poles throughout cell division and to microtubule minus ends of the central spindle during telophase (Supplementary Fig. 1b). To identify potential binding partners of ASPM, we isolated strep-GFP-ASPM and associated proteins from mitotic cells and analyzed them by mass spectrometry. In addition to the known partner calmodulin7,10, this analysis revealed several centrosomal and spindle pole proteins, including the p60 and p80 subunits of katanin (Fig. 1a,b; Supplementary Table 2). The ASPM-katanin interaction is supported by a recent proteomics study of the katanin family27.
Figure 1. ASPM and katanin form a complex at spindle poles.
(a) Mass spectrometry results of StrepTactin pull down from strep-GFP-ASPM knock-in HEK293T cells synchronized in mitosis (see Supplementary Table 2 for a complete list of specific hits). 16 out of 111 peptides of ASPM are unique to the long isoform, although only the short isoform could be detected by Western blotting in (b).
(b) StrepTactin pull down assay with lysates of strep-GFP-ASPM knock-in HEK293T cells. A red asterisk indicates a non-specific band detected by ASPM antibody in cell extract, overlapping with the specific band visible in the pull down.. See Supplementary Fig.2a for the demonstration of the specificity of anti-katanin antibodies.
(c) Immunostaining for p80 and DNA (DAPI) in GFP-ASPM knock-in HeLa cells during mitosis. Scale bar, 5 μm.
(d-f) Immunofluorescence staining and quantification of ASPM and p80 intensities at spindle poles in control or the indicated knockout HeLa cells. For ASPM intensity, n=105 spindle poles, control; n=124, p80 knockout; for p80 intensity, n=74 spindle poles, control; n=77, ASPM knockout. Scale bars, 5 μm. Data represent mean ± SD. ***, P<0.001, Mann-Whitney U test. Unprocessed original scans of blots are shown in Supplementary Figure 8. Source data for 1f can be found in Supplementary Table 5
During mitosis, ASPM and katanin co-localize at spindle poles, with the strongest overlap observed from prophase to metaphase (Fig. 1c). Knockouts of ASPM and p80 reduced the localization of each other to spindle poles by ~50% (Fig. 1d-f; Supplementary Fig.2a,b). This effect was specific, because the spindle pole intensities of two other major spindle pole proteins, NuMA and the dynactin subunit p150Glued, which localize to the minus ends of spindle microtubules28,29, were largely unaffected upon ASPM and p80 knockout (Supplementary Fig.2c-e). This result suggests that ASPM and p80 knockouts do not affect the number of microtubule minus ends at spindle poles.
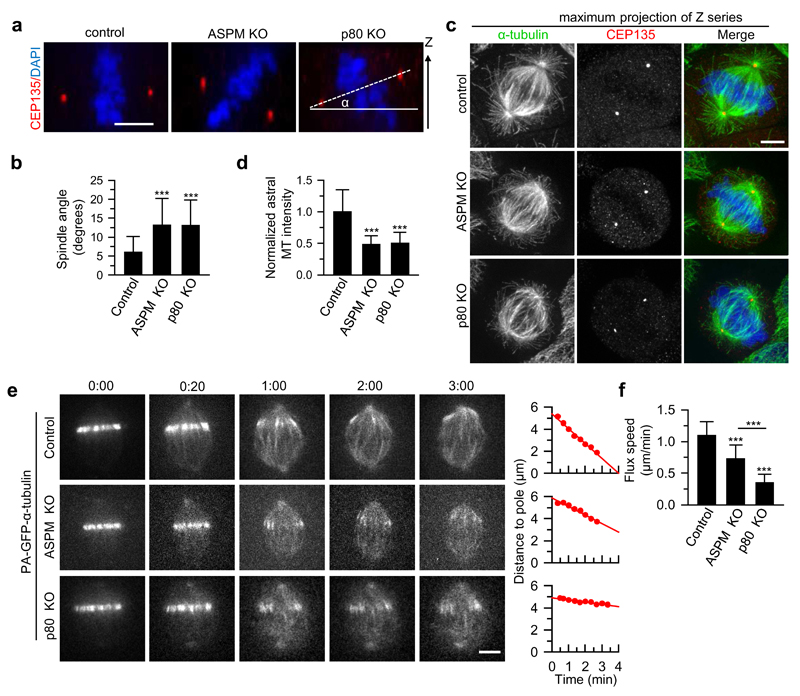
Knockouts of ASPM and p80 showed the previously described spindle defects, such as a strong increase in multipolar spindles in the p80 knockout and spindle misorientation in the ASPM knockout8,19,30 (Fig. 2a,b; Supplementary Fig. 2f-i). In addition, the knockout of p80 also perturbed spindle orientation (Fig. 2a,b). The spindle misorientation upon ASPM and p80 knockout was likely due to the dramatic reduction of astral microtubules30 (Fig. 2c,d). Since the knockout of ASPM did not display an increase in multipolar spindles, while both ASPM and p80 knockouts caused spindle orientation defects, these data suggest that the ASPM/katanin complex does not function in restricting centrosome numbers but may regulate spindle pole organization or dynamics.
Figure 2. The ASPM/katanin complex regulates spindle orientation and poleward flux.
(a) Orthogonal view (x-z) of metaphase HeLa cells stained for CEP135 and DAPI in control, ASPM knockout and p80 knockout HeLa cells. The solid line depicts the horizontal plane; the dashed line indicates the orientation of the spindle with respect to the horizontal plane. Scale bar, 5 μm.
(b) Quantification of spindle angles as shown in (a). n=41 cells, control; n=41, ASPM knockout; n=39, p80 knockout.
(c) Immunostaining for α-tubulin, CEP135 and DNA (DAPI) in control, ASPM knockout and p80 knockout HeLa cells. Maximum intensity projections of Z series with ~40 stacks and 200 nm step are shown. Scale bar, 5 μm.
(d) Quantification of astral microtubule intensity shown in c. n=44 cells, control; n=45, ASPM knockout; n=40, p80 knockout. Intensity was normalized to the average intensity in control cells.
(e) Live cell images of spindle flux in the indicated U2OS cells stably expressing PA-GFP-α-tubulin. Time displayed as min:sec. Plots of the distance between photoactivated GFP stripe and the spindle pole over time are shown on the right. The flux speed was derived from the plot slopes. Scale bar, 5μm.
(f) Flux speed in control and knockout cells. n=26 cells, control; n=29, ASPM knockout; n=26, p80 knockout.
Data represent mean ± SD. ***, P<0.001, Mann-Whitney U test. Source data for panels 2b,d and f can be found in Supplementary Table 5.
To address this possibility, we imaged poleward microtubule flux in control, and in ASPM or p80 knockout U2OS cells expressing photoactivatable (PA)-GFP-α-tubulin (Supplementary Fig. 2a,b). The knockout of ASPM and p80 strongly reduced the flux speed in metaphase spindles, with a more severe defect in p80 knockout cells (Fig. 2e,f; Supplementary Video 1). The strong decrease in spindle flux in ASPM and katanin knockout cells did not lead to major changes in spindle length (Supplementary Fig. 2f,h,j), indicating that the reduction in flux might be compensated by altered microtubule dynamics at kinetochores31,32.
