Abstract
Programmed double-strand breaks (DSBs), which initiate meiotic recombination, arise through the activity of the evolutionary conserved topoisomerase homolog Spo11. Spo11 is believed to catalyze the DNA cleavage reaction in the initial step of DSB formation, while at least a further 11 factors assist in Saccharomyces cerevisiae. Using chromatin-immunoprecipitation (ChIP), we detected the transient, noncovalent association of Spo11 with meiotic hotspots in wild-type cells. The establishment of this association requires Rec102, Rec104, and Rec114, while the timely removal of Spo11 from chromatin depends on several factors, including Mei4 and Ndt80. In addition, at least one further component, namely, Red1, is responsible for locally restricting Spo11's interaction to the core region of the hotspot. In chromosome spreads, we observed meiosis-specific Spo11-Myc foci, independent of DSB formation, from leptotene until pachytene. In both rad50S and com1Δ/sae2Δ mutants, we observed a novel reaction intermediate between Spo11 and hotspots, which leads to the detection of full-length hotspot DNA by ChIP in the absence of artificial cross-linking. Although this DNA does not contain a break, its recovery requires Spo11's catalytic residue Y135. We propose that detection of uncross-linked full-length hotspot DNA is only possible during the reversible stage of the Spo11 cleavage reaction, in which rad50S and com1Δ/sae2Δ mutants transiently arrest.
Keywords: spo11-Y135, cleavage complex, “tight” binding, DSB repair, Spo11 cleavage model
Meiosis is a specialized form of cell division essential for sexually reproducing organisms to generate gametes. Following a single round of DNA replication, two successive rounds of chromosome segregation reduce the diploid chromosome complement by half. In yeast, as in most organisms, meiotic recombination is required to sort the two sets of homologs and connect the corresponding chromosome pairs, so that they can be accurately segregated in meiosis I.
In Saccharomyces cerevisiae, meiotic recombination results from the formation and repair of transient DNA double-strand breaks (DSBs). SPO11 was one of the first meiotic recombination genes to be identified (Esposito and Esposito 1969; Klapholz et al. 1985). The meiosis-specific protein, Spo11, shares homology with the catalytic subunit (TOP6A) of an archeal type II topoisomerase from Sulfolobus shibatae (Bergerat et al. 1997). Spo11 is thought to catalyze the formation of programmed meiotic DSBs by generating a phosphodiester link between its catalytic tyrosine, Y135, and a DNA 5′ terminus. rad50S, mre11S, and com1/sae2 mutants are thought to be blocked at this stage of the cleavage reaction (Alani et al. 1990; Keeney and Kleckner 1995; McKee and Kleckner 1997; Nairz and Klein 1997; Prinz et al. 1997). In fact, the covalent linkage of Spo11 with DNA ends has been directly observed in rad50S and com1 mutants (Keeney and Kleckner 1995), but not in wild-type cells. (Below we will refer to the com1Δ/sae2Δ mutant as com1Δ for simplicity.)
In S. cerevisiae, neither meiotic DSBs nor Holliday junctions nor mature recombination products are formed in the absence of Spo11, and homologous chromosomes do not synapse (Cao et al. 1990; Schwacha and Kleckner 1994; for review, see Keeney 2001). Furthermore, it was recently shown that the targeting of Spo11 to a given DNA region (by virtue of its fusion with a DNA-binding domain) is sufficient to significantly increase the level of meiotic recombination at that site (Peciña et al. 2002). Spo11 appears to be the evolutionary conserved key protein in the initiation of recombination. Molecular and genetic studies in other fungi and higher eukaryotes have demonstrated that Spo11 orthologs are universally required for the initiation of meiotic recombination. Furthermore, Schizosaccharomyces pombe (Lin and Smith 1994), Drosophila melanogaster (McKim and Hayashi-Hagihara 1998), Caenorhabditis elegans (Dernburg et al. 1998), Arabidopsis thaliana (Grelon et al. 2001), Coprinus cinereus (Celerin et al. 2000), Sordaria macrospora (Storlazzi et al. 2003), Mus musculus (Baudat et al. 2000; Metzler-Guillemain and de Massy 2000; Romanienko and Camerini-Otero 2000) and also Homo sapiens (Romanienko and Camerini-Otero 1999; Shannon et al. 1999) maintain and require Spo11 orthologs.
Spo11 requires the help of at least 11 further proteins for DSB formation, namely Mer1, Mer2/Rec107, Mre2/Nam8, Mei4, Rec102, Ski8/Rec103, Rec104, Rec114, Rad50, Mre11, and Xrs2. Although many of these proteins were identified in a pioneering genetic screen over 10 years ago (Malone et al. 1991), the molecular roles of most of them remain elusive. Rec102, Ski8, and Rec104 have been shown to interact both genetically and physically with Spo11 (Uetz et al. 2000; Kee and Keeney 2002). Mre11, Rad50, and Xrs2 form the MRX complex, which is implicated in various vegetative DSB repair pathways, telomere maintenance, and damage signaling. The structure and biochemistry of the MRX complex have been well studied, revealing various nuclease activities and the ability to capture and hold together up to two DNA ends (Chen et al. 2001; for reviews, see Haber 1998; D'Amours and Jackson 2002). Very recently, a systematic study of two-hybrid interactions of various Spo11 cofactors revealed interactions between Mei4, Mer2, and Rec114 as well as between Rec104 and Rec114 (Arora et al. 2004). Binding of Rec102 to chromatin requires Spo11 and Rec104, while full levels of association of Rec104 with chromatin require Spo11, Rec102, Ski8, and Rec114 (Kee et al. 2004). The key question, which factors are responsible to position the Spo11 nuclease correctly, has, however, not been addressed so far in yeast. In Sordaria, Ski8 is required for formation of foci and lines by a Gfp-tagged Spo11 (Tesse et al. 2003).
Three further meiosis-specific proteins, namely, Mek1, Red1, and Hop1, are required for full levels of DSB formation. Mek1 is a serine/threonine kinase that regulates the activities of Red1 and Hop1 (Hollingsworth and Ponte 1997). Recently, Red1 has been found to bind to meiotic axis association sites, as defined by the mitotic cohesin Scc1/Mcd1 (Blat et al. 2002). Red1 is required at the base of meiotic loops to promote full levels of DSB formation, possibly by contributing to loop architecture, and also supports the subsequent loading of Dmc1, which may direct strand invasion to the homologous chromosome.
Ndt80 is a meiosis-specific transcription factor essential for wild-type levels of crossing over, disassembly of the synaptonemal complex (SC), and general cell cycle progression past prophase I (Xu et al. 1995; Chu and Herskowitz 1998). Specifically, ndt80Δ mutants exhibit prolonged persistence of DSB fragments probably due to prolonged Spo11 activity (Allers and Lichten 2001). Furthermore, joint molecules accumulate and crossover, but not gene conversion, products are markedly diminished, suggesting that the processing of a fraction of joint molecules into crossovers depends on a gene regulated by Ndt80 (Allers and Lichten 2001).
At this time, relatively little is known about the interaction of Spo11 with its target sequences on meiotic chromatin, namely, the recombination hotspots. The permanent attachment of Spo11 with hotspots in rad50S cells was employed for the genome-wide mapping of DSB sites in S. cerevisiae (Gerton et al. 2000). Only limited information on the nuclear distribution of Spo11 on chromosomes is available from M. musculus (Romanienko and Camerini-Otero 2000), D. melanogaster (Liu et al. 2002), and S. macrospora (Liu et al. 2002), while a correlation with other chromosomal features, such as the colocalization with repair proteins, is still missing.
We used a functional Spo11-Myc fusion protein to characterize the association of Spo11 with meiotic chromatin and with meiotic recombination hotspots by cytological and molecular analysis (chromatin immunoprecipitation [ChIP]) (Hecht et al. 1996; Tanaka et al. 1997), respectively. We report that Spo11 transiently associates with meiotic hotspots in wild-type cells and accumulates over time in rad50S and com1Δ mutants. In addition, we have characterized all the known essential cofactors, with respect to their influence on Spo11's hotspot association, which are independent of DSB formation. Furthermore, we observed an intermediate of the cleavage reaction in rad50S and com1Δ cells, which we termed “tight” binding and which we propose to correspond to the Spo11 cleavage complex.
Results
Spo11-Myc transiently associates with meiotic hotspots in an exclusively noncovalently bound form in wild-type cells
To study the interaction of Spo11 both with chromosomes on nuclear spreads and with hotspot sequences, we tagged the protein at the C terminus with 18 copies of the Myc epitope (Zachariae et al. 1996). Spo11-Myc is expressed from its original promoter at its native chromosomal locus and is the only source of Spo11 in all the tagged strains used in this study. Homozygous Spo11-Myc strains exhibit wild-type sporulation frequencies and spore viability is normal at 16°C, 30°C, and 34°C. Meiotic gene conversion frequencies at the his4X, his4B heteroalleles at three different temperatures (16°C, 30°C, and 34°C) were determined by single spore analysis in the Spo11-Myc strains (for further details of this analysis see Supplemental Material). No differences in conversion rates were detected for the tagged versus the wild-type strains at 16°C and 34°C and only a slight increase by a factor of 1.2 or 1.7 was measured in the tagged strains at 30°C. DSB assays were performed and confirmed that the Spo11 Myc tag does not alter the genetic requirements for DSBs (data not shown). We therefore conclude that Spo11-Myc can carry out all the functions of Spo11, which are essential for DSB formation, meiotic recombination, and spore viability.
