Abstract
A stochastic lateral signaling interaction between two developing Caenorhabditis elegans AWC olfactory neurons causes them to take on asymmetric patterns of odorant receptor expression, called AWCOFF and AWCON. Here we show that the AWC lateral signaling gene tir-1 (previously known as nsy-2) encodes a conserved post-synaptic protein that specifies the choice between AWCOFF and AWCON. Genetic evidence suggests that tir-1 acts downstream of a voltage-gated calcium channel and CaMKII (UNC-43) to regulate AWC asymmetry via the NSY-1(ASK1) p38/JNK MAP (mitogen-activated protein) kinase cascade. TIR-1 localizes NSY-1 to post-synaptic regions of AWC, and TIR-1 binds UNC-43, suggesting that it assembles a synaptic signaling complex that regulates odorant receptor expression. Temperature-shift experiments indicate that tir-1 affects AWC during a critical period late in embryogenesis, near the time of AWC synapse formation. TIR-1 is a multidomain protein with a TIR (Toll-interleukin-1 receptor) domain that activates signaling, SAM repeats that mediate localization to post-synaptic regions of axons, and an N-terminal inhibitory domain. TIR-1 and other TIR proteins are implicated in vertebrate and invertebrate innate immunity, as are NSY-1/ASK1 kinases, so this pathway may also have a conserved function in immune signaling.
Keywords: Olfactory development, C. elegans, calcium-calmodulin-dependent protein kinase II (CaMKII), cell signaling, innate immunity
The developing nervous system generates a spectacular diversity of cell types. One mechanism for creating diversity is lateral signaling, in which interactions between developing neurons cause equivalent cells to take on different identities (Greenwald 1998). The left and right Caenorhabditis elegans AWC olfactory neurons develop distinct sensory properties through an unusual stochastic lateral signaling interaction. The two AWC neurons are initially equivalent, but after a signaling interaction one AWC neuron, AWCON, expresses the str-2::GFP transgene and detects the attractive odor butanone, and the other AWC neuron, AWCOFF, detects the odor pentanedione and does not express str-2 (Troemel et al. 1999; Wes and Bargmann 2001). This decision is random but coordinated, such that exactly one AWCON and one AWCOFF neuron are generated per animal (Troemel et al. 1999). If one AWC neuron is killed in the embryo, the surviving neuron always becomes AWCOFF, indicating that an interaction or competition between AWC neurons is necessary for the AWCON state. The asymmetry between the left and right AWC neurons is stabilized after hatching and is maintained throughout life.
Although several examples of left–right asymmetry are found in the C. elegans nervous system, AWC asymmetry is the only known example that exhibits antisymmetry, defined as stochastic assignment of the two cell states. The left and right Q-neuroblast fates, which are specified by Wnt signaling and Hox genes (Harris et al. 1996; Maloof et al. 1999; Whangbo and Kenyon 1999), and the left and right ASE-neuron fates, which are specified by a gene regulatory cascade (Pierce-Shimomura et al. 2001; Chang et al. 2003; Johnston and Hobert 2003), are invariant and apparently defined by prepatterns from cell lineage. The AWC neurons arise from separate cell lineages and reside on opposite sides of the head, but the axons of the two AWC neurons make direct contact with each other in the nerve ring (White et al. 1986). Axon guidance mutants disrupt AWC asymmetry, suggesting that axon contact could mediate the AWC interaction. The Notch pathway acts in many lateral signaling events, but mutations in the Notch pathway do not affect AWC, suggesting the existence of an alternative mechanism for generating stochastic asymmetry (Troemel et al. 1999).
Calcium signaling plays an instructive role in defining AWC asymmetry. Gain-of-function mutations in unc-43, which encodes the C. elegans calcium-calmodulin-dependent protein kinase II (CaMKII), lead to the generation of two AWCOFF cells, whereas loss-of-function mutations in CaMKII lead to the development of two AWCON cells. In addition, mutations in an N/P-type voltage-gated calcium channel (the α1 subunit unc-2 or the α2δ subunit unc-36) and a calcium-activated potassium channel (nsy-3/slo-1) disrupt AWC asymmetry (Troemel et al. 1999; Davies et al. 2003). Downstream of UNC-43, the mitogen-activated protein kinase kinase kinase (MAPKKK) NSY-1 (nsy, neuronal symmetry gene), activates the MAPKK SEK-1 to execute the AWCOFF decision (Sagasti et al. 2001; Tanaka-Hino et al. 2002). nsy-1 encodes a homolog of human ASK1, the apoptosis signal-regulated kinase, which can activate both JNK and p38 MAPKs (Ichijo et al. 1997). Genetic epistasis and genetic mosaic analyses suggest a model in which lateral signaling inhibits the Ca++/CaMKII/MAPKKK signaling cassette in one AWC neuron, causing it to become AWCON. The Ca++/CaMKII/MAPKKK signaling cassette represents a new pathway that is conserved in mammalian neurons (Takeda et al. 2004). Interestingly, Ca++ signaling can regulate the neurotransmitter phenotype of vertebrate spinal cord neurons, suggesting that calcium affects neuronal specification in a variety of contexts (Borodinsky et al. 2004). Differences in external Ca++ levels have also been implicated in vertebrate left–right asymmetry (Raya et al. 2004).
In addition to their functions in AWC, nsy-1 and sek-1 act with the p38 MAP kinase pmk-1 in C. elegans innate immunity, the ability to resist bacterial pathogens such as Pseudomonas aeruginosa (Kim et al. 2002; Aballay et al. 2003). The mammalian NSY-1 ortholog ASK1 also functions in innate immunity, as it induces programmed cell death in Mycobacterium avium-infected macrophages (Bhattacharyya et al. 2003). The mechanism of ASK1 activation in innate immunity is unknown, but C. elegans unc-43 null mutants have normal innate immunity, suggesting that there are different modes of activation of NSY-1 in AWC development and immune responses (Kim et al. 2002).
Here we describe the function of an AWC asymmetry gene, tir-1/nsy-2, that was identified based on a mutant with str-2::GFP expression in both AWC cells (2 AWCON phenotype) (Troemel et al. 1999; Sagasti et al. 2001). Recent RNA interference (RNAi) studies have implicated tir-1 in C. elegans innate immunity (Couillault et al. 2004; Liberati et al. 2004); our results suggest that this function arises from regulation of the conserved NSY-1/ASK1 MAPKKK signaling cassette. We show that TIR-1 localizes NSY-1/ASK1 to AWC synapses, implicating the synapse as the site of AWC lateral signaling.
Results
nsy-2/tir-1 encodes a conserved protein with Armadillo, SAM, and TIR domains
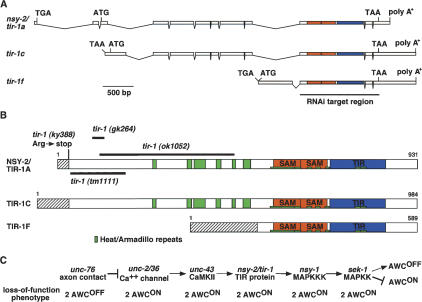
The 2 AWCON mutation nsy-2(ky388) was mapped to a small interval on the third chromosome, and then rescued with a cosmid containing only one full-length open reading frame, F13B10.1 (Fig. 1A). This open reading frame has been previously characterized by RNAi under the name tir-1 (Couillault et al. 2004; Liberati et al. 2004). It encodes a family of at least six different proteins with alternative N termini from alternative promoters. The ky388 allele was associated with a premature stop codon in an early exon of one alternative transcript of F13B10.1, called tir-1a (Fig. 1B). Three other alleles of tir-1 have been generated by reverse genetics, but none has previously been characterized. tir-1(tm1111), deleting early exons of tir-1a, tir-1c, and tir-1e, caused a stronger 2 AWCON phenotype than nsy-2(ky388) (Fig. 1B, Table 1). RNA interference of the F13B10.1 gene caused a 2 AWCON phenotype similar to that of nsy-2(ky388) and tir-1(tm1111) (Fig. 2B,C). Further reduction of tir-1(tm1111) function through RNAi did not lead to a more severe phenotype (data not shown). We conclude that ky388 likely represents a reduced function allele of tir-1, and that a general reduction of tir-1 function through RNAi can also generate the 2 AWCON phenotype. In agreement with other groups working on this locus (J. Ewbank, F. Ausubel, and J. Hodgkin, pers. comm.), the gene nsy-2 has been renamed tir-1. Although the reduction-of-function allele tm1111 inactivates all three long forms of tir-1, none of the EMS-induced alleles affect the shorter tir-1b, tir-1d, and tir-1f forms. Therefore, the null phenotype of this complex locus is not yet defined. All alleles of tir-1 caused milder defects in AWC development than null alleles of unc-43 or nsy-1; moreover, all alleles of tir-1 caused milder defects in innate immunity than null alleles of nsy-1 (Supplementary Fig. 1). Either some residual tir-1 activity persists in all alleles, or tir-1 is redundant with other genes during AWC development and innate immunity.
