Abstract
Plant peroxisomal proteins catalyze key metabolic reactions. Several peroxisome biogenesis PEROXIN (PEX) genes encode proteins acting in the import of targeted proteins necessary for these processes into the peroxisomal matrix. Most peroxisomal matrix proteins bear characterized Peroxisomal Targeting Signals (PTS1 or PTS2), which are bound by the receptors PEX5 or PEX7, respectively, for import into peroxisomes. Here we describe the isolation and characterization of an Arabidopsis peroxin mutant, pex7-1, which displays peroxisome-defective phenotypes including reduced PTS2 protein import. We also demonstrate that the pex5-1 PTS1 receptor mutant, which contains a lesion in a domain conserved among PEX7-binding proteins from various organisms, is defective not in PTS1 protein import, but rather in PTS2 protein import. Combining these mutations in a pex7-1 pex5-1 double mutant abolishes detectable PTS2 protein import and yields seedlings that are entirely sucrose-dependent for establishment, suggesting a severe block in peroxisomal fatty acid β-oxidation. Adult pex7-1 pex5-1 plants have reduced stature and bear abnormally shaped seeds, few of which are viable. The pex7-1 pex5-1 seedlings that germinate have dramatically fewer lateral roots and often display fused cotyledons, phenotypes associated with reduced auxin response. Thus PTS2-directed peroxisomal import is necessary for normal embryonic development, seedling establishment, and vegetative growth.
INTRODUCTION
Peroxisomes are organelles housing diverse and vital processes. Plant peroxisomes contain enzymes for photorespiration (Liepman and Olsen, 2001; Reumann, 2002) and are the primary, if not exclusive, site of fatty acid β-oxidation (Gerhardt, 1992; Kindl, 1993; Graham and Eastmond, 2002). In addition, plant peroxisomes are necessary for jasmonic acid (JA) biosynthesis (Wasternack and Hause, 2002) and are implicated in the conversion of indole-3-butyric acid (IBA) into the active auxin indole-3-acetic acid (IAA; Zolman et al., 2000; Bartel et al., 2001).
Peroxisome matrix proteins must be imported from the cytosol because peroxisomal proteins are encoded by nuclear genes. Import of matrix proteins is accomplished by an array of PEROXIN (PEX) proteins that function in matrix protein import or general peroxisome assembly. Although >30 pex mutants have been identified in yeast and mammals, in plants, only pex2 (Hu et al., 2002), pex5 (Zolman et al., 2000), pex6 (Zolman and Bartel, 2004), pex10 (Schumann et al., 2003; Sparkes et al., 2003), pex14 (Hayashi et al., 2000), and pex16 (Lin et al., 1999) Arabidopsis mutants have been isolated. However, plausible Arabidopsis homologues for many of the PEX genes remaining to be characterized have been identified through sequence homology (Mullen et al., 2001; Charlton and López-Huertas, 2002).
Two sequences sufficient to signal matrix protein import into peroxisomes have been identified. One is the extreme C-terminal amino acid sequence Ser-Lys-Leu (SKL), or a conserved variant, designated Peroxisomal Targeting Signal type 1 (PTS1; Gould et al., 1989; Mullen, 2002; Neuberger et al., 2003; Reumann, 2004). The PEX5 receptor recognizes and binds cytosolic PTS1-containing proteins. Mutations in human PEX5 can cause the peroxisome biogenesis disorders Zellweger syndrome and neonatal adrenoleukodystrophy (Dodt et al., 1995). The PEX5-PTS1 complex binds a PEX14-PEX13 receptor complex at the peroxisome membrane (Albertini et al., 1997) and is translocated into the peroxisome matrix (Dammai and Subramani, 2001) in a process dependent on PEX2, PEX10, and PEX12 (Dodt and Gould, 1996; Chang et al., 1999). In the peroxisome matrix, PEX5 releases its cargo and is recycled to the cytosol (Dammai and Subramani, 2001) in a process dependent on PEX1, PEX4, PEX6, and PEX22 (Collins et al., 2000).
An alternate sequence is the N-terminal Peroxisomal Targeting Signal type 2 (PTS2) composed of Arg-Leu-(X)5-His-Leu or a number of variants within the first ∼30 amino acids (Osumi et al., 1991; Flynn et al., 1998; Reumann, 2004). PEX7 is the receptor for PTS2-containing proteins (Rehling et al., 1996). Mutations in human PEX7 can cause the peroxisome biogenesis disorders Refsum disease (van den Brink et al., 2003) and rhizomelic chondrodysplasia punctata (Braverman et al., 1997; Motley et al., 1997; Purdue et al., 1997). In Saccharomyces cerevisiae, Pex7p interacts with Pex18p and Pex21p, two functionally redundant proteins necessary for PTS2 protein import (Purdue et al., 1998). In Neurospora crassa and Yarrowia lipolytica, Pex18p and Pex21p are replaced with the single Pex7p docking protein Pex20p (Sichting et al., 2003; Einwächter et al., 2001).
