Abstract
Relevant mouse models of E2a–PBX1-induced pre-B cell leukemia are still elusive. We now report the generation of a pre-B leukemia model using E2a–PBX1 transgenic mice, which lack mature and precursor T-cells as a result of engineered loss of CD3ε expression (CD3ε–/–). Using insertional mutagenesis and inverse-PCR, we show that B-cell leukemia development in the E2a–PBX1 × CD3ε–/– compound transgenic animals is significantly accelerated when compared to control littermates, and document several known and novel integrations in these tumors. Of all common integration sites, a small region of 19 kb in the Hoxa gene locus, mostly between Hoxa6 and Hoxa10, represented 18% of all integrations in the E2a–PBX1 B-cell leukemia and was targeted in 86% of these leukemias compared to 17% in control tumors. Q-PCR assessment of expression levels for most Hoxa cluster genes in these tumors revealed an unprecedented impact of the proviral integrations on Hoxa gene expression, with tumors having one to seven different Hoxa genes overexpressed at levels up to 6600-fold above control values. Together our studies set the stage for modeling E2a–PBX1-induced B-cell leukemia and shed new light on the complexity pertaining to Hox gene regulation. In addition, our results show that the Hoxa gene cluster is preferentially targeted in E2a–PBX1-induced tumors, thus suggesting functional collaboration between these oncogenes in pre-B-cell tumors.
Keywords: E2a–PBX, B-cell leukemia model, homeobox, oncogenic collaborators, transgenics
Leukemias are often induced by translocations that deregulate the expression of genes involved in the regulatory network underlying hematopoiesis (Rabbitts 1994; Look 1997). The mode of action of the resultant fusion proteins to interfere with normal genetic programs is poorly understood. Several types of leukemias have been modeled in mice by genetic engineering, providing a powerful tool to identify oncogenic complements (Berns et al. 1999). The translocation t(1;19) occurs in 5% of all childhood acute lymphoblastic leukemias (ALL). Those ALLs are typically classified as pre-B ALLs since they express cytoplasmic but not surface immunoglobin. The specific association of t(1;19) with pediatric pre-B ALL (25% of all cases) makes it an attractive disease to model (Carroll et al. 1984; Williams et al. 1984). As a result of this translocation, two transactivation domains in E2A are fused to the C-terminal portion of PBX1 (Kamps et al. 1990; Nourse et al. 1990). In gene transfer studies the fusion protein E2a–PBX1 showed transforming activity in several cell types, including fibroblasts and lymphoid and myeloid cells (Kamps and Baltimore 1993; Kamps et al. 1996; Thorsteinsdottir et al. 1999; Sykes and Kamps 2004). Despite the several in vivo murine models that have been described for E2a–PBX1, none as yet reflects the human B-cell disease. Retroviral overexpression of E2a–PBX1 in hematopoietic chimeras resulted in a myeloid disorder (Kamps and Baltimore 1993; Thorsteinsdottir et al. 1999). Transgenic animals in which E2a–PBX1 expression is driven toward lymphoid cells by the immunoglobulin heavy-chain promoter developed an aggressive T-cell leukemia within 5 mo (Dedera et al. 1993). The underlying cause for this difference in disease induced by E2a–PBX1 in human and mice is unknown. In order to model the appropriate B-cell leukemia and avoid the development of T-cell tumors, we engineered lymphoid-specific transgenic mice for E2a–PBX1 and crossed these animals with mice mutant for the CD3ε chain, which fail to develop mature T-cells (Malissen et al. 1995). Successfully, our compound transgenic mice developed B-cell leukemia of the pre-B-cell phenotype, which in addition could be significantly accelerated by proviral insertional mutagenesis, providing a model to identify collaborator genes for E2a–PBX1-induced B-cell leukemia. Analysis of the integration sites in the genomic DNA of transgenic B-cell tumors revealed a very high incidence of MMLV integrations within the Hoxa locus. In addition, expression analysis of the Hoxa cluster showed that the integrations resulted in activation of several Hoxa genes in the majority of specimens that were analyzed. Thus the generation and exploitation of this E2a–PBX1-induced B-cell leukemia model provided unbiased in vivo evidence for genetic interaction between Hoxa genes and E2a–PBX1 in B-cell leukemia.
Results
Generation and analysis of nonleukemic E2a–PBX1 transgenic mice
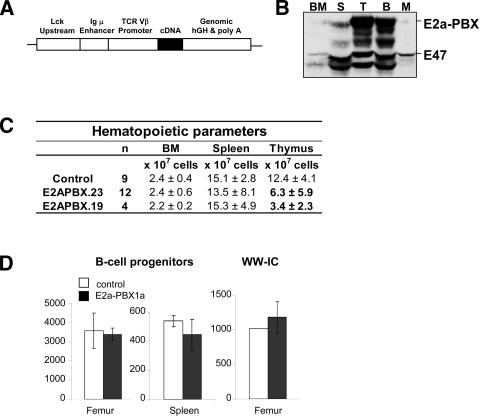
Two transgenic mouse lines (nos. 19 and 23) in which the E2a–PBX1 oncogene was placed under control of lymphoid-specific promoter/enhancer elements found in the pLIT3 expression vector (Fig. 1A; Hough et al. 1994) were generated. Western blot analysis confirmed the expression of the fusion protein E2a–PBX1 in cells from the bone marrow (BM), spleen, and thymus. Transgene levels in the thymocytes appeared higher than in splenocytes and BM (Fig. 1B; not shown but similar for line 19). Protein levels of E2a–PBX1 were also verified in purified B-cells from the transgenic lines and appeared comparable to those detected in T-cells (Fig. 1B, cf. lanes identified as B vs. T). Sorted myeloid cells expressed low to undetectable levels of E2a–PBX1 (Fig. 1B).
Figure 1.
Description of E2a–PBX1 mice generated for these studies. (A) Schematic representation of the transgenic construct. (B) Western blot analysis determining expression of E2a–PBX1 fusion protein in bone marrow (BM), spleen (S), thymus (T), sorted B-cells (B), and myeloid cells (M) from transgenic line 23. B-cells were sorted based on the surface marker B220 and myeloid cells on Mac1. Expression of E2a–PBX1 was detected using an antibody against the E47 chain of the E2A component of E2a–PBX1; see E47 label for endogenous expression of E47. (C) Table showing the cellularity (mean ± SD) determined for the indicated hematopoietic organ. (D) Numbers of IL-7-responding B-cell progenitors (pre-B) per femur and spleen of E2a–PBX1 transgenic mice (line 23, n = 3) and control littermates (n =3) estimated by colony-forming cell (CFC) assay in semisolid medium. The right graph represents the numbers of functionally primitive B-cells (so-called Whitlock-Witte initiating cell or WW-IC) found in the individual organs of E2a–PBX1 transgenics and littermate controls.