As an assay for the interaction of Spo11 with a particular hotspot, we adapted a ChIP method (Hecht et al. 1996; Tanaka et al. 1997) for use with meiotic recombination hotspots. In this technique, we stabilized DNA–protein associations by exposure to formaldehyde (FA), after which DNA was extracted, fragmented, and immunoprecipitated using a Myc-antibody against our Spo11-Myc tag. After immunoprecipitation, preferential association between Spo11 and particular DNA sequences was sensitively detected by multiplex PCR (mPCR) and by quantitative real-time PCR (qPCR) with the appropriate primer pairs (PPs) (Fig. 1A). qPCR, in addition to yielding quantitative results, has the further advantage that the PCR is performed in separate reactions, thereby excluding any interference between the different PPs. To assay the interaction of Spo11-Myc with a recombination hotspot, DNA surrounding the natural YCR048w recombination hotspot was analyzed by qPCR (Fig. 1A). One of the PPs was located in close proximity to the YCR048w hotspot region (PP5, Fig. 1A), while the other PP was 3 kb away (PP6, Fig. 1A). As an additional negative control to PP6, we designed a PP located in a region devoid of DSBs (YCR011C), in other words, a cold region (originally defined by the absence of localized DSBs in a rad50S mutant [Baudat and Nicolas 1997]). mPCR was used to analyze the interaction of Spo11-Myc with DNA in the vicinity of the artificially created his4XLEU2 hotspot (Fig. 1A; Xu and Kleckner 1995).
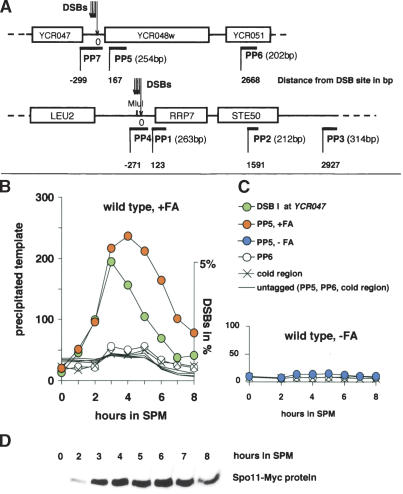
Figure 1.
Hotspot association of Spo11 persists longer in the population than DSBs. (A) Overview of the hotspots and primer pairs used. All distances are drawn to scale. (PP1–7) Primer pairs 1–7 (size of PCR product in base pairs); below each primer pair the distance to the most proximal DSB of a cluster is indicated (according to Liu et al. 1995; Xu and Kleckner 1995). Size of PP4 product: 358 bp; size of PP7 product: 362 bp. (B) Quantitative real-time PCR (qPCR) of immunoprecipitated DNA from samples of a meiotic time course experiment were taken at 1-h intervals, using primer pairs PP5 located 167 bp and PP6 located 2668 bp distal to the DSB site of the YCR048w hotspot and a cold region primer pair located in a cold DSB domain (YCR011C). The IP subsequent to FA cross-linking reveals preferential precipitation of PP5 from the 2-h time point onward. The meiotic DSBs at the YCR048w hotspot from the same time course are shown in the same graph (green). The relation between DNA and ChIP scales is arbitrary in this graph. (C) No enrichment of precipitated DNA in the same time course in the absence of FA. (D) Significant expression of Spo11-Myc from the 2-h time point onward.
In wild type, ChIP followed by qPCR identified a transient interaction of Spo11 with the YCR048w hotspot proximal sequence (PP5, Fig. 1A) during most of meiotic prophase (Fig. 1B). No signals were observed when we used an untagged wild-type strain, indicating that the enrichment was dependent on Spo11's epitope tag Myc (Fig. 1B). The interaction of Spo11 with the hotspot DNA was only detectable after cross-linking the DNA in vivo with Spo11 using FA, thus providing no indication for the presence of a stable, covalent Spo11–DNA intermediate in wild type (Fig. 1C). It is, however, formally possible that there is a covalent intermediate that is not abundant enough or too short lived to be detected.
When we analyzed Spo11 expression, the formation of DSBs, and Spo11–hotspot association by ChIP in parallel, all three signals appeared simultaneously, suggesting that steps leading to the formation of stable breaks occur rapidly after the initiation of Spo11 expression (Fig. 1B,D). Quantification of precipitated DNA revealed that the wave of the ChIP signal peaked ∼1–2 h later than the wave of the DSB signal and also decreased later. In other words, after the DSB peak, Spo11 molecules increasingly occupy uncleaved hotspots. This phenomenon can be explained either by Spo11 binding to hotspots after repair or by late-arriving Spo11 molecules binding to novel hotspots without cleaving them.
Spo11 foci turn over in wild-type cells, accumulate in rad50S mutants, and form independently of DSB formation
Meiosis-specific Spo11-Myc foci were detected on meiotic chromatin of spread yeast nuclei and their abundance varied according to the meiotic stage. Unexpectedly, Spo11 foci were abundant in some pachytene cells, a stage in which the formation of DSBs is believed to have concluded. During pachytene, but not in the earlier stages, most Spo11 foci touched or overlapped with Zip1 (the central element component of the SC) (Fig. 2A). No foci were detected in spread nuclei beyond the pachytene stage.
Figure 2.
Spo11-Myc foci form independently of DSBs and colocalize with Rad50 and Mre11 in rad50S cells. (A) A spread nucleus from early zygotene (z) and one from pachytene (p) are displayed side by side. Black and white panels show Spo11, Zip1, and DAPI separately, while the color panel presents Spo11 (red) and Zip1 (green) superimposed. The abundance of foci in both stages fluctuates considerably, suggesting dynamics in which foci are formed during zygotene and removed during pachytene. (B) The graphs show the appearance of Spo11-Myc foci during meiotic time courses in wild type, rad50Δ, rad50S, rec102Δ, rec104Δ, and rec114Δ cells. One-hundred cells were scored at each time point. The red symbols represent the fraction of nuclei with >20 foci, empty symbols the fraction containing less than five foci, and the blue symbols the fraction of cells past MI (determined by DAPI staining). (C) Typical spread meiotic nuclei from spo11-Y135F, rec102Δ, and rec114Δ mutant strains, stained with antibodies to Spo11-Myc (red) and to Zip1 (green). (D) Nuclear localization of Spo11 is relaxed in rec102Δ and rec104Δ mutants. Quantification of in situ staining of whole cells of rec102Δ and rec104Δ mutants at 6 h in meiosis stained with antibodies to Spo11-Myc (red) and to Rec8-Ha (green). One-hundred cells with nuclear Rec8 were counted in each experiment. Rec8 is an early meiosis-specific protein localizing to the nucleus, which serves to exclude errors from faulty staining or from nonmeiotic cells. Staining was classified as being “exclusively”, “predominantly”, or “equally” nuclear compared with the cytoplasm. These three classes are illustrated in the right panel, with the red and the green channel slightly horizontally dislocated. Under “absent” we counted cells with no detectable Spo11, but normal Rec8 staining. “AnaI” and “AnaII” denote meiotic anaphase stages, in which Spo11 is nuclear, even in the two mutants. (E) Spo11 and Rad50, and Spo11 and Mre11 foci colocalize in rad50S meiotic prophase nuclei. Cells from a 6-h time point were spread and stained with antibodies against Spo11-Myc (red), Rad50 (green), and Mre11 (green) and with DAPI. Regions of overlap between Spo11 and Rad50, or between Spo11 and Mre11, appear yellow in the merged images. A significant fraction but never all of the foci colocalize.
