Abstract
Polarized growth in filamentous fungi requires the integrity of the microtubule (MT) cytoskeleton. We found that growing MTs in Aspergillus nidulans merge at the center of fast growing tips and discovered that a kinesin motor protein, KipA, related to Tea2p of Schizosaccharomyces pombe, is required for this process. In a ΔkipA strain, MT plus ends reach the tip but show continuous lateral movement. Hyphae lose directionality and grow in curves, apparently due to mislocalization of the vesicle supply center (Spitzenkörper) in the apex. Green fluorescent protein (GFP)-KipA accumulates at MT plus ends, whereas a KipA rigor mutant protein, GFP-KipAG223E, coated MTs evenly. These findings suggest that KipA requires its intrinsic motor activity to reach the MT plus end. Using KipA as an MT plus-end marker, we found bidirectional organization of MTs and determined the locations of microtubule organizing centers at nuclei, in the cytoplasm, and at septa.
INTRODUCTION
The ability to generate cell polarity is a key feature of both prokaryotic and eukaryotic cells. In eukaryotes, it is achieved and maintained through the localized assembly of signaling complexes, the rearrangement of the cytoskeleton, the interaction of the cytoskeleton with the cortex, and the delivery of proteins to the sites of membrane growth and cell extension. One early step for the generation of asymmetry is the positioning of cell surface landmark proteins, which direct the cytoskeleton and thereby the flow of vesicles toward this site (Nelson, 2003).
The general principles of cell polarization are relatively well understood in the single-cell yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. In S. pombe, mutants with T-shaped cells were isolated and named tip elongation aberrant (tea) (Snell and Nurse, 1994; Verde et al., 1995). Two of the corresponding genes, tea1 and tea2, were cloned and analyzed in detail (Mata and Nurse, 1997; Browning et al., 2000). Tea1p is a kelch-domain protein that localizes to the growing plus ends of MTs in the cell and is thereby delivered to the cortex (Behrens and Nurse, 2002). In addition to being transported in an MT-dependent manner, the protein affects the organization of the MT cytoskeleton: tea1 mutants have a higher frequency of cells in which MTs curl around the ends of the cell (Mata and Nurse, 1997). tea2 encodes a kinesin-like protein (Browning et al., 2000) that is responsible for the correct localization of Tea1p and localizes itself to the cell poles and the MT plus ends. One possible explanation for the Tea1p localization defect is the disturbance of the MT cytoskeleton observed in tea2 mutants. However, it was proposed that Tea2p is loaded onto MTs in the middle of the cell and then travels toward the MT plus ends by using its intrinsic motor activity (Browning et al., 2003). Thus, it could be that Tea1p is a cargo of the Tea2p motor. On the other hand, Tea2p anchorage at the cell ends depends on Tea1p (Browning et al., 2000, 2003). After being transported and delivered to the cortex, Tea1p needs to be anchored at the membrane. A candidate protein for that is Mod5p, which was discovered through an insertional mutagenesis approach (Snaith and Sawin, 2003). Mod5p contains a signal for carboxy-terminal prenylation and is thus a prime candidate for membrane association. Although localization of Tea1p depends on this protein, the restriction of Mod5p localization itself to the cell tip requires the presence of Tea1p. How growth is directed to the site of Tea1p is not yet well understood.
Despite the growing body of information on cell polarity in S. pombe, little is known about the molecular mechanisms of cell polarity in filamentous fungi, in which polar growth is by far more prominent (Harris and Momany, 2004). In contrast to S. pombe, tip extension is not linked to the cell cycle, and growth direction can be changed. Fast hyphal growth in filamentous fungi requires a continuous supply of enzymes and cell wall components. This is achieved by MT-dependent vesicle transport mediated by motor proteins such as conventional kinesin (Seiler et al., 1997, 1999; Riquelme et al., 2000; Requena et al., 2001). An accumulation of vesicles, named the vesicle supply center or “Spitzenkörper” (SPK), is located in the hyphal tip close to the cortex. Although the exact nature of the vesicles is still unknown, it is likely that the SPK serves as a transit station for secreted enzymes, e.g., for cell wall biosynthesis. The position of the SPK has been associated with the direction of hyphal growth (Girbardt, 1957; Grove and Bracker, 1970; Bartnicki-Garcia et al., 1995; Riquelme et al., 1998). Other genes and factors required for establishment and maintenance of polarity in filamentous fungi have been identified, and some of the genes have been analyzed at the molecular level (Momany et al., 1999; Shaw et al., 2002; Lin et al., 2003; Seiler and Plamann, 2003). However, it is not yet possible to develop a comprehensive model of how the components act and interact.
We analyzed the genome of Aspergillus nidulans for kinesin-like proteins and identified 11 candidates, which were phylogenetically grouped into nine of the currently 11 different kinesin families (Rischitor et al., 2004). In this study, we have characterized the kinesin-like protein KipA and found that deletion of kipA affects the maintenance of growth directionality. We propose that the polarity defect is due to a lack of temporal anchorage of MTs to the growing cortex at the cell tip.
MATERIALS AND METHODS
Strains, Plasmids, and Culture Conditions
Supplemented minimal (MM) and complete media for A. nidulans and standard strain construction procedures were used, as described by Hill and Käfer (2001). Expression of tagged genes under control of the alcA promoter was regulated by carbon source: repression on glucose, derepression on glycerol, and induction on threonine or ethanol (Waring et al., 1989). A list of A. nidulans strains used in this study is given in Table 1. Standard laboratory Escherichia coli strains (XL-1 blue, Top 10 F′) were used. Plasmids and cosmids are listed in Table 2 or described below.
Table 1.
A. nidulans strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| SRF200 | pyrG89; ΔargB::trpCΔB; pyroA4; veA1 | Karos and Fischer, 1999 |
| GR5 | pyrG89; wA3; pyroA4; veA1 | Waring et al., 1989 |
| RMS011 | pabaA1, yA2; ΔargB::trpCΔB; trpC801, veA1 | Stringer et al., 1991 |
| SPR26 | pyrG89; ΔkipB::argB; pyroA4; veA1 | Rischitor et al., 2004 |
| SPR36 | ΔkipB::argB; pyroA4; veA1; ΔkinA::pyr4 | Rischitor et al., 2004 |
| SRL1 | GR5 transformed with pRL19 [ΔkipA::pyr4] | See text |
| SRS27 | SRF200 transformed with pRS31 and pDC1 [GFP-nuclei] | Suelmann et al., 1997 |
| SRS29 | SRF200 transformed with pRS54 and pDC1 [GFP-mitochondria] | Suelmann and Fischer, 2000 |
| SJW02 | wA3; pyroA4; ΔargB::trpCΔB; alcA(p)::GFP::tubA; veA1 [GFP-MTs] | J. Warmbold, Marburg, Germany |
| SJW100 | SJW02 transformed with pJW18 [GFP-MTs and red nuclei] | J. Warmbold, Marburg, Germany |
| SNR3 | pyrG89, yA2; ΔargB::trpCΔB; veA1; ΔkinA::pyr4 | Requena et al., 2001 |
| SSK13 | pabaA1; wA3; ΔkipA::pyr4; veA1 | SRL1 × RMS011 |
| SSK28 | pabaA1; wA3; ΔkipB::argB; pyroA4, ΔkipA::pyr4; veA1 | SSK13 × SPR26 |
| SSK44 | pabaA1; wA3; ΔargB::trpCΔB; ΔkipA::pyr4; veA1 | SSK13 × RMS011 |
| SSK61 | SSK44 transformed with pRS31 and pDC1 [ΔkipA and GFP-nuclei] | See text |
| SSK67 | pabaA1; ΔkipA::pyr4; alcA(p)::GFP::tubA; veA1 [ΔkipA and GFP-MTs] | SSK44 × SJW100 |
| SSK69 | SSK44 transformed with pRS54 and pDC1 [ΔkipA and GFP-mitochondria] | See text |
