Abstract
A major goal in the study of vertebrate mitosis is to identify proteins that create the kinetochore-microtubule attachment site. Attachment sites within the kinetochore outer plate generate microtubule dependent forces for chromosome movement and regulate spindle checkpoint protein assembly at the kinetochore. The Ndc80 complex, comprised of Ndc80 (Hec1), Nuf2, Spc24, and Spc25, is essential for metaphase chromosome alignment and anaphase chromosome segregation. It has also been suggested to have roles in kinetochore microtubule formation, production of kinetochore tension, and the spindle checkpoint. Here we show that Nuf2 and Hec1 localize throughout the outer plate, and not the corona, of the vertebrate kinetochore. They are part of a stable “core” region whose assembly dynamics are distinct from other outer domain spindle checkpoint and motor proteins. Furthermore, Nuf2 and Hec1 are required for formation and/or maintenance of the outer plate structure itself. Fluorescence light microscopy, live cell imaging, and electron microscopy provide quantitative data demonstrating that Nuf2 and Hec1 are essential for normal kinetochore microtubule attachment. Our results indicate that Nuf2 and Hec1 are required for organization of stable microtubule plus-end binding sites in the outer plate that are needed for the sustained poleward forces required for biorientation at kinetochores.
INTRODUCTION
By conventional electron microscopy (EM), the vertebrate kinetochore is a multilayered structure with its inner core containing the DNA-binding proteins CENP-A and CENP-B and the structural proteins CENP-C, H, and I. The inner core is linked by a 40-nm fibrous gap to a dense outer plate that is ∼40 nm thick. (Jokelainen, 1967; Rieder, 1982; McEwen et al., 1993). Extending outward ∼150 nm from the outer plate is a fibrous corona that includes the motor proteins CENP-E and cytoplasmic dynein (Cooke et al., 1997; Yao et al., 1997; Cleveland et al., 2003). Together, the outer plate and fibrous corona are referred to as the kinetochore “outer domain.” Also associated with the outer domain are spindle checkpoint proteins (Mad1, Mad2, Bub1, BubR1, Bub3, Mps1, and Cdc20) that inhibit anaphase until kinetochores acquire sufficient numbers of microtubules and achieve bipolar metaphase alignment of their chromosomes (reviewed in Cleveland et al., 2003).
Kinetochore microtubules (kMTs) are spindle microtubules whose minus ends are typically anchored at the spindle poles and whose plus ends terminate end-on at the kinetochore. Human kinetochores typically bind 15-22 microtubule plus ends that usually terminate within the outer plate and rarely extend into the fibrous gap. (Wendell et al., 1993; McEwen and Heagle, 1997; McEwen et al., 2001). Chromosome movement depends on the assembly dynamics of kMT plus ends within their kinetochore attachment sites (Rieder and Salmon, 1998; Maddox et al., 2003).
Although it has been known for >30 years that kMTs terminate in the outer kinetochore plate, until recently very little was known about the identities of proteins required to anchor polymerizing and depolymerizing plus-ends of kMTs into the outer plate. However, identification of protein complexes required for kMT attachment has progressed rapidly in the budding yeast S. cerevisiae. One such complex is the Ndc80 complex, which is made up of four components: Ndc80p, Nuf2p, Spc24p, and Spc25p (reviewed in McAinsh et al., 2003). Yeast cells with deleted or mutant members of the complex exhibit loss of kinetochore-microtubule attachment without global loss of kinetochore structure (i.e., Ndc10 complex is not disrupted) (He et al., 2001; Wigge and Kilmartin, 2001). Since its discovery in budding yeast, homologues of the Ndc80 complex have been identified in Schizosaccharomyces pombe, Caenorhabditis elegans, Xenopus, chicken, and human (Howe et al., 2001; Nabetani et al., 2001; Wigge and Kilmartin, 2001; DeLuca et al., 2002; Martin-Lluesma et al., 2002; Bharadwaj et al., 2004; Desai et al., 2003; Hori et al., 2003; McCleland et al., 2003, 2004). The four members of the Ndc80 complex copurify as a tight complex from budding yeast, Xenopus, and human cells (Janke et al., 2001; Wigge and Kilmartin, 2001; Bharadwaj et al., 2004; McCleland et al., 2003). In human cells, the Ndc80 complex components are named Hec1, Nuf2, Spc24, and Spc25, where Hec1 is the Ndc80 homolog based on its original identification as a protein Highly Enhanced in Cancer cells (Chen et al., 1997).
Evidence that the Ndc80 complex has a role in the formation and stability of kMT attachments in vertebrates has come from studies depleting or inhibiting protein function in cells by RNA interference (RNAi), gene disruption, or antibody microinjection. Such cells fail to align their chromosomes, and they lack sustained poleward forces at kinetochores as indicated by abnormally long spindles and unstretched centromeres between sister kinetochores (DeLuca et al., 2002; Bharadwaj et al., 2004; Hori et al., 2003; McCleland et al., 2003, 2004).
Although there is consensus that Ndc80 is essential for normal mitotic progression, its precise role in kMT attachment and kinetochore structure has been controversial. Reduction of Hec1 or Nuf2 by RNAi to low levels (5-10% of control) in HeLa cells results in cells blocked in prometaphase for prolonged periods with very low levels of the spindle checkpoint proteins Mad1 and Mad2 at kinetochores. The loss of Mad1 and Mad2 has been cited as evidence for kMT formation in cells depleted of Ndc80 complex components (Martin-Lluesma et al., 2002; Cleveland et al., 2003; Cheeseman et al., 2004). However, recent studies have indicated that retention of Mad1 and Mad2 at kinetochores depends directly on the Ndc80 complex independent of kMT formation (DeLuca et al., 2003; Meraldi et al., 2004).
Normally, kMTs are stable to cold- or calcium-induced depolymerization because of their kinetochore plus-end attachments; in contrast, most spindle MTs with free plusends are not stable before anaphase (Brinkley and Cartwright, 1975; Weisenberg and Deery, 1976). Based on cellular responses to these treatments, which induce microtubule depolymerization, the role of Hec1 and Nuf2 in kinetochore fiber formation has been controversial. One study reported that depletion of Hec1 by RNAi in HeLa cells did not result in loss of kMTs after calcium treatment, suggesting the presence of stable microtubule attachments to kinetochores (Martin-Lluesma et al., 2002). Another study demonstrated that microtubules were not stable to cold-induced depolymerization in HeLa cells depleted of Nuf2 by RNAi (which also results in a depletion of Hec1), indicating no kMTs were present (DeLuca et al., 2002, 2003). The role of Spc24 and Spc25 in kMT formation has also been debatable. Xenopus tissue culture cells injected with anti-Spc24 or anti-Spc25 antibodies were unable to retain stable kMTs after treatment with cold calcium buffer, implying a defect in kinetochore-microtubule attachment (McCleland et al., 2004). However, a study in HeLa cells found that depletion of Spc24 or Spc25 by RNAi did not result in loss of microtubule polymer by either cold or calcium induced depolymerization (Bharadwaj et al., 2004).
To further understand the role of Hec1 and Nuf2 in kinetochore fiber formation, we first resolve the controversy over whether these proteins are essential for kMT attachment in HeLa cells by a quantitative assay for cold stability, by EM, and by live-cell imaging of kinetochore movements in cells substantially depleted of these proteins by RNAi. Furthermore, we localize Hec1 to the outer plate of the kinetochore using immuno-EM and also show that depletion of Hec1 and Nuf2 by RNAi results in a major structural disruption of the outer plate. Finally, we demonstrate that Hec1 and Nuf2 are spatially separated from the outer domain protein CENP-E and do not exhibit the microtubule-dependent changes in assembly typical of other outer domain components. Together, these results place Hec1 and Nuf2 in a distinct region of the kinetochore at the site of MT plus end attachment within the outer plate, suggesting an essential role for Hec1 and Nuf2 in spatially organizing the sites for kinetochore-MT binding.