In young nonleukemic transgenic mice, there was a two- to fourfold decrease in the number of thymocytes, while numbers of peripheral T-cells in the BM and spleen were normal (Fig. 1C). Analysis of subpopulations of mature lymphoid and myeloid cells in BM, spleen, and thymus showed no alteration of a particular cell subset (data not shown). Furthermore, numbers of IL7-responding B-cell progenitors in BM and spleen, and earlier B-cell populations determined by Whitlock-Witte initiating cell (WW-IC) assays were similar between the transgenic mice and control littermates (Fig. 1D). Despite the fact that E2a–PBX1 is expressed in B-cells, no abnormalities were observed in the B-cell compartment at 2 mo of age (data not shown). In conclusion, except for a reduced number of total T-cells in the thymus, both lines of E2a–PBX1 transgenic mice lack detectable hematopoietic anomalies at 2 mo.
E2a–Pbx1 transgenic mice develop lymphoid leukemia
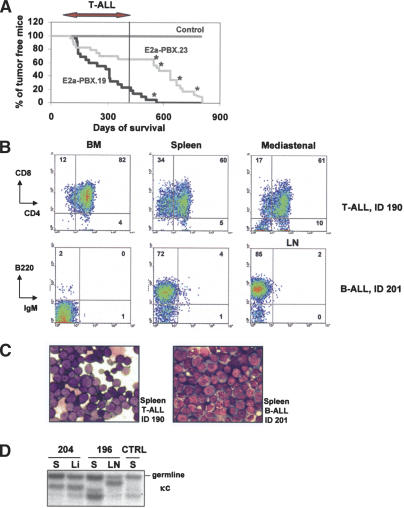
Cohorts of 24 mice from the two transgenic lines (previously backcrossed in C57Bl/6J mice) were monitored for the development of leukemia. Mice of both lines began to develop leukemia at 3–4 mo of age. Surprisingly, a significant proportion of both cohorts presented an extended life span, which appeared longer for mice from line 23 than for those from line 19 (median survival 463 ± 229 vs. 298 ± 146, respectively) (Table 1; Fig. 2A). Some mice from line 19 survived up to 600 d, and mice from line 23 even up to 800 d (Fig. 2A). A few mice died of old age. For this reason the penetrance for tumor development was determined as 88% for line 23 and 97% for line 19 (Table 1).
Table 1.
The survival of E2a–PBX1 transgenic mice and characterization of their leukemia
| Mouse lines | MMLV | n | Survival | Penetrancea | B | T | B/M | M | non-H | Unknown |
|---|---|---|---|---|---|---|---|---|---|---|
| E2aPBX.23 | - | 24 | 463 ± 229 | 88% | 13% | 33% | 4% | 4% | 12% | 29% |
| E2aPBX.19 | - | 24 | 298 ± 146 | 97% | 4% | 50% | 0% | 8% | 3% | 29% |
| E2aPBX.23/CD3e-/- | - | 15 | 403 ± 156 | 87% | 40% | 0% | 0% | 0% | 13% | 38% |
| E2aPBX.23/CD3e-/- | + | 18 | 162 ± 31 | 100% | 33% | 0% | 33% | 16% | 0% | 11% |
| Control.23/CD3e-/- | + | 23 | 191 ± 50 | 100% | 48% | 13% | 4% | 0% | 0% | 25% |
Penetrance = 100% - % of mice that died of no hematopoietic (non-H) etiology. (B) = B-ALL, (T) = T-ALL, (M) = myeloid leukemia.
Figure 2.
Description of T-cell and B-cell leukemias in E2a–PBX1 transgenics. (A) Survival curve of E2a–PBX1 transgenic mice from lines 19 and 23. Mice that die before 400 d suffer from T-ALL. After that period, some mice succumbed to B-ALL or a mixed B-cell/myeloid (B/M) phenotype (indicated with *) or myeloid leukemia or died of old age. (B) FACS profiles of a T-ALL and B-ALL that developed in E2a–PBX1 transgenic mice, using antibodies against the T-cell markers CD4 and CD8 and B-cell markers B220 and surface IgM. The profiles for different hematopoietic organs display infiltration of the CD4+/CD8+ T-cells in spleen and BM (upper panel, mouse ID 190, Supplementary Table S1) and infiltration in spleen and LN for the B-ALL (lower panel, mouse ID 201, Supplementary Table S1). (C) Cytological analysis of representative (same mice as in B) Wright-Giemsa-stained T-ALLs and B-ALLs. (D) Southern blot analysis of DNA isolated E2a–PBX1-induced leukemias to study rearrangement of the κ light-chain (κc) gene. Genomic DNA was digested with EcoRI and BamHIII. ID 204 and 196 are B-cell leukemias with rearrangement of the κ chain, as indicated by the DNA fragments that are smaller than those corresponding to the unrearranged allele (germline in D). Mouse ID204 appears to have the same leukemic clone in spleen and liver (Li), whereas mouse ID196 seems to have two different clones in spleen and LN.
Interestingly, mice from line 23 that succumbed before they reached the age of 400 d almost all suffered from T-cell leukemia (8/10 T-cell [T] and 2/10 not determined), while a proportion of mice that died at an older age suffered from either B-cell leukemia (3/14), myeloid (M) leukemia (1/14), a mixed B/M (1/14), or T/M (1/14), or causes that could be related to old age (3/14) (see Supplementary Table S1 for complete details). For selected B-cell leukemia from line 23, FACS studies demonstrated the expression of the B-cell marker B220 and lack of expression of surface IgM, suggesting the pre-B-cell origin for these tumors (Fig. 2B). Furthermore, Southern blot analysis showed that the κ light chain was rearranged for at least one of these tumors (Fig. 2D, ID204).
The different latency time for the development of T-cell versus B-cell leukemia are reflected in the survival curve shown in Figure 2A. A lag period of almost 100 d occurred between the last mouse to succumb to T-cell leukemia and the onset of B-cell or B/myeloid leukemia. This may suggest a higher sensitivity of T-cells for E2a–PBX1 compared to B-cells and myeloid cells. Only one of the longest survivors for line 19 died of B-cell leukemia at an age that such leukemia began to develop in line 23 (see Supplementary Table S1; Fig. 2A).
In line 19, 12 mice suffered from T-cell leukemia, and similar to line 23 these mice died within 400 d. Two mice developed myeloid leukemia, one B-cell leukemia, while the remaining 10 mice died of unknown aetiology (see Table 1 for summary and Supplementary Table S1 for details of individual leukemia). The majority of mice from transgenic line 19 died prior to the occurrence of B-cell tumors. B and T leukemic cells appeared as medium-sized blasts with small nuclear-to-cytoplasmic ratios and were infiltrated into BM, spleen, thymus, lymph nodes, and frequently nonhematopoietic organs, such as liver and lung (see Fig. 2B–D for a selected example and Supplementary Table S1 for complete details of individual leukemic mice).