Biochemical characterization of the ASPM-katanin interaction
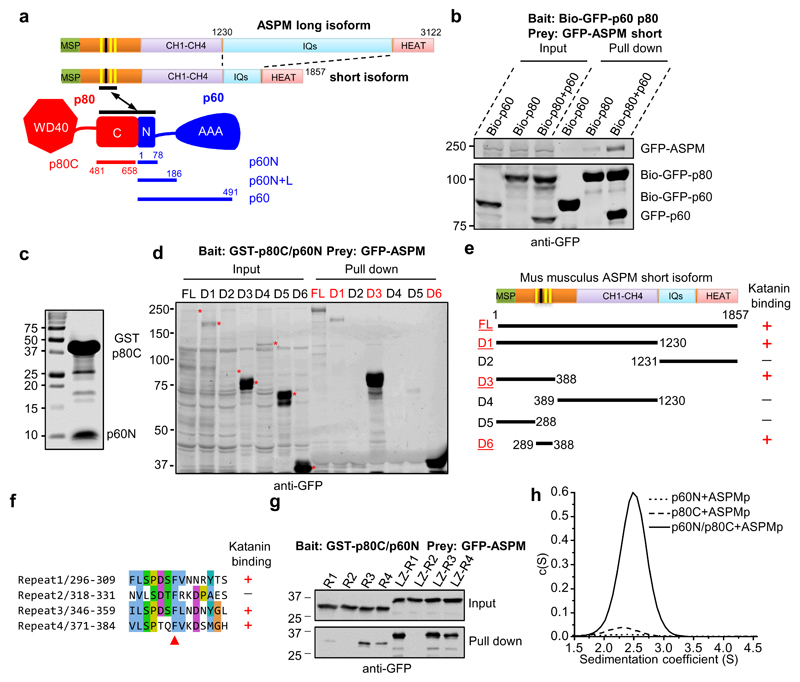
To gain insight into the functional connection between ASPM and katanin, we investigated their interaction in detail and found that both the p60 and p80 subunits of katanin are required for ASPM binding (Fig. 3a,b). ASPM contains an N-terminal major sperm protein (MSP) domain, a long unstructured region, four calponin homology (CH) domains that can bind microtubules9, multiple isoleucine-glutamine (IQ) motifs, and a C-terminal HEAT repeat region. Deletion mapping showed that the heterodimer formed between p60N and p80C interacts with ASPM through its unstructured region that contains four conserved peptide repeats (Fig. 3a-e; Supplementary Fig. 3a). Further mapping revealed that the p60N/p80C heterodimer interacted with three out of the four ASPM repeats and preferentially associated with the third repeat (Fig. 3f,g). Sedimentation velocity experiments with purified components verified the direct interaction between p60N/p80C and the third repeat of ASPM and confirmed that this interaction requires both the p60N and p80C subunits (Fig. 3h). These results suggest that the ASPM-binding site on katanin is created upon the formation of the p60N/p80C heterodimer.
Figure 3. The interaction between ASPM and katanin requires a conserved repeat sequence of ASPM and the p60N/p80C heterodimer.
(a) Scheme of the domain organization of ASPM, p60 and p80. Three (yellow) out of four linear repeats in ASPM interact with p60N/p80C. The numbering is based on the mouse protein.
(b) Streptavidin pull down assay with lysates of HEK293T cells expressing individual subunits of katanin or the p60/p80 heterodimer together with the short isoform of ASPM. A substrate peptide for the biotin ligase BirA (Bio-tag, MASGLNDIFEAQKIEWHE) was inserted at the N-terminus of GFP-tagged bait proteins. Biotinylation was accomplished by co-expressing the tagged proteins together with BirA.
(c) Coomassie blue staining of a gel with GST-p80C/p60N purified from E. coli, which was used for the GST pull down assays in panels d and g and Fig. 4f,g.
(d, e) Western blotting and a summary of the mapping of the interaction between ASPM and p60N/p80C: lysates of HEK293T cells expressing full length ASPM or fragments thereof were used for GST pull down assays with GST-p80C/p60N.
(f) Alignment of the four repeats present in the mouse ASPM protein. Red arrowhead, a conserved phenylalanine residue, corresponding to F352 in the third repeat.
(g) GST-p80C/p60N pull down assay with lysates of HEK293T cells expressing different GFP-tagged ASPM peptide repeats in monomeric or dimeric forms. LZ, leucine zipper from GCN4, which was used as a dimerization domain. The p60N/p80C heterodimer did not bind to the second repeat of ASPM and preferentially associated with the third repeat when the repeats were tested as monomers.
(h) Continuous sedimentation coefficient distribution profiles c(S) showing that p60N/p80C complex formation is a prerequisite for ASPMp peptide binding. TAMRA-labeled ASPMp peptide was detected at 555 nm. When mixed with either p60N or p80C alone, the unbound ASPMp peptide, because of its small size, did not produce any signal between 1.5 and 4.5 S. In contrast, upon the addition of the ASPMp peptide to the p60N/p80C complex, a peak at 2.5 S appeared, demonstrating binding of the peptide to the complex.
Unprocessed original scans of blots are shown in Supplementary Figure 8.
Structure of the ASPM/katanin complex
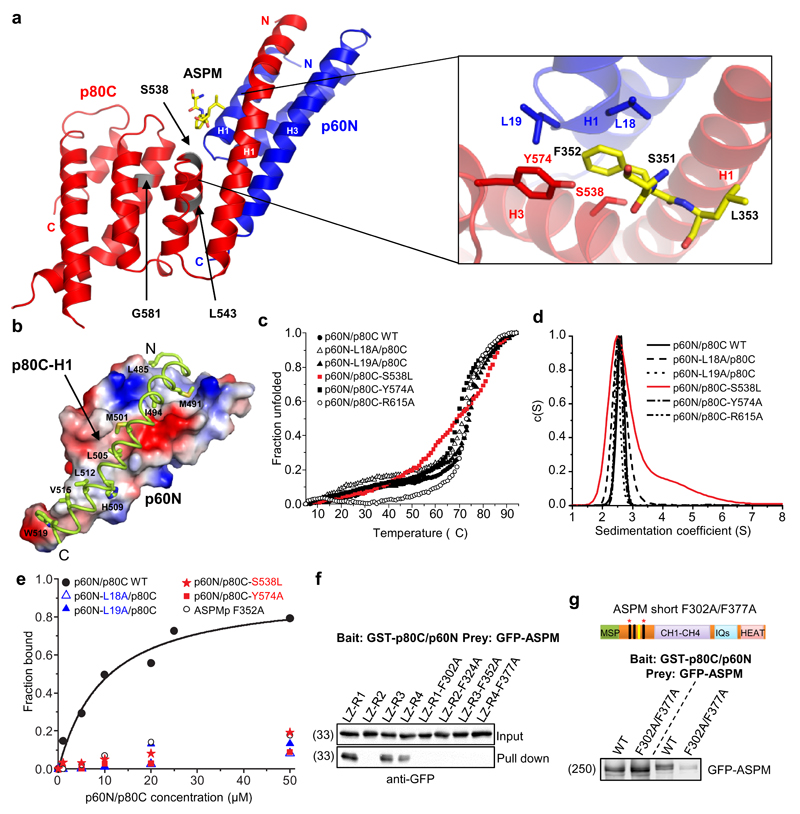
Next, we reconstituted a minimal 1:1:1 stoichiometric complex composed of p60N, p80C and a peptide corresponding to the third repeat of ASPM (ASPMp), and solved the tripartite p60N/p80C/ASPMp complex structure to 1.5 Å resolution by X-ray crystallography (Fig. 4a; Supplementary Table 3). The p60N subunit forms a three-helix bundle, which is consistent with an NMR structure obtained with a shorter version of p60N33. The p80C subunit in turn consists of seven helices that adopt an extended helix-turn-helix motif. The interaction between p60N and p80C is mediated by both hydrophobic and polar amino acid residues located in the H1 helix of p80C and helices H1 and H3 of p60N (Fig. 4b). Inspection of the p60N/p80C/ASPMp complex structure in combination with co-immunoprecipitation, sedimentation velocity experiments and circular dichroism spectroscopy suggested that the three microcephaly-associated mutations located within p80C, S538L, L543R and G581D18, affected the stability of the protein, but not its capacity to bind to p60N (Fig. 4c,d; Supplementary Fig. 3b).