| SSK72 | wA3; ΔkipA::pyr4; ΔkinA::pyr4; veA1 | SSK44 × SPR36 |
| SSK73 | wA3; ΔkipB::argB; pyroA4, ΔkipA::pyr4; veA1; ΔkinA::pyr4 | SSK44 × SPR36 |
| XX3 | pyrG89; nudA1, chaA1, veA1 | Xiang et al., 1994 |
| SSK80 | pabaA1; wA3; ΔkipA::pyr4; nudA1, veA1 | SSK44 × XX3 |
| SSK92 | GR5 transformed with pSK82 [GFP-KipA] | See text |
| SSK99 | SSK92 transformed with pJH19 and pI4 [GFP-KipA and red nuclei] | See text |
| SSK100 | SSK92 transformed with pPND1 and pI4 [GFP-KipA and mRFP1-MTs] | See text |
| SSK114 | SRF200 transformed with pPR700, homologous integration [GFP-KipArigor] | See text |
| SSK116 | SRF200 transformed with pPR700, ectopic integration [GFP-KipArigor (partial KipA protein)] | See text |
Important characteristics of the strains that might not be obvious from their genotypes are indicated in brackets.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pCR2.1-TOPO | TA-cloning vector [for cloning of PCR fragments] | Invitrogen (NV Leek, The Netherlands) |
| pDC1 | A. nidulans argB selectable marker plasmid | Aramayo et al., 1989 |
| pI4 | A. nidulans pyroA selectable marker plasmid | Osmani et al., 1999 |
| pPND1 | kipB(N-terminus) in pCMB17 apx; GFP replaced by mRFP1 | Rischitor et al., 2004 |
| PNRSTE1 | 1.9-kb pyr4 with flanking BamHI and NotI sites in pCR2.1-TOPO | Requena et al., 2001 |
| PCMB17apx | alcA(p)::GFP, for N-terminal fusion of GFP to proteins of interest; contains N. crassa pyr4 | V. Efimov (Piscataway, NJ) |
| pRS31 | gpd(p)::stuA(NLS)::GFP in pBluescript KS- | Suelmann et al., 1997 |
| pRS54 | gpd(p)::citrate synthase (N)::GFP in pBluescript KS- | Suelmann and Fischer, 2000 |
| pJW18 | alcA(p)::stuA(NLS)::DsRed and argB as selectable marker in pBluescript KS- | Toews et al., 2004 |
| pJH19 | gpd(p)::stuA(NLS)::DsRed and argB as selectable marker in pBluescript KS- | Toews et al., 2004 |
| pSK82 | 1-kb kipA fragment in pCMB17apx | See text |
| pPR700 | As pSK82 except for G223E mutation yielding kipArigor | See text |
Molecular Techniques
Standard DNA transformation procedures were used for A. nidulans (Yelton et al., 1984) and E. coli (Sambrook and Russel, 1999). For PCR experiments, standard protocols were applied using a capillary Rapid Cycler (Idaho Technology, Idaho Falls, ID) for the reaction cycles. DNA sequencing was done commercially (MWG Biotech, Ebersberg, Germany). Genomic DNA was extracted from A. nidulans with the DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). RNA was isolated with TRIzol (Invitrogen, Paisley, Scotland, UK) according to the manufacturer's protocols. DNA and RNA analyses (Southern and Northern hybridizations) were performed as described by Sambrook and Russel (1999).
Disruption and Tagging of kipA
Two regions of kipA were amplified by polymerase chain reaction (PCR) by using genomic DNA as template and primers KipA1 (5′-CAACGCACCGAATACTCAGC-3′) and KipA2-BamHI (5′-AGGATCCGAGTCGTAGACTC-3′) for the upstream region (positions -288 to +815 relative to the A of the start codon) and KipA3-NotI (5′-AGCGGCCGCCTTATGGTATGACC-3′) and KipA4 (5′-GTGTTCAGAGTCTCTGATGC-3′) for the downstream region (positions +872 to +1625 relative to the A of the start codon). The products were cloned into pCR2.1-TOPO generating pEW30 and pEW27, respectively. A 1.1-kb BamHI fragment from pEW30 and a 750-base pair NotI fragment from pEW27 were then cloned upstream and downstream of the pyr4 marker in pNRSTE1, generating pRL19. This plasmid was cut with KpnI and XbaI, which cut in the polylinker on either side of the insert, thus generating a fragment containing pyr4 flanked by kipA sequences. This fragment was transformed into the uracil-auxotrophic A. nidulans strain GR5. Its homologous integration by double crossover should lead to replacement of 56 base pairs of kipA by pyr4, with consequent disruption of the motor domain. Among 60 transformants that were analyzed by PCR, five displayed the desired homologous integration at the kipA locus. Four of them had no additional integration events as shown by Southern blot analysis. One ΔkipA strain (SRL1) was crossed to RMS011, and the resulting strain SSK13 (recognized as ΔkipA by Southern blot) was crossed again to RMS011 to generate the ΔkipA strain SSK44 and to SPR26 to generate the ΔkipA ΔkipB double-mutant strain SSK28 (identified by Southern blotting with probes for both kipA and kipB).
To create an N-terminal fusion construct, a 1-kb fragment of kipA (starting from the ATG) was amplified from genomic DNA with the primers KipAfor-AscI (5′-GGCGCGCCCGGGATGTCTACACTGCCGCAGC-3′) and KipA-PacI (5′-TTAATTAATAGATTTCGAGATAGCTGACG-3′) and cloned into pCR2.1-TOPO, yielding pSK79. The AscI-PacI fragment from pSK79 was then subcloned into the corresponding sites of pCMB17apx (kindly provided by V. Efimov, Piscataway, NJ), yielding pSK82. Homologous recombination of this plasmid into the kipA locus should result in an N-terminal GFP fusion of the entire kipA gene under control of the alcA promoter plus a truncated 5′ region under control of the kipA promoter. Among 20 transformants of strain GR5, two displayed the kipA deletion phenotype under repressing conditions (growth on glucose) and a wild-type phenotype under inducing conditions (growth on glycerol or threonine), one of which was saved as strain SSK92. PCR analysis confirmed that the construct was integrated at the kipA locus. These results indicated that the tagged KipA is fully active in vivo.
Creation of a kipA Rigor Mutant Allele
We changed glycine residue 223 to glutamate by site-directed mutagenesis by using the oligonucleotides KipA-G223E fwd (5′-GGTATGACCGGAACTGAGAAAACCTTTTCTATGC-3′) and KipA-G223E rev (5′-GCATAGAAAAGGTTTTCTCAGTTCCGGTCATACC-3′), plasmid pSK82 as template, and the QuikChange XL site-directed mutagenesis kit (Stratagene, Heidelberg, Germany); this yielded plasmid pPR700. We transformed strain SRF200 and searched for a transformant in which pPR700 was integrated at the kipA locus. Among 150 transformants, two (one named SSK114) displayed the kipA deletion phenotype under both repressing and inducing conditions. PCR analysis confirmed that the construct was integrated at the kipA locus in both transformants. The PCR fragments were sequenced to confirm the mutagenesis event. The low frequency of mutant colonies can be explained by the need for homologous recombination to occur downstream of the changed triplet in a 133-base pair region. Two strains (one named SSK116) with ectopic integrations of pPR700 were selected for phenotypic analyses as a control.
Light Microscopy
For live cell imaging, cells were grown in glass-bottom dishes (WPI, Berlin, Germany) in 2 ml of medium, either MM + 2% glycerol + pyridoxine and/or arginine or MM + 2% ethanol (or threonine) + pyridoxine and/or arginine. Cells were incubated at 30°C for 15 h, and images were captured at room temperature by using an Axiophot microscope (Carl Zeiss, Jena, Germany), a Planapochromatic 63× or 100× oil immersion objective lens, and a 50-W Hg lamp. Fluorescence was observed using standard fluorescein isothiocyanate, 4,6-diamidino-2-phenylindole (DAPI), and rhodamine filter sets. Images were collected and analyzed with a Hamamatsu Orca ER II camera system and the Wasabi software (version 1.2). Time-lapse series were obtained with an automated Wasabi program that acquires series of images with 2- or 3-s intervals, 0.5- or 0.75-s exposure time, and ∼100 exposures in a sequence. Image processing and measurements were done with Image-ProPlus (Media Cybernetics, Silver Spring, MD) and Adobe Photoshop.