Figure 1.
tir-1 encodes a TIR protein. (A) Genomic structure of three variants of F13B10.1 gene. Sequence analysis of cDNA clones corresponding to the F13B10.1 gene revealed six splice forms with alternative 5′ ends, called tir-1a–f. Shown here are tir-1a, tir-1c, and tir-1f, which were identified in this work. tir-1b and tir-1d are similar to tir-1f, and tir-1e is similar to tir-1a and tir-1c (WormBase, http://www.wormbase.org). Boxes and lines represent exons and introns, respectively. ATG and TAA indicate the positions of the putative translational start and stop codons. tir-1a, tir-1c, and tir-1f do not have splice leader sequences at the 5′ end, but have in-frame stop codons preceding the putative translational start codon. The regions that encode SAM and TIR domains are shown in orange and blue, respectively. The region that was targeted by RNAi is underlined. (B) Structure of TIR-1 proteins. Heat/Armadillo repeats, SAM domains, and TIR domain are shown in green, orange, and blue, respectively. Heat/Armadillo repeats are located at amino acids 256–266, 343–354, 360–390, 416–427, 456–464, 484–503, 554–622, 624–634, 649–660, 669–680, 693–703, 774–786, and 789–801 (Mink et al. 2001; Liberati et al. 2004). Hatched boxes indicate isoform-specific regions. (C) Epistasis relationships of nsy mutants. tir-1 may act downstream of unc-43 and upstream of nsy-1.
Table 1.
Analysis of AWC phenotypes
| Strain | 2 AWCOFF (%) | 1 AWCOFF/1 AWCON (%) | 2 AWCON (%) | n |
|---|---|---|---|---|
| Wild type (N2) | 2 | 98 | 0 | 241 |
| Single mutants | ||||
| unc-43(n1186lf) | 1 | 1 | 98 | 180 |
| unc-43(n498gf) | 83 | 17 | 0 | 71 |
| tir-1(ky388ts) | 0 | 42 | 58 | 286 |
| tir-1(tm1111lf) | 0 | 26 | 74 | 80 |
| tir-1(ok1052) | 36 | 50 | 14 | 133 |
| tir-1(gk264) | 0 | 99 | 1 | 175 |
| odr-3::tir-1::DsRed transgenea | 7 | 93 | 0 | 97 |
| odr-3::tir-1(OE) transgeneb | 94 | 6 | 0 | 617 |
| nsy-1(ky542lf) | 0 | 1 | 99 | 156 |
| odr-3::nsy-1(gf) transgeneb | 80 | 20 | 0 | 206 |
| Double mutants | ||||
| tir-1(ky388ts); odr-3::tir-1::DsRed transgene | 0 | 99 | 1 | 107 |
| unc-43(n1186lf); odr-3::tir-1(OE) transgeneb | 90 | 9 | 1 | 248 |
| unc-43(n498gf); tir-1(ky388ts) | 1 | 40 | 59 | 145 |
| unc-43(n498gf); tir-1(tm1111lf) | 1 | 30 | 69 | 115 |
| nsy-1(ky542lf); odr-3::tir-1(OE) transgeneb | 0 | 3 | 97 | 182 |
| tir-1(ky388ts); odr-3::nsy-1(gf) transgene | 85 | 13 | 2 | 257 |
| tir-1(tm1111lf); odr-3::nsy-1(gf) transgeneb | 76 | 24 | 1 | 267 |
Animals were grown at 20°C.
The odr-3::tir-1::DsRed transgene was injected at 5 ng/μl.
Results were combined from two transgenic lines, which showed similar penetrance of phenotypes. The odr-3::tir-1 transgene was injected at 50 ng/μl.
Figure 2.
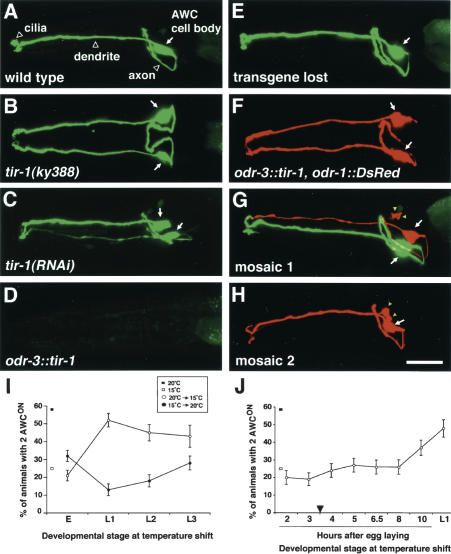
str-2::GFP expression in tir-1 mutants and mosaic animals. (A–H) Confocal projections of animals expressing an integrated str-2::GFP transgene; animals in E–H are from a strain with an unstable transgenic array bearing odr-3::tir-1 and odr-1::DsRed. GFP and DsRed were false-colored green and red, respectively. (A) Wild-type animals express str-2::GFP in one AWC neuron (1 AWCON/1 AWCOFF). (B,C) tir-1(ky388) (B) and tir-1(RNAi) (C) animals express str-2::GFP in both AWC neurons (2 AWCON). (D) Animals overexpressing the transgene odr-3::tir-1 did not express str-2::GFP in either AWC neuron (2 AWCOFF). (E–H) Phenotypes from mosaic analysis. (E) Animals that lost the odr-3::tir-1, odr-1::DsRed array had wild-type str-2::GFP expression. (F) Nonmosaic animals expressing odr-1::DsRed in both AWC neurons had a 2 AWCOFF phenotype. (G,H) Mosaic animals expressing odr-1::DsRed in one AWC neuron. Most tir-1(gf) AWC neurons (DsRed+) bearing the transgene odr-3::tir-1 were AWCOFF, while wild-type AWC neurons (DsRed–) were either AWCON (G) or AWCOFF (H) with equal frequency. Arrows indicate the AWC cell body. odr-1::DsRed is also visible in AWB, a smaller cell (yellow arrowheads). Bar, 10 μm. Anterior is at left and ventral is down in lateral or ventrolateral views in A and C–H; ventral view in B.(I) tir-1(ky388) animals were shifted between restrictive temperature (20°C) and permissive temperature (15°C) at various points during development. For the downshift (from 20°C to 15°C), a significant increase in 2 AWCON animals was observed between the embryo and L1 shift (P < 0.001). For the upshift (from 15°C to 20°C), a significant difference in the 2 AWCON phenotype was observed between the embryo and L1 shift (P < 0.001). (E) Embryo 2 h after egg laying; (L1) first larval stage; (L2) second larval stage; (L3) third larval stage. (J) Embryos were shifted from 20°C to 15°C at different times after egg laying. A significant increase in the 2 AWCON mutant phenotype occurred between the shifts at the embryonic stage 8 h after egg laying and L1 (∼15 h after egg laying) (P = 0.001). When shifted at L1, tir-1 mutants exhibited a 2 AWCON phenotype similar to that of mutants raised continuously at 20°C (P = 0.1). The AWC neurons are born ∼1 h after egg laying. Axon outgrowth starts ∼3.5 h after egg laying (arrowhead). The percentage of animals with 2 AWCON phenotypes in tir-1(ky388) animals raised continuously at 15°C or 20°C is indicated at the left of each graph. Error bars denote the standard error of proportion.