Generation of E2a–PBX1 mice with enhanced occurrence of B-cell leukemia
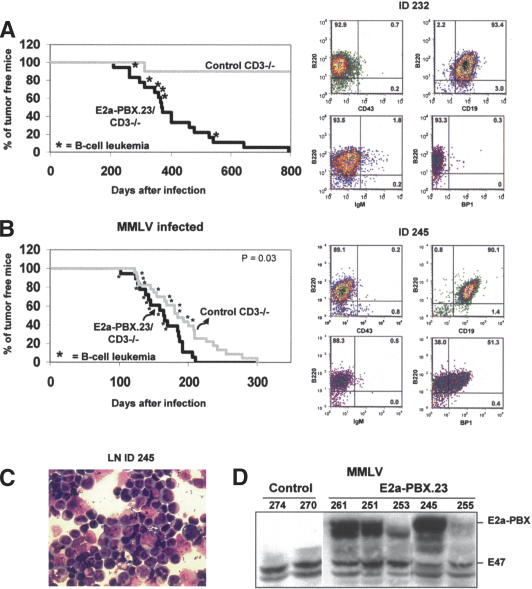
Elimination of T-cell development at an early stage as observed in mice lacking a functional CD3ε gene was reasoned as a possible means to reduce the frequency of T-cell tumors in our E2a–PBX1 transgenics, thereby allowing sufficient time for the development of B-cell leukemia in these mice. Based on this assumption, E2a–PBX1 transgenic mice from line 23 (lowest incidence of T-cell and highest occurrence of B-cell leukemia) (Table 1) were crossed to homozygosity with mice mutant for the CD3ε chain and monitored for disease occurrence over 2 yr. As expected, T-cell differentiation was inhibited in these mice, and none of them succumbed to T-cell leukemia, while at least 40% developed B-cell leukemia (see Table 1 and Supplementary Table S2 for details). Unexpectedly, the occurrence of B-cell leukemia in these compound E2a–PBX1.23 × CD3ε–/– transgenic mice was more accelerated than that seen in our single E2a–PBX1.23 transgenics (cf. occurrence of B-cell leukemia for line 23 as detailed in Figs. 2A, 3A). The phenotype of several B-cell leukemias that developed in these compound transgenics was B220+/CD43–/CD19 +/IgM– (Fig. 3A, right panels) and they expressed Pax5 (Supplementary Tables S2, S3), indicating a pre-B-cell phenotype according to the Hardy B-cell classification (Hardy et al. 1991). Consistent with the B-cell origin of these tumors, all specimens had immunoglobulin heavy (IgH) chain rearrangements, most of which showed rearrangement of the κ light chain (Supplementary Table S2).
Figure 3.
Genetic engineering of E2a–PBX1 mice with high frequency of pre-B leukemia. (A) Survival curve of E2a–PBX1 × CD3ε–/– compound transgenic mice and control CD3ε–/– littermates (left panel) and FACS profile of a representative B-ALL (right panel). (B) Survival curve of compound E2a–PBX1 × CD3ε–/– transgenic mice and their control CD3ε–/– littermates injected with MMLV (left panel) and FACS profile of a B-ALL is shown to the right.(C) Cytological evaluation of a representative Wright-Giemsa-stained MMLV-induced transgenic B-ALL. (D) Western blot analysis for E2a–PBX1 expression on cells from spleen or lymph node from E2aPBX1 × CD3ε–/– and control/CD3ε–/– mice. Expression of E2a–PBX1 as detailed in Figure 1B. Two signals of different size were detected for E2a–PBX1, possibly showing the expression of both E2a–PBX1a (the longer form) and E2a–PBX1b (shorter transcript as result of alternative splicing) in the transgenic leukemic cells.
Slow-transforming retrovirus accelerates pre-B-cell leukemia formation in E2a–PBX1 × CD3ε–/– transgenic mice
In order to identify pre-B-cell-relevant collaborators, proviral insertional mutagenesis was performed using the E2a–PBX1/CD3ε–/– mouse model described above. Neonatal E2a–PBX1/CD3ε–/– mice (n = 18) and their control littermates (n = 23) were intraperitonally injected with MMLV. The minimal latency time to leukemia occurrence was 101 and 122 d for transgenic and control animals, with mean survival of 162 ± 31 d versus 191 ± 50 d, respectively (P = 0.03) (Fig. 3B). Analysis by FACS and/or rearrangement of the IgH and κ light chain showed that 66% of the transgenic mice suffered from B-cell leukemia (Table 1), of which half had a combined myeloid phenotype (see fourth line in Table 1 for summary and Supplementary Table S3 for details). In addition, three mice died of myeloid leukemia (16%) that expressed low to undetectable levels of E2a–PBX1 (Table 1; e.g., in Fig. 3D, upper signal [E2a–PBX1] is weak in ID253). Expression of the E2a–PBX1 transgene in the B-cell tumor cells was confirmed by Western blot analysis (Fig. 3D).
The majority of the mice in the control group (i.e., CD3ε–/– littermate) that could be analyzed died of B-cell leukemia (48%). Surprisingly, two mice developed an unexpected T-cell leukemia (ID277 and 281) (Supplementary Table 3), and one showed a combined B-cell and T-cell phenotype (ID280).
Identification of collaborator oncogenes to E2a–PBX1 in pre-B-cell leukemia
To detect whether MMLV was integrated in previously reported common integration sites (CIS) such as Notch1, Pim1, and Bmi1, Southern blot analysis was performed using DNAs isolated from B-cell leukemia derived from transgenic (n = 16) and control (n = 21) animals. Using an experimental strategy including the selection of adequate probes and restriction enzymes, which allows the detection of a high proportion of rearranged oncogenes at these loci, we could not detect such rearrangements in any of these tumors (data not shown). This included Notch1, a previously reported oncogenic collaborator to E2a–PBX1 in T-cell leukemias (Feldman et al. 2000), likely indicating that oncogenic complementation is cell type specific.
To identify unknown collaborators, inverse PCR was performed on genomic DNA isolated from 13 different B-cell tumors (seven expressing E2a–PBX1 and six controls). Multiple amplification products were subcloned and sequenced for each tumor. In total, 72 different loci with MMLV integrations were identified (all integrations are shown in Supplementary Table S4). Of these, six loci were the site of MMLV integration in more than one tumor and were considered as CIS, identifying them as candidate cancer genes (Table 2; Suzuki et al. 2002). These included two novel CIS, namely, Pde4d, a phosphodiesterase and a hypothetical phospholipase (i.e., A930027K05RIK).
Table 2.