To determine SPK position, strains were grown on a coverslip for 24 h at room temperature in MM containing 17% gelatin, and images were captured with an Axiovert microscope (Carl Zeiss). For motion analysis, sequences of up to 30 frames of single germtubes were taken at 1- and 2-min intervals, and SPK position and hyphal growth were observed by phase contrast microscopy.
Indirect immunofluorescence of MTs and γ-tubulin was performed as described by Willins et al. (1995), with the following modifications. Cells were grown for 8 h at 37°C in MM containing appropriate supplements and fixed for 30 min at 37°C in 7.4% formaldehyde in PME buffer (50 mM PIPES, 5 mM EGTA, and 1 mM MgSO4, pH 6.9), followed by three washes with PME buffer. The cells were then digested with 4 mg/ml glucanase, 11 mg/ml driselase (both from InterSpex, San Mateo, CA), and 2 mg/ml yeast lytic enzyme (ICN, Eschwege, Germany) for 1 h at room temperature. Before adding antibodies, the cells were blocked with 3% bovine serum albumin in Tris-buffered saline (20 mM Tris and 700 mM NaCl, pH 7.6) for 1 h. Cells were then incubated with monoclonal anti-α-tubulin antibody (Sigma-Aldrich, Taufkirchen, Germany; clone DM1A, 1:400 dilution) or a monoclonal anti-γ-tubulin antibody (Sigma-Aldrich; clone GTU88, dilution 1:400) followed by Cy3-conjugated anti-mouse-IgG secondary antibody (Molecular Probes, Leiden, The Netherlands; 1:1000 dilution). For staining of nuclei, mounting medium with DAPI (Vector Laboratories, Burlingame, CA) was used.
RESULTS
Cloning of kipA
We analyzed the genome of A. nidulans in the databases at Cereon Genomics (Cambridge, MA) and at the Whitehead Institute (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/) for kinesin-like proteins and used the motor domain of A. nidulans conventional kinesin to identify 11 members of the kinesin family (Rischitor et al., 2004). In the studies reported here, we have characterized one of these proteins, KipA. The sequence of kipA was confirmed by sequencing 3.9 kb from a kipA-containing cosmid (pEW35, isolated from the PUI library, kindly provided by B. Miller, Moscow, ID). To determine the intron-exon borders, we amplified corresponding cDNAs by reverse transcription-PCR. Comparison of genomic and cDNA sequences revealed the presence of one 49-base pair intron (Figure 1A). The predicted KipA protein is 889 amino acids in length with a molecular mass of 96.25 kDa and a calculated isoelectric point of 9.9. The motor domain starts after 142 amino acids. Comparison of motor domains placed KipA and several other fungal kinesins in the CENP-E subfamily (Figure 1B). Sequence identity outside the motor domains is low. For example, the motor domain of KipA, which includes apparent ATP- and MT-binding sites, displayed 47% identity to S. pombe Tea2p, whereas the full-length proteins displayed only 29% identity. Nonetheless, the architecture of the fungal proteins seems similar, because they all contain the motor domain at the N terminus, N-terminal extensions, and regions with a high probability for coiled-coil formation in the C-terminal part (Figure 1C). In comparison, the human kinesin CENP-E is much larger (2663 aa), and the coiled-coil regions are much more extended, which are typical features of CENP-E kinesins from mammals. These differences suggest that the fungal kinesins comprise a distinct group. However, the functions of the proteins seem to be different. In S. cerevisiae, Kip2p is necessary for proper nuclear distribution, whereas in S. pombe, Tea2p is required for polarized growth (Huyett et al., 1998; Miller et al., 1998; Browning et al., 2000). In mammals, CENP-E is involved in the checkpoint control of mitosis (Weaver et al., 2003).
Figure 1.
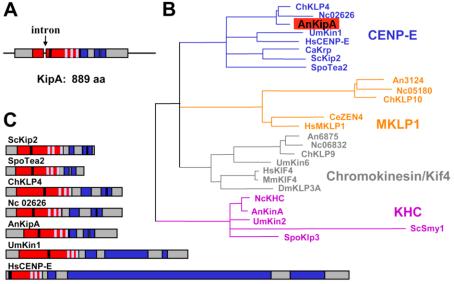
Characterization of KipA. (A) Scheme of the genomic locus and the KipA protein (accession no. AJ622826). Arrow, intron; red, motor domain; blue, coiled-coil regions; black, ATP binding site; light blue, the microtubule-binding-pocket. (B) A most likely phylogenetic tree of kinesins belonging to four different subfamilies was built by comparison of their motor domains by using Tree-puzzle (http://www.tree-puzzle.de/) and a maximum-likelihood algorithm. For evaluation of statistical significance of the topology, 25,000 replicating puzzling steps were performed. The substitution model Whelan-Goldman 2000 was used because it produced a consensus tree, as shown, which is in good agreement with the published data (Schoch et al., 2003). The kinesins shown are from the indicated organisms: An, A. nidulans; Nc, Neurospora crassa; Spo, S. pombe; Ce, Caenorhabditis elegans; Mm, Mus musculus; Um, Ustilago maydis; Sc, S. cerevisiae; Ch, Cochliobolus heterostrophus; Hs, Homo sapiens; Ca, Candida albicans; Dm, Drosophila melanogaster. The A. nidulans and N. crassa sequences were taken from the annotated genome sequence database at the Whitehead Institute. (C) Comparison of the protein architecture of KipA and related kinesins. Domains are indicated as in A.
Role of KipA in Growth Directionality in A. nidulans
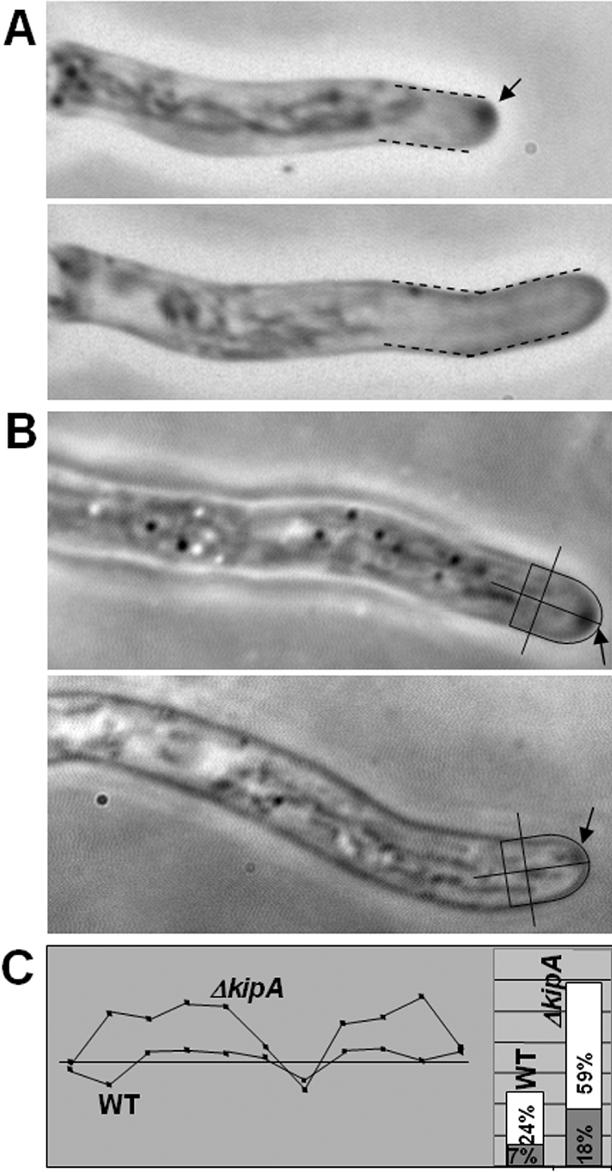
To explore the function of KipA in A. nidulans, we constructed ΔkipA disruptant strains (see Materials and Methods). The rates of colony and hyphal extension of the mutants showed no difference from those of wild type (for hyphal extension, 3.8 ± 0.9 μm/min). We also did not detect any differences with respect to asexual or sexual sporulation. However, the maintenance of growth directionality was altered. Whereas wild-type hyphae extend only at their tip and grow in an approximately straight line, the mutants displayed meandering and curved hyphae (Figure 2, A and B). The branching pattern was not significantly different from that of wild type. This morphological phenotype co-segregated with the knockout marker in crosses between ΔkipA and wild-type strains (our unpublished data). Because mutation of the KipA-related protein Kip2p in S. cerevisiae causes a misdistribution of nuclei during budding, we tested for a role of KipA in organelle movement in A. nidulans. Nuclei and mitochondria were visualized by GFP staining (strains SRS27, SRS29, SSK61, and SSK69) and analyzed by time-lapse videomicroscopy as described previously (Suelmann et al., 1997; Suelmann and Fischer, 2000). We could not find any difference with respect to nuclear migration (speed and directionality) or positioning. The mitochondrial network also displayed a wild-type appearance (our unpublished data). Vesicle transportation toward the hyphal tip should also not be reduced, because hyphal growth speed was not affected. Hence, the maintenance of growth directionality seems to be the main role of KipA. Another polarity phenotype became apparent when we analyzed spore germination. After the first germ tube from an A. nidulans conidium has reached a certain length, a second hypha emerges from the spore. In wild-type, the origin of the second hypha seems to be determined by the position of the original germ tube; thus, it emerged from the side of the spore opposite to the germ tube (bipolar) in 76% of the spores, whereas other positions were found only 24% of the time. In contrast, the second hypha emerged in seemingly random positions in 84% of the spores in the ΔkipA mutant (Figure 2D).