A quantitative analysis of Spo11-Myc foci on wild-type spreads revealed that foci appeared at the 2-h time point (Fig. 2B) and declined after the 5-h time point. The average number of Spo11-Myc foci peaked at the 4-h time point (25 ± 20, n = 20) versus 3 h (20 ± 25, n = 20) and 5 h (14 ± 14, n = 20). At all of these time points only few nuclei do show >60 foci, suggesting that a high foci content may be a short stage, which not every cell may go through because the half-life of individual foci may be quite short relative to their asynchrony within a nucleus. The time of Spo11-Myc foci appearance coincides with the association of Spo11-Myc with meiotic hotspots (as determined by ChIP) and with the appearance of DSBs. These observations are consistent with the idea that Spo11 foci correspond to sites of DSB initiation in wild-type cells. In rad50S cells, where DSBs accumulate with Spo11 covalently attached to the 5′-DSB end (de Massy et al. 1995; Keeney and Kleckner 1995; Liu et al. 1995; Gerton et al. 2000), Spo11 foci accumulated over time to higher levels than in wild-type or any other mutants (Fig. 2B). The average number of Spo11-Myc foci at the 6-h time point is 77 (±18, n = 20) and the numbers are much more homogenous than in wild type. In rad50Δ cells, where meiotic DSBs are not introduced, Spo11 foci appeared to be like those in wild type (Fig. 2B). In addition, the spo11-Y135F mutation, which completely abolishes Spo11's catalytic activity, does not affect Spo11 foci formation (Fig. 2B,C), indicating that focus formation is independent of the generation of DSBs. It does, however, depend on the presence of the Rec102 protein. Absence of Rec104 causes a strong reduction, and absence of Rec114 nearly no reduction of Spo11 foci numbers (Fig. 2C). While in wild-type cells in situ staining showed Spo11 almost exclusively in the nucleus, a significant increase in cytoplasmic Spo11 label was found in rec102Δ and rec104Δ mutants (Fig. 2D). ski8Δ mutants, which are normal for formation of Spo11 foci on spread nuclei (Fig. 2B), show an intermediate decrease of nuclear Spo11 label in in situ staining (Fig. 2D).
Spo11-Myc foci colocalize with Rad50 and Mre11 foci in rad50S cells
If Spo11 foci marked sites of DSB formation, they might colocalize with other proteins required for break formation. We chose to analyze the colocalization of Spo11-Myc with Mre11 and Rad50, as both proteins are required for DSB formation as well as DSB repair (Alani et al. 1990; Nairz and Klein 1997), hence suggesting that they may be present at active DSB sites. As Mre11 and Rad50 are not detectable in wild-type cells, a rad50S strain background was used for this analysis. In rad50S spreads, Mre11 and Rad50 showed extensive colocalization with Spo11-Myc (Fig. 2E), consistent with the hypothesis that Spo11-Myc foci correspond to sites of DSB formation. An average of 30% (20.1 ± 6.0) of Spo11 foci (67.4 ± 22.5) colocalized with Rad50 foci (75.8 ± 35.6) n = 21, t = 5 h under most stringent criteria (when random overlaps were estimated to be 2.1 ± 0.31 by simulation, p > 0.99). Colocalization between Spo11 and Mre11 at t = 5 h was slightly lower, 24% (14.7 ± 6.8) of Spo11 foci (60.3 ± 21.5) colocalized with Mre11 foci (58.3 ± 19.2) n = 22 (random overlaps 1.2 ± 0.25, p > 0.99). No colocalization was observed between Spo11-Myc foci with Zip1, Hop1, Red1, and Mnd1 in wild type (data not shown), suggesting that the binding sites of these proteins on chromosomes are timely or spatially distinct from those of Spo11.
REC102, REC104, and REC114 are required for the association of Spo11 with hotspots
We analyzed Spo11's association with recombination hotspots in null mutants for all the genes known to be essential for DSB formation (for reviews, see Smith and Nicolas 1998; Keeney 2001), with the exception of MER1 and NAM8/MRE2 because their role in DSB formation lies exclusively in the maturation of MER2-mRNA (Engebrecht et al. 1991; Nakagawa and Ogawa 1997). The mutants fell into two main categories, one in which Spo11 failed to interact with hotspot DNA and one in which interaction was observed (Fig. 3). In strains lacking REC102, REC104, or REC114, Spo11 did not precipitate detectable hotspot DNA (Fig. 3D–F). In the ski8Δ, mer2, mei4Δ, red1, hop1, mre11Δ, rad50, and xrs2 mutants, hotspot DNA above background levels could be amplified following Spo11-Myc ChIP (Fig. 3). Each experiment was repeated at least three times by mPCR at the his4LEU2 hotspot (data not shown) and by qPCR at the YCR048w locus with identical results. To demonstrate that cells expressed Spo11-Myc normally during these experiments, Spo11 protein levels were analyzed in parallel by Western blotting, confirming that Spo11-Myc was clearly expressed in all the mutant strains. In addition, nuclear division was followed by DAPI staining and DSB assays were performed to show that the Spo11 Myc tag did not alter the requirements for break formation in these mutants (data not shown). Except for the hop1 mutant, where we did not observe any breaks, probably because the low level of DSBs expected in the hop1 mutant fell below our threshold of detection (10% of wild type [Mao-Draayer et al. 1996]), the rest of the mutants were as expected (data not shown).
Figure 3.
Genetic control of Spo11–hotspot interaction qPCR of precipitated DNA from wild type and mutants. Filled circles represent the hotspot signal (PP5), while empty circles represent sites farther away from the hotspot (PP6). × (cold region) represents a sequence from YCR011c, from a chromosome region without detectable DSBs. (A) Wild-type curve. (B) Dependence of the signal on the tag. (C) Enhanced levels of hotspot precipitation in the mutant carrying a catalytically inactive Spo11 (spo11-Y135F-Myc). (D–F) Absence of detectable hotspot association in rec102Δ, rec104Δ, and rec114Δ mutants. Spo11 expression was normal in all strains shown in this figure, but is displayed for the three mutants that produce no detectable ChIP signal. (G–I) Enhanced levels of Spo11–hotspot interaction in mre11Δ, rad50, and xrs2. (J) In red1 cells, PP6 (a fragment almost 3 kb away from the hotspot) was precipitated at similar levels as the hotspot itself. Graphs without any symbol represent DNA from untagged strains throughout the figure, showing that the PP6 signal is also dependent on the tag. (K) Hotspot association of Spo11 in the hop1 mutant was slightly reduced. (L,M) Meiosis-specific enrichment of hotspot DNA, similar to wild type in ski8Δ and mer2. (N,O) In mei4Δ and ndt80Δ mutants, no relevant decrease in the amount of precipitated hotspot DNA was observed by qPCR up to the 10-h time point. (SPM) Sporulation medium.
A Spo11 signal, roughly double that of wild type, was observed in the rad50, mre11Δ, and xrs2 mutants and in the spo11-Y135F mutant (Fig. 3C,G–I). This suggests that the association of Spo11 with the hotspot DNA either persists for longer or that more Spo11 molecules bind to the hotspot DNA in these mutants.
We next tested hop1 and red1 mutants for Spo11-Myc hotspot association, as they cause a severe reduction in the initiation of recombination (Hollingsworth and Byers 1989; Rockmill and Roeder 1990). For the hop1 mutant, the ChIP signal was reduced but still preferentially associated with the hotspot (Fig. 3K). As a control, we repeated the same experiment in an isogenic strain lacking the Myc tag, which confirmed that the signal depended on Spo11. For red1, an increased precipitation of PP6 (Fig. 3J), which is not immediately adjacent to the hotspot, as well as of PP5 was observed. The PP from the cold region also detected almost equal amounts of meiosis-specific signal, while in an untagged red1 strain, all PPs amplified equally low amounts of DNA corresponding to background levels. The observed signal from PP6 and the cold region, therefore, depends on Spo11-Myc. This phenomenon suggests that Spo11 is not restricted to the hotspot region in red1 mutants. Red1 may, therefore, be required to limit Spo11 to the core hotspot area, perhaps by preventing additional Spo11 from binding to the flanking regions.
Ndt80 and Mei4 are required for terminating Spo11's association with the hotspot
qPCR revealed that Spo11's association with the hotspot did not decrease until 8 h after transfer to sporulation medium in mei4Δ cells, which neither activate Spo11 nor produce any DSBs (Fig. 3N). At this time, the culture had progressed normally through meiosis. Sixty-six percent of cells had passed the first and 50% the second meiotic division. In ndt80Δ mutants, Spo11 associated normally with the tested hotspots and remained attached throughout the arrest (Fig. 3O). qPCR showed that the level of Spo11 binding reached values at or slightly above the peak found in wild-type cells in both mei4Δ and ndt80Δ mutants.
Similar to the red1Δ mutant, ndt80Δ and mei4Δ mutants show an increased interaction of Spo11 with non-hotspot DNA (Fig. 3J,N,O). In the latter mutants, however, there is still a clear preference for the sequence close to the hotspot (PP5) over PP6, and the cold region DNA remained at background levels (Fig. 3N,O). This suggests that the observed intermediate enrichment of PP6 in mei4Δ and ndt80Δ may reflect the overloading of hotspots with Spo11 rather than a relaxation in the stringency of Spo11 localization. Slightly elevated PP6 levels were also seen in spo11-Y135F, mre11Δ, and xrs2Δ mutants, which show high levels of Spo11 binding (Fig. 3C,G,I).