List of the common insertion sites recovered from transgenic (tg) and control (ctrl) tumors
| Insertions
|
||||||
|---|---|---|---|---|---|---|
| CIS | Protein family | Total | tg | ctrl | P-valuea | Other leukemia models |
| Hoxa | Transcription factor | 9 | 8 | 1 | 0.04 | M in BXH2; M, B, T in AKxD |
| Pde4d | Phosphodiesterase | 3 | 1 | 2 | 0.56 | New |
| Pou2f2 | Transcription factor | 2 | 1 | 1 | 1 | B-cell AKxD |
| Pdm16 | Transcription factor | 3 | 1 | 2 | 1 | B-cell Cdkn2a-/- |
| Osbp13 | Oxysterol-binding protein | 2 | 1 | 1 | 1 | Myeloid BXH2 |
| A930027K05Rik | Hypothetical phospholipase | 2 | 1 | 1 | 1 | New |
The P-value is based on the Pearson χ2 test, using exact parameters.
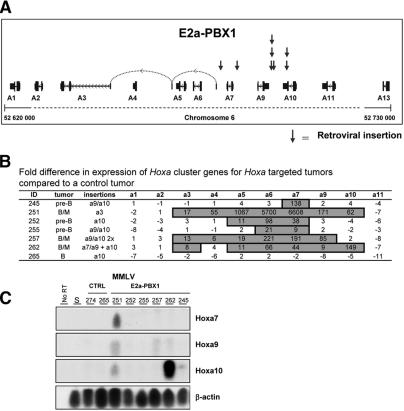
Interestingly, the only CIS that was preferentially targeted in E2a–PBX1-induced B-ALL was recovered from a 19-kb region located between Hoxa6 and Hoxa10 (P = 0.04) (Table 2). This locus accounted for ∼18% (8/45) of all integrations in these tumors. This contrasts with only one integration from 38 (3%) in the same locus in the control B-cell leukemias. The eight integrations derived from the E2a–PBX1 × CD3ε–/–/MMLV leukemias were found in six of the seven leukemias analyzed. Since two of these leukemias contained two different integrations in this locus, at best only 14% (1/7) of the E2a–PBX1 B-cell tumors lacked MMLV integration in the Hoxa locus. This value compares favorably to 83% of the control B-cell leukemia not showing integrations in this region.
The eight integrations in the Hoxa locus of the transgenic tumors were distributed as follows: Four were located in the intergenic region between Hoxa9 and Hoxa10, two in the 3′-UTR of Hoxa10, one between Hoxa7 and Hoxa9, and one telomeric to the 5′-UTR of Hoxa3 (which is uniquely found between Hoxa6 and Hoxa7) (see Fig. 4A; sequences adjacent to the Hoxa integration site are shown in Supplementary Table S5). The only insertion in the control (nontransgenic) tumor was telomeric to Hoxa10.
Figure 4.
Analysis of MMLV integrations in the Hoxa locus of E2a–PBX1 B-cell leukemia. (A) Schematic representation of MMLV integrations localized in the Hoxa locus in E2a–PBX1 transgenic mice as determined by I-PCR. Integrations are indicated by the vertical arrows above the representation of the Hoxa cluster. Large vertical bars indicate Hoxa gene exons, small vertical bars indicate the UTRs, and solid lines with arrows pointing to the left indicate the orientation of the gene and the introns. (B) Table showing Q-PCR results as fold difference in expression levels of Hoxa cluster genes in the Hoxa-locus-targeted tumors compared to a control non-Hoxa-targeted B-cell tumor (ID274). Based on previous studies with these Q-PCR probes (Thompson et al. 2003), values of 8, representing three PCR cycles, and more are considered as substantial increases of expression (boxed numbers). (C) Semiquantitative RT–PCR analyses of Hoxa7, Hoxa9,and Hoxa10 expression in E2a–PBX1 B-cell leukemias that contain MMLV integration(s) in the Hoxa locus (lanes 5–10) or control B-cell leukemias (lanes 3,4) of which one (ID265) also contains a proviral integration in this locus. The globally amplified cDNA was transferred to nylon membranes and hybridized to probes specific for the indicated genes as detailed in Lessard et al. (1998). (S) RNA extracted from spleen cells of a healthy E2a–PBX1 transgenic mouse; (no RT) no reverse transcription before the PCR amplification.
Hoxa gene activation in E2a–PBX1 B-cell leukemia: integration versus expression
To test whether the integrations of MMLV in the Hoxa locus resulted in aberrant gene expression, Q-PCR was performed for 10 of the 11 known Hoxa genes using total RNA isolated from each of the seven tumors described above (six transgenic and one control B-cell leukemia). From this analysis, several observations were made: (1) All six E2a–PBX1 B-cell leukemias analyzed that contain MMLV integration(s) in the Hoxa locus presented a marked increase (e.g., up to 6600-fold) in expression of one to several Hoxa genes when compared to control B-cell leukemias that do not have apparent integration in this locus (e.g., control B-cell tumor ID274 was used as the baseline for the data presented in Fig. 4B but values with other B-cell leukemias are comparable). (2) Hoxa6 and Hoxa7 were the genes most frequently overexpressed with 83% and 100% of the tumors expressing levels that were nine to 6600 times above values measured in control tumors. On average, levels of expression of Hoxa6 and Hoxa7 in E2a–PBX1 transgenic B-cell leukemia were 2–3 logs above values obtained in control B-cell leukemia (Fig. 4B). Interestingly, neither Hoxa6 nor Hoxa7 was overexpressed in the control B-cell tumor, which had an integration in the Hoxa locus (Fig. 4B). (3) In all leukemias analyzed, there was a very poor correlation between the integration site of MMLV (see third column in Fig. 4B) and the expression of Hoxa genes. For example, leukemias ID245, ID252, ID255, and ID265 all have integration in or near Hoxa10, and none of these leukemias overexpress Hoxa10 or Hoxa9. In contrast, integration close to the 5′-UTR of Hoxa3 results in the activation of seven genes from Hoxa3 to Hoxa10. (4) The single Hoxa-targeted control tumor (ID265) did not overexpress any of the Hoxa genes (Fig. 4B), possibly indicating the insignificant nature of this particular integration in the absence of E2a–PBX1.
For Hoxa7, Hoxa9, and Hoxa10, we verified the results obtained by Q-PCR using a semiquantitative RT–PCR approach that exploits global amplification of cDNA as detailed previously (Sauvageau et al. 1994; Lessard et al. 1998, 1999; Lessard and Sauvageau 2003). The results from these experiments confirmed that leukemia ID262 expressed the highest levels of Hoxa10 and that ID251 was the only other tumor that overexpressed this gene (Fig. 4C, third panel). Hoxa9 expression was the highest in ID251 followed by ID257 and ID262, again in agreement with the Q-PCR results shown in Figure 4B. Finally, Hoxa7 could only be detected in ID251 with this methodology, again in agreement with the Q-PCR results indicating much higher expression levels of this gene in this sample versus all other leukemias analyzed.
Together, these results suggest a common activation of Hoxa7 in all E2a–PBX1-induced B-cell leukemias that contained MMLV integrations in the Hoxa locus. In addition, several of these leukemias showed high expression levels of other Hoxa cluster genes.