MATERIALS AND METHODS
Live Cell Imaging
A GFP-CENP-B full-length construct (kindly provided by Dr. Kevin Sullivan, Scripps Research Institute, San Diego, CA) was transiently transfected into HeLa cells using Fugene6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN). Twenty-four hours after transfection with GFP-CENP-B, cells were transfected with Nuf2 small, interfering RNAs (siRNAs; DeLuca et al., 2002), Hec1 siRNA (Martin-Lluesma et al., 2002), or mock-transfected with transfection reagent alone. Forty-eight hours after siRNA transfection, cells were mounted in Rose chambers and filled with dye-free L-15 media. Cells were then imaged using a Nikon 100× Planapochromat oil immersion lens (Garden City, NY) on a spinning disk confocal fluorescence microscope (DeLuca et al., 2003).
Immunofluorescence
Cells were fixed and prepared for immunofluorescence as described in DeLuca et al. (2002). For nocodazole experiments, cells were incubated with DMEM containing 20 μM nocodazole for 3 h before fixation. Quantification of CENP-E kinetochore fluorescence was carried out as described in Hoffman et al. (2001). CENP-E antibodies were used at a dilution of 1:750 (kindly provided by Dr. Tim Yen, Fox Chase Cancer Center, Philadelphia, PA). Hec1 antibodies (Abcam, Cambridge, United Kingdom) were used at a dilution of 1:1000. CREST serum (Antibodies, Davis, CA) was used at 1:2000 dilution. Tubulin antibodies (Serotec, Raleigh, NC) were used at a dilution of 1:350. Sodium azide/deoxyglucose incubations were carried out as described in Howell et al. (2001). For colocalization analysis, we acquired z-series stacks of confocal images at 0.2-μm intervals (Metamorph software; Universal Imaging, Downington, PA) through cells and processed the image stacks to obtain 3D projections after digital image deconvolution using a Delta Vision image processing workstation (Applied Precision, Seattle, WA).
Electron Microscopy
Coverslips containing treated cells were fixed and flat embedded in Epon, and 80-nm-serial sections were cut, stained, and imaged at 6300× or 8000× as described in McEwen et al. (1997). All calculations and graphs were computed using Microsoft Excel (Redmond, WA). For the immuno-EM, the cells were permeabilized by incubation in 0.2% Triton in PHEM (60 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.9, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2) for 1 min and immediately fixed in 0.5% gluteraldehyde in PHEM for 5 min at room temperature. After fixation, the cells were placed in 0.5 mg/ml NaBH4 in phosphate-buffered saline (PBS) for 5 min to reduce free aldehydes, washed three times in PBS, blocked with KB (10 mM Tris, pH 7.7, 150 mM NaCl) supplemented with 5% bovine serum albumin (BSA) for 30 min at room temperature, and washed two times with incubation buffer (KB supplemented with 0.1% Aurion-BSA-c; Aurion, Wageninger, the Netherlands). Monoclonal antibody Hec1 was diluted 1:500 in incubation buffer and incubated for 1 h at room temperature. After antibody incubation, the cells were washed three times in incubation buffer and treated with 0.8 nm secondary goat anti-mouse gold (Aurion) at 4°C overnight, then washed again with incubation buffer (four times, 5 min) and PHEM (two times, 5 min). The cells were then postfixed with 2.5% gluteraldehyde in PHEM for 30 min, washed with PHEM, and silver-enhanced (Aurion R-GENT SE-EM). They were then dehydrated through a graded ethanol series, flat-embedded in Epon, and processed as previously described in McEwen et al. (1997).
For the RNAi experiments, cells were chosen based on the published phenotype for Nuf2 and Hec1 RNAi depletion: poor chromosome alignment with chromosomes dispersed throughout the cells at varying degrees. For the RNAi plus nocodazole experiments, mitotic cells were chosen at random. The appearance of kinetochores was not used to exclude data once cells were chosen, but in all cases, cells showing signs of apoptosis were avoided (for both control and siRNA-transfected cells, with and without nocodazole treatment). In particular, we scanned cells to make sure there were no signs of chromosome fragmentation; condensed, ruptured, or otherwise abnormal mitochondria; or cell lyses (e.g., large clear zones in the cytoplasm). The number of kinetochores per micron was determined by counting the number of kinetochores identified in serial sections over a 2-4-μm depth (i.e., 25-50 80-nm-thick sections) through an area rich in chromosomes and generally near the center of the cell. This measure was adopted in order to analyze several cells from each treatment rather than to expend a considerably greater amount of effort to determine the total number of kinetochores in one or two cells. Determining the number of kinetochores per chromosome analyzed is unsatisfactory because the boundary between individual chromosomes is ill defined, particularly in the metaphase plate of control cells (e.g., see Figure 2A). This rough estimate of kinetochore density was sufficient to demonstrate to a striking and consistent difference in the number of kinetochores detected in Nuf2-depleted and control cells.
Figure 2.
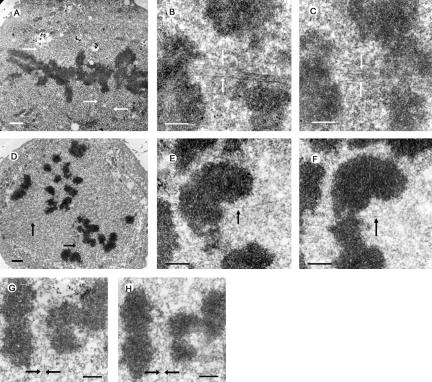
The effect of Nuf2 depletion on kMT binding. Electron micrographs of HeLa cells 48 h after mock-transfection (A-C) or transfection with hNuf2-siRNA (D-H). Arrows indicate typical examples of the robust (A-C) and deficient (D-H) bundles of microtubules associated with kinetochores of mock- and siRNA-transfected cells. (A) Low magnification view of a single serial section of a mock-transfected control cell. The well-formed metaphase plate persisted through all serial sections of the cell. (B and C) Serial section views of a kinetochore from a control cell. All kinetochores that were followed through serial sections had robust kinetochore fibers. (D) Low-magnification view of a single serial section from a transfected cell. Although some sections gave the appearance of partial metaphase alignment, this was not well maintained through the serial sections and virtually no kinetochore fibers were found. (E and F) Serial section views of a kinetochore from a transfected cell. It was more difficult to find kinetochores in transfected cells than in controls cells (see Table 1), and only a few of the kinetochores examined had more than 6-8 kMTs bound. (G and H) Serial section views from a transfected cell showing lateral interactions between spindle MTs and the centromere region of a chromosome. Scale bars: (A and D) 1 μm; (B, C, E, F, G, and H) 400 nm.
RESULTS
Nuf2 and Hec1 Are Essential for Stable kMT Attachments
To resolve the controversial issue regarding the role of Hec1 and Nuf2 in kinetochore fiber formation, we subjected Hec1 siRNA-transfected cells to cold-induced microtubule depolymerization and quantified the fluorescence of the remaining microtubules. We have previously shown that reduction of Hec1 or Nuf2 protein by RNAi in HeLa cells results in reduced protein levels of both (DeLuca et al., 2003), and cells depleted of Nuf2 and Hec1 undergo a long prometaphase block followed by cell death (DeLuca et al., 2002; Martin-Lluesma et al., 2002). Although RNAi of Nuf2 or Hec1 reduces protein levels in a cell population on average to 10-15% (detected by Western blot), the level in many individual cells based on immunofluorescence analysis is 5-10% of normal (DeLuca et al., 2002). Forty-eight hours after Hec1 siRNA transfection, we cooled cells rapidly to 4°C for 10 min before extraction and fixation. Cells were stained for tubulin and Hec1 and whole cell fluorescence values for each were quantified. The amount of cold-stable microtubule polymer remaining in Hec1 siRNA-transfected cells was dependent on the amount of Hec1 at kinetochores (Figure 1A, ♦), and the amount extrapolated to near zero at zero Hec1. As expected, control metaphase cells exhibited high levels of tubulin fluorescence and Hec1 (▪). Furthermore, kinetochores in Hec1-depleted cells were clearly not associated with the few cold-stable fibers that remained after cold treatment, as detected by immunofluorescence (Figure 1B). This is in contrast to control mitotic cells, where kinetochores could clearly be traced to microtubules (Figure 1B). These results demonstrate that few cold-stable microtubules persist in HeLa cells substantially depleted of Nuf2 and Hec1 and that kinetochores in these cells are not attached to the remaining microtubules.