`Tight' binding of Spo11 to hotspots occurs while formation of stable DSBs are delayed in rad50S cells
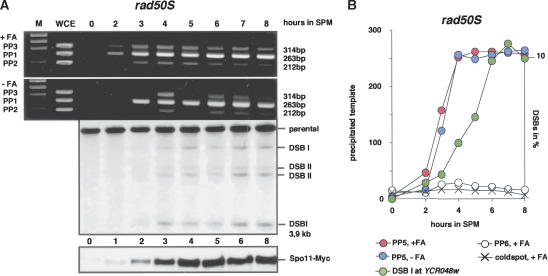
As expected, selective precipitation of hotspot DNA by ChIP in rad50S cells was independent of FA cross-linking (Fig. 4A). Spo11 is known to accumulate in a covalently attached form in this mutant (de Massy et al. 1995; Keeney and Kleckner 1995; Liu et al. 1995; Gerton et al. 2000). FA-cross-linked samples produced a slightly earlier signal than untreated ones, suggesting that Spo11 may bind first by weak interaction, followed by a tighter protein–DNA bond, which survives precipitation in the absence of cross-linking in the rad50S mutant.
Figure 4.
Formaldehyde-independent hotspot association of Spo11 precedes DSB formation in rad50S. (A) The left upper panel (IP after FA cross-linking) shows specific enrichment of the PP1 product. The panel below (IP w/o FA) shows a very similar enrichment, although the signal appears slightly later. (WCE) Whole-cell extract. The Western blot shows significant expression of Spo11-Myc from the 2-h time point onward. The Southern blot shows signal corresponding to DNA fragments generated by meiotic DSBs at the his4XLEU2 hotspot from the same time course. (B) qPCR of rad50S ChIP-DNA (IP–FA and +FA cross-linking) at the YCR047c hotspot of an experiment independent of that shown in A. Corresponding DSB signals at YCR047c were quantified and superimposed to compare intensity and timing. The scale for DSBs was chosen so that the final level of DSBs corresponds to the final level of bound Spo11. Note the delayed increase of DSBs in the rad50S strain compared with cross-linking-independent association of Spo11-Myc to the hotspot region.
Whereas in wild type, FA-dependent ChIP signal and DSB signal rise in parallel (Fig. 1B), DSBs are clearly behind FA-dependent and FA-independent ChIP signals in rad50S cells (see 3-h time point in Fig. 4A,B representing two independent experiments and different hotspots). We would like to emphasize that we compare DSB formation and ChIP signal of the same time course relative to each other, excluding variability coming from the speed with which a particular population undergoes meiosis. That FA-independent ChIP signals preceded DSB signals in rad50S mutants suggested that fairly stable associations between Spo11 and the hotspots exist before equivalent numbers of stable DSBs form. We will refer to such binding as “tight” binding. Because “tight” binding was not detectable in wild-type cells, the phenomenon might be caused by the accumulation of a reaction product as a consequence of the rad50S defect.
Are ChIP signals too early in rad50S or are DSBs late? DSB signals appeared at least 1 h later in the rad50S experiment than in the wild-type time course (Figs. 4A, 1B), while ChIP signals appeared approximately on time. Because also Spo11 expression (Figs. 1, 4) as well as Rec8 expression (data not shown) proceeded on schedule in the rad50S population, this strongly suggests that stable DSBs appear with a delay in rad50S.
A possible interpretation of these results is that the Spo11 cleavage reaction may go through a “tight” complex in which the DNA strands are largely intact. This intermediate may correspond to the predicted reversible stage (Keeney 2001), in which cleavage and rejoining occur reversibly or to an even earlier stage, when cleavage has not yet ensued. Somehow, rad50S cells accumulate this intermediate transiently.
Full-length hotspot DNA is detected in precipitates of Spo11 in rad50S and in com1Δ cells in the absence of FA
The proposed transient accumulation of the “tight” complex predicts that in the absence of FA, full-length hotspot DNA fragments may be detected in the Spo11 precipitates. The full-length fragments should occur only transiently, before stable breaks are formed in rad50S cells. In order to detect full-length hotspot DNA fragments selectively, we designed the PPs PP4 and PP7 (see Fig. 1A). The corresponding primers of each pair are located on opposite sides of the DSB array of the hotspot site (Liu et al. 1995; Xu and Kleckner 1995), thus producing a signal only from intact template. Indeed, both PPs detected precipitated full-length hotspot DNA during the predicted window in the absence of FA (Figs. 5A, 6A). As a control, we show that PP5, which is located entirely to one side of the YCR048w-DSB array, produced a permanent signal from the same extracts (Figs. 5A, 4B), consistent with detecting the sum of reversibly and irreversibly covalently bound Spo11.
Figure 5.
“Tight” binding is an intermediate in the cleavage reaction of both rad50S and com1Δ mutants. All precipitations were carried out without prior FA treatment. (A) Quantification by qPCR of precipitated DNA from rad50S cells. Blue and yellow diamonds represent PP7, which selectively detects intact hotspot, but for the yellow symbols the salt concentration in the extraction buffer was raised to 400 mM NaCl. Orange and yellow circles represent PP5, which detects hotspots associated with Spo11 independently of their cleavage. Again, for the yellow symbols salt concentration in the extraction buffer was raised to 400 mM NaCl. Green circles show the fraction of cleaved hotspot molecules. (B) Same as in A, except for com1Δ cells.
Figure 6.
“Tight” binding of Spo11 requires its catalytic tyrosine. (A) All precipitations were carried out without prior FA treatment in rad50S cells. (Upper left panel) Multiplex analysis of ChIP using PP4 (see Fig. 1A), which detects selectively uncut hotspot DNA. Note the transient enrichment at the 2- and 4-h time points. (Next panel down) PP1 (see Fig. 1A), detecting both cleaved and intact precipitated hotspot DNA. This yields permanent enrichment of hotspot DNA. (Right two panels) Same analysis as to the left but from spo11-Y135F, rad50S double mutants. No cleaved DNA, but also no uncleaved DNA is precipitated. (WCE) Whole-cell extract; (M) molecular weight marker. (B) No indication of localized nicks in rad50S and com1Δ cells. Digested genomic DNA of rad50S and com1Δ cells from time points showing “tight” binding was separated under neutral conditions in the first, and under denaturing conditions in the second dimension, then blotted and hybridized with a probe from the YCR048w hotspot. Filled-in arrowheads mark the expected position of the 2.5-kb DSB fragment. Empty arrowheads mark the position of an expected signal from a SSB at YCR048w. No such signal can be detected. The smear from the parental fragment toward smaller sizes is likely caused by random SSBs introduced during the preparation. The four panels below provide quantification of the above signals. One profile was created from the diagonal (black, DSBs) and one from the SSB smear (red). The profiles were normalized so that corresponding peaks would appear on top of each other.
We found a way to selectively eliminate the recovery of full-length hotspot fragments without interfering with the recovery of cleaved hotspot by raising the NaCl concentration in the extraction buffer (and thereafter) to 400 mM (Fig. 5A, yellow symbols). The transient signal from PP7 is eliminated under high salt conditions. Interestingly the PP5 signal obtained under high salt conditions now coincides with the DSB curve, as expected, if it now detected only irreversibly bound hotspot DNA. These results show that PP5 signal is the sum of a transient signal also detected by PP7, plus the signal from irreversibly bound DNA, which parallels DSB formation. However, as we don't know how NaCl eliminates the PP7 signal, this experiment does not address the nature of “tight” binding.
If the “tight” complex accumulated as a precursor of the cleavage reaction due to the failure of rad50S mutants to efficiently exit this stage, the com1Δ mutation, which causes a repair defect similar to that of rad50S (McKee and Kleckner 1997; Prinz et al. 1997), might also accumulate the cleavage complex. If, on the other hand, “tight” binding resulted from a failure of the rad50-K81I point mutant to separate the Spo11 activating function from its function in DSB repair completely, no accumulation was expected in com1Δ cells. In contrast to Rad50, Com1 has not been implicated in Spo11 activation. Figure 5A and B shows that “tight” binding is observed in com1Δ mutants and that qPCR of precipitates from rad50S and com1Δ cells yields almost identical results. This observation suggests a mechanistic link between the newly observed phenomenon of “tight” binding and the accumulation of covalently bound Spo11 in com1Δ and rad50S cells. It excludes that the defect might be specific to only the particular point mutation rad50-K81I.
`Tight' binding in rad50S and com1Δ mutants requires Spo11's catalytic tyrosine and does not involve a stable nick
During the proposed reversible stage the cleavage complex is thought to interact with the DNA termini via Spo11's Y135. To address whether the “tight” complex requires the catalytic tyrosine for its formation, we analyzed precipitated DNA from the spo11-Y135F rad50S double mutant (Bergerat et al. 1997), using the full-length specific PP4 (Fig. 1). “Tight” binding was undetectable in this strain (Fig. 6A). The identical result was obtained using PP7 followed by qPCR (data not shown). FA-dependent hotspot precipitation (i.e., weak association of Spo11 with the hotspot) was not diminished by mutation of the catalytic tyrosine (Fig. 3C). This result shows that Spo11's catalytic tyrosine is required for “tight” binding. It is consistent with cleavage being required for “tight” binding, as expected during the reversible stage. However, it does not completely rule out that Spo11's DNA-binding capacity is affected and responsible for the loss of signal.