Discussion
In this study we modeled the human pre-B-cell leukemia induced by the fusion protein E2a–PBX1 in genetically engineered mice. Our transgenic mice, which show comparable expression levels of E2a–PBX1 in B and T lymphoid cells, suffered both from T-cell and B-cell leukemias. The predominance of T-cell leukemias and their shorter latency than the B-cell counterparts indicates a higher sensitivity of murine T-cells to E2a–PBX1. This is in accordance with previously generated E2a–PBX1 transgenic mice, using the Ig heavy-chain enhancer, that develop aggressive T-cell leukemias within 5 mo in the absence of B-cell leukemias (Dedera et al. 1993). The discrepancy between B-cell disease in humans and preference for T-cell disease in genetically engineered mice caused by E2a–PBX1 may be related to the copies of E2A present and its function in B-cell and T-cell differentiation. In human patients with t(1;19), one functional copy is left, while the other is presumably inactivated by the translocation. Transgenic mice, however, still carry two E2A copies in addition to the E2a–PBX gene. Decreased dose of E2A has been shown to affect B-cell differentiation. In E2A+/– mice, a higher proportion of pro-B-cells are cycling, related to lower p21waf/cip1 levels, suggesting an antiproliferative function for E2A (Herblot et al. 2002). Therefore, in contrast to our E2a–PBX1 transgenic mice, it is possible that B-cell development is perturbed in human E2a–PBX1 leukemia due to decreased quantities of E2A as result of one translocated locus. The importance of E2A allelic integrity in E2a–PBX1 B-cell leukemia can be investigated using a similar genetic approach as described in this paper for CD3ε.
Here we present a transgenic B-cell model for E2a–PBX1 in which T-cell leukemogenesis is circumvented by generation of transgenic mice in a CD3ε chain mutant background. A large proportion of the mice that were generated from this approach suffered from pre-B and B-cell leukemias (see Supplementary Table S2). Interestingly, the compound E2a–PBX1 × CD3ε–/– transgenic mice developed B-cell leukemias at much earlier time points (most leukemia occurred between 263 and 370 d of life) than B-cell leukemias that were observed in our parental transgenic line (after 560 d). The nature of this observation remains unclear but may include an immune T-cell-dependent response to the E2a–PBX1 B-cell leukemia that could not happen in the CD3ε–/– background. Other explanations such as greater backcrossing into C57Bl/6J are unlikely since all of our mice were from backcross ≥6.
Acceleration of tumorigenesis by MMLV integration provides a valuable tool to identify collaborator genes to E2a–PBX1 in this B-cell leukemia model. Interestingly, we noted an important proportion of the MMLV-induced leukemias in E2a–PBX1/CD3ε–/– that carried markers for both B-cells (B220) and myeloid cells (Mac1). These tumors could originate from a common myeloid and B-cell progenitor. Biphenotypic, B-cell, and macrophage progenitors have been described to be derived from fetal liver cells (Kee et al. 1994). The occurrence of myeloid leukemias exclusively in transgenic animals is also of interest, especially considering that E2a–PBX1 expression was very low to undetectable in these leukemias. We suspect that myeloid precursor cells are particularly sensitive to transformation induced by E2a–PBX1 (Kamps and Baltimore 1993; Kamps and Wright 1994; Thorsteinsdottir et al. 1999), although non-cell-autonomous effects cannot be ruled out at this stage. The development of rare T-cell leukemias only in control CD3ε mutant mice might be explained by the presence of a few early T-cell progenitors or common lymphoid progenitor cells in CD3ε mutant mice and the tropism of MMLV for these cells.
Our insertional mutagenesis screen was carried out in order to identify loci that could be overtly targeted in E2a–PBX1 B-cell tumors. As detailed above, this “small-scale” functional screen was strongly biased for a small region of 19 kb in the Hoxa cluster that represented up to 18% of all integrations analyzed in E2a–PBX1 B-cell tumors, including two tumors bearing two integrations in this locus. Q-PCR analysis for Hoxa gene expression in these tumors highlighted the collaborative nature of these integrations since up to several Hoxa genes were overexpressed as a result of these integrations in the transgenics, compared to no overexpression in the only control B-cell tumor with a MMLV integration in the Hoxa locus. A more refined analysis of our data suggests an association between the identity of the B-cell leukemia analyzed and the selection of Hoxa gene that was overexpressed as a result of insertional mutagenesis. For example, all three pre-B leukemias described in Figure 4B (ID245, ID252, ID255) showed overexpression of Hoxa7 and, except for one, Hoxa6. Interestingly, all bipheno-typical B/myeloid leukemias also co-overexpressed Hoxa9 (n = 3/3) and Hoxa10 (n = 2/3), two genes associated with myeloid leukemia in human or mouse (Nakamura et al. 1996; Thorsteinsdottir et al. 1997; Kroon et al. 1998). Subgroup analysis as done here must be interpreted cautiously because the numbers of specimens per group are very small. A larger-scale study would be necessary to provide definitive conclusions on this point. However, the preponderance for targeting of the Hoxa locus is noteworthy.
The insertion site Hoxa7/Hoxa9 has been determined earlier as a CIS in other (non-MMLV) models. These include the myeloid leukemias from BXH2 mice (Suzuki et al. 2002), Cas-Br-M-MuLV-induced myeloid leukemias (Joosten et al. 2002), and in myeloid, B-cell, or T-cell leukemias in the AKxD model (Suzuki et al. 2002). A systematic assessment of Hoxa gene expression level was not performed in these mouse models, precluding the comparison of our expression data with those previously reported in other models. Our study reveals the site Hoxa7/Hoxa9 for the first time as a CIS for MMLV, which is significantly correlated with the presence of the E2a–PBX1 transgene (P = 0.04). Interestingly, and although not the major point of these studies, our results also highlight the complexity of Hoxa gene expression in the context of proviral integration.
Identification of Hox genes as major CIS in E2a–PBX1-induced B-cell leukemia merits attention since HOX proteins represent important functional DNA-binding partners to PBX1 (Lu et al. 1995) and have been shown to bind DNA cooperatively also with E2a–PBX1 (Penkov et al. 2000; Schnabel et al. 2000; Knoepfler et al. 2001). We have previously showed that in myeloid cells, Hoxa9 is a strong collaborator to E2a–PBX1 (Thorsteinsdottir et al. 1999). However, Cleary and colleagues previously reported that a truncated E2a–PBX1 transgene lacking the homeodomain of PBX1, essential for cooperative DNA binding with HOX proteins, is also transforming in T-cells. This suggests that the transforming activity of E2a–PBX1 in T-cells is independent of cooperativity with HOX proteins (Monica et al. 1994). Thus, it may be that E2a–PBX1 activates numerous transforming pathways in primary cells and that some of these are not critical for transformation of T-cells but may be necessary for B-cell transformation. Specific studies will be required to address this important question.