Mammals and plants appear to lack PEX18, PEX20, and PEX21 orthologues. Mammalian PEX7 function instead depends on interaction with an isoform of PEX5 (PEX5L; Braverman et al., 1998) containing a PEX7-binding domain (Matsumura et al., 2000; Dodt et al., 2001). Whereas most PEX5 mutants are defective in PTS1 protein import, a PEX5 mutant Chinese hamster ovary (CHO) cell line is specifically deficient in PTS2 protein import because of inability to bind PEX7 (Matsumura et al., 2000). Similarly, Arabidopsis PEX7 interacts with PEX5 in vitro (Nito et al., 2002), and in vitro import of a PTS2 protein into Cucurbita pepo glyoxysomes is enhanced by PTS1 protein addition (Johnson and Olsen, 2003). Indeed, a role for PEX7-PEX5 interaction in Arabidopsis PTS2-protein import has been proposed (Sparkes and Baker, 2002). The dependence of PEX7 on PEX5 interaction in some organisms suggests a convergence of function carried to the extreme in Caenorhabditits elegans, which lacks PEX7 entirely and uses a PTS1 on proteins targeted by PTS2 in other organisms (Gurvitz et al., 2000; Motley et al., 2000).
Insertional null mutations in Arabidopsis pex2 (Hu et al., 2002) and pex10 (Schumann et al., 2003; Sparkes et al., 2003) confer embryo lethality, revealing the necessity of peroxisome function for early plant development. Plants heterozygous for the AtPex10 knockout allele bear ∼20% abnormal seeds that fail to germinate (Sparkes et al., 2003); aborted seeds remain white rather than proceeding to green and contain embryos arrested at the late globular or heart stages of development (Schumann et al., 2003). As peroxisomes are necessary for JA biosynthesis (Wasternack and Hause, 2002) and are implicated in the conversion of IBA into IAA (Bartel et al., 2001), deficiencies in JA and IAA have been suggested as possible causes for pex10 mutant lethality (Sparkes et al., 2003). In addition, a double mutant defective in two peroxisomal acyl-CoA oxidase genes is embryo lethal (Rylott et al., 2003), suggesting that β-oxidation is an essential function for which peroxisomes are required during embryogenesis.
Here we describe pex7-1, an Arabidopsis mutant defective in IBA response and PTS2 protein import into peroxisomes. In addition, we show that the previously isolated PTS1-receptor mutant pex5-1 (Zolman et al., 2000) is deficient in PTS2-, but not PTS1-protein import. Further, a pex7-1 pex5-1 double mutant is severely deficient in peroxisome functions, requiring exogenous sucrose for growth, and displaying frequent embryonic abnormalities suggestive of reduced auxin levels or response.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
pex7-1 was isolated from the Col-0 T-DNA insertion line SALK_005354 (Alonso et al., 2003) obtained from the ABRC and isolated using a PCR-based method. Amplification with the oligonucleotide PEX7-2 (5′-CTTCTCGAAGATTCAATTCAACGAT-3′) and the modified LBb1 (Alonso et al., 2003) T-DNA left border primer LB1-Salk (5′-CAAACCAGCGTGGACCGCTTGCTGCA-3′) yielded a ∼200-base pair product from mutant DNA, whereas PEX7-1 (5′-CTCGAATTTAGATTTCTCTCTCACTTTTA-3′) combined with PEX7-2 yielded a 252-base pair product in the presence of wild-type DNA, enabling genotypic determination. Combining either PEX7-1 or PEX7-2 with LB1-Salk yields a PCR product; both products were sequenced to determine the T-DNA/genomic DNA junctions, which are present at 95/96 and 98/99 base pairs upstream of the PEX7 start codon. This result indicates an insertional event that resulted in T-DNA left border sequences oriented in both directions, with the T-DNA replacing a 3-base pair deletion from 96 to 98 base pairs upstream of the PEX7 start codon. The pex5-1 mutant in the Col-0 background contains an ethyl methanesulfonate-induced missense mutation described previously (Zolman et al., 2000).
Phenotypic analyses were performed on mutants backcrossed to Col-0 at least once. Seeds were surface-sterilized, stratified overnight at 4°C in 0.1% agar, and grown on plant nutrient medium (PN; Haughn and Somerville, 1986) with 0.5% sucrose (PNS) unless otherwise noted. For root elongation assays, 5 μM IBA was added to medium from a 100 mM ethanol stock, and plants were grown for 8 d in continuous light under yellow filters (25-45 μE m-2 sec-1) to reduce the destruction of indolic compounds by short-wave-length light (Stasinopoulos and Hangarter, 1990). Lateral roots were counted under a dissecting microscope after 9 d of growth at 22°C under yellow light filters. For sucrose-dependence assays, seeds were prepared as above and plated on PN and PNS, grown 1 d under continuous white light, then wrapped with foil, and grown 5 additional days in darkness. Plants grown to maturity were transferred to soil (MetroMix 200, Scotts, Marysville, OH) after ∼14 d of growth and placed under continuous white light (Sylvania Cool White fluorescent bulbs, Danvers, MA) at 22-25°C.