To confirm the identification of F13B10.1 as nsy-2/tir-1 and determine the site of gene action, we generated an odr-3::tir-1::DsRed transgene that drives expression of a tagged tir-1a cDNA strongly in AWC neurons and weakly in four other classes of sensory neurons (Roayaie et al. 1998) (the tir-1a cDNA affected by the ky388 mutation was used in all molecular studies). The transgene rescued the str-2 expression defect in tir-1(ky388) mutant when injected at 5 ng/μL, resulting in a high percentage of animals with the wild-type 1 AWCON phenotype (Table 1). This result suggests that tir-1 acts in AWC to affect asymmetric odorant receptor expression.
tir-1 encodes predicted cytoplasmic proteins with HEAT-Armadillo repeats, two sterile α motif (SAM) domains, and one Toll/Interleukin-1 receptor (TIR) domain (Fig. 1A,B). HEAT-Armadillo repeats are scaffolding domains found in many proteins (Huber et al. 1997; Chook and Blobel 1999; Cingolani et al. 1999). The ∼70-amino acid SAM domain, which can homodimerize, was first described in proteins in the yeast MAP kinase mating pathway, and is also present in receptor protein kinases, serine/threonine kinases, transcription factors, and adaptor proteins (Ponting 1995; Schultz et al. 1997; Kyba and Brock 1998; Ramezani-Rad 2003). The TIR domain is a 200-amino acid sequence shared by the transmembrane Toll-like receptors (TLRs) and the interleukin-1 receptor, as well as cytoplasmic adaptor proteins that act downstream of TLRs in the innate immune response (Medzhitov 2001). TIR domains often homodimerize and heterodimerize with other TIR domains. A single TIR-1-like protein is present in C. elegans, Drosophila, mouse, and human (Mink et al. 2001); the mammalian ortholog is known as SARM, for sterile α and HEAT-Armadillo motifs (O'Neill et al. 2003). The function of SARM is unknown.
Two of the deletion alleles of tir-1 resulted in in-frame deletions in the predicted protein. One of these mutants, gk264, had a wild-type phenotype, and one mutant, ok1052, had a mixed phenotype with both 2 AWCON and 2 AWCOFF animals (Fig. 1B; Table 1). Based on its molecular nature as well as structure-function analysis (see below), tir-1(ok1052) is likely to encode a misregulated protein product.
Overexpression of odr-3::tir-1, achieved by injecting an odr-3::tir-1 transgene at a high concentration (50 ng/μL), generated a 2 AWCOFF phenotype opposite to the tir-1 reduction-of-function phenotype (Fig. 2D; Tables 1, 2). This result indicates that the level of tir-1 activity can specify AWC odorant receptor expression in an instructive manner, with a low level of tir-1 activity defining the AWCON state and a high level of activity defining the AWCOFF state. Thus tir-1 is likely to be directly involved in AWC signaling.
Table 2.
Overexpression of TIR-1 and mosaic analysis
| Transgene expressed in both AWC neuronsa
|
|||||||||
| Transgene | 2 AWCOFF (%) | 1 AWCOFF/1 AWCON (%) | 2 AWCON (%) | n | |||||
| odr-3::tir-1 #1 | 90 | 9 | 1 | 631 | |||||
| odr-3::tir-1 #2 | 94 | 6 | 0 | 527 | |||||
| odr-3::tir-1 #3 | 95 | 5 | 0 | 727 | |||||
| Mosaic animals with one wild-type and one mutant AWC neurona
|
|||||||||
| 1 AWCOFF/1 AWCON (%)b
|
χ2 analysis for execution model
|
||||||||
| 2 AWCOFF (%) | Wild-type AWCON/tir-1(gf) AWCOFF | Wild-type AWCOFF/tir-1(gf) AWCON | n | χ2 | P | ||||
| odr-3::tir-1 #1 | 53 | 40 | 7 | 60 | 2.46 | 0.25-0.5 | |||
| odr-3::tir-1 #2 | 48 | 50 | 2 | 92 | 0.35 | 0.5-0.9 | |||
| odr-3::tir-1 #3 | 61 | 37 | 2 | 51 | 3.88 | 0.1-0.25 | |||
An odr-1::DsRed transgene was used to identify AWC cells with the extrachromosomal array. Upper table shows results from animals with the array in both AWC cells (two DsRed+ cells). Lower table shows results from mosaic animals, which contained the array in only one AWC cell (one DsRed+ and one DsRed- cell). Three independent lines were analyzed.
The wild-type cell was DsRed-, and the tir-1(gf) cell was DsRed+.
tir-1 may act between unc-43 and nsy-1 in AWC signaling
tir-1 reduction-of-function mutants share the 2 AWCON phenotype with null mutants for unc-43/CaMKII and null mutants in the nsy-1/ASK1 MAP kinase pathway (Table 1). To characterize the relationship between tir-1 and other genes, we made double mutants between gain-of-function (gf) and loss-of-function (lf) mutations in the pathway.
tir-1 mutations were epistatic to unc-43 mutations (Table 1). The dominant unc-43(n498) mutant had a 2 AWCOFF phenotype, whereas unc-43(gf) tir-1(lf) double mutants had a 2 AWCON phenotype, like tir-1(lf) single mutants. The unc-43(n1186) null mutant had a 2 AWCON phenotype, whereas unc-43(lf) tir-1(gf) mutants had the 2 AWCOFF phenotype of tir-1(gf) single mutants (Table 1). These results suggest that tir-1 acts downstream of unc-43.
nsy-1 mutations were epistatic to tir-1 mutations. tir-1(gf) nsy-1(lf) double mutants had a 2AWCON phenotype, like nsy-1(lf) single mutants. An overexpressed odr-3::nsy-1 transgene with a deletion in its regulatory N terminus caused a 2 AWCOFF phenotype (Sagasti et al. 2001), and tir-1(lf) nsy-1(gf) mutants exhibited the same 2 AWCOFF phenotype, unlike tir-1(lf) mutants (Table 1). These results suggest that nsy-1 acts downstream of tir-1.
These epistasis results are consistent with a model in which tir-1 acts downstream of unc-43 and upstream of nsy-1 in a linear genetic pathway to regulate AWC asymmetry (Fig. 1C). None of the alleles of tir-1 eliminated all potential splice forms, so there is a remaining possibility that tir-1 function is partly parallel to nsy-1. However, all genetic results were qualitatively and quantitatively consistent with this interpretation.
tir-1 can act cell-autonomously to execute the AWCOFF decision
Although the rescue of tir-1 using the odr-3 promoter suggests a site of action in AWC, it does not distinguish whether the protein acts in the AWCON cell, the AWCOFF cell, or both. These possibilities can be distinguished in mosaic animals in which tir-1 activity is altered in only one of the two AWC neurons. The partial penetrance of tir-1 mutations was problematic for mosaic analysis, so the gain-of-function phenotype of tir-1 overexpression was used to ask whether TIR-1 acts in AWCON or AWCOFF. An extrachromosomal array containing odr-1::DsRed and odr-3::tir-1 transgenes was introduced into a str-2::GFP integrated line. Nonmosaic animals expressed DsRed fluorescence in both AWC cells, but str-2::GFP fluorescence in neither AWC cell (the 2-AWCOFF phenotype) (Fig. 2F). Spontaneous loss of the extrachromosomal array resulted in mosaic animals, recognizable as animals in which only one AWC neuron expressed DsRed fluorescence. The single DsRed-expressing AWC cell did not express str-2::GFP in 95% of the animals (Fig. 2G,H; Table 2), a percentage consistent with a cell-autonomous repression of str-2::GFP by the odr-3::tir-1 transgene in that neuron.
The AWC neuron in the mosaics that did not express DsRed should be genotypically wild-type. Approximately 40% of the AWC cells that lacked DsRed fluorescence expressed str-2::GFP (Fig. 2G; Table 2). The expression of str-2::GFP in about half of the cells suggests that both AWC neurons participated in the initial lateral signaling interaction: If no interaction had taken place, the wild-type AWC neuron should always have taken on a default AWCOFF state. These results indicate that TIR-1 acts cell autonomously to execute the AWCOFF cell decision, downstream of the initial interaction that diversifies the two AWC neurons.
tir-1 acts embryonically
The tir-1(ky388) allele is temperature-sensitive for its 2 AWCON phenotype, with 25% of animals being AWCON at 15°C, 58% at 20°C, and 81% at 25°C. To determine the time of tir-1 gene action, and help infer the time of AWC lateral signaling, animals were shifted between 20°C and 15°C at different developmental stages, and adults were scored for str-2::GFP expression in AWC.