In conclusion, we have used a genetic approach to generate a relevant B-cell leukemia model for E2a–PBX1 and used this model to identify the Hoxa locus as the preferred MMLV integration site in these leukemias. This model can now be fully exploited in both fundamental and clinically relevant studies.
Materials and methods
Transgenic animals
The backbone of the pLIT3 vector has been described elsewhere. In short, a fragment of E2a–PBX1 cDNA was cloned down-stream of a TCR Vβ promoter, immunoglobulin (Ig) enhancer elements, and sequences from the proximal promoter of the lck gene. The human growth hormone (hGH) gene with a frame-shift mutation in the coding region was inserted 3′ of the transgene cDNA to provide introns that enhance the expression. Transgenic animals were generated using standard techniques. The transgene was injected into pronuclei of (C57Bl/6J × C3H) F2 hybrid zygotes. Two to four hours after injection, surviving eggs were transplanted in oviducts of pseudopregnant CD-1 host females. Transgenic progeny were identified by dot blot or Southern blot of tail DNA using a probe for hGH. Mice mutant for the CD3ε chain (Malissen et al. 1995) were kindly provided by Dr. M. Malissen (Centre d'Immunologie de Marseille-Luminy, Marseille, France).
For the induction of leukemias in transgenic mice, 1-d-old newborn transgenic and control littermates were intraperitonally injected with 105 infectious units of MMLV suspended in 50 μL of DMEM (van Lohuizen et al. 1991).
In vitro clonogenic progenitor assays
For myeloid clonogenic progenitor assays, cells were plated in 35-mm dishes in semisolid medium, containing 1% methylcellulose in α medium supplemented with 10% fetal calf serum (FCS), 5.7% bovine serum albumin, 10–5 β-mercaptoethanol, 1 U/mL erythropoietin (Epo), IL-3, IL-6, steel, 2 mM glutamine, and 200 mg/mL transferrin. BM and spleen cells from transgenic mice and control littermates were plated at concentrations of 3 × 104 and 106 cells/mL, respectively. Colonies were scored on day 12–14 of incubation and identified according to standard criteria. For pre-B-cell clonogenic progenitor assays, 3 × 105 bone marrow cells or 106 spleen cells were plated in 1% methylcellulose in α medium supplemented with 30% FCS, 10–4 β-mercaptoethanol, 2 mM glutamine, and 0.2 ng/mL IL-7. Colonies were scored on day 8.
In vitro long-term culture assays for B-cells
The frequency of primitive lymphoid B-cells was determined with a limiting dilution analysis in the Whitlock-Witte culture system (Whitlock and Witte 1987). Total BM cells from transgenic and control mice were cultured for 3 wk on S17 stromal cells in RPMI medium containing 5% preselected FCS, 5 × 10–5 β-ME, and 50 μg/mL gentamicin at concentrations ranging from 3 × 102 to 2 × 105 bone marrow cells per well. After 3 wk, each well was scored for the presence of nonadherent B-cells. The B-cell phenotype was confirmed by staining cells from multiple wells with B220 and Mac1 antibody, and analyzed by FACS. The frequency of the WW-ICs was calculated by applying Poisson statistics to the proportion of negative wells at different dilutions.
Flow cytometry
For the phenotypic analysis of healthy transgenic mice and for analysis of the tumor cell type of sick mice, fresh BM, spleen, and thymus cells were isolated and stained for two- or three-color analysis. Antibodies used to detect T-cell, B-cell, and myeloid cell populations are CD4-FITC, CD8-PE, B220-PE, B220-PE-cy5, CD19-APC, BP1-PE, CD43-FITC, Mac1-bio, IgM-bio (all from BD Bioscience Pharmingen), IgD-FITC (SBA), and Gr1-FITC labeled, which was kindly provided by P. Lansdorp (British Columbia Cancer Agency, Vancouvar, BC, Canada). Biotinylated antibodies were detected with PE, APC (Pharmingen), or PE-cy7 (e-Bioscience) conjugated streptavidin. Fluorescence was analyzed using the FACS Calibur (BD Bioscience) or the LSR II (BD Bioscience), using DIVA software. FACS data were analyzed with WinMDI or FCS Express software (De Novo Software).
For the detection of the E2a–PBX1 protein in B-cells and myeloid cells, fresh spleen cells and BM cells were stained with antibodies B220-PE and Mac1-biotinylated, followed by SA-PE, respectively, and subsequently sorted on a Moflo FACS sorter (Cytomation), using the summit software.
DNA analysis
High-molecular-weight DNA was isolated from fresh tumor cells (BM, spleen, thymus, and lymph nodes, when enlarged) using DNAzol (InVitrogen, Life Technologies), digested with the appropriate restriction enzymes, and analyzed by Southern blotting. For analysis of immunoglobulin rearrangement, a 2.4-kb fragment of κC was used, kindly provided by Dr. G. Gilliland (Brigham and Women's Hospital, Boston, MA). For detection of MMLV integrations in MMLV-induced leukemias by Southern, a 380-bp sacI–PstI fragment of the 3′-untranslated region of MMLV was used (kindly provided by Dr. P. Jolicoeur, IRCM, Montreal, Québec, Canada).
Reverse-transcriptase PCR
RNA was isolated using trizol, according to the manufacturer's protocol (InVitrogen). One microgram of total RNA from tumors induced by MMLV from E2a–PBX1 transgenic and control littermates or human leukemic cells was used for the RT–PCR reaction as described by Sauvageau et al. (1994) with minor modifications. Gene 32 protein was omitted from the PCR reaction and replaced by water. RT–PCR products were divided in five fractions and run on a 2% agarose gel followed by blotting on a membrane (Zetaprobe). Membranes were probed for Hoxa gene expression, and reused after stripping. A probe for β-actin was used to adjust for loading differences.
Quantitative RT–PCR
Total RNA was isolated by Trizol, DNase-I-treated, and cDNA was prepared (MMLV-RT, random primers) according to the manufacturer's instructions (InVitrogen). Q-PCR was carried out using TaqMan-probe-based chemistry (Applera). Oligonucleotides for all 10 murine Hoxa genes were designed against nucleotide sequences deposited in murine genome databases (GenBank, http://www.psc.edu/general/software/packages/genbank/genbank.html; RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq; and EMBL, http:/www.ebi.ac.uk/embl) using Primer Express (Applera). Reactions, analysis, and validation of the Hox amplicons were carried out as previously described (Thompson et al. 2003).