Mutant Rescue
The PEX7 cDNA APZ50H10R was obtained from the Kazusa Stock Center (Asamizu et al., 2000), sequence verified (Lone Star Labs, Houston, TX), and a 1.3-kb SmaI/PvuII fragment was gel-purified and cloned into a SmaI-digested 35SpBARN vector (LeClere and Bartel, 2001) to generate 35S-PEX7. The subcloning boundaries were sequenced with vector-derived oligonucleotides to verify the insert orientation, and 35S-PEX7 was electroporated into Agrobacterium tumefaciens GV3101 (Koncz and Schell, 1986) and transformed into Col-0 and pex7-1 plants using the floral dip method (Clough and Bent, 1998). T1 seeds were grown on PNS supplemented with 7.5 μg/ml glufonisate-ammonium (BASTA; Crescent Chemical, Augsburg, Germany); resistant plants were rescued to unsupplemented PN after 8 d and later transferred to soil. The presence of the construct in T1 plants was confirmed by PCR-amplification of genomic DNA with a vector-derived oligonucleotide and an appropriate gene-specific primer. T1 plants confirmed to carry an intact construct were grown to maturity, and T2 progeny were examined for IBA resistance.
Visualization of Matrix Protein Import Using Green Flourescent Protein
A PTS2-tagged green fluorescent protein (GFP) was created by amplifying the 5′ 147 base pairs of Arabidopsis PED1 thiolase (Hayashi et al., 1998) clone U09045 (Yamada et al., 2003) that encode the N-terminal region containing the PTS2 signal sequence and the signal sequence cleavage site (Johnson and Olsen, 2003). Oligonucleotides PTS2Xba (5′-CCAGAAGGATCTAGAAAAAATGGAGAAAGCGATCG-3′, modifications to add a XbaI site and adenines 5′ of the start codon are underlined) and PTS2Bam (5′-CAACAGGATCCCATAGAGAGAGGTCCTCTG-3′, modifications to add a BamHI site and allow in-frame subcloning are underlined) were used for PCR amplification of U09045 template DNA with Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The resultant 175-base pair bluntended product was gel-purified and subcloned into EcoRV-cut pBluescript KS+ (Stratagene) and sequenced to verify the absence of mutations. From this construct, the 155-base pair XbaI/BamHI fragment was gel-purified and cloned into XbaI/BamHI-digested CD3-326 (Davis and Vierstra, 1998) downstream of the 35S promoter and in frame with the gene encoding enhanced GFP. From this construct, a 2-kb EcoRI/HindIII product that included the 35S promoter and PTS2-GFP fusion was gel-purified and cloned into EcoRI/HindIII-digested pBIN19 plant transformation vector (Bevan, 1984) to yield PTS2-GFP. Cytoplasmic eGFP (unmodified CD3-326) and PTS2-GFP were transformed into Agrobacterium and then Col-0, pex7-1, and pex5-1 plants as described above. Col-0, pex5-1, and pex7-1 T1 seeds were screened on PNS with 13 μg/ml kanamycin sulfate from a water-dissolved stock, and kanamycin-resistant Col-0 and pex5-1 T1 plants were rescued to PN plates after 9 d. The existence of a kanamycin resistance gene in the T-DNA insert in pex7-1 T1 plants necessitated isolating transformants by screening for GFP fluorescence on a Leica MZFLIII dissecting scope (Deerfield, IL), and GFP-expressing pex7-1 plants were rescued as above. To monitor GFP localization, T2 seedlings were mounted on slides, and GFP fluorescence in root tips of 3-d-old seedlings grown in white light suspended in 0.1% agar and root hairs of 7-d-old seedlings grown in white light on PNS were examined by using a Zeiss Axioplan 2 fluorescence microscope (Thornwood, NY) equipped with a narrow-band GFP filter set (41020, Chroma Technology, Rockingham, VT).
Col-0 plants expressing PTS1-GFP, a CD3-326 construct mutagenized to introduce the PTS1 signal sequence SKL to the extreme C-terminus of eGFP (the construct GFP-SKL from Zolman and Bartel, 2004), were crossed to pex7-1 and pex5-1 plants, and IBA-resistant F2 progeny were genotyped using PCR for the pex7-1 and pex5-1 mutations. Lines segregating GFP fluorescence were identified and used for analysis as described above.
Western Blot Analysis
Protein was extracted from entire 2-d-old Col-0, pex7-1 and pex5-1 seedlings grown aseptically in water under white light and 3-d-old Col-0, pex7-1, pex5-1, and pex7-1 pex5-1 seedlings grown in 0.5% sucrose under white light. Germination is slightly delayed by sucrose, so the different chronological ages represent the equivalent developmental stages. Twenty-five seedlings with emerged embryonic roots (radicles) were selected from each genotype, frozen, ground with a pestle, and suspended in one volume of extraction buffer (0.1 M Tris-HCl, pH 6.8, 20% glycerol, 4% SDS). Samples were briefly centrifuged to pellet debris, and supernatants were heated for 10 min at 80°C. Protein extracts were subjected to SDS-PAGE on NuPAGE 10% Bis-Tris gels (Invitrogen, Carlsbad, CA) with Cruz size standards (Santa Cruz Biotechnology, Santa Cruz, CA).