Figure 4. Structure of the p60N/p80C/ASPMp complex.
(a) Ribbon representation of the p60N/p80C/ASPMp complex structure and close-up view of the ASPMp binding site with interacting residues shown in stick representation. The position of the three microcephaly-associated mutations in the p80C subunit, S538L, L543R and G581D, are highlighted in grey.
(b) Close-up view of the hydrophobic interactions between p60N (shown as electrostatic surface representation; positively-, negatively- and hydrophobic residues are depicted in blue, red and grey, respectively) and helix H1 of p80C (shown as a ribbon representation with hydrophobic residues in stick representation).
(c) Normalized thermal unfolding profiles of p60N/p80C complex variants assessed by circular dichroism spectroscopy. All mutants display stabilities comparable to the wild-type protein except for the S538L mutant (red), which is 6 °C less stable. We could not purify the L543R and G581D mutants from E. coli, most likely because these mutations affect the fold and/or stability of the p60N/p80C complex.
(d) Continuous sedimentation coefficient distribution profiles c(S) of p60N/p80C showing that mutations p60N-L18A, p60N-L19A, p80C-Y574A and p80C-R615A did not affect p60N/p80C heterodimer formation. The microcephaly-associated mutation S538L (red) did show signs of aggregation.
(e) Binding curves obtained from sedimentation velocity experiments for the interaction between the TAMRA-labeled ASPM peptide and p60N/p80C. The ASPM peptide interacts with p60N/p80C with an apparent dissociation constant, Kd, of 9 μM. The individual substitutions F352A of ASPM, L18A or L19A of p60N, or S538L or Y574A of p80C abolished the interactions between the TAMRA-ASPM peptide and p60N/p80C.
(f) GST-p80C/p60N pull down assays with lysates of HEK293T cells expressing either the WT ASPM peptide repeats or their mutated versions, in which the conserved phenylalanine residue (red arrowhead in Fig. 3f), was substituted for alanine (FA).
(g) GST-p80C/p60N pull down assays with lysates of HEK293T cells expressing either the WT full length ASPM or the F302A/F377A mutant, in which the conserved phenylalanine residues of repeats 1 and 4 of ASPM have been simultaneously mutated to alanines. Unprocessed original scans of blots are shown in Supplementary Figure 8.
In the p60N/p80C/ASPMp complex structure, the ASPMp peptide bound to the site formed at the interface between p60N and p80C, a result that explains why both subunits are needed for tripartite complex formation (Fig. 4a). Residues S351, F352 and L353 of ASPMp were clearly visible in the electron density of the structure (Fig. 4a; Supplementary Fig. 3c). We found that F352 of ASPMp inserts into a hydrophobic pocket formed by L18 and L19 of p60N, and by Y574 of p80C. Individual substitutions of F352 of ASPMp, L18 or L19 of p60N, or Y574 of p80C to alanine abolished the binding between ASPMp and the p60N/p80C heterodimer although they had no effect on the formation and stability of the p60N/p80C heterodimer (Fig. 4c-e). These residues thus have critical roles for tripartite complex formation. The microcephaly-associated p80 S538L substitution also blocked the binding of ASPMp (Fig. 4e), suggesting that the loss of ASPM association might partially underlie the microcephaly phenotype. Finally, mutation of the conserved phenylalanine residues in the first and fourth repeat of ASPM (F302A/F377A) strongly reduced the binding between full-length ASPM and katanin (Fig. 4f,g), demonstrating that tandem ASPM repeats cooperate in katanin binding. Taken together, these results demonstrate that formation of the ASPM/katanin complex depends on residues located at the interface of the p60/p80 heterodimer and on conserved residues in the linear motifs of ASPM.
ASPM associates with microtubule minus-ends in vitro and in cells
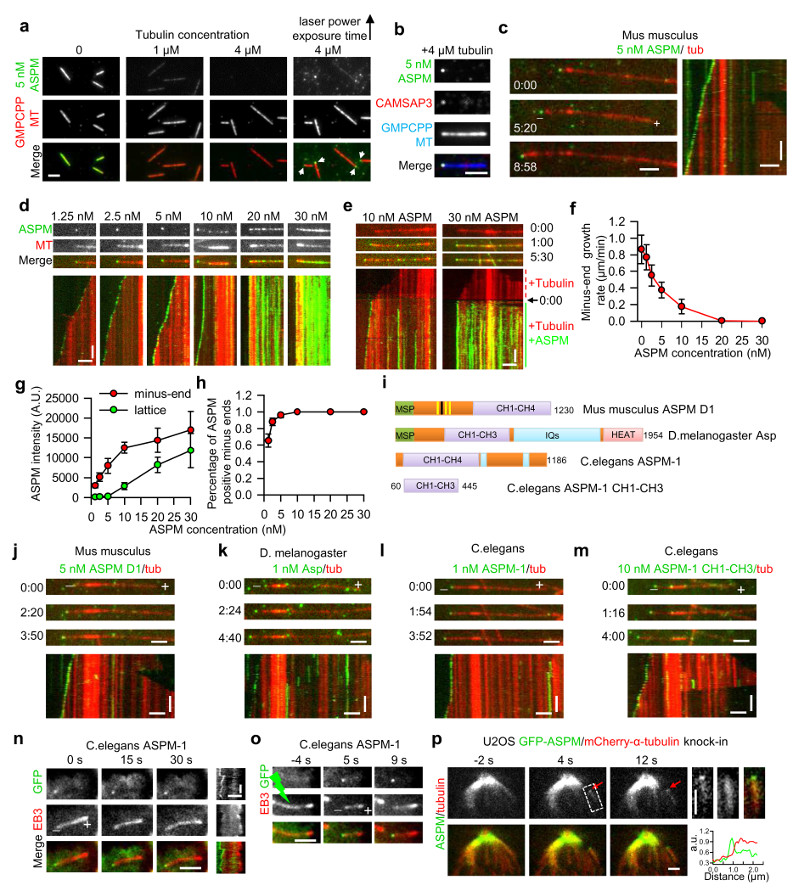
To understand how ASPM and katanin work together, we set out to reconstitute their activities in vitro. To this end, we purified the short isoform of mouse ASPM in complex with calmodulin from overexpressing HEK293T cells (GFP-ASPM; Fig. 3a and Supplementary Fig. 7a). ASPM decorated GMPCPP-stabilized microtubules in the absence of free tubulin; however, when tubulin was added even at a concentration insufficient to induce polymerization, ASPM preferentially localized to microtubule minus ends, as determined by co-localization with the microtubule minus-end marker CAMSAP334 (Fig. 5a,b). At tubulin concentrations supporting microtubule polymerization, ASPM strongly accumulated at growing microtubule minus ends and slowed down their growth. This effect was more pronounced at higher ASPM concentrations, when both the enrichment of ASPM and its binding frequency at the minus ends were higher (Fig. 5c-h). At ASPM concentrations above 20 nM, a complete block of minus-end outgrowth was observed (Fig. 5d-f); at such higher concentrations, ASPM also regained microtubule lattice binding (Fig.5d,e,g).