Inverse PCR
Tumor DNA (5 μg) was digested to completion with SstII or BamHI (InVitrogen) in a total volume of 40 μL. After precipitation with ethanol, the digested DNA was self-circularized by dilution and ligation using T4 DNA ligase (40 U; InVitrogen) in a total volume of 400 μL at 16°C for 16 h. Circular DNA was precipitated with ethanol and dissolved in 40 μL of TE buffer. PCR was performed in 50 μL using 2 μL of the DNA template, 20 nmol of each dNTP, 10 pmol of each forward and reverse primer, 1× buffer, and 3.5 U of enzyme mix of the Expand Long Template PCR System (Roche). PCR was carried out on a Perkin Elmer thermocycler with the following parameters: 92°C for 2 min, followed by 10 cycles of 92°C for 10 sec, 63°C for 30 sec, 68°C for 15 min, then 20 cycles of 92°C for 10 sec, 63°C for 30 sec, 68°C for 15 min with a 20-sec auto-extension, and a final extension step at 68°C for 30 min. A small aliquot of the PCR reaction (5 μL) was used for Southern analysis using a probe specific for the U3M sequence of the LTR of MMLV to confirm that the PCR products were derived from proviral sequence. PCR products were separated on a 1% agarose gel, purified using the QIAEX II Gel Extraction Kit (QIAGEN), and directly cloned using the TOPO TA Cloning Kit for Sequencing (InVitrogen). The primers used for IPCR were S3′F (5′-TGCTTCTATGCG GACCACACAGG-3′) and S3′R (5′-GCTAAGGGGAGTAG GAGCACCC-3′) for SstII 3′ cloning; S5′F(5′-GGACCTAACCA AGTTGGGGGCC-3′) and S5′R (5′-GCAGAGCAGAAGGTAA CCCAACG-3′) for SstII 5′ cloning; S3′F and B3′R (5′-CCGCT ACTAGACACAACCAGCAC-3′) for BamHI 3′ cloning; and B5′F (5′-GAATACTGGTACCCTGCCAGTCC-3′) and S5′R for BamHI 5′ cloning.
DNA sequencing
DNA sequencing was performed using the CEQ Dye Terminator Cycle Sequencing with a Quick Start kit (Beckman-Coulter) on a CEQ 2000 Sequencer (Beckman-Coulter) either at the Molecular Biology service of the IRCM (Montreal) or by Genome Québec. The sequencing primers used were 5′-SeqC (5′-GTTC CATCTGTTCCTGACCT-3′) for 5′ clones and 3′-SeqC (5′-GC TCAATAAAAGAGCCCACA-3′) for 3′ clones).
Analysis of MMLV integration site sequences
The retroviral insertion site sequences were filtered for the presence of repetitive DNA and candidate genes were identified by homology searches using the BLAT alignment tool from UCSC Genome Browser (Mouse Genome Sequencing Consortium, Mouse Assembly May 2004 freeze). The definition of retroviral common insertion sites (CIS) was based on previously described statistical analysis (Mikkers et al. 2002). The correlation of the Hoxa locus as a target for MMLV in transgenic versus control mice was tested using the Pearson χ2 test, using exact parameters.
URLs
More information about the tumor-derived clones and the exact position of MMLV integrations can be found at the Mouse Retroviral Tagged Cancer Gene Database (http://rtcgd.ncifcrf.gov).
GenBank accession numbers
The accession numbers for the flanking sequences of the retroviral insertions in the E2a–PBX1 and control (nontransgenic) leukemias are AY745893 through AY745976, excluding AY745916.
Protein analysis
Preparation of cellular extracts from BM, spleen, and thymus derived from healthy or sick transgenic mice for E2a–PBX1 or from control littermates and Western blot analyses were performed as described (Krosl and Sauvageau 2000). Briefly, proteins were separated by gel electrophoresis using 10% polyacrylamide–SDS and transferred to Immobilon P membranes (Milipore). Membranes were blocked with 5% nonfat milk in TBST (20 mM Tris-Cl at pH 7.6, 140 mM NaCl, 0.05% Tween-20) and probed with the E47 antibody (Santa Cruz, Biotechnology Inc.) to detect E2a–PBX1. Bound antibodies were detected using horseradish peroxidase-conjugated anti-rabbit antibodies (Sigma) followed by enhanced chemiluminescence (ECL; Amersham).
Acknowledgments
We thank Nadine Mayotte for helping to generate the E2a–PBX1 transgenic mice and Melanie Frechette for the animal care. Special thanks are owed to Nathalie Tessier, Eric Massicotte, and Martine Dupuis from the flow cytometry service of IRCM for sorting and assistance in FACS analysis. We also thank Miguel Chagnon for his help with the statistical analysis. This project was supported by a grant to G.S. from the National Cancer Institute of Canada and in part by the National Institutes of Health. Members of the leukemia cell bank of Quebec are also acknowledged for their support and expertise on leukemia. M.S. is a fellow of the Fond de Recherches en Sante du Quebec, and A.T. is supported by a fellowship from the American Cancer Society for Beginning Investigators (ACSBI) under the administration of the Union Internationale Contre le Cancer (UICC). G.S. is a recipient of a Canada Research Chair in molecular genetics of stem cells and a scholar of the Leukemia Lymphoma Society of America.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1268505.
References
- Berns A., Mikkers, H., Krimpenfort, P., Allen, J., Scheijen, B., and Jonkers, J. 1999. Identification and characterization of collaborating oncogenes in compound mutant mice. Cancer Res. 59: 1773s–1777s. [PubMed] [Google Scholar]
- Carroll A.J., Crist, W.B., Parmley, R.T., Roper, M., Cooper, M.D., and Finley, W.H. 1984. Pre-B cell leukemia associated with chromosome translocation 1;19. Blood 63: 721–724. [PubMed] [Google Scholar]
- Dedera D.A., Waller, E.K., LeBrun, D.P., Sen-Majumdar, A., Stevens, M.E., Barsh, G.S., and Cleary, M.L. 1993. Chimeric homeobox gene E2A–PBX1 induces proliferation, apoptosis, and malignant lymphomas in transgenic mice. Cell 74: 833–843. [DOI] [PubMed] [Google Scholar]
- Feldman B.J., Hampton, T., and Cleary, M.L. 2000. A carboxy-terminal deletion mutant of Notch1 accelerates lymphoid oncogenesis in E2A–PBX1 transgenic mice. Blood 96: 1906–1913. [PubMed] [Google Scholar]
- Hardy R.R., Carmack, C.E., Shinton, S.A., Kemp, J.D., and Hayakawa, K. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herblot S., Aplan, P.D., and Hoang, T. 2002. Gradient of E2A activity in B-cell development. Mol. Cell. Biol. 22: 886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough M.R., Takei, F., Humphries, R.K., and Kay, R. 1994. Defective development of thymocytes overexpressing the costimulatory molecule, heat-stable antigen. J. Exp. Med. 179: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M., Vankan-Berkhoudt, Y., Tas, M., Lunghi, M., Jenniskens, Y., Parganas, E., Valk, P.J., Lowenberg, B., van den, A.E., and Delwel, R. 2002. Large-scale identification of novel potential disease loci in mouse leukemia applying an improved strategy for cloning common virus integration sites. Oncogene 21: 7247–7255. [DOI] [PubMed] [Google Scholar]