Protein was transferred to Hybond enhanced chemiluminescence nitrocellulose membranes (Amersham Pharmacia Biotech, Piscataway, NJ) for 1 h at 24 V. Membranes were blocked in 5% powdered milk in Tween Tris-buffered saline (TTBS) buffer (Ausubel et al., 1995) for at least 2 h, and then incubated with rocking at 4°C for at least 13 h in a 1:1000 dilution of rabbit antiplant thiolase antibody (Kato et al., 1996) or a 1:200 dilution of a rabbit anti-Arabidopsis PEX5 antibody (Zolman and Bartel, 2004) in 5% milk TTBS. Membranes were washed four times in 5% milk TTBS and then incubated for at least 1 h at room temperature in a 1:500 dilution of a horseradish peroxidase-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology). Membranes were washed as before and visualized using LumiGLO reagent (Cell Signaling, Beverly, MA).
PEX7 Expression Analysis
Seven-day-old Col-0, pex7-1, pex5-1, and pex7-1 pex5-1 seedlings grown under white light on PNS were frozen in liquid nitrogen and ground in a chilled mortar and pestle. RNA was extracted using RNeasy Plant Mini Kits (Qiagen, Valencia, CA). After OD260 quantification and electrophoresis to ensure the absence of RNA degradation, each sample was treated with DNaseI (Amplification Grade, Roche Applied Science, Indianapolis, IN) and 0.3 μg was reverse-transcribed in a 20 μL volume using random hexamer primers and SuperScript III polymerase (Invitrogen). Controls lacking reverse transcriptase were performed for each sample.
Quantitative real-time reverse-transcription PCR using an ABI Prism 7000 Sequence Detection System was performed on triplicate 25-μL reactions containing 2 μL cDNA, 0.5 μM each primer, and 0.2 μM probe in Taqman universal PCR Master Mix (Applied Biosystems, Foster City, CA). Primers for PEX7 were PEX7-QRTF (5′-TGGCTGTGCTTAATGGTCATG-3′) and PEX7-QRTR (5′-CCTCCTATGCGGCGAGAA-3′) and a 5′ 6-FAM-labeled, 3′ MGBNFQ (minor groove-binder/nonfluorescent quencher) probe (5′-ATATGCGGTGAGGAAGG-3′). Primers for the APRT control were APRT-F (5′-CATATCTGTTGTTGCAGGTGTTGA-3′) and APRT-R (5′-CCAATAGCCAACGCAATAGGA-3′) and a 5′ 6-FAM-labeled, 3′ MGBNFQ-quenched probe (5′-TAGAGGTTTCATTTTTGGCC-3′). APRT amplification was used to normalize PEX7 levels using a comparative CT method (ABI Prism 7700 Sequence Detection System User Bulletin no. 2, http://www.appliedbiosystems.com). Control reactions without reverse transcriptase amplified later than experimental samples.
RESULTS
Peroxisome Defects in the Arabidopsis pex7-1 Mutant
Sequence similarity searches have been used to identify At1g29260 as Arabidopsis PEX7 (Schumann et al., 1999). The single-copy gene encodes a protein ∼40 and ∼59% identical to PEX7 from S. cerevisiae and humans, respectively, and is largely comprised of WD40 domains (Figure 1, A and B). To determine the role of PEX7 in plant development, we isolated the pex7-1 mutant from the Salk Institute T-DNA insertion collection (Alonso et al., 2003) and identified a single kanamycin resistance-linked T-DNA 95 base pairs upstream of the start codon, placing it within the 5′ untranslated region.
Figure 1.

Organization of Arabidopsis PEX7 and PEX5 proteins. (A) Schematic showing domain architecture of Arabidopsis PEX7 and PEX5 proteins. The bracket above PEX5 is the region shown in C, and the asterisk indicates the Ser to Leu mutation indicated in C. (B) PEX7 is the single Arabidopsis (At) homolog of human (Hs) PEX7 and is highly similar to tomato (Le) GenBank accession AY186749 and rice (Os) TIGR gene temporary ID 8351.t01315 (OsPEX7). (C) Partial alignment of Arabidopsis PEX5 with orthologues from rice, human, yeast (Sc), and roundworm (Ce). pex5-1 contains a missense mutation in a conserved serine (Zolman et al., 2000) needed for PEX7 interaction and PTS2-containing protein import into peroxisomes in Chinese hamster (Matsumura et al., 2000). Note the absence of this domain in CePEX5; C. elegans does not contain PEX7 or a PTS2 import pathway (Gurvitz et al., 2000; Motley et al., 2000). Sequences were aligned with the MegAlign program (DNA Star, Madison, WI) using the ClustalW method; residues identical in three or more sequences are shaded black, residues chemically similar in three or more sequences are shaded gray. WD-40 domains determined by Pfam (Bateman et al., 2002); PPR and TPR domains after Zolman et al. (2000).
Like previously described Arabidopsis mutants with peroxisomal defects (Zolman et al., 2000, 2001a, 2001b; Zolman and Bartel, 2004), pex7-1 displays reduced sensitivity to exogenously supplied IBA (Figure 2A). In addition, the mutant is resistant to an analogous synthetic proto-auxin, 2,4-dichlorophenoxybutyric acid (2,4-DB; unpublished data), which is converted in peroxisomes to the active derivative 2,4-dichlorophenoxyacetic acid (2,4-D; Hayashi et al., 1998). The IBA resistance of pex7-1 is rescued by overexpression of wild-type PEX7 (Figure 2B), indicating that the phenotype observed results from reduced PEX7 function.