Figure 5. ASPM associates with microtubule minus-ends in vitro and in cells.
(a) Images of mouse GFP-ASPM (green) binding to stable GMPCPP-stabilized microtubules (red) in the presence of tubulin at different concentrations. White arrowheads indicate ASPM specifically associated with one microtubule end. Scale bar, 2 μm.
(b) Images showing colocalization of GFP-ASPM (green) and SNAP-Alexa647-CAMSAP3 (red) at the minus end of a GMPCPP-stabilized microtubule (blue). Scale bar, 2 μm.
(c, d, j-m) Images and corresponding kymographs of GFP-full length ASPM (c,d), ASPM D1 (j), fly Asp (k), worm ASPM-1 full lenth (l) and CH1-CH3 (m) (green) tracking dynamic microtubule minus ends (red). Scale bars: horizontal, 2 μm; vertical, 1 min.
(e) Images and corresponding kymographs of GFP-tagged full length ASPM tracking (10 nM) or blocking (30 nM) dynamic microtubule minus ends (red) in a flow-in experiment. 0:00, the moment of flow-in. Scale bars: horizontal, 2 μm; vertical, 1 min.
(f) Quantification of minus-end growth rate in the presence of ASPM at different concentrations. n = 49, 41, 37, 29, 28, 30 and 30 microtubules for ASPM concentration from 0-30 nM,
(g,h) ASPM intensity on microtubule minus ends and lattice, and percentage of ASPM-positive minus ends; At ASPM concentration from 1.25-30 nM, n=22, 12, 11, 12, 13 and 13 microtubules (end intensity and accumulation frequency measurement); n=20, 20, 19, 15, 19 and 20 microtubules (lattice intensity). Minus-end intensity of ASPM was averaged only over the time points at which ASPM accumulation was present.
(i) A scheme of the domain organization of mouse ASPM D1, fly Asp and worm ASPM-1.
(n,o) Full length worm ASPM-1 associates with minus ends of free microtubules (n) or minus ends freshly generated by photoablation (o) in interphase MRC5 cells. Green lightning bolt indicates the site of photoablation. Scale bars, horizontal, 2 μm; vertical, 10 s.
(p) Images of a GFP-ASPM and mCherry-α-tubulin double knock-in cell before and after photoablation. Enlarged images at 4 s and the corresponding line scan of the two channels are shown on the right. Scale bars, 2 μm.
Time represented as min:sec. Data represent mean ± SD. Source data for panels f,g and h can be found in Supplementary Table 5.
To assess which parts of ASPM confer microtubule minus-end binding specificity, we performed deletion mapping of the mouse protein and found that its minus-end binding does not require the C-terminal HEAT- and IQ repeats (Fig. 5i,j; Supplementary Fig. 7a); shorter fragments could not be assayed due to their propensity to aggregate. Fly Asp and worm ASPM-1 also accumulated at growing microtubule minus-ends in vitro (Fig. 5k,l; Supplementary Fig. 7a), and we found that the first three conserved CH domains (CH1-CH3) of worm ASPM-1 were sufficient for this activity (Fig. 5m; Supplementary Fig. 7a). Mass spectrometry analysis of different ASPM preparations revealed no other microtubule-associated proteins (MAPs, Supplementary Table 4), supporting the conclusion that minus-end tracking is an autonomous activity of ASPM.
The specific minus-end binding of worm ASPM-1 and its CH1-CH3 fragment was confirmed in interphase mammalian cells (Fig. 5n, o; Supplementary Fig. 4a,b). Mouse ASPM could not be analyzed in a similar way because it strongly accumulated in the nucleus during interphase. However, spindle microtubule photoablation experiments showed that besides weak lattice binding, the endogenous GFP-tagged ASPM localized to freshly generated minus-ends within 7.0±2.4 s after ablation, and moved to the spindle pole with the speed of 3.1±1.4 μm/min (mean±SD, n=13, Fig. 5p). This dynamic behavior is similar to that of the NuMA-dynein complex after photoablation of spindle microtubules28,29. We conclude that ASPM can autonomously localize to microtubule minus ends and that this activity is evolutionarily conserved. Furthermore, at high concentrations ASPM can completely block minus-end growth and associate with microtubule lattice.
The katanin p60/p80 heterodimer binds and bends microtubule ends in cells and in vitro
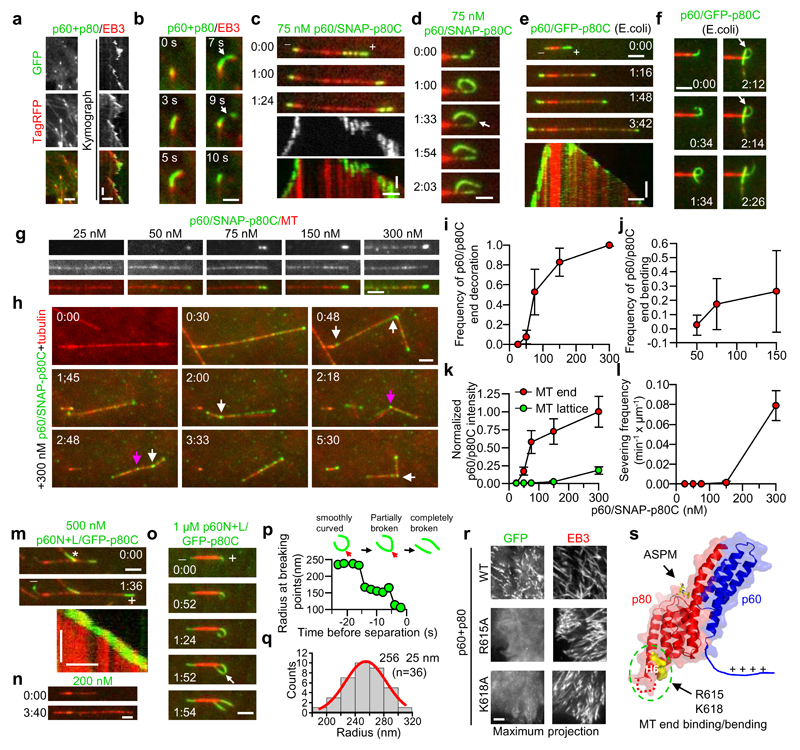
Next, we investigated whether katanin can also interact with microtubule ends. In cells, overexpressed p60/p80 frequently localized to growing microtubule tips labeled with the plus-end marker EB3 and caused their bending and breakage (Fig. 6a, b; Supplementary Video 2). This microtubule end localization required simultaneous expression of the p60 subunit and the p80 C-terminus, but was independent of the WD40 domain of p80 (Supplementary Fig. 5a-c). In vitro, the p60/p80C complex purified from HEK293T cells (Supplementary Fig. 7b) readily severed stabilized microtubules at 75 nM (see below), but, similar to p60 alone in vitro26, this activity was strongly inhibited in the presence of free tubulin. However and in agreement with cellular data, 75 nM purified p60/p80C heterodimer could track, decorate, bend and break growing microtubule plus- and minus ends (Fig. 6c,d; see Fig. 6i,j for end binding and bending frequency). These activities were not mediated by additional factors that were co-purified from HEK293T cells, as we detected no other MAPs in our purifications (Supplementary Table 4). Furthermore, the p60/p80C complex purified from E. coli (Supplementary Fig. 7b) also showed robust microtubule end-binding, -bending and -breaking activities (Fig. 6e,f).