One alternative way to explain precipitation of full-length template by Spo11 in the absence of FA is to postulate covalent interaction of only one sister strand with Spo11, while the opposite sister strand may remain intact. Spo11 is thought to create meiotic DSBs by cleaving both single strands at the same position in a coordinated manner. In rad50S and com1Δ cells, even moderate interference with this coordination could result in the accumulation of a nicked molecule with Spo11 covalently bound to the 5′-end of the nick. This molecule would contain both a covalent link for FA-independent precipitation and an intact DNA strand suitable as a template for PP7. We directly tested this possibility by two-dimensional DNA electrophoresis, in which separation in the second dimension was performed under denaturing conditions to separate the nicked half strand from the intact one. As “tight” binding in rad50S and com1Δ cells was as strong as covalent binding at the 3-h time point (Fig. 5A,B), the signal originating from the nicked fragment should equal that of the DSB, if nicking were to explain “tight” binding. Figure 6B shows such an analysis for the 2-, 3-, and 4-h time points of com1Δ cells (same experiment as in Fig. 5B) as well as the 3-h time point of rad50S cells (same experiment as in Fig. 5A). While DSBs are readily detectable, no signal corresponding to a localized single strand break could be observed. This result agrees with previous two-dimensional DNA separation at several different hotspots in rad50S cells, in which localized single strand breaks were also not found (de Massy et al. 1995; Liu et al. 1995; Xu and Kleckner 1995). These experiments were, however, mostly undertaken for isolated time points. As we were looking for an early intermediate, it was important to measure “tight” binding in parallel to the two-dimensional analysis and to perform the two-dimensional analysis when “tight” binding was maximal (3 h for this experiment). The absence of localized nicks leads us to conclude that “tight” binding in com1Δ or rad50S mutants cannot be explained by a relaxation of the coordination between the two single-strand DNA cleavages in vivo.
Discussion
Activation of Spo11 is controlled separately from its physical interaction with meiotic hotspots
Spo11 has been shown to be a major determinant in the initiation of meiotic recombination in all tested eukaryotic organisms, and convincing evidence exists that it catalyzes DNA cleavage in a topoisomerase-like reaction (Bergerat et al. 1997; Keeney et al. 1997; Keeney 2001). The simplest model could therefore assume that hotspot cleavage ensues when Spo11 interacts physically with the hotspot. However, several observations lead us to conclude that the mere interaction of Spo11 with a hotspot is not sufficient to cleave it and that an additional activation step is required. By recording the kinetics of Spo11's association with hotspots by ChIP in wild-type cells, we found that the wave of Spo11–hotspot interaction lasts longer than the wave of DSBs. Spo11 must, therefore, associate with intact hotspots in wild-type cells. The presence of Spo11 foci on pachytene bivalents, which are thought not to contain DSBs, further supports this finding. In mouse Spo11 is also found on synapsed chromosomes (Romanienko and Camerini-Otero 2000), consistent with a possible conserved role at this stage. Spo11's cleavage activity seems to be restricted to an early phase or to a certain number of hotspots, after which Spo11 keeps accumulating for some period on hotspots, without cleaving them.
Which of all the known factors essential for DSB formation govern Spo11–hotspot binding and which ones control activation? Measuring the physical interaction of Spo11 with meiotic hotspot DNA by ChIP, we established that only three factors are essential for this interaction, namely, Rec102, Rec104, and Rec114. Of these, Rec102 and Rec104 seem to be required for any interaction of Spo11 with chromatin, because rec102Δ and rec104Δ mutants display a strongly reduced number of Spo11 foci on spread meiotic nuclei. In fact Rec102 and Rec104 are required for nuclear accumulation of Spo11, because Spo11, which by in situ staining is found almost exclusively in the nucleus in wild-type cells, is also present in the cytoplasm in rec102Δ and in rec104Δ cells. Because even in rec102Δ and in rec104Δ mutants Spo11 is still enriched inside the nucleus in most cells, we propose that Rec102 and Rec104 mediate nuclear anchoring of Spo11 by chromatin binding, rather than mediating nuclear import. Interestingly, chromatin staining of Rec102 and Rec104 is abolished in spo11Δ mutants and their cytoplasmic localization increases dramatically (Kee et al. 2004). Thus, chromatin association of each of the three proteins depends on the other two, suggesting that they bind to chromatin as a preformed complex. Consistent with this interpretation, physical interactions between them were observed by coimmunoprecipitation and in two-hybrid experiments (Kee and Keeney 2002; Jiao et al. 2003; Arora et al. 2004).
In contrast nearly normal levels of Spo11 foci were observed in rec114Δ, yet no binding of Spo11 to meiotic hotspots occurred. This identifies Rec114 as specifically required for recognizing hotspots and for recruiting Spo11 to them. However, DBSs in rec114Δ mutants are not delocalized but rather completely abolished, revealing an additional role in activation. Rec114 has an ortholog in S. pombe called rec7 (Molnar et al. 2001) and may therefore be part of an evolutionarily conserved Spo11-targeting system. When Spo11 is fused to the DNA-binding domain of Gal4, DSBs can be observed at Gal4-binding sites (Peciña et al. 2002). If Rec102, Rec104, and Rec114 had no role in activation of Spo11, they should become dispensable for breaks at the Gal4-binding sites. However, they still remain essential. We suggest that activation, but not localization, of Spo11 requires the complete initiation complex, and that the roles of Rec102, Rec104, and Rec114 in activation are mainly structural. We found no essential role for Ski8 in localizing Spo11 to hotspots or for focus formation, but an intermediate reduction in nuclear localization. Therefore we think Ski8's main role in yeast is the activation of the Spo11 nuclease. Since Spo11 is required for Ski8's nuclear localization (Arora et al. 2004), Ski8 may be part of a preinitiation complex containing also Spo11, Rec102, and Rec104 and possibly others. In Sordaria, Ski8 is essential for focus formation of Spo11 on DNA besides Spo11 activation (Tesse et al. 2003), but only the activation function seems to be conserved in evolution.
One of the activators of Spo11 is Mre11. Localization of Mre11 requires all other DSB genes, including Spo11, but with the exception of Rad50 (Borde et al. 2004). This shows that Mre11 itself has little or no affinity for the hotspot and may bind only to the almost complete preinitiation complex. That Spo11, in contrast, only requires three loading factors, argues for a much closer interaction between Spo11 and the targeting factors, probably in the form of a subcomplex.
Not only the creation, but also the termination, of Spo11's associations with hotspots is dependent on other proteins. In addition to rad50S and com1Δ mutants, which accumulate covalently attached Spo11 on the 5′-DNA termini of cleaved hotspots (Keeney and Kleckner 1995; Liu et al. 1995), mei4Δ and ndt80Δ mutants also show defects in terminating Spo11–hotspot interaction. As the dissociation of Spo11 from the hotspot does not depend on successful cleavage, mei4Δ mutants are the only example in which Spo11 is at least strongly retarded at uncleaved hotspots in the absence of any cell cycle arrest. Ndt80 is a transcription factor, the absence of which causes pachytene arrest (Xu et al. 1995), DSB and unresolved Holliday junction accumulation, and a decrease in crossover formation (Allers and Lichten 2001). Persistence of the Spo11 signal at hotspots in the ndt80Δ mutant may be due to recurrent association and cleavage of hotspots and/or to a failure to dissociate Spo11 after cleavage.
`Tight' binding occurs before stable DSB formation and may define an intermediate of the cleavage reaction
ChIP experiments reproducibly ascertained that FA-independent association of Spo11 with the hotspot precedes detection of stable DSBs in com1Δ and rad50S mutants. We termed this interaction of Spo11 with the hotspot, which is specific to these mutants, “tight” binding. In wild-type time courses “tight” binding was never observed (Figs. 1, 4). If “tightly” bound Spo11 is not recovered due to high salt concentration in the extraction buffer, the PP5 curve for Spo11 binding aligns with the DSB curve (Fig. 5A,B, yellow and green symbols), as originally expected for rad50S and com1Δ cells. Subtraction of the high salt curve from the low salt curve reveals that the additionally “tightly” bound Spo11 detected by PP5 in low salt extracts peaks at the 3-h time point, before the bulk of DSBs are detected. Direct detection of “tightly” bound Spo11 with primers specific for full-length hotspot confirms its transient nature and also peak at the 3-h time point before DSB formation (Figs. 5, 6A).
We detect DSBs in com1Δ and rad50S cells ∼30–60 min later than in wild type using only internal parameters of the cultures as a reference, such as Rec8 and Spo11 expression. “Tight” binding appears at exactly the time when a temporary deficit in DSBs is observed. We therefore propose that “tight” binding in com1Δ and rad50S mutants may correspond to an intermediate of the cleavage reaction, which accumulates transiently before stable DSBs form.