Figure 2.
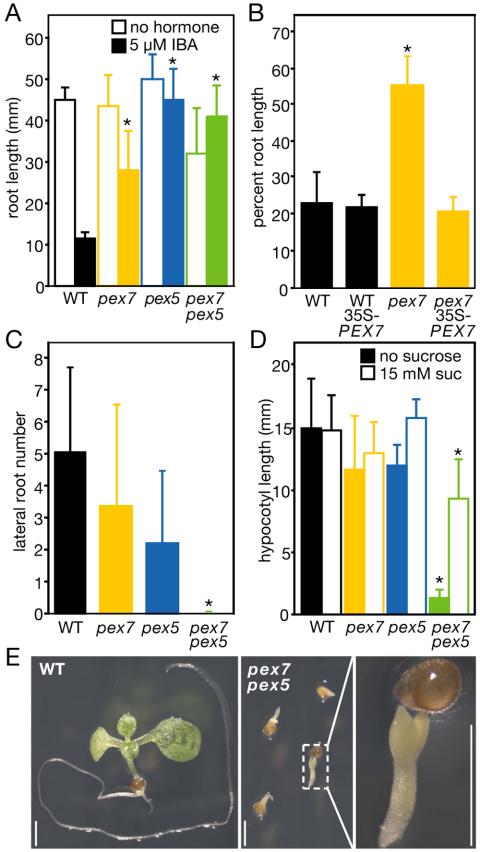
pex7-1, pex5-1, and pex7-1 pex5-1 are deficient in peroxisomal processes. (A) Root elongation on IBA. Root lengths of 8-d-old plants grown on sucrose-supplemented medium with or without addition of 5 μM IBA under yellow light filters are shown. (B) Percent root elongation on 5 μM IBA versus hormone-free medium. Plants were grown as in A. (C) Lateral root number. Lateral roots of 9-d-old plants grown on PNS under yellow light filters were counted under a dissecting microscope. (D) Hypocotyl elongation in darkness with and without sucrose. Plants were grown one day in white light and then were grown in darkness for 5 additional days. Bars, means + SDs; n ≥ 6 in A, n ≥ 10 in B-D. *Significantly different from wild type (p < 0.01; one-tailed t test assuming unequal variance). (E) pex7-1 pex5-1 does not develop after germination without sucrose in the light. Wild-type (left) or pex7-1 pex5-1 (right) plants were grown for 7 d under white light on medium lacking sucrose. Scale bars, 1 mm.
Some peroxisomal mutants produce fewer lateral roots than wild type even without exogenous auxin (Zolman et al., 2001b; Zolman and Bartel, 2004). Any lateral root production defect in pex7-1 on unsupplemented medium is weak (Figure 2C), though reproducible. In addition, pex7-1 forms fewer lateral roots than wild type after IBA treatment (unpublished data). pex7-1 is not generally defective in auxin responses, because it responds normally to IAA both in primary root elongation inhibition and induction of lateral roots (unpublished data). Whereas defects in β-oxidation of fatty acids stored in seeds renders many peroxisome-defective mutants completely sucrose dependent for seedling establishment (Hayashi et al., 1998, 2000; Zolman et al., 2001a, 2001b; Footitt et al., 2002; Fulda et al., 2004; Zolman and Bartel, 2004), the pex7-1 hypocotyl is not markedly shorter than wild type when grown in darkness without added sucrose (Figure 2D). This lack of sucrose dependence suggests that any defect in peroxisomal β-oxidation of endogenous fatty acids in the pex7-1 mutant may be modest. Adult pex7-1 plants are not discernibly different from wild type in growth rate or morphology (unpublished data).
To test whether matrix proteins are imported normally into pex7-1 peroxisomes, we expressed PTS1- and PTS2-tagged versions of GFP in the pex7-1 mutant. As expected, PTS1-tagged GFP (Zolman and Bartel, 2004) is efficiently imported into peroxisomes in pex7-1 (Figure 3). The presence of PTS1-GFP in a normal punctate pattern in pex7-1 roots indicates that peroxisome abundance and gross morphology are unaffected by PEX7 deficiency. In contrast, PTS2-tagged GFP is not efficiently imported into pex7-1 peroxisomes; although some faint punctate fluorescence is still observed, PTS2-GFP confers largely diffuse fluorescence similar to cytoplasmically localized GFP (Figure 3). To confirm that this defect observed with an engineered substrate reflected endogenous proteins, we used Western blotting to indirectly examine import of 3-ketoacyl-CoA thiolase (thiolase) into peroxisomes. As the PTS2 signal is removed from thiolase following import, the consequent molecular weight shift is diagnostic of peroxisomal import. We detected primarily mature processed thiolase in wild-type seedlings, but some residual unprocessed thiolase remains in pex7-1 seedlings (Figure 4A), especially in seedlings grown in the absence of sucrose (Figure 4B). These results imply that peroxisomal deficiencies observed in pex7-1 result from poor import of PTS2-containing proteins.