Figure 6. The katanin p60/p80 complex binds and bends microtubule ends in cells and in vitro.
(a) Images and kymographs of the colocalization of GFP-p60/p80 and EB3-TagRFP in MRC5 cells. Scale bars: horizontal, 2 μm; vertical, 10 sec.
(b) Images of a single microtubule plus end bound to GFP-p60/p80 and EB3-TagRFP. White arrow, breaking of a bent microtubule end. Scale bar, 1 μm.
(c-f, m-o) Images and corresponding kymographs illustrating the behavior of p60/SNAP-Alexa647-p80C (c,d), p60/GFP-p80C (e,f, purified from E. coli) and p60N+L/ GFP-p80C (m-o) on dynamic microtubule ends in vitro; white arrow, breaking of a bent microtubule. Scale bars: horizontal, 2 μm; vertical, 1 min. White asterisk in panel m denotes the signal from another microtubule.
(g) Images of p60/SNAP-Alexa647-p80C localizing to dynamic microtubule plus ends and lattices at different concentrations. Scale bar, 2 μm.
(h) Images of p60/SNAP-Alexa647-p80C severing dynamic microtubules at 300 nM in a flow-in experiment. White and purple arrows indicate complete and incomplete severing events, respectively. The misalignment between different channels was the result of sequential imaging of freely moving microtubules. Scale bar, 2 μm.
(i-j) Frequency of p60/p80C accumulation and bending of growing plus ends at different concentrations. At 300 nM, end bending could not be reliably measured due to movement of severed microtubules. i, n=22, 23, 24, 20 and 19 microtubules (25-300 nM); j, n=23, 24 and 21 microtubules (50-150 nM).
(k) Intensities of p60/p80C on microtubule ends and lattices at different concentrations. n=100, 101, 110, 100 and 100 microtubules (25-300 nM).
(l) Severing activity of p60/p80C on dynamic microtubules at different concentrations. n=2 (25-75 nM) or 3 (150-300 nM) experiments.
(p) Quantification of the radius of p60N+L/p80C decorated microtubule during a bending and breaking event.
(q) Histogram of radii of microtubule curvature induced by p60N+L/p80C; n=36 microtubules.
(r) Maximum projection of 100 sec time lapse movies of indicated GFP-p60/p80 variants co-expressed with EB3-TagRFP in HeLa cells. Scale bar, 2 μm.
(s) Structural illustration of the positions of key residues of p60N/p80C required for microtubule end decoration.
Time in min:sec. Data represent mean ± SD. Source data for panels i, j, k, l and q can be found in Supplementary Table 5.
To get further insight into the end binding and severing activities of katanin, different concentrations of p60/p80C together with 20 μM tubulin were flowed into the reaction chamber containing pre-assembled dynamic microtubules (Fig. 6g-h; Supplementary Fig. 5d). The intensity of katanin and its binding frequency to microtubule ends had a sigmoid-like response at 50-300 nM concentration (Fig. 6i,k), suggesting cooperative binding. In contrast, p60/p80C concentrations ≥300 nM were needed to observe katanin binding along pre-existing microtubules and microtubule severing (Fig. 6h,l, white arrows; Supplementary Video 3). Complete severing within one minute was observed at 750 nM p60/p80C (Supplementary Fig. 5d). Strikingly, no microtubule severing was observed in the vicinity of the ends, although the p60/p80C intensity was much higher near the ends than along microtubules. This result suggests that the end-decorating pool of katanin is much less potent, if not incapable, of microtubule severing compared to its lattice pool.
While the p60N/p80C heterodimer did not bind to microtubule ends, the p60N+L/p80C complex (Supplementary Fig. 7b), which includes the positively charged linker (L) adjacent to p60N, but lacks the ATPase domain, displayed robust microtubule end-tracking, -bending and -breaking activity when present at concentrations of ≥0.5 µM (Fig. 6m-o; Supplementary Video 4). The ATPase domain of p60 is thus not essential for microtubule end decoration and breaking, but likely contributes to this activity by directly interacting with microtubules or by promoting katanin oligomerization. The radius of p60N+L/p80C decorated and bent microtubule ends before breakage was 256 ± 25 nm (mean ±SD, n=36; Fig. 6p,q), which corresponds to a 1.8±0.2° angle between two longitudinally arranged tubulin dimers. This degree of curvature is significantly smaller than that of free tubulin or tubulin in complex with stathmin35, but similar to the curvature of tubulin sheets at growing microtubule ends36, suggesting a plausible growing end recognition mechanism.
Based on the crystal structure of the p60N/p80C/ASPMp complex, we identified positively charged surface-exposed residue patches on p60N/p80C, which might bind to the negatively charged microtubule lattice. Mutation analysis showed that two highly conserved residues, R615 and K618 in helix H6 of p80C, but not any of the other arginine or lysine residues used as controls were essential for microtubule end binding in cells (Fig. 6r,s; Supplementary Fig. 5e-g). The highly conserved four-residue segment 607-610 located in the loop connecting helices H5 and H6 was also required for the interaction with microtubule ends (Supplementary Fig. 5f,g). The N-terminal part of p60 and the C-terminal part of p80 thus form a microtubule-binding module that recognizes and bends microtubule ends through a combination of a conserved positively charged surface patch at the tip of p80 and a positively charged linker present in the p60 subunit (Fig. 6s).