Figure 1.
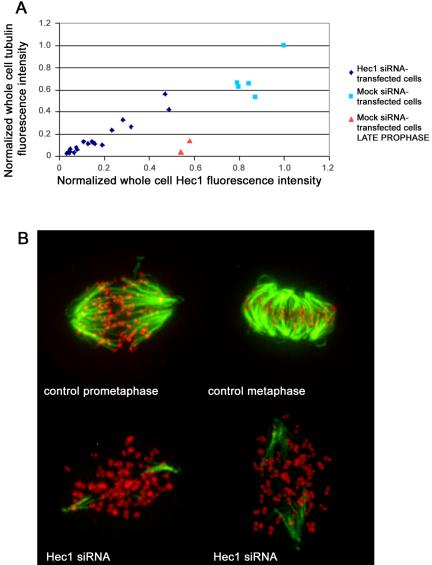
Cold stable microtubules do not persist in cells depleted of Nuf2 and Hec1. Forty-eight hours posttransfection with Hec1 siRNA, HeLa cells were incubated in an icewater bath for 10 min, followed by extraction and fixation on ice. (A) Cells were processed for immunofluorescence and stained with Hec1 and α-tubulin antibodies. Image stacks of mock-transfected and Hec1 siRNA-transfected cells were captured using a spinning disk confocal fluorescence microscope. For quantification, a 20-image z-series stack was summed. A circle that included the entire cell was chosen as the constant area for quantification for all conditions. A background circle twice the area of the original circle was used to measure the background fluorescence. The circle sizes (small circle: d = 200 pixels; large circle: d = 283 pixels) were constant for every cell analyzed. The average whole cell integrated fluorescence intensities for Hec1 and tubulin are plotted in X and Y, respectively. Cells from the test coverslip were chosen to include those with a range of Hec1 depletion levels (blue diamonds). Metaphase cells from control coverslips are represented by the squares (light blue). Cells from control coverslips in late prophase are also shown (pink triangles): these cells have high levels of Hec1, but have not yet formed a bipolar spindle and kMT attachments; thus, the levels of stable microtubule polymer is low. (B) Control and Hec1 siRNA-transfected cells were cold-treated, processed for immunofluorescence, and stained with tubulin antibodies (green) and CREST antibodies (red). Maximum projections were produced from image stacks using the “stack arithmetic” function accessed using the Metamorph software analysis system (Universal Imaging) with identical image processing and contrast enhancement.
The cold-stable assays predict that cells depleted of Hec1 and Nuf2 should have few end-on kinetochore microtubules. To test this, we used transmission EM to examine kinetochores from cells with reduced levels of Nuf2 and Hec1 by Nuf2 RNAi. Our strategy for EM analysis on how Nuf2-depletion alters kMT attachment and kinetochore structure was based on the following. We chose cells with poor chromosome alignment (chromosomes distributed along the length of the spindle) but otherwise normal in appearance (see Materials and Methods). Previously we had shown by immunofluorescence that the majority of mitotic cells 48 h posttransfection fail to align their chromosomes into a metaphase plate, and these cells have low levels of Nuf2 at kinetochores (5-10% of control levels). It is statistically likely that the cells chosen for EM analysis contained unaligned chromosomes as a result of Nuf2 and Hec1 depletion rather than being poorly transfected cells in prometaphase without substantial Nuf2 and Hec1 depletion. This is because at 48 h posttransfection, more than 90% of the living cells have very low levels of Nuf2 and approximately half of these are blocked in a prometaphase-like state (DeLuca et al., 2002). In contrast, <1% of poorly transfected cells are expected to be in prometaphase, because the mitotic index of control cells is low (∼5%) and prometaphase is relatively short in HeLa cells. Furthermore, all of the Nuf2 siRNA-transfected cells analyzed by EM showed significant differences from control cells (Table 1).
Table 1.
Summary of control and siRNA-transfected HeLa cells examined by electron microscopy
| Cell | Treatment | Kin/μm | kMT/Kin | Range kMT/Kin | N |
|---|---|---|---|---|---|
| C-1 | Control | 7.5 | 16.0 | 13-20 | 16 |
| C-2 | Control | 4.9 | 15.3 | 12-22 | 16 |
| Average | Control | 6.2 | 15.6 | 12-22 | 32 |
| TF-1 | Nuf2-siRNA | 2.7 | 6.8 | 3-11 | 6 |
| TF-2 | Nuf2-siRNA | 2.2 | 5.2 | 2-8 | 5 |
| TF-3 | Nuf2-siRNA | 2.8 | 8.3 | 4-12 | 4 |
| TF-4 | Nuf2-siRNA | 1.7 | 4.9 | 2-8 | 7 |
| TF-5 | Nuf2-siRNA | 1.3 | 4.8 | 3-7 | 6 |
| Average | Nuf2-siRNA | 2.1 | 6.0 | 2-12 | 28 |
| NC-1 | Nocodazole control | 28.6 | N/A | N/A | 32 |
| NC-2 | Nocodazole control | 38.5 | N/A | N/A | 52 |
| NC-3 | Nocodazole control | 31.7 | N/A | N/A | 43 |
| Average | Nocodazole control | 32.9 | N/A | N/A | 127 |
| NTF-1 | Nocodazole Nuf2-siRNA | 9.1 | N/A | N/A | 11 |
| NTF-2 | Nocodazole Nuf2-siRNA | 17.5 | N/A | N/A | 21 |
| NTF-3 | Nocodazole Nuf2-siRNA | 13.3 | N/A | N/A | 17 |
| NTF-4 | Nocodazole Nuf2-siRNA | 10.9 | N/A | N/A | 14 |
| NTF-5 | Nocodazole Nuf2-siRNA | 6.3 | N/A | N/A | 7 |
| NTF-6 | Nocodazole Nuf2-siRNA | 10.6 | N/A | N/A | 11 |
| Average | Nocodazole Nuf2-siRNA | 11.3 | N/A | N/A | 81 |
We first analyzed control mitotic HeLa cells, which spend most of their time with chromosomes near the metaphase plate. In these cells, portions of kinetochore fibers were visible in many of the serial section electron micrographs as distinct bundles of microtubules (Figure 2A, arrows). At slightly higher magnifications individual kinetochores were easily identified and exhibited classical kinetochore morphology with an electron-dense outer plate and a robust kinetochore fiber (Figure 2, B and C). The average number of microtubules bound per kinetochore was 15.6 with a range of 12-22 (Table 1, Figure 3B).
Figure 3.
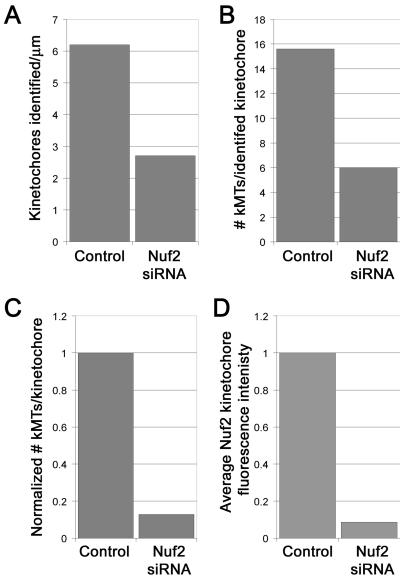
Quantification of microtubule binding to kinetochores in control versus Nuf2-depleted cells. The numbers of kinetochores identified per micrometer of cell sectioned (A), and the number of microtubules per kinetochore (B), were averaged for control (n = 2 cells; 32 kinetochores) and Nuf2 siRNA-transfected cells (5 cells; 28 kinetochores). Kinetochore-microtubule numbers in Nuf2 siRNA-transfected cells were normalized to kinetochore-microtubule numbers in control cells by assigned a value of zero attached microtubules to the “unidentified kinetochores” (C). The average reduction of kinetochore-microtubules detected by EM in C is similar to the average reduction in Nuf2 at kinetochores in siRNA-transfected HeLa cells as measured by immunofluorescence microscopy (D).