Figure 3.
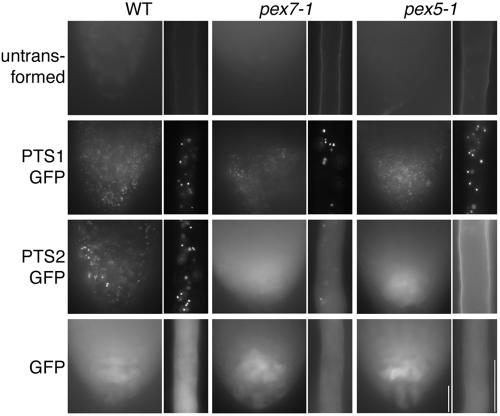
pex7-1 and pex5-1 are deficient in PTS2 protein import into peroxisomes. Three-day-old root tips from seedlings grown in 0.1% agar in white light (left panels) and 7-d-old root hair cells (right panels) are shown for wild-type, pex7-1, and pex5-1, and transformants expressing PTS1-GFP, PTS2-GFP, and cytoplasmic GFP. Note the punctate fluorescence in plants expressing PTS1-GFP and wild-type plants expressing PTS2-GFP; fluorescence is diffuse in pex7-1 and pex5-1 expressing PTS2-GFP, indicating disrupted PTS2 protein import into peroxisomes. Untransformed plant images were captured at the maximum exposure time used for any of the transformed lines. Scale bars, 200 μm.
Figure 4.
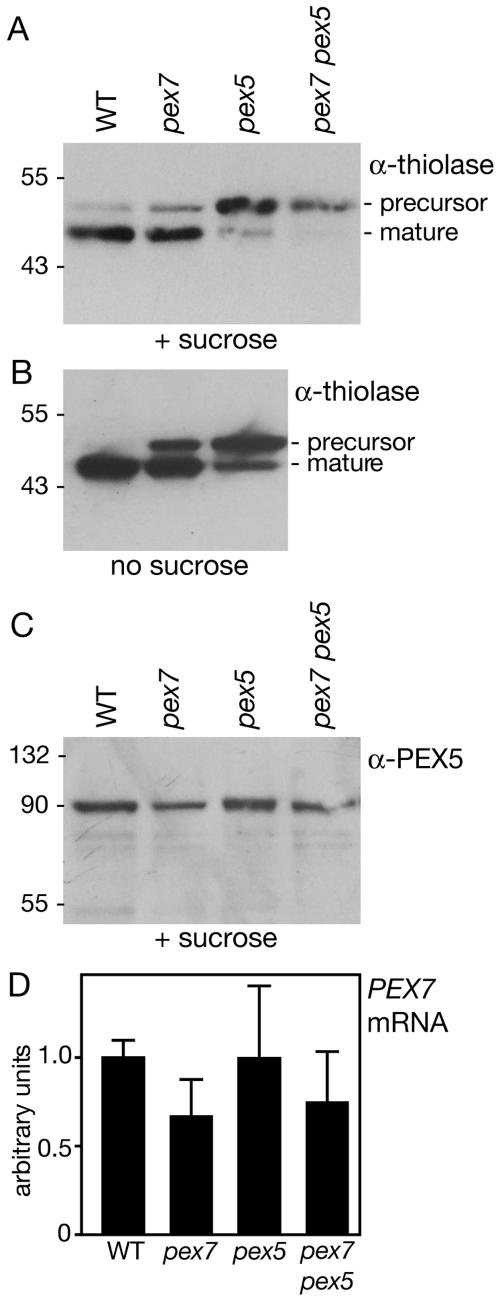
pex7-1 and pex5-1 are deficient in import of the PTS2 protein thiolase into peroxisomes. (A) Thiolase import defects. Precursor thiolase protein is translated with an N-terminal, PTS2-containing sequence that is cleaved after entry into peroxisomes to produce mature thiolase. Protein was extracted from seedlings grown on sucrose and visualized using an antithiolase antibody (Kato et al., 1996). (B) Thiolase import defects are exaggerated in plants grown without exogenous sucrose. pex7-1 pex5-1 was omitted because of developmental arrest in the absence of sucrose. (C) PEX5 protein is present in all mutants. Protein was extracted from seedlings grown on sucrose and visualized using anti-PEX5 antibody (Zolman and Bartel, 2004). Positions of molecular mass markers (in kDa) are indicated at the left in A-C. (D) PEX7 message is present in all mutants. PEX7 mRNA levels relative to an APRT control in wild type and each mutant were determined using quantitative real-time reverse-transcription PCR. Error bars, SDs of mean PEX7 levels expressed in arbitrary units.