Figure 2.

Morphological phenotypes of kipA mutants. (A and B) Differential interference contrast images of wild-type hyphae (RMS011) (A) and ΔkipA hyphae (SSK44) (B). (C) Phase-contrast images of alcA(p)-GFP-kipA strain SSK92 under inducing (threonine) and repressing (glucose) conditions. (D) Effect of kipA deletion on the establishment of polarity. Conidia of wild-type (RMS011) and ΔkipA (SSK13) strains were germinated and analyzed for the direction of second germ tube formation. For each strain, 200 germlings were counted.
Determination of growth directionality in filamentous fungi involves an organelle in the center of the apical dome of the hyphae. This organelle, named the “vesicle supply center” or SPK, provides, for example, enzymes for cell wall biosynthesis. When the SPK moves away from the center of the cell, growth occurs in the direction of the SPK location (Figure 3A). We determined the position of the SPK by phase contrast microscopy in wild type and the ΔkipA mutant (Figure 3, B and C). Its average distance from the central axis was ∼2.5-fold greater in the mutant, although the diameters of the hyphae were equivalent in mutant and wild type (2.1 ± 0.8 μm). In addition to its altered positioning, the SPK typically seemed smaller in the mutant. Because the rate of hyphal tip extension was not altered in the mutant, we presume that vesicles are simply more dispersed in the tip region.
Figure 3.
Mispositioning of the SPK in a kipA mutant. (A) Correlation between SPK position and growth direction. Top, noncentral SPK (arrow) in a hypha of ΔkipA mutant strain SSK13. Bottom, 5 min later, growth has occurred in the direction of the noncentral SPK. (B) Determination of SPK position in wild-type (top, RMS011) and ΔkipA mutant (bottom, SSK44) hyphae. The center of the tip and central axis of the hypha were defined as indicated by the lines, and SPK position was determined by measuring the distance from the center of the SPK to the central axis and expressing the value as a percentage of the radius of the hypha (i.e., the distance from the central axis to the outer edge of the hypha at the position indicated by the line perpendicular to the central axis). (C, left) Examples of tracks of SPKs in wild-type (RMS011) and ΔkipA (SSK44) strains. The shaded box represents (top to bottom) half the diameter of the hypha in each case; the straight line represents its central axis and a time period of 10 min over which measurements were made. (C, right) Summary of data for 129 measurements (17 germlings) on strain RMS011 and 121 measurements (28 germlings) on strain SSK44. The average (shaded box) and maximum (white box) distances of the SPK from the central axis are indicated. Note that, in some cases, the SPKs moved considerably further from the central axis than seen in the typical tracks shown on the left.
Stabilization of MTs by KipA and Functional Interactions with Other Motor Proteins
Recently, it was shown that addition of low amounts of the MT-destabilizing drug benomyl caused curved growth in A. nidulans and thus that growth directionality depends on the integrity of the MT cytoskeleton (Riquelme et al., 2003). To begin investigating the potential effect of KipA on MT dynamics, we studied the sensitivity of a ΔkipA strain toward benomyl. The mutant was slightly more sensitive than wild type, suggesting a MT-stabilizing effect of KipA (Figure 4A). Because MT stability also is affected by mutations of other motor proteins, we studied genetic interaction between ΔkipA and these mutations by comparing growth at 30°C. A double mutant between conventional kinesin, ΔkinA, and ΔkipA (strain SSK72) revealed an additive phenotype in which the colonies displayed a compact appearance like ΔkinA mutants (Figure 4B) and curved hyphal growth like the ΔkipA mutant (our unpublished data). Deletion of kinA stabilizes MTs (as judged by benomyl resistance) and the additional deletion of kipA did not detectably change this effect (our unpublished data). Another MT-destabilizing kinesin is the Kip3-family kinesin KipB (Rischitor et al., 2004). The ΔkipA, ΔkipB strain (SSK28) grew ∼50% slower than the wild-type, although each mutation alone had no effect on hyphal extension, suggesting partially redundant functions of the two motors (Figure 4B). Benomyl resistance was not reduced in comparison with the ΔkipB strain (our unpublished data). A triple mutant, ΔkinA, ΔkipA, ΔkipB (strain SSK73) was still viable (our unpublished data). We also tested for genetic interaction between ΔkipA and a temperature-sensitive dynein mutation (nudA1). Deletion of kipA partially suppressed the growth defect of the dynein mutant (Figure 4B). Benomyl sensitivity was similar in the nudA1 and ΔkipA, nudA1 strains (our unpublished data). In all mutant combinations that contained the ΔkipA allele, we observed the curved growth phenotype (our unpublished data). Because in all those double and triple mutants, MTs are even more stable than in wild-type, the effect of the kipA mutation on growth directionality cannot be caused primarily by an effect on MT stability.
Figure 4.

Comparison of growth and benomyl sensitivity of wild-type and mutant strains. (A) Wild-type (RMS011) and ΔkipA (SSK44) conidia were spotted at the centers of agar plates containing different concentrations of benomyl. Colony diameters were measured after 2 d at 37°C, and the value for wild type at that concentration was used as 100% (10 colonies per strain for each concentration). (B) Wild-type and mutant strains were grown for 2 d at 30°C, and the diameters of 25 colonies per strain were measured. The strains used were RMS011, SSK44, SNR3, SSK72, SPR36, SSK73, XX3, and SSK80 (see Table 1).
KipA Moves along MTs and Accumulates at Their Plus Ends
To examine KipA localization and movement, we created strains expressing a GFP-KipA fusion protein under control of the regulatable alcA promoter (see Materials and Methods). In strains in which GFP-KipA is the only source of KipA, growth under repressing conditions resulted in the kipA mutant phenotype, whereas wild-type hyphal growth was observed under inducing conditions, showing that the GFP-KipA fusion protein is biologically active (Figure 2C). In interphase cells, we observed a spotted distribution of the protein in the cytoplasm and labeling of the septa during growth under derepressing conditions (Figure 5A). Timelapse analyses revealed that the spots were moving rapidly within the cell. In the hyphal tip, GFP spots were moving with a speed of 8-12 μm/min toward the tip (Figure 5B and Movie 1). When the protein arrived at the tip, the GFP fluorescence intensity decreased rapidly. The speed and direction of spot movement were similar to the dynamics of growing MTs (Figure 5B and Movie 2). To confirm the apparent MT association, we colocalized GFP-KipA and MTs using a red fluorescent protein (mRFP1)-KipB motor domain fusion protein, which efficiently decorates all populations of MTs (Rischitor et al., 2004). We found GFP-KipA spots at the ends of MT fibers, suggesting that KipA is a plus-end-associated protein (Figure 5C, top). In addition, we observed GFP-KipA spots at places where the MT fluorescence signal changed (Figure 5C), suggesting that the MT tracks contain overlapping MT bundles.