In Nuf2 siRNA-transfected cells, chromosomes were generally not aligned into a metaphase plate. Although partial alignment of chromosomes near the spindle equator was observed in many of the serial section electron micrographs, the alignment was never as tight as the metaphase plate in controls, particularly when examining a run of serial-section series. Few substantial bundles of microtubules were observed in these cells (Figure 2D). Furthermore, in contrast to control cells, kinetochores in transfected cells were difficult to identify because few kinetochores exhibited classical kinetochore morphology, and even fewer had robust kinetochore fibers (Figure 2, E and F). To obtain a quantitative estimate of this effect, we counted the number of kinetochores identified per micrometer of cell depth (kinetochores/μm) as described in Materials and Methods. On average only 2.1 kinetochores/μm were identified in Nuf2-depleted cells, whereas 6.2 kinetochores/μm, or three times as many, were identified in control cells (Table 1, Figure 3A). The kinetochores that were detected in Nuf2-depleted cells had an average of 6.3 kinetochore microtubules bound in the classic end-on manner, with a range of 2-12 (Table 1, Figure 3B). This number most likely represents an overestimate of the number of microtubules attached to the majority of the kinetochores in Hec1- and Nuf2-depleted cells, because ∼2/3 of the kinetochores could not be identified and probably had no stably attached microtubules (Table 1, Figure 3A). Figure 3C shows the average number of microtubules per kinetochore, assuming the unidentifiable kinetochores had no attached microtubules. For the graph in Figure 3C, the average number of kinetochores identified per micrometer in control cells was used as a normalized value to determine the estimated number of attached microtubules to kinetochores in Nuf2 siRNA-transfected cells. This number is ∼9% of control values, similar to the average reduction in Nuf2 at kinetochores in the siRNA-transfected HeLa cells as measured by immunofluorescence microscopy (Figure 3D). Thus, in Nuf2- and Hec1-depleted cells, kinetochores were largely unidentifiable, and those kinetochores that were visualized failed to form robust kinetochore fibers. Finally, a small but significant number of the kinetochores exhibited lateral interactions with microtubules (Figure 2, G and H).
Kinetochores with Depleted Levels of Nuf2 and Hec1 Exhibit Erratic Motility
To investigate the nature of kinetochore-microtubule interactions in human cells with reduced levels of Nuf2 and Hec1, we imaged control and Nuf2 siRNA-transfected HeLa cells expressing a fluorescent kinetochore marker, CENPB-GFP, and tracked kinetochore motility. Kinetochores of control cells became aligned along a central axis and exhibited normal oscillations characteristic of kinetochores on bipolar, aligned chromosomes (Figure 4A, Supplementary Movie 1). Oscillatory motions were, for the most part, constrained to a single axis as pairs switched between poleward and away from the pole movements as expected (Shelby et al., 1996). In addition to oscillating, kinetochore pairs also underwent periods of stretching and relaxation (Figure 4A′), reflecting centromere tension between sister kinetochores.
Figure 4.
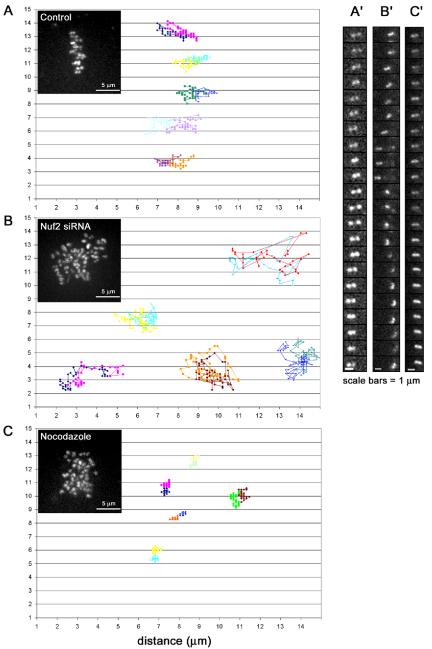
Kinetochores with depleted levels of Nuf2 and Hec1 move erratically and in a microtubule-dependent manner. HeLa cells transiently expressing CENPB-GFP were mock siRNA transfected (A and A′) or transfected with Nuf2 siRNA (B and B′). Forty-eight hours after siRNA transfection, cells were imaged on a spinning disk confocal microscope. Images were collected every 10 s over a period of 10-40 min. For microtubule depolymerization experiments, mock-transfected cells were incubated with 20 μM nocodazole for 2 h before imaging (C and C′). Individual kinetochores and their corresponding sisters were tracked using the “track points” function accessed using the Metamorph software analysis system (Universal Imaging), and points were plotted on X and Y axes (A-C). Insets show a maximum projection of a z-series stack through a cell from each corresponding condition (scale bars, 5 μm). Montages in A′-C′ are frames from the time-lapse movies at 10-s intervals showing a single representative sister kinetochore pair from each condition (scale bars, 1 μm).
Cells depleted of Nuf2 and Hec1 by Nuf2 RNAi also exhibited movements, but these movements differed from control cells in several ways. Kinetochore pairs did not exhibit sustained alignment along a central axis or bidirectional oscillations. Instead they were scattered throughout the spindle region with random orientations of the axis between sister pairs. They exhibited erratic movements, which included flipping and rotating (Figure 4, B and B′, Supplementary Movie 2). This erratic kinetochore behavior was observed for all Nuf2 siRNA-transfected cells analyzed (n = 30). Kinetochore pairs did undergo transient directed movements, but not in a consistent direction (Figure 4B). Pairs very infrequently exhibited brief centromere stretching, but in most cells, no stretching was observed.
To determine if erratic movements occur in the absence of stable kinetochore-microtubule interactions, we treated control-transfected cells with nocodazole to monitor kinetochore movements in the absence of spindle microtubules. As shown in Figure 4C, kinetochores in nocodazole-treated cells were more centrally clustered, and tracking individual kinetochore movements revealed very little motion (Figure 4, C and C′, Supplementary Movie 3). These results indicate that the transient movements observed in the Nuf2 siRNA-transfected cells are microtubule dependent and that Nuf2 and Hec1 are needed for the formation of kMTs capable of generating productive chromosome movements and the tensions associated with bipolar chromosome alignment.
The Ndc80 Complex Defines a Stable Region in the Kinetochore Outer Domain
As detected by immunofluorescence, the Ndc80 complex components localize to the outer domain of the kinetochore similar to many spindle checkpoint proteins and the motor proteins CENP-E and dynein (reviewed in Cleveland et al., 2003). There is evidence, however, that the assembly properties of the Ndc80 complex components differ from many other outer domain proteins. The components of the Ndc80 complex persist at nearly constant levels at kinetochores throughout mitosis, whereas the motors and spindle checkpoint proteins become depleted by kMT formation (Hoffman et al., 2001; DeLuca et al., 2002; Martin-Lluesma et al., 2002; Bharadwaj et al., 2004; Hori et al., 2003; McCleland et al., 2003). By fluorescence recovery after photobleaching (FRAP) analysis, Nuf2 and Hec1 are very stable components of the kinetochore, with recovery times of >30 min (Hori et al., 2003), whereas many other outer domain components are quite dynamic. For example, Cdc20, Bub3, BubR1, Mps1, and 50% of Mad2 turnover with half-times of 20 s or less (Howell et al., 2000, 2004; Kallio et al., 2002; Shah et al., 2004).