The AWC neurons are born at 300 min (∼1 h after egg laying) and their axons extend at 450 min (∼3.5 h after egg laying) (Sulston et al. 1983). When shifted from 20°C to 15°C early in embryogenesis (2 h after egg laying), tir-1 mutants exhibited a mild 2 AWCON phenotype similar to that of mutants raised continuously at 15°C (Fig. 2I). The time of the temperature shift corresponds approximately to the birth of the AWC neurons. This result indicates that tir-1 is probably not required in very early cell lineages. An upshift from 15°C to 20°C at the same time in embryogenesis was sufficient to rescue significant tir-1 function compared to continuous growth at 20°C (P < 0.001) (Fig. 2I), suggesting that embryonic expression of TIR-1 was sufficient for development. In contrast, temperature shifts in the first larval stage (L1) resulted in phenotypes consistent with the embryonic temperature, even when animals were raised for several days at the new temperature. Similar results were obtained following L2 or L3 larval stage temperature shifts (Fig. 2I). These results suggest that tir-1 is required embryonically for AWC asymmetry.
To further define the time of tir-1 gene action, embryos were shifted from 20°C to 15°C at different times after egg laying (Fig. 2J). The 2 AWCON phenotype significantly increased during the shifts at the late embryonic stage (8 h after egg laying) and L1 (∼15 h after egg laying) (P = 0.001). These results suggest that tir-1 function must be present by late embryogenesis, the time at which synapses form in the nerve ring.
TIR-1 is localized to post-synaptic regions of axons
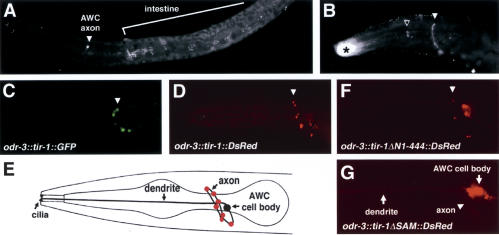
An anti-TIR-1 antibody was able to detect protein in whole-mount immunostaining of animals that expressed odr-3::tir-1 or F13B10.1 rescuing transgenes, although the endogenous expression of TIR-1 was undetectable. Staining revealed punctate expression in the nerve ring, the region where AWC axons reside (Fig. 3A,B). The subcellular localization of TIR-1 in AWC was examined more closely using a transgene in which a TIR-1::GFP fusion protein was expressed under the odr-3 promoter. TIR-1::GFP was localized in a punctate pattern along the AWC axon, and mostly excluded from the cell body and dendrites, like TIR-1 detected by the antiserum (Fig. 3C). A similar punctate localization pattern of TIR-1 was present in animals bearing a rescuing odr-3::tir-1::DsRed transgene (Fig. 3D,E). Both TIR-1::GFP and TIF-1::RFP can restore wild-type tir-1 function. With the qualification that they were obtained with overexpressed or tagged proteins, these results suggest that TIR-1 protein is localized to a subcompartment in AWC axons.
Figure 3.
Subcellular localization of TIR-1. (A) Animals bearing the rescuing cosmid F13B10 were stained with TIR-1 antisera. TIR-1 staining is also visible in the intestine. (B) Animals expressing odr-3::tir-1 immunostained with TIR-1 antisera. White arrowhead indicates specific staining; open arrow and asterisk indicate nonspecific staining of the pharynx and buccal cavity, also seen with preimmune sera. (C) Confocal projections of animals expressing odr-3::tir-1::GFP false-colored green. (D) Confocal projections of animals expressing odr-3::tir-1::DsRed false-colored red. Cross-mark indicates autofluorescence. (E) Schematic representation of TIR-1::DsRed distribution pattern shown in D. (F,G) Animals expressing odr-3::tir-1ΔN1-444:: DsRed (F) or odr-3::tir-1ΔSAM::DsRed (G).
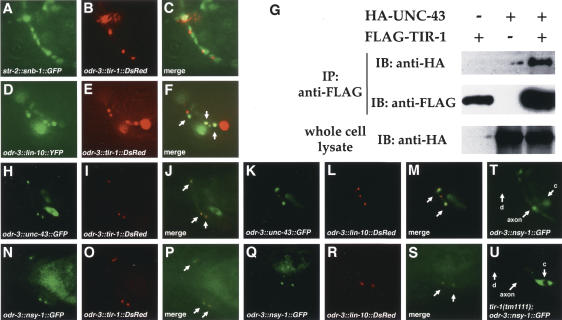
The punctate localization of TIR-1 in axons is reminiscent of the localization of synaptic proteins. Each AWC neuron sends synapses to AIA, AIB, and AIY neurons and the contralateral AWC neuron, and receives a few synapses from ASI and the contralateral AWC neuron (White et al. 1986). Presynaptic and post-synaptic regions are adjacent and interspersed along the AWC axon. To ask whether TIR-1 is localized to presynaptic or post-synaptic sites in the AWC axon, the odr-3::tir-1::DsRed transgene was coexpressed with str-2::synaptobrevin (snb-1)::GFP, a GFP-tagged synaptic vesicle protein that serves as a presynaptic marker (Nonet et al. 1998), or odr-3::lin-10::YFP, a YFP-tagged PDZ-domain protein that localizes to post-synaptic densities in C. elegans neurons (Rongo et al. 1998). TIR-1::DsRed was strikingly colocalized with LIN-10::YFP (Fig. 4D–F), but was usually adjacent to SNB-1::GFP rather than overlapping with it, despite the greater overall expression of SNB-1::GFP (Fig. 4A–C). These results suggest that TIR-1 may be localized to post-synaptic regions of AWC.
Figure 4.
Colocalization of TIR-1 with LIN-10, UNC-43, and NSY-1 in the AWC axon. (A–C) Animals expressing odr-3::tir-1::DsRed and str-2::snb-1::GFP in unc-13(e450) background. SNB-1::GFP puncta are better-resolved in unc-13 mutants (Richmond et al. 1999). (D–F) odr-3::tir-1::DsRed and odr-3::lin-10::YFP. Colocalization of LIN-10::YFP with TIR-1::DsRed in F appears yellow (arrows). (G) Interaction of TIR-1 with UNC-43. Transfected HEK 293 cells were immunoprecipitated (IP), followed by immunoblotting (IB) with indicated antibodies. Whole-cell lysate was ∼10% of input. In parallel experiments, UNC-43 also associated with NSY-1 (Sagasti et al. 2001; our unpublished results); this interaction was not appreciably altered in the presence of TIR-1 (data not shown). TIR-1 did not coimmunoprecipitate SEK-1 (data not shown). (H–J) odr-3::unc-43::GFP and odr-3::tir-1::DsRed. (K–M) odr-3::unc-43::GFP and odr-3::lin-10::DsRed. (N–P) odr-3::nsy-1::GFP and odr-3::tir-1::DsRed. (Q–S) odr-3::nsy-1::GFP and odr-3::lin-10::DsRed. Arrows indicate colocalization of UNC-43::GFP and TIR-1::DsRed (J), UNC-43::GFP and LIN-10::DsRed (M), NSY-1::GFP and TIR-1::DsRed (P), or NSY-1::GFP and LIN-10::DsRed (S). (T,U) Expression of odr-3::nsy-1::GFP in wild type (T) or tir-1(tm1111) (U). (c) Cell body; (d) dendrite.
TIR-1 interacts with UNC-43 and mediates synaptic localization of NSY-1
Since tir-1 acts genetically between unc-43 and nsy-1, we examined the possibility that these proteins could associate with one another directly. Plasmids encoding Flag-tagged TIR-1 and HA-tagged UNC-43 were transiently transfected into HEK 293 cells, the cell lysate was immunoprecipitated with an anti-Flag antibody, and the immunoprecipitated complex was analyzed on Western blots with HA or Flag antibodies (Kawasaki et al. 1999). TIR-1 and UNC-43 could be coimmunoprecipitated when coexpressed in HEK 293 cells, suggesting that TIR-1 physically associates with UNC-43 (Fig. 4G). A weak interaction between Flag-tagged TIR-1 and T7-tagged NSY-1 could be detected, but this interaction appeared inefficient compared to the TIR-1/UNC-43 interaction (data not shown).