The ASPM/katanin complex severs microtubules and blocks minus-end growth in vitro
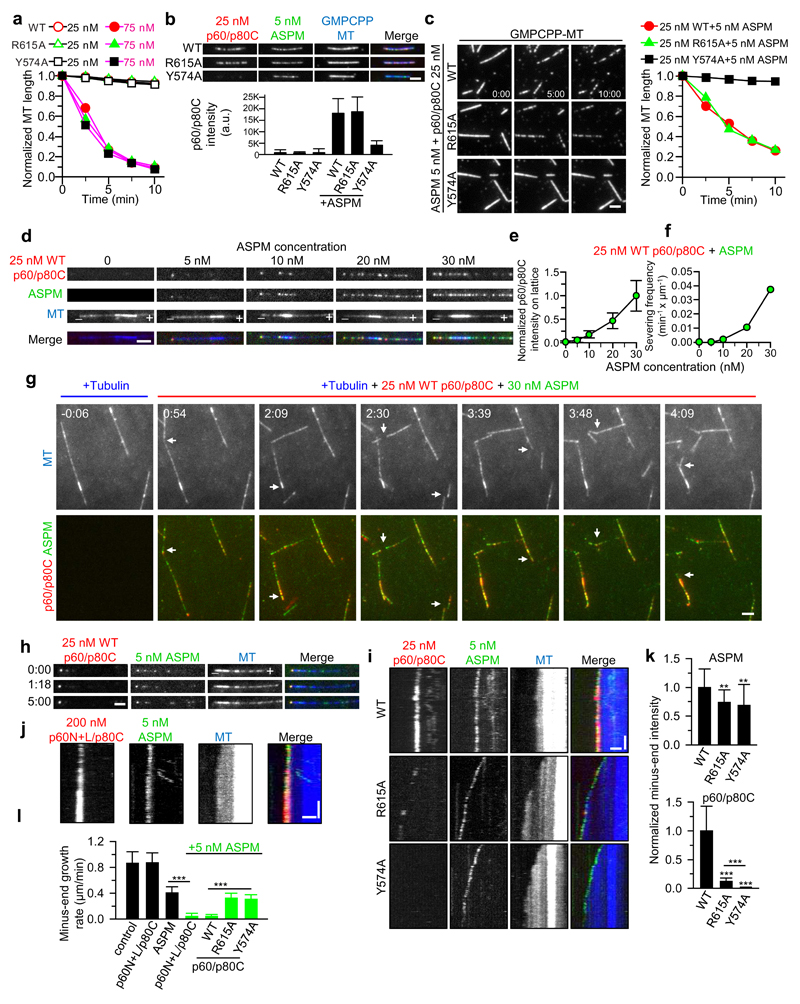
Having established that both ASPM and the p60/p80C heterodimer specifically interact with microtubule ends at low concentration and associate with microtubule lattice at high concentration, we next tested their combined activities using either wild type p60/p80C or the two p80C mutants, R615A and Y574A, which are deficient in binding to dynamic microtubule ends or ASPM, respectively (Supplementary Fig. 6a,b; Fig. 4e). In the absence of free tubulin, all three p60/p80C variants could efficiently sever microtubules when present at concentrations of ≥75 nM but not at 25 nM (Fig. 7a; Supplementary Fig. 6c). Upon the addition of 5 nM ASPM, both the wild type and the R615A p60/p80C variants, but not the ASPM binding-deficient Y574A mutant, displayed strong microtubule recruitment and severing at 25 nM concentration (Fig. 7b,c). ASPM can thus enhance the activity of katanin by recruiting it to microtubules. Under these conditions, the R615A mutant could sever microtubules as robustly as wild type p60/p80C, again suggesting that the microtubule end binding and severing activities of katanin are biochemically distinct.
Figure 7. The ASPM/katanin complex severs microtubules and blocks minus-end growth in vitro.
(a) Quantification of microtubule severing activity of 25 nM (black lines) and 75 nM (purple lines) p60/SNAP-Alexa647-p80C (wild type (WT) and mutants). For microtubule images, see Supplementary Fig. 6c. Data are the mean of 2 experiments.
(b) Images and quantification of the recruitment of 25 nM p60/SNAP-Alexa647-p80C (red) to GMPCPP-stabilized microtubules (blue) in the presence of 5 nM GFP-ASPM (green). From left to right, n=47, 47, 49, 49, 45 and 52 microtubules.
(c) Images and quantification of microtubule severing activity of 25 nM p60/SNAP-Alexa647-p80C (WT and mutants) in the presence of 5 nM GFP-ASPM. Data are the mean of 2 experiments
(d) Images showing recruitment of WT p60/SNAP-Alexa647-p80C onto dynamic microtubule minus ends and lattices by GFP-ASPM at different concentrations.
(e) Quantification of microtubule lattice intensity of WT p60/SNAP-Alexa647-p80C at different ASPM concentrations. From left to right, n=65, 63, 71, 65 and 64 microtubules.
(f) Quantification of severing activity of WT p60/SNAP-Alexa647-p80C at different ASPM concentrations. Data are the mean of 2 experiments.
(g) TIRFM time lapse images of severing of dynamic microtubules by 25 nM WT p60/SNAP-Alexa647-p80C and 30 nM GFP-ASPM. White arrows indicate severing events.
(h) Images showing recruitment of WT p60/SNAP-Alexa647-p80C to dynamic microtubule minus ends by 5 nM GFP-ASPM.
(i,j) Kymographs showing localization of GFP-ASPM and WT p60/SNAP-Alexa647-p80C, R615A and Y574A mutants (i) or p60N+L/SNAP-Alexa647-p80C (j) to dynamic microtubule minus ends. Vertical scale bar, 1 min.
(k) Quantification of ASPM and p60/SNAP-Alexa647-p80C intensities on minus ends. From left to right, ASPM, n=32, 19 and 19 microtubule; p60/p80C, n=33, 19 and 19 microtubules.
(l) Quantification of minus-end growth rate in conditions with 200 nM p60N+L/p80C alone, 5 nM ASPM alone, and 5 nM ASPM together with 200 nM p60N+L/p80C or 25 nM p60/p80C (WT, R615A and Y574A). From left to right, n=49, 41, 38, 55, 58, 96, 84 microtubules. The data for control were re-plotted from Fig. 5f.
Horizontal scale bars in all panels, 2 μm. Data represent mean ± SD. **, P<0.01; ***, P<0.001, Mann-Whitney U test. Time in min:sec. Source data for panels a, b, c, e, f, k and l can be found in Supplementary Table 5.
We then asked whether ASPM can promote microtubule severing by katanin in the presence of free tubulin and found that while 25 nM p60/p80C alone did not bind or sever dynamic microtubules, increasing concentrations of ASPM caused its recruitment to microtubules and their severing (Fig. 7d-g; Supplementary Video 5). Upon the addition of 30 nM ASPM, 25 nM p60/p80C displayed a severing activity that was approximately half of that observed with 300 nM p60/p80C alone (Fig. 6l and 7f), which is close to an order of magnitude of improvement. The R615A but not the Y574A mutant of p60/p80C was able to sever dynamic microtubules at 25 nM concentration in the presence of 30 nM wild type ASPM (Supplementary Fig. 6d-g), while no increase in severing was observed with 30 nM ASPM F302A/F377A, which is deficient in katanin binding (Supplementary Fig. 6h-j). These data demonstrate that katanin-mediated microtubule severing is promoted by the direct interaction with ASPM but does not require katanin’s microtubule end-binding activity.
Next, we investigated whether katanin can enhance the minus-end blocking by ASPM. 5 nM wild type ASPM but not the F302A/F377A mutant that is deficient in katanin binding, was sufficient to recruit 25 nM p60/p80C onto microtubule minus ends (Fig. 7d; Supplementary Fig. 6k-m), and this coincided with a dramatic suppression of their growth (Fig. 7h,i,k,l). In contrast, the R615A and the Y574A mutants showed only weak or no ASPM-mediated minus-end binding activity, respectively, and neither of the mutants could block minus-end elongation (Fig. 7i,k,l). The intensity of the ASPM signal at microtubule minus ends was higher in the presence of wild type p60/p80C than with the two mutant heterodimers (Fig. 7k), explaining the strong inhibition of minus-end growth. ASPM and katanin thus enhance each other’s minus-end accumulation, in line with their mutual interdependence for spindle pole localization (Fig. 1d-f).