A subtle defect in DSB formation in com1Δ and rad50S mutants has been observed before (Borde et al. 2000). In both mutants, late replicating regions, which also activate their hotspots later, show reduced DSB formation. The slight delay in DSB formation offers an explanation for this result. If there was only a limited time window in which Spo11 could be activated, early hotspots might become late, while late hotspots may become too late to be activated.
The rad50S and com1Δ defects link DSB formation, `tight' binding, and release of covalently bound Spo11
The fact that both rad50K81I and com1Δ mutations cause transient accumulation of “tight” binding as well as terminal accumulation of irreversibly bound Spo11 to cleaved hotspots decreases the possibility that these defects are independent of each other. What could be the primary defect responsible for both phenomena?
We have tried to address experimentally the possibility that coordination of the two single-strand breaks (SSBs) required for a DSB might be defective in rad50S and com1Δ mutants. An accumulation of an intermediate composed of one full-length strand and a broken one with Spo11 covalently attached to a 5′-DNA end would explain both the strong interaction and recovery of full-length template. However, consistent with earlier observations (de Massy et al. 1995; Liu et al. 1995; Xu and Kleckner 1995), no SSBs were detectable in 2D-gels (Fig. 6B). In contrast to earlier studies, we analyzed the DNA from time points in which “tight” binding was detected in parallel by ChIP. This suggests to us that not enough SSBs exist in vivo to explain the observed precipitation of full-length template.
Assuming that “tightly” bound Spo11 indeed represents an intermediate in the Spo11 cleavage reaction, its transient accumulation certainly represents an earlier defect than the failure to remove covalently bound Spo11. Usually an accumulation of an intermediate points to a problem of exiting a particular stage. A unifying proposal to explain both the well-known and the novel rad50S (and com1Δ) phenotype might, therefore, state that transient accumulation of an intermediate may be caused by failure in these mutants to exit that stage, while permanent covalent linkage of Spo11 to 5′-ends might be the consequence of an “alternative” exit from that stage, which makes the earlier intermediate transient. In this respect, it may be interesting to note that many events, even simple protease treatment (Liu and Wang 1979), can result in covalent topoisomerase–DNA complexes, suggesting that the “alternative” exit could be a rather crude event. Formally, it is therefore possible that covalently linked Spo11 presents a problem to meiotic cells, because it is an alternative product that may not occur in the same form during the wild-type reaction. On the other hand, it is equally possible—but not proven to date—that a fully functional MRX complex could cope with Spo11-blocked 5′-DNA termini, irrespective of whether they arose through the wild-type or an alternative pathway.
The precise chemical nature of the “tight” complex between Spo11 and the hotspot DNA remains elusive, as it would require characterization of the complex in vitro. However, the fact that the catalytic hydroxyl group of Y135 of Spo11 is essential for detection of “tight” binding leads us to favor the idea that a transesterification reaction may play a role in formation or detection of the “tight” complex. But a mechanism simply based on differential hotspot binding properties of Spo11 and Spo11-Y135F cannot be ruled out at this point.
Materials and methods
Yeast strains and meiotic time courses
All strains used in this study are isogenic to SK1 (Kane and Roth 1974) and are listed in Table 1. Spo11 and Spo11-Y135F proteins were tagged at the C terminus with 18 Myc epitopes at their original chromosomal loci by one-step gene targeting (Zachariae et al. 1996; Wach et al. 1997). Sporulation, formation of DSBs, and viability were identical to their untagged parental strains. Tagging was confirmed by PCR and Southern and Western analysis. Ski8, Rec104, and Rec114 were deleted using PCR-mediated gene replacement (Wach et al. 1994) and confirmed by Southern analysis. MER2, HOP1, RAD50, and XRS2 were disrupted using plasmids pR990 (Shirleen Roeder, New Haven, CT), pNH37-2 (Breck Byers, Seattle, WA), pNKY83 (Nancy Kleckner, Cambridge, MA), and pEI40 (Francis Fabre, Paris, France) and confirmed by Southern analysis. All mutants and their tagged variants showed no detectable DSBs and <1% spore viability.
Table 1.
The strains used in this study
| Strain | Genotype |
|---|---|
| YFK0047 | NKY1238xNKY1240 |
| YFK1478 | MAT a/α, ho::LYS2, lys2, SPO11::MYC18::TRP1, REC8::HA3::URA3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, ura3 |
| YFK1965 | MAT a/α, ho::LYS2, lys2, SPO11::MYC18::TRP1, (REC8::HA3::URA3)/REC8::HA3::URA3, his4XLEU2-URA3, ade2::hisG/ADE2, trp1::hisG, ura3, his3::hisG, rad50S::URA3, leu2::hisG (rad50S = rad50-Ki81) |
| YFK1667 | MAT a/α, ho::LYS2, lys2, leu2::hisG, trp1::hisG, ura3, spo11-Y135F::MYC18::TRP1, his4XLEU2-Mlul::BamH1-URA3 |
| YFK2116 | MAT a/α, ho::LYS2, lys2, his4XLEU2-URA3, leu2, ura3, trp1::hisG, spo11-Y135F::MYC18::TRP1, rad50S::URA3 |
| YFK2075 | MAT a/α, ho::LYS2, SPO11::MYC18::TRP1, ura3, leu2::hisG, trp1::hisG, his4BLEU2-Mlu1/his4XLEU2-URA3 |
| YFK2048 | MAT a/α, ho::LYS2, lys2, his4XLEU2-URA3, ura3, leu2::hisG, trp1::hisG/TRP1, ARG4/arg4-nsp |
| YFK1507 | MAT a/α, ho::LYS2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, arg4-nsp, rad50::URA3 |
| YFK1938 | MAT a/α ho::LYS2, lys2, xrs2::LEU2, leu2::hisG, ura3, trp1:hisG, his4XLEU2-Mlul::BamHI-URA3, SPO11::MYC18::TRP1 |
| YFK1878 | MAT a/α ho::LYS2, his4XLEU2-Mlul::BamHI-URA3, ura3, leu2, trp1, SPO11::MYC18::TRP1, mre11Δ::KAN MXc |
| YFK1974 | MAT a/α ho::LYS2, lys2, SPO11::MYC18::TRP1, REC8::HA3::URA3, his4XLEU2-URA3, trp1::hisG, ura3, leu2::hisG, ade2::hisG, mer2::ADE2 |
| YFK1510 | MAT a/α ho::LYS2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, arg4-nsp/ARG4, red1::Leu2b |
| YFK1513 | MAT a/α ho::LYS2,SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, arg4-nsp, hop1::LEU2 |
| YFK1528 | MAT a/α ho::LYS2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, arg4-nsp, rec114Δ::KANMX |
| YFK1516 | MAT a/α ho::LYS2, lys2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, trp1::hisG, his4XLEU2-Mlu1::BamH1-URA3/ his4XLEU2-URA3, rec102Δ::URA3a |
| YFK1527 | MAT a/α ho::LYS2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, arg4-nsp, rec104Δ::KANMX |
| YFK1532 | MAT a/α ho::LYS2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, trp1::hisG, arg4-nsp, ski8Δ::KANMX |
| YFK1444 | MAT a/α ho::LYS2, his4XLEU2-Mlul::BamHI-URA3, ura3, leu2, trp1/TRP1, SPO11::MYC18::TRP1, mei4Δ::URA3a |
| YFK1535 | MAT a/α ho::LYS2, lys2, SPO11::MYC18::TRP1, REC8::HA3::URA3, ura3, leu2::hisG, his4XLEU2-URA3, arg4-nsp, trp1::hisG, ndt80Δ::LEU2a |
All markers are homozygous unless stated otherwise.
Deletion strain constructs generously provided by aN. Kleckner (Harvard University, Cambridge, MA); bS. Roeder (Yale University, New Haven, CT), and cA. Nicolas (Institut Curie, Paris, France).
Strains were grown and sporulated as previously described (Nairz and Klein 1997). In short, for a meiotic time course, diploid cells were grown to a concentration of 4 × 107 cells/mL in yeast extract/peptone/acetate medium and incubated at the same cell concentration in 2% potassium acetate at 30°C–33°C with vigorous shaking. Meiotic divisions were monitored by fluorescence microscopy after staining with 0.2 μg/mL DAPI.
Cytology
Yeast meiotic spreads were performed as described (Nairz and Klein 1997; Loidl et al. 1998). Spo11-Myc was detected using 9E10 (mouse anti-Myc, 1:50) and CY3-conjugated goat anti-mouse antibody (1:100, Dianova). Rabbit anti-Zip1 antibody was raised against a purified Zip1–GST fusion protein and affinity purified against the same protein. The purified Zip1 antibody was used (1:50) and detected by FITC-conjugated goat anti-rabbit serum (1:200, Sigma). Rabbit anti-Mre11, 1:5000, a gift from J. Petrini (Madison, WI), and rabbit anti-Rad50, 1:200, a gift from N. Kleckner, were used for immunostainings. Colocalization was determined using IPLAB software, and regarding only the centers of the signals, on the average ∼15 pixel (∼0.01 μm2). Only overlapping signals were counted. Random overlaps were estimated by running 250 simulations, using focus size and number as well as nucleus area and sample size as parameters.