To determine whether TIR-1, UNC-43, and NSY-1 are present in the same subcellular compartment in AWC, odr-3::tir-1::DsRed was coexpressed with odr-3::unc-43::GFP or odr-3::nsy-1::GFP in wild-type animals. All of these transgenes encoded functional proteins. Tagged UNC-43 and NSY-1 proteins were not as tightly localized as TIR-1 protein, but their expression was notably enriched in the TIR-1-containing puncta in the AWC axon (Fig. 4H–J,N–P). UNC-43 has previously been shown to be enriched in post-synaptic domains (Rongo and Kaplan 1999), and UNC-43::GFP and NSY-1::GFP were also colocalized with LIN-10::DsRed expressed in AWC (Fig. 4K–M,Q–S). These results suggest that TIR-1, UNC-43, and NSY-1 reside together at post-synaptic regions of AWC.
Since genetic results suggested that tir-1 might regulate nsy-1, the subcellular localization of NSY-1 in AWC was examined in wild-type and tir-1(tm1111) animals. In wild type, NSY-1::GFP was localized in a punctate pattern in the AWC axon with some diffuse expression in the AWC cell body (Fig. 4T). In tir-1(tm1111), NSY-1::GFP expression was faint and delocalized in the AWC axon, and expression in the AWC cell body was stronger than in wild type (Fig. 4U, as seen in >20 animals). Thus tir-1 stimulates the synaptic localization of NSY-1 protein in the AWC axon.
TIR-1 contains separable functional domains for signaling, localization, and regulation
To define the functional domains of TIR-1, a series of TIR-1::DsRed deletion transgenes were analyzed for their activity and subcellular localization in AWC. These experiments focused on three regions of the protein: the conserved TIR and SAM domains and the N-terminal domain that was deleted in the tir-1(ok1052) allele. Each transgene was tested for its ability to rescue tir-1 mutant when expressed at low levels, and its ability to generate a dominant phenotype in a wild-type background when expressed at high levels.
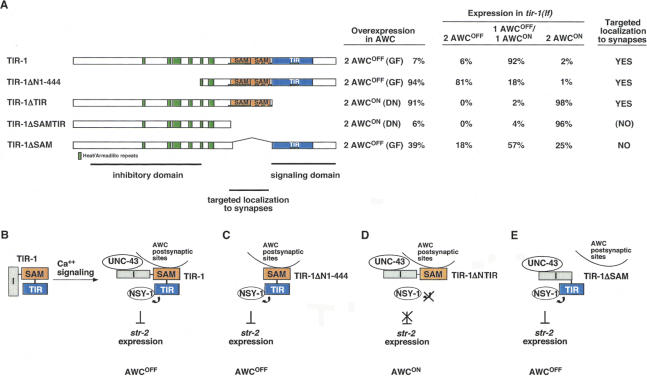
Like full-length TIR-1 protein, the TIR-1ΔN1-444 protein lacking the first 444 amino acids of the protein was localized to AWC synapses (Fig. 3F). This deleted protein suppressed the 2 AWCON phenotypes in tir-1(lf) mutants, but consistently generated a 2 AWCOFF gain-of-function phenotype instead of the wild-type 1 AWCON phenotype (Fig. 5A). TIR-1ΔN1-444 also caused a stronger gain-of-function 2 AWCOFF phenotype in wild-type animals than full-length TIR-1 expressed at similar levels, suggesting that the N-terminal region is an inhibitory or regulatory domain (Fig. 5). This result is consistent with the mixed phenotype caused by tir-1(ok1052), an in-frame deletion of the same domain (Fig. 1B; Table 1), and supports the identification of tir-1(ok1052) as a partial gain-of-function mutation.
Figure 5.
Identification of TIR-1 functional domains. There are three inferred functional domains in the TIR-1 protein: a regulatory region at the N terminus, SAM domains for targeted localization to synapses, and a TIR domain for activation of signaling. (A) Structure-function analysis of TIR-1. (Far left) Diagram of mutated TIR-1 proteins tested for biological activity. (Left) Overexpression of transgenes in wild-type animals. “Gain-of-function” transgenes were injected into wild-type at 5–7 ng/μL. “Dominant-negative” transgenes were injected into wild-type at 35 ng/μL. Two lines were examined for each transgene; in all cases, both lines gave similar results. For overexpression experiments, n =63–141 animals. (Center) Rescue of tir-1(tm1111lf) animals. Two lines were examined for each transgene; in all cases, both lines gave similar results. For rescue experiments, n =73–107 animals. (Right) Subcellular localization of tagged protein. TIR-1ΔSAMTIR was expressed poorly, so it is possible that it is present at synapses at a level too low to be detected with the RFP marker. (B) Model for TIR-1 functions. The N-terminal inhibitory domain (I) could mask the TIR signaling domain. Ca++ signaling through UNC-43 stimulates TIR-1-dependent activation of NSY-1 at the synapse. (C) When the N-terminal inhibitory domain is deleted, TIR-1ΔN1-444 can activate NSY-1 independently of Ca++ signaling, leading to a strong gain-of-function 2 AWCOFF phenotype. (D) When the TIR signaling domain is deleted, TIR-1ΔTIR competes with wild-type TIR-1 for UNC-43 or other activators at the synapse, leading to a 2 AWCON dominant-negative effect. (E) TIR-1ΔSAM, deleting the synaptic localization domain, is not targeted to the synapse and has a mild gain of function phenotype when overexpressed.
TIR-1ΔTIR, in which the TIR domain is deleted, was also localized to AWC synapses. However, this protein failed to rescue the defects in tir-1 mutants, and caused a strong dominant-negative 2 AWCON phenotype in a wild-type background (Fig. 5A). These results indicate that the TIR domain is essential for TIR-1 function, and that other domains of TIR-1ΔTIR can inhibit endogenous TIR-1 either by homodimerizing with it (Couillault et al. 2004) or by interacting nonproductively with essential partners such as UNC-43 and NSY-1.
TIR-1ΔSAMTIR, deleting both SAM and TIR domains, failed to rescue tir-1 mutants and caused a weak dominant-negative phenotype in a wild-type background. This protein was poorly localized to synapses (Fig. 5A). Deletion of only the SAM domains in TIR-1ΔSAM led to a protein that had both a reduced ability to rescue the tir-1 mutant, and a weak gain-of-function phenotype when expressed in a wild-type background (Fig. 5A). This mixed partial loss-of-function/partial gain-of-function phenotype suggests loss of an important regulatory activity. TIR-1ΔSAM was not targeted to synapses (Fig. 3G). These results suggest that the SAM domain localizes TIR-1 to synapses, contributing both to its normal function and to the dominant-negative function of TIR-1ΔTIR. Together, these results identify the N-terminal domain of TIR-1 as a regulatory or inhibitory domain, the TIR domain as a signaling domain, perhaps for activation of NSY-1, and the SAM repeats as a synaptic localization domain essential for the full activity of TIR and N-terminal domains.
Discussion
Developmental signaling generates two distinct AWC olfactory neurons with overlapping sensory specificities. Our experiments define a new component of the AWC signaling pathway, the Heat/Armadillo-SAM-TIR protein TIR-1. TIR-1 is localized to post-synaptic regions of the axon, and stimulates the localization of the MAP-KKK NSY-1 to post-synaptic regions, suggesting that AWC lateral signaling and diversification of odorant receptor expression are initiated at the synapse.
The AWC neurons form synapses on each other, and in the two animals reconstructed by White et al. (1986), most synapses were made from AWCR to AWCL. It is possible that this synaptic asymmetry is directly related to the asymmetry in receptor expression between AWCON and AWCOFF. Temperature-shift experiments indicate that tir-1 acts in the AWCON/AWCOFF decision during the last few hours of embryogenesis, the time at which synapses are made in the nerve ring, and TIR-1, NSY-1, and UNC-43 are all present in post-synaptic regions (Rongo and Kaplan 1999; this work). The striking synaptic localization of TIR-1 and the timing of tir-1 gene action suggest that synapse formation may be coupled to AWC asymmetry. Strong loss-of-function mutations in the genes unc-13, unc-18, and unc-104, which disrupt classical synaptic transmission, have no effect on AWC asymmetry (Troemel et al. 1999; A. Sagasti and C.I. Bargmann, unpubl.). None of these mutations result in a complete block in spontaneous and evoked release, but all cause much stronger defects in synaptic function than the mutations in unc-2, unc-36, and unc-43 that result in strong AWC phenotypes. Therefore, the developmental effects of calcium signaling on AWC asymmetry are unlikely to be explained by a block in classical synaptic transmission. Instead, these results suggest that the AWC synapse has a distinct lateral signaling function that alters odorant receptor expression.