Figure 5.
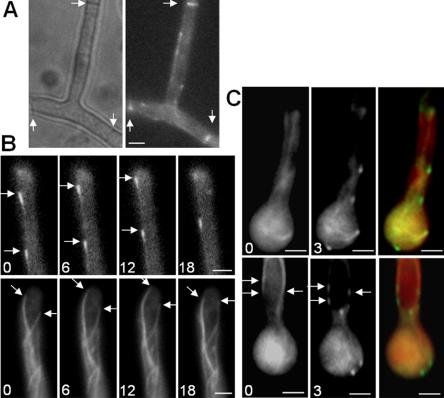
GFP-KipA localization in interphase cells. Strains SSK92 (A and B, top sequence), SJW02 (B, bottom sequence), and SSK100 (C) were grown on glycerol to derepress expression of the GFP-tagged proteins without overexpressing them. (A) Localization of GFP-KipA. Left, phase contrast; right, GFP; arrows, the location of septa. (B) Top sequence, movement of GFP-KipA spots (arrows) toward the tip of the hypha. Times are indicated in seconds. Bottom sequence, GFP-labeled MTs in a hyphal tip. The MT on the left grows, pauses at the tip, and retracts after a catastrophe event. In comparison, the GFP-KipA signal disappears without retracting after reaching the tip (see Movies 1 and 2). (C) Localization of GFP-KipA at the plus ends of mRFP1-decorated MTs. Top and bottom rows show two different germlings. Left, mRFP1 fluorescence; MTs are visible as fibers. Middle, 3 s after the first exposure, the fluorescence channel was changed to visualize GFP-KipA. Right, overlay of the colored images. Arrows indicate positions in the MT bundle at which the mRFP1 fluorescence intensity decreases (left) and at which GFP-KipA is concentrated (middle).
When the expression level of GFP-KipA was increased, we observed a comet-like staining pattern. The length of the comets increased with increasing protein concentrations until the entire MT was stained (Figure 6A). To determine whether the comets were the result of one-dimensional diffusion of the protein from the plus end, we deenergized the cells by addition of 100 μM CCCP (carbonylcyanide-m-chlorophenylhydrazone) and thus presumably transformed KipA into a rigor state. After 30-min incubation, MT plus ends were still visible, but in addition filaments were rather evenly decorated, and a GFP-KipA concentration gradient toward the plus end was not obvious (Figure 6B). Thus, the comet-like appearance can be explained by hypothesizing that the motor actively moves along MTs.
Figure 6.
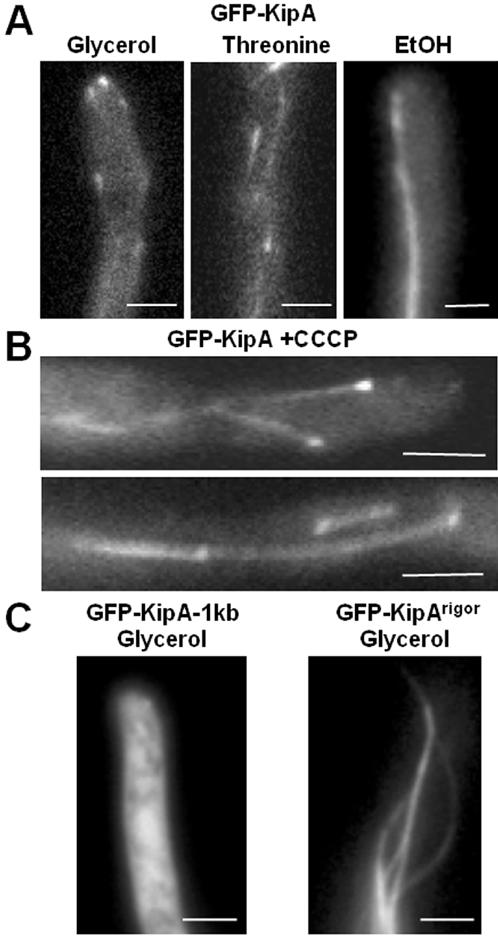
Localization of GFP-KipA and the rigor mutant GFP-KipAG223E. (A) GFP-KipA labels MT plus ends when expressed at low levels and the entire MTs when overexpressed. Strain SSK92 was grown at 25°C on glycerol or threonine for 1 d or on ethanol for 2 d. (B) GFP-KipA staining of MTs and MT plus ends 20 min after addition of CCCP to strain SSK100 growing on glycerol medium at 25°C. Comet-like structures were not observed. (C) GFP-KipAG223E stains entire MTs. Ectopic integration of the construct led to staining of the cytoplasm (left), whereas homologous integration led to production of full-length GFP-KipAG223E and coating of the entire MTs. The images were taken after strains SSK116 and SSK114 were grown for 1 d on glycerol.
To test further whether an intrinsic KipA motor activity is required for its MT plus-end localization, we created a KipA mutant version in which glycine 223 was changed to glutamate (G223E). This amino acid change is located in the predicted ATP-binding domain (P-loop) and has been shown to transform kinesin motor proteins into a “rigor” state (Nakata and Hirokawa, 1995; Browning et al., 2003). In strain SSK114, GFP-KipAG223E was the only source of KipA. Under both inducing (threonine) and repressing (glucose) conditions, we observed the kipA mutant phenotype (our unpublished data), and a GFP signal was detectable during growth either on threonine or under derepressing (glycerol) conditions. During growth on glycerol, MTs were evenly decorated, and no accumulation of the protein at the MT plus ends was detectable (Figure 6C, right). As a control, we analyzed strain SSK116, in which the plasmid had integrated ectopically. Induction of the promoter produced cytoplasmic GFP staining (Figure 6C, left), as expected given that the GFP-fusion protein contains only a truncated motor domain that lacks the MT binding regions.
In mitotic cells, GFP-KipA localized to the spindle pole bodies and the midzone of the spindle (Figure 7), as well as to the plus ends of astral MTs (our unpublished data).
Figure 7.
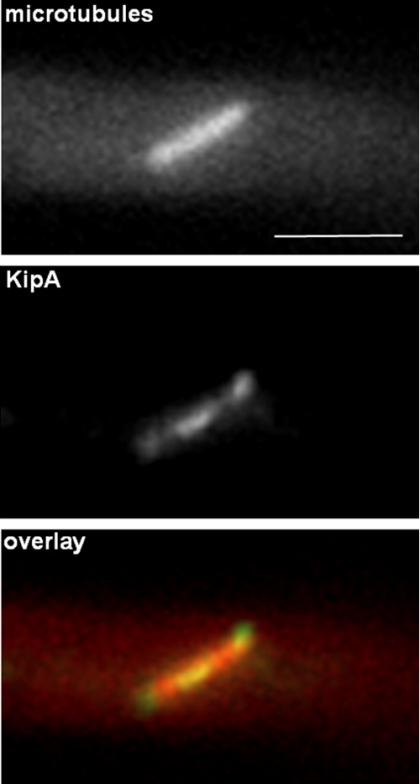
GFP-KipA localization during mitosis. Strain SSK100 was grown on glycerol for one day at 25°C and observed for mRFP1-KipB (top) and GFP-KipA (middle). The bottom panel shows the overlaid images.