Similarly, 200 nM p60N+L/p80C heterodimer could be recruited to microtubule minus ends by 5 nM ASPM and caused minus-end growth inhibition (Fig. 7j,l). However, by itself this truncated version of katanin, which lacks the ATPase domain, did not localize to microtubule ends at 200 nM (Fig. 6n). Taken together, these in vitro data indicate that ASPM and katanin mutually stimulate each other’s activities: ASPM enhances the microtubule lattice severing activity of katanin; conversely, katanin promotes the ASPM-dependent blockage of microtubule minus-end growth.
Dissociation of katanin from ASPM impairs spindle orientation and poleward flux
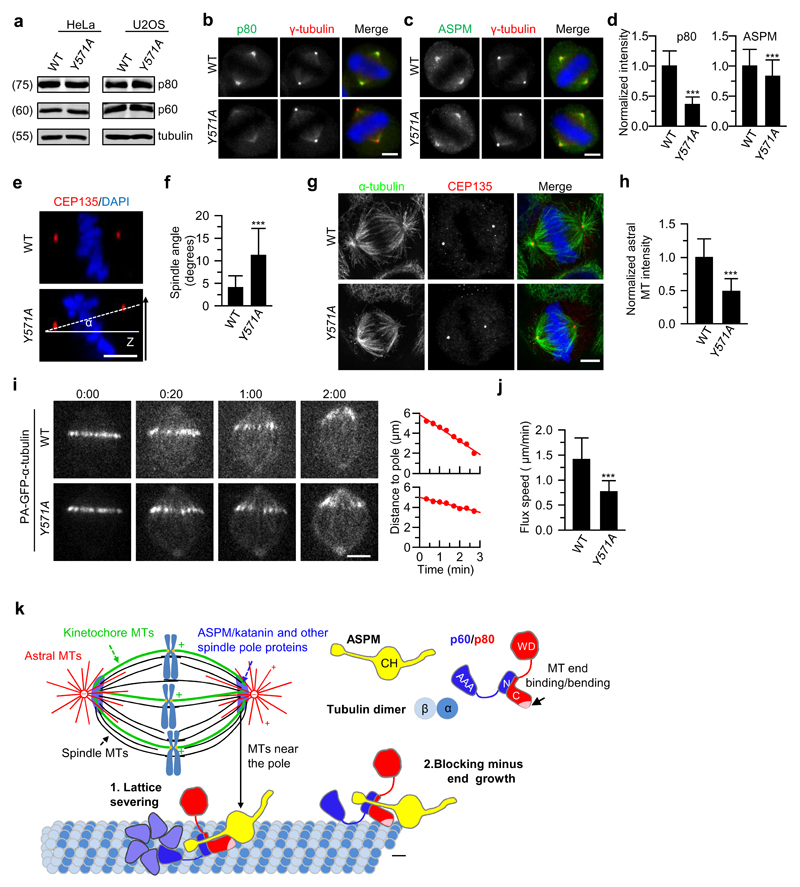
Finally, to investigate the physiological role of the interaction between ASPM and katanin, we introduced in HeLa and U2OS cells a point mutation corresponding to the mouse p80 Y574A (Y571A in human) using CRISPR/Cas9 technology. The protein levels of p60 and p80 were comparable in wild type and p80Y571A cells (Fig. 8a), which is consistent with our biophysical data showing that this mutation did not affect formation of a stable p60/p80 heterodimer (Fig. 4c,d). In line with the biochemical data and in vitro reconstitution assays, the intensity of spindle pole localization of katanin p80 Y571A was reduced to a level similar to that of the ASPM knockout (Fig. 8b,d). We also observed a decrease in ASPM intensity at the spindle pole in p80Y571A cells (Fig. 8c,d), confirming the mutual dependence of ASPM and katanin for pole localization. Furthermore, p80Y571A cells displayed defects in spindle orientation, astral microtubule density and poleward microtubule flux, similar to the ASPM knockout (Fig. 8e-j). Together, these data fully support our conclusion that the ASPM/katanin complex plays an important role in regulating microtubule dynamics at spindle poles.
Figure 8. Dissociation of katanin from ASPM impairs spindle orientation and poleward flux.
(a) Western blotting with the indicated antibodies of extracts of wild type (WT) and p80Y571A HeLa and U2OS cells. Katanin protein levels in WT and p80Y571A mutant cell lines are similar.
(b-d) Immunofluorescence staining and quantification of p80 and ASPM intensities at spindle poles in WT and p80Y571A HeLa cells. For p80 intensity, n=204 spindle poles, WT; n=220, p80Y571A; for ASPM intensity, n = 244, WT; n=240, p80Y571A. Scale bars, 5 μm.
(e,f) Orthogonal view (x-z) of metaphase HeLa cells stained for CEP135 and DAPI in WT, p80Y571A HeLa cells (e) and quantification of spindle angles (f; n=40 cells, WT and p80Y571A). Scale bar, 5 μm.
(g) Immunostaining for α-tubulin, CEP135 and DNA (DAPI) in WT and p80Y571A HeLa cells. Maximum intensity projections of Z series with ~40 stacks and 200 nm step were shown here. Scale bar, 5 μm.
(h) Quantification of astral microtubule intensity shown in g. n=39 (WT) and 41 (p80Y571A) cells. Intensity was normalized to the average value in the WT cells.
(i) Live cell images of spindle flux in WT and p80Y571A U2OS cells stably expressing PA-GFP-α-tubulin. Time displayed as min:sec. Plots of the distance between photoactivated GFP stripe and the spindle pole over time are shown on the right. The flux speed was derived from the plot slopes. Scale bar, 5 μm.
(j) Flux speed in WT and p80Y571A U2OS cells. n=24 (WT) and 28 (p80Y571A) cells.
(k) Model for regulation of microtubule minus-end dynamics at spindle poles by the ASPM/katanin complex. (1) Katanin recruited to microtubule lattice by ASPM can oligomerize through the p60 ATPase domain and sever microtubules. (2) Minus-end binding of the ASPM/katanin complex promotes its accumulation near the pole. Through its end-binding activity, katanin enhances blocking of microtubule minus-end growth by ASPM, which likely primes the minus-ends for depolymerization by depolymerases.
Data represent mean ± SD. ***, P<0.001, Mann-Whitney U test. Unprocessed original scans of blots are shown in Supplementary Figure 8. Source data for panels d, f, h and j can be found in Supplementary Table 5.
Discussion
The results of this study demonstrate that in mammalian cells, ASPM and katanin form a physiological complex that plays a key role in regulating spindle poles. They uncover the structural basis of the formation of the katanin p60/p80 heterodimer and its interaction with ASPM. The data further provide insight into selected mutations associated with microcephaly by showing that disease-relevant residue substitutions in the p80 subunit of katanin interfere with the activity of the protein by either destabilizing its fold or by disrupting the binding capacity of katanin for ASPM. Most importantly, our work reveals that the ASPM/katanin complex acts on both microtubule minus ends and the lattice, and shows that the combined activities of the two proteins can regulate microtubule dynamics at spindle poles.