Spore viability, gene conversion
Spore viability was assayed by tetrad dissection and ascus frequencies were always determined to estimate spore yield. Gene conversion was assayed between the his4X and his4B heteroalleles (Cao et al. 1990) before and after sporulation (3 d) on selective medium. Viability was determined on YPD plates in parallel. Meiotic gene conversion frequencies were calculated as meiotic convertants per viables minus the vegetative background.
DSB assay
Physical analysis of DSBs was performed on DNA prepared by alkaline lysis of spheroplasted yeast cells. Briefly, cells were sporulated and 20-mL aliquots were taken at 1-h intervals, pelleted, washed once in H2O, and frozen in liquid nitrogen for later use. After thawing, cells were resuspended in 200 μL SCE/zymolyase/2-ME and incubated shaking for 60 min at 37°C. Upon >90% spheroplasting, 200 μL SDS solution was added, and after careful mixing by inversion, cells were incubated for 5 min at 65°C, gently shaking. Two-hundred microliters of 5M-KOAc was added followed by an incubation for 20 min on ice. Cell debris was removed by a 15-min spin at 5000 rpm. Five hundred microliters of supernatant was transferred to a new tube; 200 μL 5M-NH4OAc and 1 mL isopropanol were added. DNA was pelleted during a 30-sec spin at 5000 rpm and supernatant was carefully removed. The pellet was dissolved in 100 μL TE and again precipitated with 200 μL isopropanol. The supernatant was removed, and the pellet was washed in 80% ethanol and finally dissolved in 50 μL TE. RNAse A (0.5 μg) was added for 1 h at 37°C before restriction analysis. Neither proteinase K digestion nor phenol/chloroform/isoamylalcohol extraction were performed. Following digestion of the genomic DNA with the appropriate restriction enzyme, the DNA was separated on a 0.8% TAE-buffered agarose gel and transferred to a Hybond-Nylon membrane (Amersham) by capillary transfer. Probes were gel-purified by the Freeze'N Squeeze kit (Bio-Rad) according to protocol and labeled, either radioactively by random priming using α-32P dATP or nonradioactively by using the ECF random prime labeling kit (Amersham). Hybridization was carried out overnight at 60°C or 65°C. Direct quantification of the signal was undertaken on a PhosphorImager (Typhoon 8600, green fluorescence, 500–650 PMT) and evaluated using ImageQuant (both Molecular Dynamics). An intensity profile was generated for each lane. Peaks corresponding to the bands were identified and separated from the background and finally integrated. Overexposure of parental bands was carefully avoided. We examined DSBs on both the artificial his4LEU2 hotspot (Cao et al. 1990) and the natural hotspot YCR048w (Goldway et al. 1993).
Native/denaturing 2D electrophoresis
The genomic DNA used in this assay was from the same batch as the genomic DNA used in the DSB assay. Native/denaturing 2D electrophoresis was performed as described (Allers and Lichten 2001). A square 0.7% agarose Seakem GTG agarose gel in 1× TAE was prepared. The first dimension was run for 17 h at 4°C, 35 V (1 V/cm). After rinsing the gel twice with dH2O, the DNA was denatured for 30 min in 250 mM NaOH/5 mM EDTA, followed by equilibration in 50 mM NaOH/1 mM EDTA (2 × 20 min). The gel was rotated 90° and run for 19 h at 4°C, 20 V (0.6 V/cm) in 50 mM NaOH/1 mM EDTA, followed by 30 min equilibration in transfer buffer (0.6 M NaCl/0.5 M NaOH). Then it was transferred to a Hybond-N+ membrane (Amersham) by vacuum blotting for 1.5 h. A AvrII–NheI restriction fragment of YCR048w (Peciña et al. 2002) was labeled with 32P-dCTP and hybridized as described (Borde et al. 2000). Quantification of the signal was undertaken on a Storm PhosphorImager (Amersham).
Western blotting
To obtain yeast extracts, cells (3 mL of 4 × 107 cells/mL) were resuspended in 1 mL of ice-cold H20 and lysed by adding 150 μL of YEX Lysis Buffer (1.85 M NaOH, 7.5% β-mercaptoethanol, natural pH) and incubated for 10 min on ice. One hundred fifty microliters of cold 50% trichloroacetic acid was added and incubated for a further 10 min on ice. The precipitated proteins were pelleted, resuspended in 100 μL 1× GSD-loading buffer and neutralized by adding 15 μL of unbuffered 1 M Tris. Separation and blotting were performed according to standard procedures. Myc-tagged proteins were detected using the 9E10 mouse anti-Myc (1:300) and HRP-conjugated goat anti-mouse (1:100,000, Pierce) antibodies. Detection was performed by the ECL plus Western blotting detection system (Amersham).
ChIP assay
Every ChIP experiment was carried out at least three times with similar results. The ChIP protocol is based on Hecht et al. (1996) and Tanaka et al. (1997). Samples of meiotic cells (50 mL of 4 × 107 cells/mL) were collected at the indicated time points and incubated with 1% FA for 15 min at room temperature for protein–DNA cross-linking. After addition of 125 mM glycine and a further incubation for 5 min at room temperature, cells were harvested and washed three times with ice-cold 1× TBS at pH 7.5 (20 mM TrisHCL at pH 7.5, 150 mM NaCl). The cells were resuspended in 500 μL of lysis buffer (50 mM HepesKOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 1 mM PMSF, 10 μL Aprotinin, 1 tablet of complete inhibitor cocktail [Roche] in 50 mL solution) and lysed with acid-washed glass beads for 10 min in a VIBRAX-IKA at full output. The chromatin was fragmented to an average size of 500 bp by sonication (4 × 25 sec, Bandelin, Sonoplus HD70). To obtain whole-cell extract (WCE), a 50-μL pre-IP sample was removed and microcentrifuged at full speed for 10 sec to pellet the cell debris (supernatant = WCE). The rest of the samples were also centrifuged at 12,000 rpm (4°C) for 20 sec to pellet the cell debris. IP was performed by adding the 450-μL extract to a pellet of magnetic dynabeads M-450 (pan mouse IgG, Dynal), corresponding to 50 μL or 2 × 107 beads, which were preincubated with the 9E11 (monoclonal mouse anti-Myc) antibody overnight at 4°C. Precipitates were washed twice with lysis buffer, twice with lysis buffer plus 360 mM NaCl, twice with washing buffer (10 mM TrisHCl at pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA), and finally once with 1× TE at pH 7.5, using the magnetic device supplied by Dynal. After reversal of cross-linking by heating in TE-1% SDS overnight at 65°C, the Spo11 protein was digested with pronase (12 μL of 20 mg/mL stock) for 3 h at 40°C and after adding 3 μL glycogen (10 mg/mL), the remaining DNA was purified by phenol/chloroform/isoamylalcohol extraction. RNA digestion (10 μg RNase) was carried out for 1 h at 37°C. The DNA was finally resuspended in 30 μL 1× TE at pH 8.0.
mPCR was carried out as described (Tanaka et al. 1997) using PPs located 123 bp (263-bp product, PP1), 1591 bp (212-bp product, PP2), and 2927 bp (314-bp product, PP3) distal to the DSB I site of the his4XLEU2 hotspot (Fig. 1A). PP4 (358 bp) was designed to span the DSB I region, in which almost all breaks of the DSB I site occur (Xu and Kleckner 1995). PCR primers were designed as 20 mers with ∼55% GC content (sequences are available upon request). Every mPCR reaction contained three pairs of primers at a final concentration of 25 pmol each. Apart from the primers and 1–3 μL immunoprecipitate, 2 μL Taq polymerase (1 U/μL), 5 μL 10× PCR buffer, and 5 μL of 25 mM MgCl2 (all Fermentas) were added to a final volume of 50 μL. PCR steps were 4 min, 94°C; followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C; and finally 7 min, 72°C. The PCR products were separated on a 2.5% agarose gel and visualized with 0.1 μg/mL ethidium bromide.