PTS2-containing Protein Import Is Defective in the PTS1 Receptor Mutant pex5-1
Arabidopsis contains a single PEX5 gene (At5g56290; Brickner et al., 1998; Zolman et al., 2000) encoding a protein ∼20 and ∼28% identical to S. cerevisiae Pex5p and human PEX5L, respectively (Figure 1). PEX5 proteins contain N-terminal pentapeptide repeat (PPR) domains involved in PEX14 docking at the peroxisome (Nito et al., 2002) and C-terminal tetratricopeptide repeat (TPR) domains necessary for PTS1 protein cargo binding (Gatto et al., 2000). In addition, among the TPR repeats is a sequence conserved between human and plant PEX5 proteins and the PEX7-binding domains of the yeast peroxisome docking proteins Pex18p and Pex21p (Einwächter et al., 2001). A fragment of human PEX5L including this region is sufficient for interaction with human PEX7 (Dodt et al., 2001).
The previously described pex5-1 mutant displays reduced sensitivity to exogenous IBA and a slight growth defect in darkness without exogenous sucrose (Zolman et al., 2000). pex5-1 contains a serine to leucine mutation in the presumptive PEX7-binding region (Figure 1). Western blot analysis indicates that the pex5-1 mutant protein accumulates to wild-type levels (Figure 4C). Intriguingly, the analogous serine is mutated to phenylalanine in a CHO cell line defective in PTS2, but not PTS1, protein import (Matsumura et al., 2000), suggesting that the Arabidopsis pex5-1 mutant might similarly have defects in PTS2 import.
To directly examine PTS1 and PTS2 function in the pex5-1 mutant, we observed localization of PTS1- and PTS2-targeted GFP derivatives. Interestingly, we found that pex5-1 is fully competent in PTS1-GFP import (Figure 3). In marked contrast, however, PTS2-GFP is not detectably imported into peroxisomes in pex5-1 (Figure 3), consistent with the hypothesis that the primary defect in the pex5-1 mutant is in PTS2, rather than PTS1, import. Indeed, PTS2-targeted thiolase is imported less efficiently in pex5-1 than in wild type or pex7-1 (Figure 4, A and B).
Severe Developmental Deficiencies and Blocked PTS2-Protein Import in pex7-1 pex5-1
pex7-1 and pex5-1 were cross-pollinated to produce a pex7-1 pex5-1 double mutant. Like both parents, the double mutant displays reduced sensitivity to exogenous IBA (Figure 2A). Unlike either single mutant, however, pex7-1 pex5-1 is completely dependent on exogenous sucrose for growth not only in darkness (Figure 2D), but also in light (Figure 2E). Even when provided with sucrose, the double mutant grows more slowly than wild type, as evidenced by reduced root and hypocotyl elongation (Figure 2, A and D). Moreover, the possible lateral root defects observed in the single mutants are exacerbated in the double mutant; 9-d-old pex7-1 pex5-1 seedlings lack lateral roots (Figure 2C). Consistent with these morphological defects, 3-d-old double mutant seedlings apparently lack processed thiolase (Figure 4A), suggesting a severe block in import of thiolase, and presumably other PTS2 proteins, into peroxisomes.
In addition to these seedling defects, adult pex7-1 pex5-1 plants are dramatically less robust and less fecund than wild type or either parent (Figure 5, C and D). Examination of siliques of different ages reveals apparently normal fertilization indicated by the presence of plump developing seeds in green siliques, but defects become increasingly apparent during the course of seed maturation (Figure 5A). A majority of mature double mutant seeds are shrunken (Figure 5, B and D). Seed development is dependent upon PEX7 dosage; plants homozygous for pex5-1 and heterozygous for pex7-1 generate an intermediate number of shrunken seeds (Figure 5D). In contrast, pex5-1 is fully recessive for seed morphology in the pex7-1 background, as pex7-1/pex7-1 PEX5/pex5-1 plants produce normal seeds (Figure 5D). Among pex7-1 pex5-1 seeds with a wild-type appearance, fewer than half germinate when grown on media supplemented with sucrose, whereas only occasional abnormally shaped seeds germinate (Figure 5E).
Figure 5.
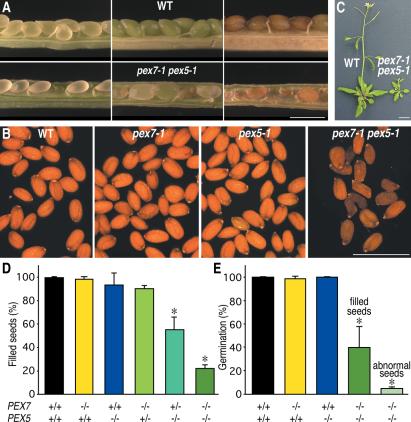
pex7-1 pex5-1 double mutant phenotypes. (A) Seed development is aberrant in pex7-1 pex5-1. One valve was removed from wild-type and pex7-1 pex5-1 siliques to reveal developing seeds of increasing age from left to right. Scale bar, 1 mm. (B) Mature pex7-1 pex5-1 seeds are shrunken. Scale bar, 1 mm. (C) pex7-1 pex5-1 adult plants have reduced stature. Plants were grown under white light on PNS for 14 d, then transferred to soil, and grown an additional 17 d. Scale bar, 1 cm. (D) Percent seeds with normal filled morphology. Mature seeds were assayed for plump appearance. Bars, means + SDs from progeny of three plants of the indicated genotype; n ≥ 60 seeds per plant. (E) Percent germination in single and double mutants. pex7-1 pex5-1 were sorted by seed morphology in D. Seedlings were grown on medium supplemented with 45 mM sucrose and assayed for germination (radicle emergence) after 9 d. Bars, means + SDs (or SE of measurement for pex5-1) from progeny of three plants of the indicated genotype (progeny of two pex5-1 plants); n ≥ 23 seeds per plant, except pex7-1 pex5-1 filled seeds where n ≥ 10 seeds per plant. *Significantly different from wild type (p < 0.02; one-tailed t test assuming unequal variance).