We propose that ASPM and katanin can cooperate in two ways (Fig. 8k). First, due to its high local concentration at the poles resulting from preferential minus-end association, the ASPM/katanin complex could bind to and sever microtubules and thus directly contribute to microtubule disassembly at the poles. Second, ASPM can localize to the outmost microtubule minus ends and prevent their growth. The minus-end growth inhibiting activity of ASPM can be potentiated by the p60N+L/p80C module of katanin. Blocking minus-end growth in turn might make microtubules better substrates for depolymerases of the kinesin-13 family, which are involved in driving spindle flux by disassembling microtubule minus ends32,37–39. Along these lines, the kinesin-13 MCAK can efficiently accumulate at the ends of stable microtubules40, but requires additional factors to bind and destabilize the ends of growing microtubules41,42.
The demonstration that ASPM and katanin are microtubule end-binding proteins helps to understand their cellular activities. The function of fly Asp in centrosome-pole attachment, for example, might be explained by its dual affinity for microtubule minus ends and centrosomes12, and worm ASPM-1 might rely on its ability to recognize microtubule minus ends to target the dynein-NuMA complex to spindle poles7. The R233Q mutation in the C. elegans p80 homologue, corresponding to our rationally introduced R615A mutation in mouse p80, has a synthetic interaction with the P99L mutant of the p60 homologue in controlling meiotic spindle length24, supporting the in vivo significance of the end-binding and -bending activity of katanin. These biochemical properties, which are distinct from microtubule severing, might also be the basis of katanin’s role in regulating microtubule plus-end dynamics in interphase34,43.
It has been hypothesized that during embryonic development, mitotic defects in neural precursors lead to microcephaly44. Here, we showed that ASPM and katanin knockout cells, as well as cells expressing a point mutant of katanin that cannot bind to ASPM, have defects in spindle orientation. It is possible that this abnormality is one of the factors contributing to perturbed cortical development. To conclude, our results connect two proteins known to be involved in microcephaly into the same biochemical pathway and suggest that proper regulation of microtubule minus ends during cell division is a critical factor for human brain development.
Supplementary Material
Acknowledgements
We thank A. Prota for the help in refining the crystal structures, M. Boxem, C. Janke and M. Mikhaylova for the gift of materials, and the beamline scientists at beamlines X06DA of the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland) for technical assistance with the X-ray data collection. This work was supported by the European Research Council Synergy grant 609822 and Netherlands Organization for Scientific Research (NWO) CW ECHO grant (711.011.005) to A.A., the EMBO long-term and Marie Curie IEF fellowships to L.R. and grants from the Swiss National Science Foundation (31003A_166608 to M.O.S. and 31003A_163449 to R.A.K.), a NWO VIDI grant (723.012.102) to A.F.M.A and as part of the National Roadmap Large-scale Research Facilities of the Netherlands (project number 184.032.201) to A.F.M.A. and A.J.R.H. The structural data reported in this paper are available in PDB (PDB code 5LB7).
Footnotes
Author Contributions
K.J, L.R, S.H., R.A.K, M.O.S. and A.A. designed experiments, analyzed data and wrote the paper. A.A. coordinated the project. K.J and S.H performed cellular and in vitro reconstitution experiments, L.R. performed biophysical experiments, L.R and G.C performed crystallography experiments, Q.L., A.F.M.A and A.J.R.H performed and analyzed mass spectrometry experiments.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Morris-Rosendahl DJ, Kaindl AM. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH) Mol Cell Probes. 2015;29:271–281. doi: 10.1016/j.mcp.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–624. doi: 10.1016/s0960-9822(03)00530-x. [DOI] [PubMed] [Google Scholar]
- 3.Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- 4.Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakefield JG, Bonaccorsi S, Gatti M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol. 2001;153:637–648. doi: 10.1083/jcb.153.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Voet M, et al. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- 8.Higgins J, et al. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito A, Goshima G. Microcephaly protein Asp focuses the minus ends of spindle microtubules at the pole and within the spindle. J Cell Biol. 2015;211:999–1009. doi: 10.1083/jcb.201507001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoborg T, Zajac AL, Fagerstrom CJ, Guillen RX, Rusan NM. An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J Cell Biol. 2015;211:987–998. doi: 10.1083/jcb.201509054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripoll P, Pimpinelli S, Valdivia MM, Avila J. A cell division mutant of Drosophila with a functionally abnormal spindle. Cell. 1985;41:907–912. doi: 10.1016/s0092-8674(85)80071-4. [DOI] [PubMed] [Google Scholar]
- 12.do Carmo Avides M, Glover DM. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 1999;283:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]
- 13.Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol Biol Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 15.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 17.Hartman JJ, et al. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 18.Mishra-Gorur K, et al. Mutations in KATNB1 Cause Complex Cerebral Malformations by Disrupting Asymmetrically Dividing Neural Progenitors. Neuron. 2014;84:1226–1239. doi: 10.1016/j.neuron.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu WF, et al. Katanin p80 Regulates Human Cortical Development by Limiting Centriole and Cilia Number. Neuron. 2014;84:1240–1257. doi: 10.1016/j.neuron.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNally KP, Bazirgan OA, McNally FJ. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J Cell Sci. 2000;113(Pt 9):1623–1633. doi: 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- 21.Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 22.McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNally KP, McNally FJ. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol Biol Cell. 2011;22:1550–1560. doi: 10.1091/mbc.E10-12-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNally K, et al. Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol Biol Cell. 2014;25:1037–1049. doi: 10.1091/mbc.E13-12-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;147:1397–1407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey ME, Sackett DL, Ross JL. Katanin Severing and Binding Microtubules Are Inhibited by Tubulin Carboxy Tails. Biophys J. 2015;109:2546–2561. doi: 10.1016/j.bpj.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung K, et al. Proteomic Analysis of the Mammalian Katanin Family of Microtubule-severing Enzymes Defines KATNBL1 as a Regulator of Mammalian Katanin Microtubule-severing. Mol Cell Proteomics. 2016 doi: 10.1074/mcp.M115.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elting MW, Hueschen CL, Udy DB, Dumont S. Force on spindle microtubule minus ends moves chromosomes. J Cell Biol. 2014;206:245–256. doi: 10.1083/jcb.201401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikirzhytski V, et al. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J Cell Biol. 2014;206:231–243. doi: 10.1083/jcb.201401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gai M, et al. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep. 2016;17:1396–1409. doi: 10.15252/embr.201541823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the "Pacman-flux" machinery that moves chromosomes. J Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 33.Iwaya N, et al. A common substrate recognition mode conserved between katanin p60 and VPS4 governs microtubule severing and membrane skeleton reorganization. J Biol Chem. 2010;285:16822–16829. doi: 10.1074/jbc.M110.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang K, et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev Cell. 2014;28:295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Brouhard GJ, Rice LM. The contribution of alphabeta-tubulin curvature to microtubule dynamics. J Cell Biol. 2014;207:323–334. doi: 10.1083/jcb.201407095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janosi IM, Chretien D, Flyvbjerg H. Modeling elastic properties of microtubule tips and walls. Eur Biophys J. 1998;27:501–513. doi: 10.1007/s002490050160. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM. Patronin mediates a switch from kinesin-13-dependent poleward flux to anaphase B spindle elongation. J Cell Biol. 2013;203:35–46. doi: 10.1083/jcb.201306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers GC, et al. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 39.Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 41.Montenegro Gouveia S, et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Tanenbaum ME, et al. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 43.Zhang D, et al. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat Cell Biol. 2011;13:361–370. doi: 10.1038/ncb2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taverna E, Gotz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.