Dispersion of MTs in the Hyphal Tips of ΔkipA Mutants
To study the behavior of MTs in detail, we compared wild-type and ΔkipA strains expressing GFP-labeled α-tubulin. We found no difference in the number and length of MTs in the cytoplasm in the two strains, and, in both cases, the MTs showed dynamic behavior and reached the tip cortex. In the mutant, the MTs paused at the cortex for an average of 7.8 ± 3.4 s, with a maximum of 15 s (43 MTs in 18 germlings), in comparison with an average of 13.5 ± 5.5 s, with a maximum of 27 s, in the wild-type (44 MTs in 16 germlings). In addition to the different times of pausing, we found a difference with respect to the arrangement of MTs in the apical dome of the tip. In wild-type cells MTs converged in the center of the tip (Figure 8A and Movie 2). This phenomenon was also evident when we examined the movement of GFP-KipA, in that the GFP signal could be tracked until it reached the center of the hyphal tip (Figure 8B). We also found that the tracks along which GFP-KipA moved were used several times (Figure 8, B and C, and Movie 3). In comparison, MTs in the ΔkipA strain did not always converge to a single point. Instead, they diverged at the tip and ended at different places in the cortex (Figure 8, D and E, and Movie 4). Another difference was observed when we analyzed the MTs after they reached the center of the tip. In wild-type cells, MTs typically remained at their position until a catastrophic event caused depolymerization. In contrast, in the kipA mutant, MTs frequently changed their positions even after they reached the cortex (Figure 8, D and E, and Movie 5). These results suggest that there is a MT-cortex interaction that depends on plus-end-localized KipA.
Figure 8.
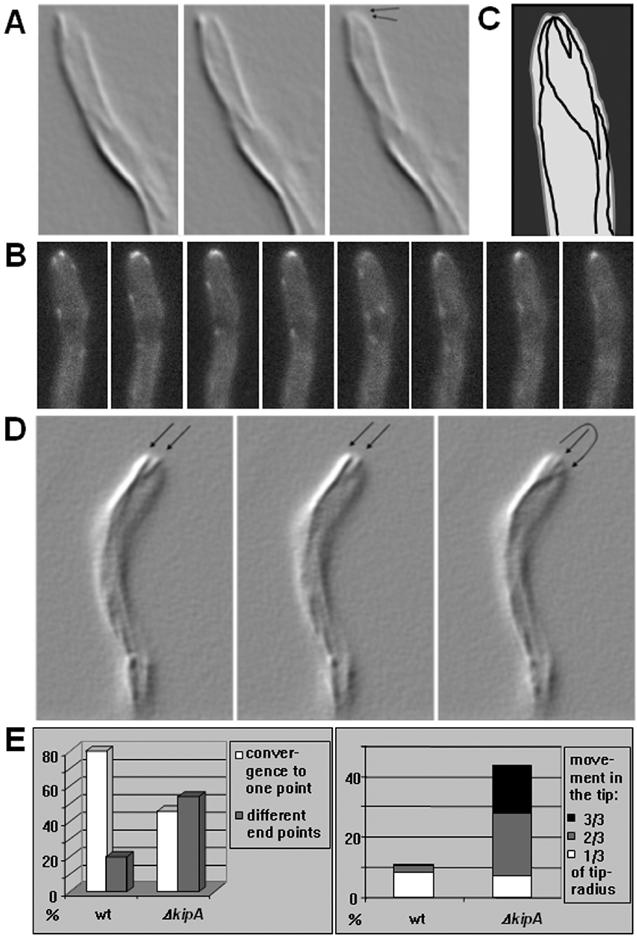
MT organization in growing hyphal tips of wild type and a ΔkipA mutant (see Movies 2-5). (A-C) Wild-type strain SJW02 was grown on glycerol at 25°C. (A) GFP-labeled MTs are displayed in shadow mode in images captured at 3-s intervals. (B) GFP-KipA images captured at 2-s intervals. The disappearance of tip signal in image 5 and its reappearance in image 6 suggests that the maintenance of KipA at the tip requires continuous delivery from the plus ends. (C) Scheme of the traces of GFP-KipA. During an observation time of 4 min, 18 MT plus ends used the same tracks indicated with lines. (D) ΔkipA strain SSK67 was grown on glycerol at 25°C and imaged as in A. (E) Quantification of the effects. Left, data from 157 MTs in 27 germlings of strain SJW02 and from 151 MTs in 29 germlings of strain SSK67. Right, in the same dataset, the movements of MT plus ends away from their meeting point were measured and grouped into short, medium, and long movements.
Microtubule Organization in A. nidulans
MT plus-end-associated proteins have been used in various organisms to unravel the polarity and organization of the MT cytoskeleton (Straube et al., 2003; Piehl et al., 2004). Because KipA seems to be an effective marker protein for MT plus ends in A. nidulans, we used it as a tool to study MT organization and the number and location of MT-organizing centers (MTOCs). We monitored GFP-KipA traces in growing hyphal tips and in subapical compartments of interphase cells and determined the numbers of MTs growing toward the tip or oriented with their plus ends in the opposite direction (Movies 6-8). In the tip regions of 42 apical compartments, we found 285 tip-directed MTs (Figure 8B) and seven that polymerized away from the tips (Figure 10A, top diagram, and Movie 1). In contrast, in the area behind the tip region and in subapical compartments, the numbers of retrograde- and anterograde-growing MTs were equal (Figure 10A, top diagram). To determine whether the spindle pole bodies at the nuclei served as the only MTOCs in the cytoplasm, as in S. cerevisiae, we labeled nuclei with a DsRed fusion protein in a strain that also expressed GFP-KipA (SSK99). We defined an MTOC as a point from which several GFP spots originated and moved in different directions. In 42 hyphae, 118 MTs were analyzed. In young, unseptated germlings, ∼50% of the MTs originated from the nucleus and ∼50% from apparent MTOCs in the cytoplasm, which were often localized close to a nucleus. In some cases, an MTOC was located between the leading nucleus and the hyphal tip. In older germlings, >40% of the MTs emanated from septa (Figure 9, A and B, and Movies 6-8). We did not observe MTs passing through the septa. Immunostaining of α-tubulin and in vivo observations of MTs confirmed that MTs both originate and end at septa (Figures 9C and 10A, bottom diagram, and Movie 9). To confirm the presence of multiple MTOCs, we analyzed the distribution of γ-tubulin, which is a characteristic protein of MTOCs (Oakley et al., 1990; Job et al., 2003). γ-Tubulin was detected in interphase at nuclei but also weakly in the cytoplasm and at septa (Figure 9D). In agreement with this, we found two other SPB-associated proteins, ApsB and AlpA (a protein of the Dis1/TOG family of MT-associated proteins) as dots in the cytoplasm and at septa (our unpublished data). These observations on MT organization in A. nidulans are summarized in Figure 10B.
Figure 10.
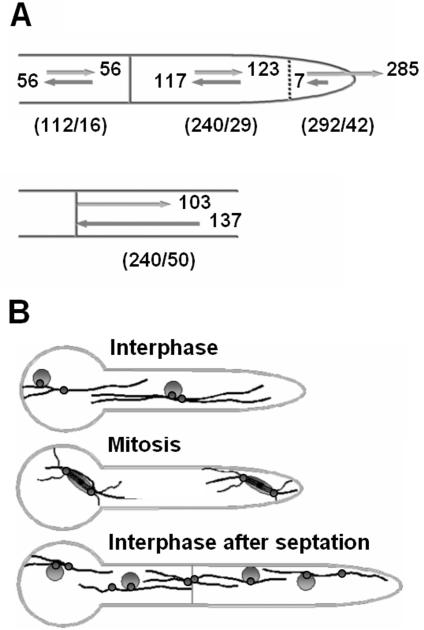
MTOCs and MT organization during the cell cycle. (A) Quantitation of the orientation of the MTs. The numbers of MTs counted and the numbers of germlings are denoted below the hyphae in parentheses. Top scheme, MTs have mixed polarities in various compartments. In the very tip (indicated with a dashed line), most MTs are oriented with their plus ends toward the tip. Bottom scheme, MTs growing away from the septum and MTs ending at the septum. (B) Summary of MTOC organization during early growth of a germling. Black lines, MTs; small circles, MTOCs; large circles, nuclei.
Figure 9.