We tested the assembly dynamics of the Ndc80 complex at kinetochores relative to the motor and checkpoint proteins of the outer domain in two other ways. One was treating Ptk1 cells with sodium azide/deoxyglucose to lower ATP concentration by ∼100 fold (Howell et al., 2000). We previously found under these conditions that many outer domain proteins leave kinetochores and are transported to the poles along spindle microtubules by dynein. Tested proteins include the spindle checkpoint proteins Mad2, BubR1, and Cdc20 and the motors CENP-E and cytoplasmic dynein (Howell et al., 2001). Cells treated in this manner exhibit a normal outer plate and retain full complements of kMTs by EM (Howell et al., 2001). Thus, these proteins are not required to maintain microtubule attachment at kinetochores nor do they comprise the outer plate structure. If Ndc80 complex components are required to mediate kMT attachment in the outer plate, we predict that they should be retained at kinetochores after treatment with azide/deoxyglucose. As shown in Figure 5A, Hec1 levels in the presence of azide decreased modestly to 80% of control values. In contrast, CENP-E levels were reduced to 15% of control levels as expected (Figure 5B; Howell et al., 2001).
Figure 5.
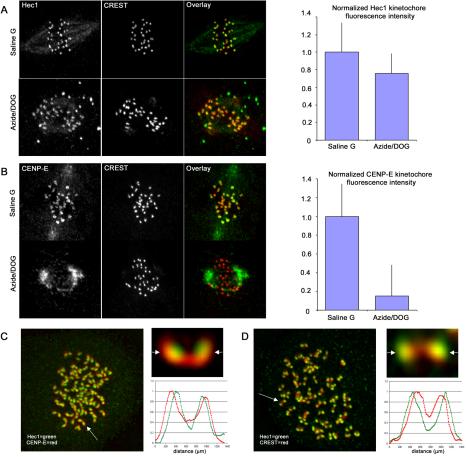
The Ndc80 complex defines a distinct region of the kinetochore outer domain. (A and B) PtK1 cells were incubated for 30 min in either control buffer (Saline G) or buffer supplemented with 5 mM sodium azide/deoxyglucose to deplete ATP before fixation for immunofluorescence. Hec1 is largely retained at kinetochores after inhibitor treatment (A), whereas CENP-E fluorescence is diminished at kinetochores and concentrated at spindle poles after inhibitor treatment (B). For quantification, ∼130 kinetochores from 8 different cells were measured for each condition. (C and D) Deconvolved immunofluorescence images from cells treated for 2 h with 20 μM nocodazole before fixation: stained for CENP-E (C) or CREST antigens in red (D), and Hec1 in green. A single kinetochore pair (top) is also shown for each: arrows indicate the planes where line scans were performed (bottom). Line scans were performed using the “Linescan” function accessed using the Metamorph software system (Universal Imaging). Data from linescans were logged into Microsoft Excel to generate graphs.
We next tested whether depolymerization of all spindle microtubules causes Hec1 to assemble into the extended “crescents” and “collars” around the centromere as seen for outer domain spindle checkpoint and motor proteins (Thrower et al., 1996; Hoffman et al., 2001). We treated cells with nocodazole for 2 h to depolymerize all microtubules and then coimmunolocalized Hec1 with CENP-E. Although some expansion of Hec1 localization was observed, it remained as more punctate fluorescence within the center of the extended “crescents” and “collars” exhibited by CENP-E (Figure 5C). Line scans through deconvolved image stacks of kinetochore pairs demonstrate this point, as the Hec1 peak fluorescence is interior relative to the peak CENP-E fluorescence at the kinetochore (Figure 5C). We found this localization pattern of Hec1 and Nuf2 relative to CENP-E at kinetochores in nonnocodazole-treated cells as well (unpublished data). Although Hec1 fluorescence is detected interior to the CENP-E fluorescence, it remains exterior to the CREST antigens, which is made obvious by the line scans in Figure 5D (Wigge and Kilmartin, 2001). Together, the above results suggest that the Ndc80 complex comprises a stable “core” region of the outer domain of the kinetochore involved in kMT attachment, but spatially distinct from motor and spindle checkpoint proteins that may be more concentrated in the corona fibers.
Hec1 Localizes to the Kinetochore Outer Plate
Previous reports have immunolocalized Hec1 and Nuf2 exterior to the CREST antigens (CENP-A, -B, and -C; Wigge and Kilmartin, 2001; DeLuca et al., 2002; Martin-Lluesma et al., 2002), and we present evidence above that Hec1 localizes to the interior of CENP-E. However, the specific localization of Ndc80 complex components at kinetochores is unknown. To determine where within the vertebrate kinetochore the Ndc80 complex resides, we carried out immuno-EM using a monoclonal Hec1 antibody in nocodazole-treated HeLa cells. Results with preextracted cells show gold particles concentrated predominantly into linear arrays at the centromere region of chromosomes, with a very low background staining elsewhere in the cell (Figure 6A). Often the outer plate was visible in regions between gold particles and examination of several such images reveals that gold labeling can be found on the inner, outer, and middle portions of the outer plate (Figure 6, B-D). In the occasional en face views we found the gold label was distributed in more of a circular area (Figure 6E). Collectively, these results give strong support to our postulate that Hec1 is located throughout the kinetochore outer plate.
Figure 6.
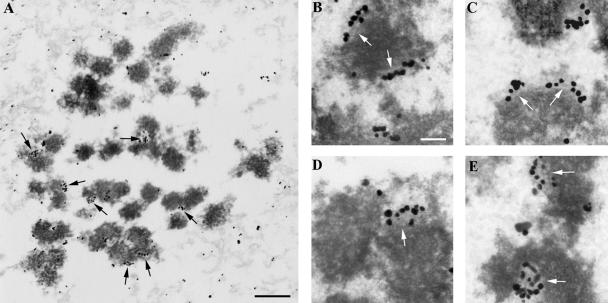
Hec1 localizes to the kinetochore outer plate. HeLa cells grown to 60-80% confluency were treated with 20 μM nocodazole for 3 h, extracted, and fixed. Fixed cells were treated with monoclonal anti-Hec1 primary antibody and anti-mouse secondary conjugated to 0.8-nm gold clusters that were then enhanced with silver reagent. (A) Low-magnification overview. Linear concentrations of silver-enhanced gold particles distinguishes kinetochore labeling (arrows) from the sparse occurrence of particles in the background. (B-D) Higher magnification examples of kinetochore labeling. Arrows indicate regions where the outer plate is visible underneath gold particles. (E) Enface or semienface views of labeled kinetochores (arrows). Note the more circular arrangement of gold particles. Scale bars: (A) 1 μm; (B-E) 200 nm.
Nuf2 and Hec1 Are Essential for Maintenance of the Kinetochore Outer Plate
Because Hec1 localizes to the kinetochore outer plate (Figure 6), and kinetochores were extremely difficult to detect by EM in Nuf2 siRNA-transfected cells (Figures 2 and 3), we tested whether Nuf2 and Hec1 have a structural role in forming and/or maintaining the kinetochore outer plate. Mock-transfected control cells and Nuf2 siRNA-transfected cells were incubated with 20 μM nocodazole for 3 h to depolymerize all spindle microtubules before fixation for EM. Incubation under these conditions is known to result in conspicuous kinetochores with prominent electron dense outer plates and a dense corona of fibrous material as expected from accumulation of motors and spindle checkpoint proteins in the absence of MTs (Rieder, 1979; McEwen et al., 1993; Thrower et al., 1996; Hoffman et al., 2001). In control cells treated with nocodazole, unbound kinetochores were readily located in regions of the cells where unaligned chromosomes accumulated (Figure 7A). Most kinetochores exhibited an unambiguous outer plate, although some were more poorly formed (Figure 7, B and C). The poorly formed kinetochores could result from orientation of the kinetochore at large angles to the microscope axis or actual structural differences between kinetochores.
Figure 7.