- Kamps M.P. and Baltimore, D. 1993. E2A–Pbx1, the t(1;19) translocation protein of human pre-B-cell acute lymphocytic leukemia, causes acute myeloid leukemia in mice. Mol. Cell. Biol. 13: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps M.P. and Wright, D.D. 1994. Oncoprotein E2A–Pbx1 immortalizes a myeloid progenitor in primary marrow cultures without abrogating its factor-dependence. Oncogene 9: 3159–3166. [PubMed] [Google Scholar]
- Kamps M.P., Murre, C., Sun, X.H., and Baltimore, D. 1990. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell 60: 547–555. [DOI] [PubMed] [Google Scholar]
- Kamps M.P., Wright, D.D., and Lu, Q. 1996. DNA-binding by oncoprotein E2a–Pbx1 is important for blocking differentiation but dispensable for fibroblast transformation. Oncogene 12: 19–30. [PubMed] [Google Scholar]
- Kee B.L., Cumano, A., Iscove, N.N., and Paige, C.J. 1994. Stromal cell independent growth of bipotent B cell—Macrophage precursors from murine fetal liver. Int. Immunol. 6: 401–407. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S., Sykes, D.B., Pasillas, M., and Kamps, M.P. 2001. HoxB8 requires its Pbx-interaction motif to block differentiation of primary myeloid progenitors and of most cell line models of myeloid differentiation. Oncogene 20: 5440–5448. [DOI] [PubMed] [Google Scholar]
- Kroon E., Krosl, J., Thorsteinsdottir, U., Baban, S., Buchberg, A.M., and Sauvageau, G. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17: 3714–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosl J. and Sauvageau, G. 2000. AP-1 complex is effector of Hox-induced cellular proliferation and transformation. Oncogene 19: 5134–5141. [DOI] [PubMed] [Google Scholar]
- Lessard J. and Sauvageau, G. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255–260. [DOI] [PubMed] [Google Scholar]
- Lessard J., Baban, S., and Sauvageau, G. 1998. Stage-specific expression of Polycomb group genes in human bone marrow cells. Blood 91: 1216–1224. [PubMed] [Google Scholar]
- Lessard J., Schumacher, A., Thorsteinsdottir, U., van Lohuizen, M., Magnuson, T., and Sauvageau, G. 1999. Functional antagonism of the Polycomb-group genes eed and Bmi1 in hemopoietic cell proliferation. Genes & Dev. 13: 2691–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look A.T. 1997. Oncogenic transcription factors in the human acute leukemias. Science 278: 1059–1064. [DOI] [PubMed] [Google Scholar]
- Lu Q., Knoepfler, P.S., Scheele, J., Wright, D.D., and Kamps, M.P. 1995. Both Pbx1 and E2A–Pbx1 bind the DNA motif ATCAATCAA cooperatively with the products of multiple murine Hox genes, some of which are themselves oncogenes. Mol. Cell. Biol. 15: 3786–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M., Gillet, A., Ardouin, L., Bouvier, G., Trucy, J., Ferrier, P., Vivier, E., and Malissen, B. 1995. Altered T cell development in mice with a targeted mutation of the CD3-ε gene. EMBO J. 14: 4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H., Allen, J., Knipscheer, P., Romeijn, L., Hart, A., Vink, E., Berns, A., and Romeyn, L. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 32: 153–159. [DOI] [PubMed] [Google Scholar]
- Monica K., LeBrun, D.P., Dedera, D.A., Brown, R., and Cleary, M.L. 1994. Transformation properties of the E2a–Pbx1 chimeric oncoprotein: Fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol. Cell. Biol. 14: 8304–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Largaespada, D.A., Lee, M.P., Johnson, L.A., Ohyashiki, K., Toyama, K., Chen, S.J., Willman, C.L., Chen, I.M., Feinberg, A.P., et al. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 12: 154–158. [DOI] [PubMed] [Google Scholar]
- Nourse J., Mellentin, J.D., Galili, N., Wilkinson, J., Stanbridge, E., Smith, S.D., and Cleary, M.L. 1990. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell 60: 535–545. [DOI] [PubMed] [Google Scholar]
- Penkov D., Tanaka, S., Di Rocco, G., Berthelsen, J., Blasi, F., and Ramirez, F. 2000. Cooperative interactions between PBX, PREP, and HOX proteins modulate the activity of the α 2(V) collagen (COL5A2) promoter. J. Biol. Chem. 275: 16681–16689. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H. 1994. Chromosomal translocations in human cancer. Nature 372: 143–149. [DOI] [PubMed] [Google Scholar]
- Sauvageau G., Lansdorp, P.M., Eaves, C.J., Hogge, D.E., Dragowska, W.H., Reid, D.S., Largman, C., Lawrence, J.H., and Humphries, R.K. 1994. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. 91: 12223–12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel C.A., Jacobs, Y., and Cleary, M.L. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19: 608–616. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Shen, H., Akagi, K., Morse, H.C., Malley, J.D., Naiman, D.Q., Jenkins, N.A., and Copeland, N.G. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32: 166–174. [DOI] [PubMed] [Google Scholar]
- Sykes D.B. and Kamps, M.P. 2004. E2a/Pbx1 induces the rapid proliferation of stem cell factor-dependent murine pro-T cells that cause acute T-lymphoid or myeloid leukemias in mice. Mol. Cell. Biol. 24: 1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A., Quinn, M.F., Grimwade, D., O'Neill, C.M., Ahmed, M.R., Grimes, S., McMullin, M.F., Cotter, F., and Lappin, T.R. 2003. Global down-regulation of HOX gene expression in PML-RARα + acute promyelocytic leukemia identified by small-array real-time PCR. Blood 101: 1558–1565. [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Sauvageau, G., Hough, M.R., Dragowska, W., Lansdorp, P.M., Lawrence, H.J., Largman, C., and Humphries, R.K. 1997. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell. Biol. 17: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Krosl, J., Kroon, E., Haman, A., Hoang, T., and Sauvageau, G. 1999. The oncoprotein E2A–Pbx1a collaborates with Hoxa9 to acutely transform primary bone marrow cells. Mol. Cell. Biol. 19: 6355–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M., Verbeek, S., Scheijen, B., Wientjens, E., van der, G.H., and Berns, A. 1991. Identification of cooperating oncogenes in E μ-myc transgenic mice by provirus tagging. Cell 65: 737–752. [DOI] [PubMed] [Google Scholar]
- Whitlock C.A. and Witte, O.N. 1987. Long-term culture of murine bone marrow precursors of B lymphocytes. Methods Enzymol. 150: 275–286. [DOI] [PubMed] [Google Scholar]
- Williams D.L., Look, A.T., Melvin, S.L., Roberson, P.K., Dahl, G., Flake, T., and Stass, S. 1984. New chromosomal translocations correlate with specific immunophenotypes of childhood acute lymphoblastic leukemia. Cell 36: 101–109. [DOI] [PubMed] [Google Scholar]