MTOCs at nuclei, in the cytoplasm, and at septa. (A) Images from movie sequences at different regions of one germling of strain SSK99 showing several GFP-KipA signals (green) originating from MTOCs at the nucleus (top row), in the cytoplasm (middle row), and at a septum (bottom row). Each sequence covered a period of ∼40 s; images were recorded at 2-s intervals, but only selected images are shown. The crosses indicate the MTOCs and the arrows point to growing MT plus ends. During each period of observation, two to three spots are moving away from the MTOC (see Movies 6-8). (B) Young germling of strain SJW02 with GFP-labeled MTs. The arrows point to two MTs emanating from an MTOC at the septum. Pictures were taken at 3-s intervals (see Movie 9). (C and D) Immunostaining of MTs (C) and γ-tubulin (D) in young germlings of strain RMS011. Nuclei were stained with DAPI. The arrows point to the septa. MTs do not continue through the septa (C). γ-Tubulin signals are visible at the nucleus and at the septum (D).
DISCUSSION
Polarized growth in filamentous fungi is a fascinating process that also provides a model for the cell biology of other highly polarized cells such as neurons in higher eukaryotes. An understanding of fungal cell extension also may help to control fungal diseases of plants and animals as well as improve the use of filamentous fungi in biotechnology. In this study, we analyzed MT organization and characterized the KipA kinesin motor protein in the filamentous fungus A. nidulans, finding an involvement of KipA in polarized growth. We found that KipA localizes to MT plus ends, and we propose that it determines growth directionality through the control of MT cortex interactions.
Multiple MTOCs Organize the MT Cytoskeleton in A. nidulans
We used GFP-KipA as a MT plus-end marker to determine the polarity and dynamic behavior of MTs in A. nidulans. In an earlier study, Han et al. (2001) used a strain with GFP-labeled MTs to address the polarity of MTs. They found that at the tip of the hypha, MTs are oriented almost exclusively with their plus ends toward the cortex. We confirmed this result and also observed that MTs converge in the hyphal tip and seem fixed at a central point until a catastrophe event. In addition, we noticed that several GFP-KipA signals seemed to use the same track in the cell. This can be explained if we assume that the MT fibers as visualized are actually bundles of several MTs with individual dynamics. A catastrophe event does not lead to complete disassembly of the entire bundle but only of some MTs. New MTs can then grow along the remaining MTs.
It is well accepted that in fungi, MTs originate from the spindle pole body, a typical fungal organelle. However, it also was found in S. pombe that several MTOCs exist in the vicinity of the nucleus in interphase cells (Tran et al., 2001). Similarly, Straube et al. (2003) found in the single-cell stage of the basidiomycete U. maydis that MT formation originated from the SPB, the bud neck, and from cytoplasmic sites. The use of the different MTOCs was cell-cycle regulated. Like-wise, in N. crassa, Minke et al. (1999) proposed the existence of two populations of MTs, only one of which was connected to the nuclei. Here, we analyzed for the first time the locations of MTOCs that were not connected to the nuclei in a filamentous fungus. We found MTOCs in the cytoplasm, some, but not all, of which were close to nuclei. In addition, very active MTOCs were detected at the septa. These MTOCs give rise to a mixed polarity of MTs in A. nidulans. Whether the different MTOC activities are structurally all alike is not yet known. There is evidence that MT nucleation can occur in the absence of γ-tubulin (Martin et al., 1997; Job et al., 2003). Likewise, we observed some cases of MT polymerization starting from the hyphal tip, where γ-tubulin was not detected. However, MT formation from the tip occurred only very rarely, indicating that there may be no true MTOC in this region.
Is KipA a Processive Motor?
We found that GFP-KipA associates with MTs and labels the MT plus ends. The extent of MT decoration, however, depended on the amount of GFP-KipA in the cell. We observed a spot-like labeling of the plus ends when the expression level was low, a comet-like appearance at intermediate expression levels, and uniform decoration of MTs upon overexpression. The comet-like appearance could be explained if we suppose that the motor actively moves along MTs and jams behind the saturated plus ends. This hypothesis was further supported by the construction of a rigor mutant version of GFP-KipA. Based on studies of similar mutants of other kinesin-family proteins, this protein is expected to be unable to hydrolyze ATP and to bind tightly to MTs while being unable to move along them. Similarly, Browning et al. (2003) showed recently that rigor mutants of Tea2p were able to attach to MTs close to the nucleus but were unable to reach the growing MT plus end.
A role of KipA as a processive motor rather than as a principle determinant of MT stability also is supported by the finding that in contrast to the behavior of a tea2 mutant in S. pombe, MTs reach the tip in a ΔkipA mutant. In addition, the benomyl sensitivity of a ΔkipA strain is only slightly increased relative to wild-type, and the double mutants constructed with ΔkipB, ΔkinA, or nudA1, which displayed decreased benomyl sensitivity (our unpublished data), still displayed the curved growth phenotype.
When we asked whether KipA has functions redundant with those of other motor proteins, we found that disruption of kipA partially suppressed the growth defect of a dynein mutation (nudA1) at restrictive temperature. This could be explained if we suppose that dynein and KipA are involved in cargo transportation. Dynein transports cargoes toward the minus end of MTs, whereas KipA seems to be a plus- end-directed motor. The imbalance of motor activities in strains in which either nudA or kipA is mutated could be overcome if both motors were absent from the cell. Besides this interaction between nudA1 and ΔkipA, we found an interaction between ΔkipA and ΔkipB. Disruption of both kinesins affected hyphal extension much more than either of the single mutations, suggesting that KipA and KipB act in different pathways but have overlapping functions. In contrast, the phenotypes of ΔkinA and ΔkipA were both expressed without detectable synergy in the double mutant, suggesting that the two motors act in independent pathways.
Roles for KipA
We found that in a ΔkipA mutant strain, MTs reach the cortex as in wild type, but their ends do not converge at a single point, and instead seem somewhat dispersed and mobile. We propose that KipA is involved in the temporal anchorage of MTs at the cortex. This situation resembles the search-capture mechanism for astral MTs during nuclear migration in S. cerevisiae. MT plus ends dynamically search for and then capture specific sites at the cortex (reviewed by Yamamoto and Hiraoka, 2003). Cortical proteins at the tip of the daughter cell, such as Kar9p and Num1p, are involved in this process (Miller and Rose, 1998; Bloom, 2000). A Num1p homologue, ApsA, is present in A. nidulans and probably serves a similar function (Fischer and Timberlake, 1995; Yamamoto and Hiraoka, 2003; Xiang and Fischer, 2004). However, mutation of num1 or apsA results in nuclear-migration defects rather than polarized-growth defects. Therefore, ApsA is probably not involved in the KipA-dependent anchorage of MTs at the growing tip. Although we did not yet identify the presumed interacting cortex protein, our results point to a new mechanism by which KipA anchors MTs at the center of a growing tip, which in turn seems to be indispensable for the positioning of the SPK (vesicle supply center), which plays a major role in determining growth directionality in filamentous fungi.
Whether KipA also is involved in processes other than polarized growth is not yet clear. However, we detected the protein at the spindle pole bodies and in the midzone of the mitotic spindle during mitosis. This localization may suggest functions of KipA in chromosome segregation like those of the CENP-E kinesin in mammals (Weaver et al., 2003). However, hyphal compartments of A. nidulans contain several nuclei, so that chromosome missegregation in some nuclei may not cause an obvious phenotype.
Acknowledgments
We are grateful to E. Winzenburg for the initial cloning of kipA and thank R. Liese, J. Scheld, and D. Stöhr for excellent technical assistance. We thank S. Kolb for the phylogenetic analysis and Dr. M. Bölker (University of Marburg) for critically reviewing the manuscript. We thank the monitoring editor for many helpful suggestions on the manuscript. We thank Dr. G. Steinberg (Max-Planck-Institute, Marburg) for helpful discussions. This work was supported by the Fonds der Chemischen Industrie, the Max-Planck-Institute for Terrestrial Microbiology, and the Deutsche Forschungsgemeinschaft.