Real-time PCR was carried out using PP5 (254-bp product) 167 bp and PP6 (202-bp product) 2668 bp distal to the DSB site of the YCR048w hotspot (Fig. 1A). PP7 (362-bp product) was designed to span the DSB region, in which all breaks of this DSB site occur (Goldway et al. 1993). As an additional negative control, a PP from YCR011c (130-bp product) was designed and referred to as a cold region (DSB free region in rad50S strains) (Baudat and Nicolas 1997). Our qPCR reactions were carried out either on a Rotorgene cycler from Genexpress or on an ABI prism 7000 (ABI). To label PCR products, we used the LightCycler-DNA Master SYBR Green I Kit from ROCHE or the SYBR Green JumpStart Taq Ready Mix from Sigma. PCR primers adjacent to the YCR047 hotspot were designed as 20 mers with ∼55% GC content and used at a final concentration of 20 pmol per PP. Sequences are available upon request. PCR steps for the Rotorgene cycler were 2 min at 94°C, followed by 30–35 cycles of 10 sec at 94°C, 5 sec at 55°C, 15 sec at 72°C, 15 sec at 81°C, and 15 sec at 84°C, of which only the 72°C was used for fluorescence measurement. PCR steps for the ABI prism 7000 were 10 min at 95°C, followed by 30–35 cycles of 15 sec at 95°C and 1 min at 60°C; the measurement step was carried out at 60°C. The melting curves, measured from 60°C to 99°C, indicated no primer–dimer formation at the used temperature. The amount of precipitated DNA (P) was calculated by comparison with a standard sample obtained from the WCE. One microliter of WCE–DNA was set to 10,000. In cases were the CHIP PCR input differed from 1 μL, the values were corrected accordingly. A value of 100 thus corresponds to 1% of the amount of WCE–DNA. We observed that the relative amount of recovered DNA varied with the detection method rather than with the immunoprecipitations, that is, different kits led to different estimates of the yield of IPs. These varied typically from 0.1% to up to 2% of total rad50S DNA.
Acknowledgments
We are indebted to Kim Nasmyth for advice and his lab for supplying the 9E11 antibody, the Myc18 tagging cassette made by W. Zachariae, and for initial help with ChIP from T. Tanaka. We thank S. Ferscha and C. Erhart for technical assistance. Special thanks to M.T. Hauser and A. Redweik for initial help with qPCR and for sharing their qPCR machine funded by the FWF-grant P14477-GEN to M.T. Hauser and to D. Chen for computer simulation of random colocalizations. We also thank N. Kleckner, S. Roeder, A. Nicolas, M. Lichten, J. Petrini, and B. Byers for supplying strains, constructs, or antibodies. Thanks to Kim Nasmyth, Jim Wang, and Maria Siomos for critical discussion of the manuscript. This work was funded by the Austrian Science Foundation FWF, grant S8203 and by the Austrofan grant from the Austrian Ministry of Science. Valérie Borde was supported by the Association pour la Recherche sur le Cancer and grant #5739 to Alain Nicolas.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.321105.
References
- Alani E., Padmore, R., and Kleckner, N. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436. [DOI] [PubMed] [Google Scholar]
- Allers T. and Lichten, M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. [DOI] [PubMed] [Google Scholar]
- Arora C., Kee, K., Maleki, S., and Keeney, S. 2004. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13: 549–559. [DOI] [PubMed] [Google Scholar]
- Baudat F. and Nicolas, A. 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Manova, K., Yuen, J.P., Jasin, M., and Keeney, S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Bergerat A., de Massy, B., Gadelle, D., Varoutas, P.C., Nicolas, A., and Forterre, P. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio, R.U., Hunter, N., and Kleckner, N. 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. [DOI] [PubMed] [Google Scholar]
- Borde V., Goldman, A.S., and Lichten, M. 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290: 806–809. [DOI] [PubMed] [Google Scholar]
- Borde V., Lin, W., Novikov, E., Petrini, J.H., Lichten, M., and Nicolas, A. 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell 13: 389–401. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani, E., and Kleckner, N. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Celerin M., Merino, S.T., Stone, J.E., Menzie, A.M., and Zolan, M.E. 2000. Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 19: 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Trujillo, K., Ramos, W., Sung, P., and Tomkinson, A.E. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Chu S. and Herskowitz, I. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1: 685–696. [DOI] [PubMed] [Google Scholar]
- D'Amours D. and Jackson, S.P. 2002. The Mre11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3: 317–327. [DOI] [PubMed] [Google Scholar]
- de Massy B., Rocco, V., and Nicolas, A. 1995. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 14: 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A.M. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Engebrecht J.A., Voelkel-Meiman, K., and Roeder, G.S. 1991. Meiosis-specific RNA splicing in yeast. Cell 66: 1257–1268. [DOI] [PubMed] [Google Scholar]
- Esposito M.S. and Esposito, R.E. 1969. The genetic control of sporulation in Saccharomyces. The isolation of temperature-sensitive sporulation-deficient mutants. Genetics 61: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton J.L., DeRisi, J., Shroff, R., Lichten, M., Brown, P.O., and Petes, T.D. 2000. Inaugural article: Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldway M., Sherman, A., Zenvirth, D., Arbel, T., and Simchen, G. 1993. A short chromosomal region with major roles in yeast chromosome III meiotic disjunction, recombination and double strand breaks. Genetics 133: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon, D., Gendrot, G., and Pelletier, G. 2001. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 1998. The many interfaces of Mre11. Cell 95: 583–586. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger, S., and Grunstein, M. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N.M. and Byers, B. 1989. HOP1: A yeast meiotic pairing gene. Genetics 121: 445–462. (Erratum appears in 122: 719.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M. and Ponte, L. 1997. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K., Salem, L., and Malone, R. 2003. Support for a meiotic recombination initiation complex: Interactions among Rec102p, Rec104p, and Spo11p. Mol. Cell. Biol. 23: 5928–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S.M. and Roth, R. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K. and Keeney, S. 2002. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K., Protacio, R.U., Arora, C., and Keeney, S. 2004. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 23: 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney S. and Kleckner, N. 1995. Covalent protein–DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. 92: 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux, C.N., and Kleckner, N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Waddell, C.S., and Esposito, R.E. 1985. The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110: 187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. and Smith, G.R. 1994. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F. and Wang, J.C. 1979. Interaction between DNA and Escherichia coli DNA topoisomerase I. Formation of complexes between the protein and superhelical and nonsuperhelical duplex DNAs. J. Biol. Chem. 254: 11082–11088. [PubMed] [Google Scholar]
- Liu J., Wu, T.C., and Lichten, M. 1995. The location and structure of double-strand DNA breaks induced during yeast meiosis: Evidence for a covalently linked DNA–protein intermediate. EMBO J. 14: 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang, J.K., Kato, N., and McKim, K.S. 2002. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Klein, F., and Engebrecht, J. 1998. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. In Nuclear structure and function (ed. M. Berrios), pp. 257–285. Academic Press, San Diego, CA. [DOI] [PubMed]
- Malone R.E., Bullard, S., Hermiston, M., Rieger, R., Cool, M., and Galbraith, A. 1991. Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 128: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Draayer Y., Galbraith, A.M., Pittman, D.L., Cool, M., and Malone, R.E. 1996. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A.H. and Kleckner, N. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S. and Hayashi-Hagihara, A. 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: Evidence that the mechanism for initiating meiotic recombination is conserved. Genes & Dev. 12: 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler-Guillemain C. and de Massy, B. 2000. Identification and characterization of an SPO11 homolog in the mouse. Chromosoma 109: 133–138. [DOI] [PubMed] [Google Scholar]
- Molnar M., Parisi, S., Kakihara, Y., Nojima, H., Yamamoto, A., Hiraoka, Y., Bozsik, A., Sipiczki, M., and Kohli, J. 2001. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz K. and Klein, F. 1997. mre11S—A yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes & Dev. 11: 2272–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T. and Ogawa, H. 1997. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells 2: 65–79. [DOI] [PubMed] [Google Scholar]
- Peciña A., Smith, K.N., Mezard, C., Murakami, H., Ohta, K., and Nicolas, A. 2002. Targeted stimulation of meiotic recombination. Cell 111: 173–184. [DOI] [PubMed] [Google Scholar]
- Prinz S., Amon, A., and Klein, F. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break induced recombination in S. cerevisiae. Genetics 146: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B. and Roeder, G.S. 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P.J. and Camerini-Otero, R.D. 1999. Cloning, characterization, and localization of mouse and human SPO11. Genomics 61: 156–169. [DOI] [PubMed] [Google Scholar]
- ____. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Schwacha A. and Kleckner, N. 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. [DOI] [PubMed] [Google Scholar]
- Shannon M., Richardson, L., Christian, A., Handel, M.A., and Thelen, M.P. 1999. Differential gene expression of mammalian SPO11/TOP6A homologs during meiosis. FEBS Lett. 462: 329–334. [DOI] [PubMed] [Google Scholar]
- Smith K.N. and Nicolas, A. 1998. Recombination at work for meiosis. Curr. Opin. Genet. Dev. 8: 200–211. [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Tesse, S., Gargano, S., James, F., Kleckner, N., and Zickler, D. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes & Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Knapp, D., and Nasmyth, K. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649–660. [DOI] [PubMed] [Google Scholar]
- Tesse S., Storlazzi, A., Kleckner, N., Gargano, S., and Zickler, D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. 100: 12865–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat, A., Pohlmann, R., and Philippsen, P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13: 1065–1075. [DOI] [PubMed] [Google Scholar]
- Xu L. and Kleckner, N. 1995. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 14: 5115–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Ajimura, M., Padmore, R., Klein, C., and Kleckner, N. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W., Shin, T.H., Galova, M., Obermaier, B., and Nasmyth, K. 1996. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274: 1201–1204. [DOI] [PubMed] [Google Scholar]