The nature of the lateral signal between AWC neurons is unknown: It could occur through a direct interaction between AWC axons or a competition for another signal. The AWC-to-AWC synapse is the only strongly asymmetric connection made by AWC, so no other input represents an obvious source of random asymmetry (White et al. 1986). However it arises, this signal is predicted to inhibit the activity of UNC-43, TIR-1, and NSY-1 in the responding AWCON cell.
Genetic epistasis results are consistent with tir-1 acting downstream of the CaMKII unc-43 but upstream of the MAPKKK nsy-1. UNC-43 and NSY-1 and their orthologs can bind one another (Sagasti et al. 2001; Takeda et al. 2004), but overexpression of tir-1 can bypass an unc-43 null mutant, indicating that no CaMKII activity is required to activate the MAPKKK pathway if sufficient TIR-1 activity is available. These results suggest the possibility of a direct role for TIR-1 in NSY-1 activation. The possibility that TIR-1 activates NSY-1 in an UNC-43-independent fashion is also supported by results in the innate immune pathway in C. elegans. tir-1, nsy-1, and sek-1 are important in innate immunity, but unc-43 has no effect on this process (Couillault et al. 2004; Liberati et al. 2004).
RNAi of tir-1 leads to decreased activity of the p38 kinase PMK-1, an endogenous kinase downstream of NSY-1, consistent with our genetic results suggesting that tir-1 may activate nsy-1 (Tanaka-Hino et al. 2002; Aballay et al. 2003; Liberati et al. 2004). RNAi of pmk-1 or the related kinase pmk-2 did not affect AWC asymmetry (data not shown). AWC asymmetry was also unaffected in mutants for pmk-3 (p38) and jnk-1 (Jnk kinase). Further experiments will be necessary to identify the final kinase(s) in the AWC signaling pathway.
TIR proteins play an ancient role in innate immunity that is conserved among vertebrates, flies, and nematodes (for review, see Qureshi and Medzhitov 2003; Takeda and Akira 2003). The NSY-1 ortholog ASK1 functions in mammalian macrophage innate immunity (Bhattacharyya et al. 2003), suggesting that the overall MAPKKK innate immunity pathway, and its regulation by TIR-1, may be highly conserved.
There are numerous possible mechanisms of TIR-1 action; one speculative model consistent with the genetic results and structure-function analysis is presented in Figure 5. We suggest that TIR-1 recruits NSY-1 to a specific signaling domain at the synapse and activates NSY-1 signaling through its TIR domain. Calcium entry and activation of CaMKII (UNC-43) at the synapse could relieve the inhibitory influence of the TIR-1 N terminus on TIR-1/NSY-1 signaling; in innate immunity, another signaling mechanism could counteract the inhibitory domain of TIR-1 to activate NSY-1. NSY-1 signaling then generates a retrograde signal to the AWC nucleus that diversifies odorant receptor expression.
Diverse patterns of receptor expression are essential for the function of the olfactory system. The precise coordination of AWC asymmetry allows the animal to detect a broad group of attractive odors, and permits discrimination between odors (Wes and Bargmann 2001). Odorant receptor expression in AWC is regulated at several different stages: early acquisition of the common AWC fate, later differentiation of AWCON and AWCOFF, and finally post-embryonic activity-dependent maintenance of receptor expression by cGMP signaling in the cilia (Troemel et al. 1999; Lanjuin et al. 2003). Each of these mechanisms provides a route of receptor regulation and the potential for fine-tuning the behavioral repertoire.
The generation of AWC neuronal diversity through the calcium/MAP kinase cascade is an unusual developmental pathway, but an increasing number of cell types use calcium to affect their development. Embryonic vertebrate spinal cord neurons use calcium signaling to switch neurotransmitter expression (Borodinsky et al. 2004). Calcium signaling affects the differentiation of T cells (Adachi and Iwata 2002), osteoblasts (Dvorak et al. 2004), and myoblasts (Chin et al. 1998; Pisaniello et al. 2003), and the formation of the asymmetric vertebrate left–right axis (Raya et al. 2004). The elucidation of the role of TIR-1 and other proteins in AWC development may help define mechanisms by which calcium signaling affects numerous cell fates.
Materials and methods
Strains
Wild-type strains were C. elegans variety Bristol, strain N2. Strains were maintained by standard methods (Brenner 1974). Mutations and integrated transgenes used in this study included kyIs140 I [str-2::GFP, lin-15(+)], nsy-1(ky542lf) II, tir-1(ky388ts) III, tir-1(gk264) III, tir-1(ok1052) III, tir-1(tm1111) III, unc-43(n1186lf) IV, unc-43(n498gf) IV, and unc-13(e450) I; kyIs130 × [str-2::snb-1::GFP, lin-15(+)]. Transgenes maintained as extrachromosomal arrays included kyEx599 [odr-3::nsy-1(gf), myo-3::DsRed], kyEx681 [odr-3::tir-1, ofm-1::GFP], kyEx749 [F13B10.1, ofm-1::GFP], kyEx750 [odr-3::tir-1, odr-1::DsRed, ofm-1::GFP], kyEx752 [odr-3::tir-1::GFP, pRF4 rol-6(su1006)], kyEx753 [odr-3::tir-1::DsRed, ofm-1::GFP], kyEx754 [odr-3::tir-1::DsRed, odr-3::lin-10::YFP, ofm-1::GFP], kyEx755 [odr-3::tir-1::DsRed, odr-3::nsy-1::GFP, ofm-1::GFP], kyEx759 [odr-3::nsy-1::GFP, odr-3::lin-10::DsRed, ofm-1::GFP], kyEx779 [odr-3::unc-43::GFP, odr-3::tir-1::DsRed, ofm-1::GFP], kyEx780 [odr-3::unc-43::GFP, odr-3::lin-10::DsRed, ofm-1::GFP], kyEx781 [odr-3::nsy-1::GFP, ofm-1::GFP], kyEx782 [odr-3::tir-1ΔN1-444::DsRed, ofm-1::GFP], kyEx783 [odr-3::tir-1ΔTIR::DsRed, ofm-1::GFP], kyEx784 [odr-3::tir-1ΔSAMTIR::DsRed, ofm-1::GFP], and kyEx785 [odr-3::tir-1ΔSAM::DsRed, ofm-1::GFP].
Mapping and cloning of tir-1
tir-1(ky388) was mapped on LGIII between two Tc1 transposable element polymorphisms, stP120 and stP19, in the DP13 strain (Williams 1995). The deficiencies sDf130 and yDf10 failed to complement tir-1, whereas the deficiency sDf121 complemented tir-1. A three-factor cross placed tir-1 to the left of lon-1.
Cosmid clones representing the tir-1 genomic region were injected into tir-1(ky388) at a concentration of ∼10 ng/μL. The cosmid F13B10 (overlapping ZK1058) rescued asymmetric expression of str-2::GFP in three of four transgenic lines (all rescued lines were ≥96% 1 AWCON/1 AWCOFF). The tir-1 (F13B10.1) genomic coding region in ky388, gk264, ok1052, and tm1111 were amplified by PCR, and PCR products were directly sequenced. The tir-1(ky388) is a G → A mutation (position 22292 of cosmid F13B10), tir-1(gk264) is an in-frame deletion of 389 bp (13346–13734 of F13B10), tir-1(tm1111) is a deletion of 771 bp (13186–13956 of F13B10), and tir-1 (ok1052) has a deletion of 1959 bp (11730–13688 of F13B10) and an insertion of 8 bp, resulting in an in-frame deletion.