Article published online ahead of print in MBC in Press on November 24, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-02-0083).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aramayo, R., Adams, T. H., and Timberlake, W. E. (1989). A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics 122, 65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S., Bartnicki, D. D., Gierz, G., Lopez-Franco, R., and Bracker, C. E. (1995). Evidence that Spitzenkörper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp. Mycol. 19, 153-159. [DOI] [PubMed] [Google Scholar]
- Behrens, R., and Nurse, P. (2002). Roles of fission yeast Tea1pp in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 157, 783-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, K. (2000). It's a kar9ochore to capture microtubules. Nat. Cell Biol. 2, E96-E98. [DOI] [PubMed] [Google Scholar]
- Browning, H., Hackney, D. D., and Nurse, P. (2003). Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5, 812-818. [DOI] [PubMed] [Google Scholar]
- Browning, H., Hayles, J., Mata, J., Aveline, L., Nurse, P., and McIntosh, J. R. (2000). Tea2pp is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 151, 15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, R., and Timberlake, W. E. (1995). Aspergillus nidulans apsA (anucleate primary sterigmata) encodes a coiled-coil protein necessary for nuclear positioning and completion of asexual development. J. Cell Biol. 128, 485-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbardt, M. (1957). Der Spitzenkörper von Polystictus versicolor. Planta 50, 47-59. [Google Scholar]
- Grove, S. N., and Bracker, C. E. (1970). Protoplasmic organization of hyphal tips among fungi: vesicles and Spitzenkörper. J. Bacteriol. 104, 989-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G., Liu, B., Zhang, J., Zuo, W., Morris, N. R., and Xiang, X. (2001). The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11, 19-24. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., and Momany, C. (2004). Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41, 391-400. [DOI] [PubMed] [Google Scholar]
- Hill, T. W., and Käfer, E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newsl. 48, 20-21. [Google Scholar]
- Huyett, A., Kahana, J., Silver, P., Zeng, X., and Saunders, W. S. (1998). The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 111, 295-301. [DOI] [PubMed] [Google Scholar]
- Job, D., Valiron, O., and Oakley, B. R. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15, 111-117. [DOI] [PubMed] [Google Scholar]
- Karos, M., and Fischer, R. (1999). Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol. Genet. Genomics 260, 510-521. [DOI] [PubMed] [Google Scholar]
- Lin, X., Momany, C., and Momany, M. (2003). SwoHp, a nucleoside diphosphate kinase, is essential in Aspergillus nidulans. Eukaryot. Cell 2, 1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. A., Osmani, S. A., and Oakley, B. R. (1997). The role of γ-tubulin in mitotic spindle formation and cell cycle progression in Aspergillus nidulans. J. Cell Sci. 110, 623-633. [DOI] [PubMed] [Google Scholar]
- Mata, J., and Nurse, P. (1997). Tea1p and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89, 939-949. [DOI] [PubMed] [Google Scholar]
- Miller, R. K., Heller, K. K., Frisèn, L., Wallack, D. L., Loayza, D., Gammie, A. E., and Rose, M. D. (1998). The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9, 2051-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K., and Rose, M. D. (1998). Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140, 377-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke, P. F., Lee, I. H., and Plamann, M. (1999). Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet. Biol. 28, 55-67. [DOI] [PubMed] [Google Scholar]
- Momany, M., Westfall, P. J., and Abramowsky, G. (1999). Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics 151, 557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata, T., and Hirokawa, N. (1995). Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 131, 1039-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W. J. (2003). Adaptation of core mechanisms to generate cell polarity. Nature 422, 766-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. R., Oakley, C. E., Yoon, Y., and Jung, K. M. (1990). γ-Tubulin is a component of the spindle pole body in Aspergillus nidulans. Cell 61, 1289-1301. [DOI] [PubMed] [Google Scholar]
- Osmani, A. H., May, G. S., and Osmani, S. A. (1999). The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274, 23565-23569. [DOI] [PubMed] [Google Scholar]
- Piehl, M., Tulu, U. S., Wadsworth, P., and Cassimeris, L. (2004). Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl. Acad. Sci. USA 101, 1584-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena, N., Alberti-Segui, C., Winzenburg, E., Horn, C., Schliwa, M., Philippsen, P., Liese, R., and Fischer, R. (2001). Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 42, 121-132. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Fischer, R., and Bartnicki-García, S. (2003). Apical growth and mitosis are independent processes in Aspergillus nidulans. Protoplasma 222, 211-215. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Gierz, G., and Bartnicki-García, S. (2000). Dynein and dynactin deficiencies affect the formation and function of the Spitzenkörper and distort hyphal morphogenesis of Neurospora crassa. Microbiology 146, 1743-1752. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Reynaga-Peña, C. G., Gierz, G., and Bartnicki-García, S. (1998). What determines growth direction in fungal hyphae? Fungal Genet. Biol. 24, 101-109. [DOI] [PubMed] [Google Scholar]
- Rischitor, P., Konzack, S., and Fischer, R. (2004). The Kip3-like kinesin KIPB moves along microtubules and determines spindle position during synchronized mitoses in hyphae of Aspergillus nidulans. Eukaryot. Cell 3, 632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D. W. (1999). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schoch, C. L., Aist, J. R., Yoder, O. C., and Turgeon, B. G. (2003). A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet. Biol. 39, 1-15. [DOI] [PubMed] [Google Scholar]
- Seiler, S., Nargang, F. E., Steinberg, G., and Schliwa, M. (1997). Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16, 3025-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., and Plamann, M. (2003). The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., Plamann, M., and Schliwa, M. (1999). Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9, 779-785. [DOI] [PubMed] [Google Scholar]
- Shaw, B. D., Momany, C., and Momany, M. (2002). Aspergillus nidulans swoF encodes an N-myristoyl transferase. Eukaryot. Cell 1, 241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith, H. A., and Sawin, K. E. (2003). Fission yeast mod5p regulates polarized growth through anchoring of Tea1pp at cell tips. Nature 423, 647-651. [DOI] [PubMed] [Google Scholar]
- Snell, V., and Nurse, P. (1994). Genetic analysis of cell morphogenesis in fission yeast - a role for casein kinase II in the establishment of polarized growth. EMBO J. 13, 2066-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube, A., Brill, M., Oakley, B. R., Horio, T., and Steinberg, G. (2003). Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol. Biol. Cell 14, 642-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, M. A., Dean, R. A., Sewall, T. C., and Timberlake, W. E. (1991). Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5, 1161-1171. [DOI] [PubMed] [Google Scholar]
- Suelmann, R., and Fischer, R. (2000). Mitochondrial movement and morphology depend on an intact actin cytoskeleton in Aspergillus nidulans. Cell Motil. Cytoskeleton 45, 42-50. [DOI] [PubMed] [Google Scholar]
- Suelmann, R., Sievers, N., and Fischer, R. (1997). Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol. Microbiol. 25, 757-769. [DOI] [PubMed] [Google Scholar]
- Toews, M. W., Warmbold, J., Konzack, S., Rischitor, P. E., Veith, D., Vienken, K., Vinuesa, C., Wei, H., and Fischer, R. (2004). Establishment of mRFP1 as fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in Escherichia coli (GATEWAY). Curr. Genet. 45, 383-389. [DOI] [PubMed] [Google Scholar]
- Tran, P. T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, F., Mata, J., and Nurse, P. (1995). Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 131, 1529-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring, R. B., May, G. S., and Morris, N. R. (1989). Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin coding genes. Gene 79, 119-130. [DOI] [PubMed] [Google Scholar]
- Weaver, B.A.A., Bonday, Z. Q., Putkey, F. R., Kops, G.J.P.L., Silk, A. D., and Cleveland, D. W. (2003). Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins, D. A., Xiang, X., and Morris, N. R. (1995). An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics 141, 1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Beckwith, S. M., and Morris, N. R. (1994). Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 91, 2100-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., and Fischer, R. (2004). Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41, 411-419. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., and Hiraoka, Y. (2003). Cytoplasmic dynein in fungi: insights from nuclear migration. J. Cell Sci. 116, 4501-4512. [DOI] [PubMed] [Google Scholar]
- Yelton, M. M., Hamer, J. E., and Timberlake, W. E. (1984). Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81, 1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]