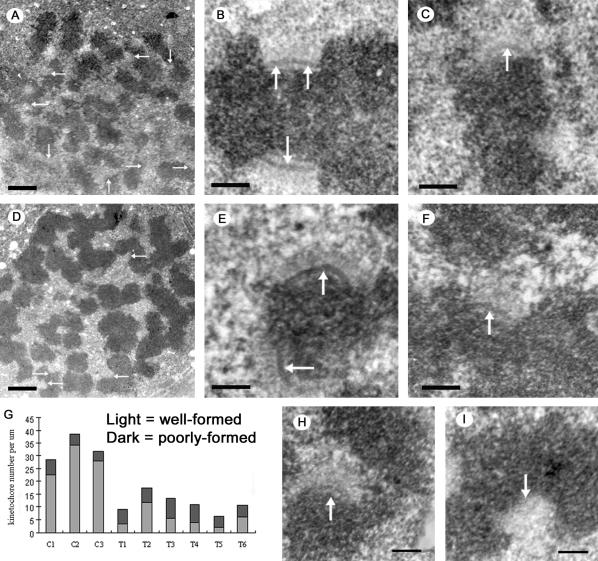
The effect of Nuf2 depletion on the formation and/or stability of the kinetochore outer plate. HeLa cells 48 h after mock-transfection (A-C), or transfection with hNuf2-siRNA (D-F, H, and I), were treated with 20 μM nocodazole for 3 h to disassemble all MTs and promote growth of the kinetochore outer domain. Cells were then fixed and prepared for EM. (A) Low-magnification view of a nocodazole-treated, mock-transfected cell. Several well-formed, unbound kinetochores are visible in the section (arrows). (B) A pair of well-formed sister kinetochores from a control cell. Note the robust outer plates (arrows). (C) Example of a poorly formed kinetochore from a control cell. The outer plate (arrow) is less dense and robust compared with the example in B. (D) Low-magnification view of a nocodazole-treated transfected cell. Unbound kinetochores (arrows) were significantly harder to locate compared with control cells. (E) A pair of well-formed sister kinetochores from a transfected cell. As in B, the outer plates (arrows) are robust. (F) Poorly formed kinetochore from a transfected cell. The outer plate is much less distinct (arrow). (G) Bar graph showing the numbers of kinetochores found in three control and six transfected cells. The light and dark shadings indicate the number of well-formed (light) and poorly formed kinetochores (dark) found per micron of depth in each of the cells. (H and I) Examples of fibrous material at the putative centromere region of chromosomes from transfected cells. Scale bars: (A and D) 1 μm; (B, C, E, F, H, and I) 200 nm.
Both robust and poorly formed kinetochores were found in nocodazole and siRNA-treated cells, but they were less numerous (Figure 7, D-F). Quantitatively, 33.2 kinetochores/μm were identified in nocodazole-treated control cells, whereas only 11.3 kinetochores/μm, or ∼1/3 of the control values, were identified in nocodazole- and siRNA-treated cells (Table 1, Figure 7G). Furthermore, a greater percentage of the kinetochores identified in the siRNA-transfected cells were poorly formed (Figure 7G). The differences were particularly pronounced in the more severely affected cells where only 6.3-10.9 kinetochores/μm could be identified, and up to 70% of those identified were the poorly formed variety (Table 1, Figure 7G). These results indicate that the kinetochore outer plate exhibits varying degrees of disorganization in cells strongly affected by hNuf2 RNAi. Consequently, small amounts of disorganization result in a poorly defined outer plate, and large amounts of disorganization result in an unrecognizable kinetochore because of the absence of a detectable outer plate. Therefore, as we found for nonnocodazole-treated cells, the predominant phenotype in Nuf2 siRNA-transfected cells is the “unidentified kinetochore.” These results strongly suggest that Ndc80 complex components are required for forming and/or maintaining the structure of the kinetochore outer plate.
Interestingly, in nocodazole-treated cells depleted of Nuf2, some chromosomes without distinct kinetochore outer plates still exhibited a fuzzy, fibrous zone extending out from what appears to be the centromere region (Figure 7, H and I). Such zones could be the fibrous components of the outer plate and corona, assembled in a somewhat disorganized manner.
The observation that corona components are still present in cells lacking high levels of Nuf2 and Hec1 is not surprising, given previous reports that have demonstrated that Nuf2 and Hec1 are not required for kinetochores to bind CENP-E, a known component of the corona region (Yao et al., 1997; DeLuca et al., 2002; Martin-Lluesma et al., 2002). To confirm quantitatively that the corona-associated protein CENP-E remains at kinetochores in cells with reduced Hec1 and Nuf2, we measured CENP-E levels at kinetochores in cells transfected with Hec1 siRNA. As shown in Figure 8, mock siRNA-transfected cells in prometaphase exhibited high levels of kinetochore-bound CENP-E. As expected from earlier work, CENP-E is still present on kinetochores of control metaphase chromosomes, but reduced to 40% of the levels at unattached kinetochores in prometaphase. In Nuf2 siRNA-transfected cells, CENP-E levels at kinetochores remained high, although slightly reduced: ∼75% of the control prometaphase value, but twice the control metaphase value (Figure 8). Thus, cells with reduced levels of Nuf2 and Hec1 are able to bind high levels of the corona protein, CENP-E.
Figure 8.
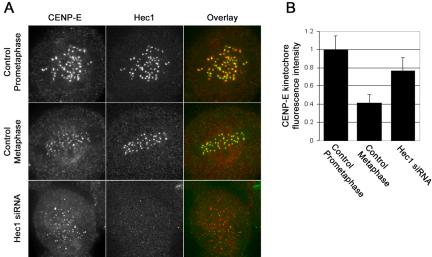
Hec1 siRNA-transfected cells retain high levels of the kinetochore corona component CENP-E. (A) Immunofluorescence images of a control (mock siRNA-transfected) prometaphase cell (top row), a control (mock-transfected) metaphase cell (middle row), and a Hec1 siRNA-transfected cell (bottom row). For each of the images shown, five sequential frames from a through-focal series were projected onto a single image. (B) CENP-E kinetochore fluorescence was measured in three cell populations: control (mock-transfected) prometaphase cells (n = 67 kinetochores), control (mock-transfected) metaphase cells (n = 27 kinetochores), and Hec1 siRNA-transfected cells (n = 165 kinetochores). Values for the graph were normalized to the average kinetochore fluorescence intensity of CENP-E at control prometaphase kinetochores.
DISCUSSION
HeLa cells with depleted levels of Nuf2 and Hec1 fail to form normal kMT attachments. Three lines of evidence support this claim. First, the amount of stable microtubule polymer remaining in these cells after cold-induced depolymerization was dependent on the level of Hec1, such that the microtubule polymer level extrapolated to near zero at zero Hec1 (Figure 1). Second, few microtubules were detected by EM in these cells (Figures 2 and 3, and Table 1). Finally, sister kinetochores in these cells were unable to generate centromere tension and the productive chromosome movements necessary for biorientation to occur (Figure 4).
It is noteworthy that some kinetochores in Nuf2 siRNA-transfected cells retained low numbers of kMTs. What accounts for the infrequent accumulation of kMTs seen by EM in the Nuf2-depleted cells? One explanation is that low levels of Nuf2 and Hec1 at kinetochores after Nuf2 or Hec1 siRNA transfection (DeLuca et al., 2003) are able to form a limited number of attachments for microtubule plus ends. It is possible that these sites are able to hold onto the plus ends at low kinetochore tension, but are unable to sustain attachment at the elevated tensions that are produced by sister kinetochores becoming attached to microtubules from opposite poles and pulling in opposite directions. These attachments are likely weak and transient because the only movements we saw for individual kinetochores were transient movements. Sister pairs were rarely stretched apart and often were rotated and flipped over, motions that do not reflect persistent microtubule attachments to kinetochores.
It is clear that kinetochores are not properly attached to microtubules in cells with depleted levels of Nuf2 and Hec1, but how is kMT attachment process perturbed? In order for productive attachment to take place, several events must occur. Microtubules must first be recruited to the kinetochore region. Initial microtubule attachments must then be converted into end-on attachments within the kinetochore outer plate. Finally, proteins within the microtubule binding sites must hold on to microtubule plus ends for substantial lengths of time as they undergo periods of polymerization and depolymerization, supporting the high tensions across kinetochore pairs associated with chromosome biorientation.