Our sequence analysis of cDNA clones corresponding to F13B10.1 generated slightly different predicted cDNAs for the previously predicted tir-1a and tir-1c isoforms, as well as a novel isoform, tir-1f. These corrected sequences have been deposited in GenBank (tir-1a, AY834226; tir-1c, AY834227; tir-1f, AY834228) and at WormBase (http://www.wormbase.org). All six forms share TIR and SAM domains, and all have unique N-terminal sequences. All three long forms share a set of HEAT/Armadillo domains as well as TIR and SAM domains, and all are taken out of frame by the tm1111 mutation. The three short isoforms tir-1b, tir-1d, and tir-1f are predicted to be intact in all mutant alleles. The cDNA clones yk171g12, yk752g04, and yk313b11 corresponding to tir-1a, tir-1c, and tir-1f, respectively, were gifts from Y. Kohara (National Institute of Genetics, Mishima, Japan).
The AWC neurons in tir-1(ky388) mutants were morphologically normal except for the pattern of str-2::GFP expression. The AWC neurons are also active, since tir-1 mutants respond to attractive odors in chemotaxis assays in a pattern consistent with their 2 AWCON phenotype (Wes and Bargmann 2001). All tir-1 mutants were generally normal in locomotion and morphology.
Plasmid construction and germline transformation
To make odr-3::tir-1, the full-length tir-1a cDNA was subcloned into pPD49.26 with an EcoRV fragment of the odr-3 promoter (Roayaie et al. 1998).
odr-3::tir-1::GFP was made by subcloning odr-3::tir-1 into pPD95.77. odr-3::tir-1::DsRed was made by replacing GFP in odr-3::tir-1::GFP with DsRed amplified from pDsRed2 (Clontech). To make odr-3::lin-10:YFP, lin-10::YFP from unc-4::lin-10::YFP (K. Shen and C. I. Bargmann, unpubl.) was subcloned to replace tir-1::GFP in odr-3::tir-1::GFP. odr-3::lin-10::DsRed was made by replacing YFP of odr-3::lin-10::YFP with DsRed. odr-3::nsy-1::GFP was made by subcloning odr-3::nsy-1 (Sagasti et al. 2001) into pPD95.77. odr-3::unc-43::GFP was made by subcloning unc-43 cDNA from yk213a11 (Y. Kohara) into pPD95.77. To make odr-3::tir-1ΔN1-444::DsRed, odr-3::tir-1ΔTIR::DsRed, odr-3:tir-1ΔSAMTIR::DsRed, and odr-3::tir-1ΔSAM::DsRed, the cDNA in odr-3::tir-1::DsRed was deleted at residues 1–1332, 2122–2793, 1675–2793, and 1675–2091, respectively. Transgenic strains were made as described (Mello and Fire 1995). odr-3::nsy-1(gf) (Sagasti et al. 2001) and odr-3::tir-1 were injected at 50 ng/μL. odr-1::DsRed (Sagasti et al. 2001) was injected at 12 ng/μL. odr-3::tir-1::GFP, odr-3::tir-1::DsRed, odr-3::lin-10::YFP, odr-3::lin-10::DsRed, odr-3::unc-43::GFP, and odr-3::nsy-1::GFP were injected at 35 ng/μL. Coinjection markers myo-3::DsRed, dominant pRF4 rol-6(su1006), and ofm-1::GFP (Miyabayashi et al. 1999) were injected at 5 ng/μL, 50 ng/μL, and 25–35 ng/μL, respectively. ofm-1::GFP is expressed in the coelomocytes.
Genetic mosaic analysis
The str-2::GFP integrated line kyIs140 I was injected with DNA for odr-3::tir-1, odr-1::DsRed, and ofm-1::GFP. Mosaic analysis and statistical analysis were performed as described (Sagasti et al. 2001). Transgenic lines were passed for six generations to allow the transgenes to stabilize before screening for mosaics. The presence of the extrachromosomal array was visible in the AWC and AWB neurons, which expressed odr-1::DsRed.
Temperature-shift experiments
tir-1(ky388) adults were allowed to lay eggs for 2 h at 20°C or 15°C, then adults were removed from the plates. Plates were placed at 20°C or 15°C for varying times, then shifted to the other temperature. Animals were allowed to develop to the adult stage, and then the percentage of animals with 2 AWCON was analyzed. Approximately 100–150 animals were scored for each shifted stage.
Immunohistochemistry
tir-1 cDNA fragments encoding amino acids 1–30, 51–274, and 589–931 were cloned into pGEX-2TK (for GST fusion, Amersham Biosciences), pRSETA, and pRSETB (for His6 fusion, Invitrogen), respectively, transformed into E. coli strain BL21-CodonPlus(DE3)-RIL (Stratagene), and induced with 0.8 mM IPTG for 3 h at 37°C; and GST and His6 fusion proteins were partially purified with Glutathione Sepharose 4B (Amersham Biosciences) and Ni-NTA resin (QIAGEN), respectively. Purified proteins were run on 10% SDS-PAGE, and bands corresponding to the correct molecular weight were excised and used to immunize rats (Covance). The titer of antisera was determined by Western blot analysis. Crude antisera (titer 1:25) against three antigens were mixed and used for immunostaining. Fixation and staining were performed as described (Finney and Ruvkun 1990).
Immunoprecipitation/Western immunoblot
Flag-TIR-1 was made by cloning tir-1 cDNA into pFlag-CMV2 (Sigma). Mammalian expression vectors were transfected into HEK 293 cells using the FuGENE 6 transfection reagent (Roche). Proteins from the cell lysates were immunoprecipitated with anti-T7 antibodies (Novagen) or anti-Flag antibodies (Sigma) as described (Kawasaki et al. 1999). Immunoprecipitates and cell lysates were separated by 10% SDS-PAGE, transferred to nitrocellulose membranes (Schleicher & Schuell), probed with anti-T7, anti-HA (Roche), or anti-Flag antibodies and horseradish peroxidase-labeled secondary antibody (Amersham Biosciences), and detected with the enhanced chemiluminescence system (Amersham Biosciences). It was not possible to test the regulation of NSY-1 kinase activity by coexpression with TIR-1 and UNC-43 in mammalian cells, because NSY-1/ASK1 kinases are activated by the unfolded protein response, and the unfolded protein response is induced by overexpression of heterologous proteins.
Acknowledgments
We thank Alvaro Sagasti for discussions and the initial characterization of tir-1; Dianne Parry, Joe Hill, and Hai Nguyen for excellent technical support; Miri VanHoven, Makoto Tsunozaki, Manuel Zimmer, Chieh Chang, and Piali Sengupta for helpful discussions and comments on the manuscript; Scott Alper for advice on immunostaining; Uta Grieshammer and Gail Martin for help with mammalian cell culture; Rick Fetter for help with confocal microscopy; Kang Shen for unc-4::lin-10::YFP; Joy Alcedo for pAD12; Malene Hansen for HT115(DE3); Naoki Hisamoto and Kunihiro Matsumoto for HA-UNC-43 and T7-NSY-1; Alan Coulson and the Sanger Center for cosmids; Yuji Kohara for EST clones; Andy Fire for C. elegans vectors; Theresa Stiernagle and the C. elegans Genetic Center for strains; Anthony Rogers and the WormBase for help with the verification of tir-1 isoforms; Shohei Mitani for tir-1(tm1111); and the C. elegans Gene Knockout Consortium for tir-1(gk264) and tir-1(ok1052). This work was supported by NIH grant DC004089. C.-F.C. was supported by the Cancer Research Fund of the Damon Runyon Foundation Fellowship, and C.I.B. is an Investigator of the Howard Hughes Medical Institute.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1276505.