Our data and others suggest that the Ndc80 complex components are not essential for the initial microtubule recruitment to the kinetochore. We have observed erratic and randomly oriented microtubule-dependent movements of kinetochores in HeLa cells depleted of Nuf2 and Hec1 (Figure 4). These results suggest that transient and/or lateral interactions between kinetochores and microtubules are not inhibited. Similarly, McCleland et al. (2004) observed erratic chromosome movements parallel to the mitotic spindle in Xenopus S3 cells injected with antibodies to Spc24 or Spc25.
Rather than functioning in microtubule recruitment to kinetochores, our results suggest a model in which Nuf2 and Hec1 are involved in formation of the end-binding sites for microtubule plus ends within the kinetochore outer plate; sites that can maintain attachment under the strong pulling forces generated at kinetochores of bioriented chromosomes (Figure 9). Support of this model is threefold: first, Hec1 and Nuf2 localize to the site of microtubule plus-end termination within the kinetochore. Second, Hec1 and Nuf2 are required for maintaining the structure of the kinetochore outer plate. And third, Hec1 and Nuf2 exhibit unique dynamic properties compared with other outer domain proteins, properties that are consistent with their role in helping form plus-end binding sites for kinetochore microtubules
Figure 9.
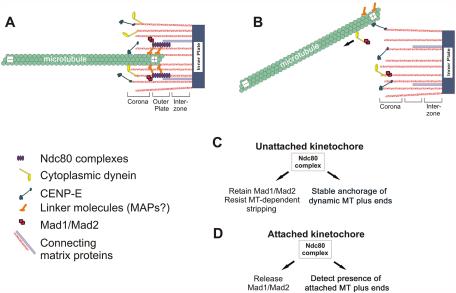
Model for Hec1 and Nuf2 function at the vertebrate kinetochore. Kinetochore structure and protein localization for (A) untreated and (B) Nuf2- or Hec1-depleted kinetochores. We suggest a role of the Ndc80 complex in preventing stripping of Mad1/Mad2 and dynein/dynactin complexes from kinetochores by microtubule interactions (C) and in signaling release of these protein complexes upon normal kinetochore microtubule formation (D). See text for details.
First, by immuno-EM, we show that Hec1 localizes specifically throughout the kinetochore outer plate and does not extend out into the fibrous corona (Figure 6). Additionally, by immunofluoresence microscopy, antibodies to Hec1 and Nuf2 localize more interiorly at the kinetochore than antibodies to CENP-E (Figure 5), which has been localized by immuno-EM to the outer regions of the outer plate into the fibrous corona (Yao et al., 1997). Thus, members of the Ndc80 complex localize to the site of microtubule plus end termination within the kinetochore outer plate (McEwen and Heagle, 1997; McEwen et al., unpublished data), which is a region of the outer domain that is distinct from CENP-E.
Second, kinetochores in cells depleted of Nuf2 and Hec1 fail to form and/or maintain outer plate structure (Figures 2 and 7), thus removing the sites for microtubule end-on attachment (Figure 9B). Presumably the Ndc80 complex is required for the formation of the microtubule binding sites in C. elegans as well; depletion of Him10 (the C. elegans Nuf2 homolog) results in loss of kMT attachment and disruption of the “ribosome exclusion zone,” which is thought to correspond to the corona in conventional EM preparations (McEwen et al., 1998; Howe et al., 2001). We propose that the Ndc80 complex is either a structural component of plus-end attachment sites within the outer plate or is responsible for their organization. Because there is no evidence that members of the Ndc80 complex directly bind to microtubules, a microtubule-associated protein or protein complex, such as the Dam1 complex identified in budding yeast (reviewed in McAinsh et al., 2003) likely mediates the interaction between microtubules and the binding sites at kinetochores through the Ndc80 complex (Figure 9A).
Notably, some chromosomes without distinct kinetochore outer plates still retained a fuzzy, fibrous region extending out from the centromere region (Figure 7, H and I), which could be the fibrous components of the outer plate and corona, although organized to a lesser degree. In nocodazole-treated cells depleted of Ndc80 components by RNAi, these loosely organized fibers must contain many outer domain proteins. This is because depletion by RNAi of Ndc80 components to ∼5% of normal results in kinetochores that retain significant levels of outer domain proteins such as CENP-F, CENP-E, dynein, dynactin, ZW10, Rod, BubR1, Mad1, and Mad2 in the absence of spindle microtubules (DeLuca et al., 2002; Bharadwaj et al., 2004; McCleland et al., 2003). These findings have important implications for vertebrate kinetochore structure, because many proteins that reside in the outer domain and fibrous corona must be capable of binding to the kinetochore independently of an intact outer plate in the absence of spindle microtubules. Therefore the different regions of the outer domain do not necessarily build on each other in a linear way, but depend on a network of interactions.
Finally, Nuf2 and Hec1 exhibit characteristics that distinguish them from other well-characterized outer domain components. First, Hec1 and Nuf2 are retained at kinetochores when ATP is depleted by sodium azide/deoxyglucose (Figure 5), which induces the outer domain proteins Mad2, BubR1, Cdc20, and CENP-E, to be transported by cytoplasmic dynein to the poles along spindle microtubules. (Howell et al., 2001). Second, Hec1 and Nuf2 do not deplete with kMT formation or form large collars encircling kinetochores after microtubule depolymerization as do the other outer domain motor and checkpoint proteins (Figure 5; Hoffman et al., 2001). Finally, a previous report has shown by FRAP analysis Nuf2 and Hec1 are stable components of the kinetochore (Hori et al., 2003). Taken together, these results suggest that Nuf2 and Hec1 within the outer plate form a stable “core” region of the kinetochore outer domain, which is distinct from many motor and spindle checkpoint protein components.
The model shown in Figure 9 is diagrammed for normal cells (Figure 9A) and for cells where the Ndc80 complex is disrupted by depletion of Hec1 and Nuf2 (Figure 9B). Substantial depletion of Hec1 and Nuf2 does not disrupt attachment of the inner plate to fibrous elements that extend throughout the interzone, outer plate, and corona regions of the kinetochore (Figure 9B). We suggest that the Ndc80 complex binds to these filaments and helps organize and concentrate plus-end attachment complexes into the dense outer plate structure seen by conventional fixation (Figure 7).
Localization of Hec1 and Nuf2 to kinetochores is dependent on the kinetochore inner plate proteins CENP-I and CENP-H, and these proteins are not lost from kinetochores in cells depleted of Nuf2 and Hec1 (Hori et al., 2003). Thus, CENP-I and CENP-H may be part of the fibrous elements that extend from the inner plate, through the interzone, and out to the Ndc80 complex in the outer plate. The fibrous elements may also include CENP-F, a kinetochore protein that is not lost with depletion of Hec1 (Martin-Lluesma et al., 2002). We show the motor proteins CENP-E and dynein/dynactin complexes localized to the outer regions of the fibrous elements since they are normally part of the fibrous corona, and they are not substantially lost from kinetochores lacking outer plates in nocodazole treated cells >90% depleted of Nuf2 and Hec1.
The Ndc80 complex is represented in Figure 9 as a single unit, due to biochemical studies that have purified Hec1 (Ndc80), Nuf2, Spc24, and Spc25 as a tight complex (Janke et al., 2001; Wigge and Kilmartin, 2001; Bharadwaj et al., 2004; McCleland et al., 2003). Depletion of Hec1 or Nuf2 results in reduced protein levels of both (DeLuca et al., 2003; Hori et al., 2003), and the same is true for Spc24 and Spc25: depletion of either protein results in depletion of both (McCleland et al., 2004). It is not yet clear, however, if loss of Spc24 and Spc25 results in loss of Nuf2 and Hec1 and vice versa (McCleland et al., 2004). This remains an important unanswered question, and its resolution will certainly further our understanding of how different members of this complex function at kinetochores.
Also shown in Figure 9 are additional connections linking the spindle checkpoint proteins Mad1 and Mad2 to both the Ndc80 complex and cytoplasmic dynein/dynactin, because there is evidence that the Ndc80 complex functions to retain Mad1/Mad2 to kinetochores (DeLuca et al., 2002; Martin-Lluesma et al., 2002; McCleland et al., 2004; Meraldi et al., 2004), whereas cytoplasmic dynein and dynactin function to strip Mad1/Mad2 from kinetochores by transport along spindle microtubules (Howell et al., 2001).
In conclusion, our data support a model in which Hec1 and Nuf2 1) are part of a stable core kinetochore complex that is required for the structure of the outer plate of the vertebrate kinetochore; 2) are required for organization of stable microtubule plus-end binding sites in the outer plate that are needed for the sustained poleward forces required for biorientation at kinetochores; and 3) regulate Mad1/Mad2 kinetochore localization.
Supplementary Material
Acknowledgments
The authors thank Dr. John Kilmartin for kindly providing the initial reagents to begin this project, Dr. Kevin Sullivan for generously providing CENPB-GFP constructs, Dr. Helder Maiato for assistance with the immuno-EM protocol, Dr. Tim Yen for CENP-E antibodies, and Drs. Chad Pearson and Daniela Cimini for helpful comments on the manuscript. We also wish to thank Mr. Curtis Joslin for recording electron micrographs, Sarah Waggoner for help preparing figures, and the Wadsworth Core Facility for Electron Microscopy. This work was supported by National Institutes of Health (NIH) GM066270 and National Science Foundation MCB0110821 to B.F.M., NIH GM24364 to E.D.S., and NIH GM66588 to J.G.D.
Article published online ahead of print in MBC in Press on November 17, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0852).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bharadwaj, R., Qi, W., and Yu, H. T. (2004). Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 279, 13076-13085. [DOI] [PubMed] [Google Scholar]
- Brinkley, B. R., and Cartwright, J., Jr. 1975. Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann. NY Acad. Sci. 253, 428-439. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I. M., Niessen, S., Anderson, S., Hyndman, F., Yates, J. R., III, Oegema, K., and Desai, A. (2004). A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18, 2255-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Riley, D. J., Chen, P. L., and Lee, W. H. (1997). HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 17, 6049-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D. W., Mao, Y., and Sullivan, K. F. (2003). Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407-421. [DOI] [PubMed] [Google Scholar]
- Cooke, C. A., Schaar, B., Yen, T. J., and Earnshaw, W. C. (1997). Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106, 446-455. [DOI] [PubMed] [Google Scholar]
- DeLuca, J. G., Howell, B. J., Canman, J. C., Hickey, J. M., Fang, G., and Salmon, E. D. (2003). Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13, 2103-2109. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., Moree, B., Hickey, J.M., Kilmartin, J.V., and Salmon, E.D. (2002). hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159, 549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., Rybina, S., Muller-Reichert, T., Shevchenko, A., Hyman, A., and Oegema, K. (2003). KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17, 2421-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Rines, D. R., Espelin, C. W., and Sorger, P. K. (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106, 195-206. [DOI] [PubMed] [Google Scholar]
- Hoffman, D. B., Pearson, C. G., Yen, T. J., Howell, B. J., and Salmon, E. D. (2001). Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12, 1995-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Haraguchi, T., Hiraoka, Y., Kimura, H., and Fukagawa, T. (2003). Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116, 3347-3362. [DOI] [PubMed] [Google Scholar]
- Howe, M., McDonald, K. L., Albertson, D. G., and Meyer, B. J. (2001). HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153, 1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B. J., Hoffman, D. B., Fang, G., Murray, A. W., and Salmon, E. D. (2000). Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150, 1233-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B. J., McEwen, B.F., Canman, J.C., Hoffman, D.B., Farrar, E.M., Rieder, C.L., and Salmon, E.D. (2001). Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B. J., Moree, B., Farrar, E. M., Stewart, S., Fang, G., and Salmon, E. D. (2004). Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14, 953-964. [DOI] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Lechner, J., Shevchenko, A. Shevchenko, A., Magiera, M. M., Schramm, C., and Schiebel, E. (2001). The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20, 777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokelainen, P. T. (1967). The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 19, 19-44. [DOI] [PubMed] [Google Scholar]
- Kallio, M. J., Beardmore, V. A., Weinstein, J., and Gorbsky, G. J. (2002). Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158, 841-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P., Straight, A., Coughlin, P., Mitchison, T. J., and Salmon, E. D. (2003). Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., DeLuca, J. G., Salmon, E. D., and Earnshaw, W. C. (2004). The dynamic kinetochore-microtubule interface. J. Cell Sci. 117, 5461-5477. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., Stucke, V. M., and Nigg, E. A. (2002). Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267-2270. [DOI] [PubMed] [Google Scholar]
- McAinsh, A. D., Tytell, J. D., and Sorger, P. K. (2003). Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19, 519-539. [DOI] [PubMed] [Google Scholar]
- McCleland, M. L., Gardner, R. D., Kallio, M. J., Daum, J. R., Gorbsky, G. J., Burke, D. J., and Stukenberg, P. T. (2003). The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17, 101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland, M. L., Kallio, M. J., Barrett-Wilt, G. A., Kestner, C. A., Shabanowitz, J., Hunt, D. F., Gorbsky, G. J., and Stukenberg, P. T. (2004). The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14, 131-137. [DOI] [PubMed] [Google Scholar]
- McEwen, B. F., and Heagle, A. B. (1997). Electron microscopic tomography: a tool for probing the structure and function of subcellular components. J. Imaging Syst. Technol. 8, 175-187. [Google Scholar]
- McEwen, B. F., Arena, J. T., Frank, J., and Rieder, C. L. (1993). Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J. Cell Biol. 120, 301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B. F., Heagle, A. B., Cassels, G. O., Buttle, K. F., and Rieder, C. L. (1997). Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 137, 1567-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B. F., Hsieh, C.-E., Mattheyses, A. L., and Rieder, C. L. (1998). A new look at the kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107, 366-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B. F., Chan, G. K., Zubrowski, B., Savoian, M. S., Sauer, M. T., and Yen T. J. (2001). CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., Draviam, V. M., and Sorger, P. K. (2004). Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7, 45-60. [DOI] [PubMed] [Google Scholar]
- Nabetani, A., Koujin, T., Tsutsumi, C., Haraguchi, T., and Hiraoka, Y. (2001). A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma 110, 322-334. [DOI] [PubMed] [Google Scholar]
- Rieder, C. L. (1979). Localization of ribonucleoprotein in the trilaminar kinetochore of PtK1. J. Ultrastruct. Res. 66, 109-119. [DOI] [PubMed] [Google Scholar]
- Rieder, C. L. (1982). The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79, 1-58. [DOI] [PubMed] [Google Scholar]
- Rieder, C. L., and Salmon, E. D. (1998). The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8, 310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J. V., Botvinick, E., Bonday, Z., Furnari, F., Berns, M., and Cleveland, D. W. (2004). Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14, 942-952. [DOI] [PubMed] [Google Scholar]
- Shelby, R. D., Hahn, K. M., and Sullivan, K. F. (1996). Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 135, 545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower, D. A., Jordan, M. A., and Wilson, L. (1996). Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskelet. 35, 121-133. [DOI] [PubMed] [Google Scholar]
- Wendell, K. L., Wilson, L., and Jordan, M. A. (1993). Mitotic block in HeLa cells by vinblastine: ultrastructural changes in kinetochore-microtubule attachment and centrosomes. J. Cell Sci. 104, 261-274. [DOI] [PubMed] [Google Scholar]
- Weisenberg, R. C., and Deery, W. J. (1976). Role of nucleotide hydrolysis in microtubule assembly. Nature 263, 792-793. [DOI] [PubMed] [Google Scholar]
- Wigge, P. A., and Kilmartin, J. V. (2001). The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152, 349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Anderson, K. L., and Cleveland, D. W. (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