References
- Aballay A., Drenkard, E., Hilbun, L.R., and Ausubel, F.M. 2003. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 13: 47–52. [DOI] [PubMed] [Google Scholar]
- Adachi S. and Iwata, M. 2002. Duration of calcineurin and Erk signals regulates CD4/CD8 lineage commitment of thymocytes. Cell Immunol. 215: 45–53. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Pathak, S., Basak, C., Law, S., Kundu, M., and Basu, J. 2003. Execution of macrophage apoptosis by Mycobacterium avium through apoptosis signal-regulating kinase 1/p38 mitogen-activated protein kinase signaling and caspase 8 activation. J. Biol. Chem. 278: 26517–26525. [DOI] [PubMed] [Google Scholar]
- Borodinsky L.N., Root, C.M., Cornin, J.A., Sann, S.B., Gu, X., and Spitzer, N.C. 2004. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature 429: 523–530. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston Jr., R.J., and Hobert, O. 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes & Dev. 17: 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin E.R., Olson, E.N., Richardson, J.A., Yang, Q., Humphries, C., Shelton, J.M., Wu, H., Zhu, W., Bassel-Duby, R., and Williams, R.S. 1998. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes & Dev. 12: 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook Y.M. and Blobel, G. 1999. Structure of the nuclear transport complex karyopherin–β2-Ran × GppNHp. Nature 399: 230–237. [DOI] [PubMed] [Google Scholar]
- Cingolani G., Petosa, C., Weis, K., and Muller, C.W. 1999. Structure of importin-β bound to the IBB domain of importin-α. Nature 399: 221–229. [DOI] [PubMed] [Google Scholar]
- Couillault C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J.F., Kohara, Y., and Ewbank, J.J. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5: 488–494. [DOI] [PubMed] [Google Scholar]
- Davies A.G., Pierce-Shimomura, J.T., Kim, H., VanHoven, M.K., Thiele, T.R., Bonci, A., Bargmann, C.I., and McIntire, S.L. 2003. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 115: 655–666. [DOI] [PubMed] [Google Scholar]
- Dvorak M.M., Siddiqua, A., Ward, D.T., Carter, D.H., Dallas, S.L., Nemeth, E.F., and Riccardi, D. 2004. Physiological changes in extracellular calcium concentration directly control osteoblast function in the absence of calciotropic hormones. Proc. Natl. Acad. Sci. 101: 5140–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M. and Ruvkun, G. 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63: 895–905. [DOI] [PubMed] [Google Scholar]
- Greenwald I. 1998. LIN-12/Notch signaling: Lessons from worms and flies. Genes & Dev. 12: 1751–1762. [DOI] [PubMed] [Google Scholar]
- Harris J., Honigberg, L., Robinson, N., and Kenyon, C. 1996. Neuronal cell migration in C. elegans: Regulation of Hox gene expression and cell position. Development 122: 3117–3131. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Nelson, W.J., and Weis, W.I. 1997. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90: 871–882. [DOI] [PubMed] [Google Scholar]
- Ichijo H., Nishida, E., Irie, K., ten Dijke, P., Saitoh, M., Moriguchi, T., Takagi, M., Matsumoto, K., Miyazono, K., and Gotoh, Y. 1997. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 275: 90–94. [DOI] [PubMed] [Google Scholar]
- Johnston R.J. and Hobert, O. 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426: 845–849. [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Hisamoto, N., Iino, Y., Yamamoto, M., Ninomiya-Tsuji, J., and Matsumoto, K. 1999. A Caenorhabditis elegans JNK signal transduction pathway regulates coordinated movement via type-D GABAergic motor neurons. EMBO J. 18: 3604–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Feinbaum, R., Alloing, G., Emerson, F.E., Garsin, D.A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M.W., et al. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297: 623–626. [DOI] [PubMed] [Google Scholar]
- Kyba M. and Brock, H.W. 1998. The SAM domain of polyhomeotic, RAE28, and scm mediates specific interactions through conserved residues. Dev. Genet. 22: 74–84. [DOI] [PubMed] [Google Scholar]
- Lanjuin A., VanHoven, M.K., Bargmann, C.I., Thompson, J.K., and Sengupta, P. 2003. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell 5: 621–633. [DOI] [PubMed] [Google Scholar]
- Liberati N.T., Fitzgerald, K.A., Kim, D.H., Feinbaum, R., Golenbock, D.T., and Ausubel, F.M. 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. 101: 6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof J.N., Whangbo, J., Harris, J.M., Jongeward, G.D., and Kenyon, C. 1999. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development 126: 37–49. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1: 135–145. [DOI] [PubMed] [Google Scholar]
- Mello C. and Fire, A. 1995. DNA transformation. Methods Cell Biol. 48: 451–482. [PubMed] [Google Scholar]
- Mink M., Fogelgren, B., Olszewski, K., Maroy, P., and Csiszar, K. 2001. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/β-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics 74: 234–244. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T., Palfreyman, M.T., Sluder, A.E., Slack, F., and Sengupta, P. 1999. Expression and function of members of a divergent nuclear receptor family in Caenorhabditis elegans. Dev. Biol. 215: 314–331. [DOI] [PubMed] [Google Scholar]
- Nonet M.L., Saifee, O., Zhao, H., Rand, J.B., and Wei, L. 1998. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 18: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill L.A., Fitzgerald, K.A., and Bowie, A.G. 2003. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol. 24: 286–290. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura J.T., Faumont, S., Gaston, M.R., Pearson, B.J., and Lockery, S.R. 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410: 694–698. [DOI] [PubMed] [Google Scholar]
- Pisaniello A., Serra, C., Rossi, D., Vivarelli, E., Sorrentino, V., Molinaro, M., and Bouche, M. 2003. The block of ryanodine receptors selectively inhibits fetal myoblast differentiation. J. Cell Sci. 116: 1589–1597. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. 1995. SAM: A novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 4: 1928–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S.T. and Medzhitov, R. 2003. Toll-like receptors and their role in experimental models of microbial infection. Genes Immun. 4: 87–94. [DOI] [PubMed] [Google Scholar]
- Ramezani-Rad M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr. Genet. 43: 161–170. [DOI] [PubMed] [Google Scholar]
- Raya A., Kawakami, Y., Rodriguez-Esteban, C., Ibanes, M., Rasskin-Gutman, D., Rodriguez-Leon, J., Buscher, D., Feijo, J.A., and Izpisua Belmonte, J.C. 2004. Notch activity acts as a sensor for extracellular calcium during vertebrate left–right determination. Nature 427: 121–128. [DOI] [PubMed] [Google Scholar]
- Richmond J.E., Davis, W.S., and Jorgensen, E.M. 1999. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2: 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K., Crump, J.G., Sagasti, A., and Bargmann, C.I. 1998. The G α protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20: 55–67. [DOI] [PubMed] [Google Scholar]
- Rongo C. and Kaplan, J.M. 1999. CaMKII regulates the density of central glutamatergic synapses in vivo. Nature 402: 195–199. [DOI] [PubMed] [Google Scholar]
- Rongo C., Whitfield, C.W., Rodal, A., Kim, S.K., and Kaplan, J.M. 1998. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94: 751–759. [DOI] [PubMed] [Google Scholar]
- Sagasti A., Hisamoto, N., Hyodo, J., Tanaka-Hino, M., Matsumoto, K., and Bargmann, C.I. 2001. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 105: 221–232. [DOI] [PubMed] [Google Scholar]
- Schultz J., Ponting, C.P., Hofmann, K., and Bork, P. 1997. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg, E., White, J.G., and Thomson, J.N. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Takeda K. and Akira, S. 2003. Toll receptors and pathogen resistance. Cell Microbiol. 5: 143–153. [DOI] [PubMed] [Google Scholar]
- Takeda K., Matsuzawa, A., Nishitoh, H., Tobiume, K., Kishida, S., Ninomiya-Tsuji, J., Matsumoto, K., and Ichijo, H. 2004. Involvement of ASK1 in Ca2+-induced p38 MAP kinase activation. EMBO Rep. 5: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Hino M., Sagasti, A., Hisamoto, N., Kawasaki, M., Nakano, S., Ninomiya-Tsuji, J., Bargmann, C.I., and Matsumoto, K. 2002. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 3: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E.R., Sagasti, A., and Bargmann, C.I. 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398. [DOI] [PubMed] [Google Scholar]
- Wes P.D. and Bargmann, C.I. 2001. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410: 698–701. [DOI] [PubMed] [Google Scholar]
- Whangbo J. and Kenyon, C. 1999. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4: 851–858. [DOI] [PubMed] [Google Scholar]
- White J.G., Southgate, E., Thomson, J.N., and Brenner, S. 1986. The structure of the nervous system of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Williams B.D. 1995. Genetic mapping with polymorphic sequence-tagged sites. Methods Cell Biol. 48: 81–96. [DOI] [PubMed] [Google Scholar]