Among pex7-1 pex5-1 mutant seeds that germinate, abnormalities in embryonic development become apparent (Figure 6). About 15% of mutant seedlings exhibit various degrees of cotyledon fusion (Figure 6), whereas such aberrant seedling morphology is seen in fewer than 1.2% of wild type or either single mutant (n ≥ 112, unpublished data). Most pex7-1 pex5-1 seedlings with fused cotyledons develop into adult plants resembling siblings with unfused cotyledons, though a small percentage arrest and perish as seedlings (unpublished data).
Figure 6.

Cotyledon fusion in the pex7-1 pex5-1 double mutant. pex7-1 pex5-1 plants were grown for 7 d on sucrose-supplemented medium and frequencies of plants with (A) wild-type, (B) asymmetric, (C) partially fused, and (D) fused cotyledons were determined. Ratios below B, C, and D represent fraction of pex7-1 pex5-1 with the indicated degree of fusion among progeny of three double mutant plants. Scale bar, 1 mm.
DISCUSSION
Using a reverse-genetic approach, we demonstrated that Arabidopsis PEX7 is necessary for PTS2 protein import into peroxisomes and that plant PEX7 function is relevant in vivo. The pex7-1 mutant is resistant to the proto-auxin IBA (Figure 2A), which is converted in peroxisomes into the active auxin IAA (Zolman et al., 2000). This phenotype was rescued by 35S promoter-driven overexpression of PEX7. The pex7-1 mutant is not notably impaired when grown without light and sucrose, suggesting that the endogenous fatty acid β-oxidation needed for seedling development in these conditions is minimally affected. The relatively weak pex7-1 phenotype may be attributable to the nature of the lesion; the T-DNA insertion in pex7-1, though within the 5′ UTR of a cDNA (Schmid et al., 2003), allows some expression of presumably fully functional PEX7 (Figure 4D). When we examined PTS2-GFP localization in pex7-1, we found little punctate fluorescence (Figure 3), indicating a severe block in PTS2 protein import. However, the apparent severity of pex7-1 in this assay could result from flooding the cells with 35S-expressed PTS2-GFP. Indeed, the endogenous PTS2-targeted thiolase import is only slightly affected in pex7-1 (Figure 4, A and B), revealing the sensitivity of IBA resistance and PTS2-GFP localization assays in detecting peroxisome defects.
Because PEX7 appears to deliver cargo to the peroxisome via interaction with PEX5 in Arabidopsis (Nito et al., 2002; Sparkes and Baker, 2002; Johnson and Olsen, 2003) as it does in mammals (Braverman et al., 1998; Matsumura et al., 2000; Dodt et al., 2001), we also examined PTS1- and PTS2-protein import in the existing pex5-1 allele (Zolman et al., 2000). Interestingly, PTS1-GFP localization was properly punctate in pex5-1, whereas PTS2-GFP was diffuse (Figure 3). Moreover, PTS2-containing thiolase was inefficiently imported into pex5-1 peroxisomes (Figure 4, A and B). There is one existing pex5 mutant of this type, a CHO cell line with a PTS2-specific defect resulting from the inability of PEX7 to bind mutant pex5 (Matsumura et al., 2000). Remarkably, the lesion in the CHO mutant pex5 is an analogous serine to that mutated in pex5-1 (Figure 1). Moreover, both are mutated to hydrophobic amino acids, leucine in pex5-1 and phenylalanine in the CHO mutant. The characterization of these two pex5 alleles with PTS2-specific defects demonstrates the critical importance of this conserved serine for PEX7 binding and suggests an ancient origin for PEX7-PEX5 interaction. Further, whereas the CHO cell line was isolated in a screen that selected for PTS2-deficient import (Matsumura et al., 2000), Arabidopsis pex5-1 was isolated from a screen for IBA resistance, which can yield a variety of peroxisomal defects (Zolman et al., 2000, 2001a, 2001b; Zolman and Bartel, 2004); this result may suggest that PTS2 proteins are particularly important for IBA metabolism in seedlings. Indeed, the peroxisome-defective mutant ped2/pex14 is deficient in IBA (Monroe-Augustus, 2004) and 2,4-DB (Hayashi et al., 2000) response as well as PTS2-protein import (Hayashi et al., 2000).
Though an N-terminal Arabidopsis PEX5 fragment lacking the region containing the pex5-1 mutation has been shown to interact with PEX7 in a yeast two-hybrid assay, the interaction was less robust than with full-length PEX5 (Nito et al., 2002). Further, the interaction was enhanced in a PEX5 fragment containing the region mutated in pex5-1 (Nito et al., 2002). Thus, the region we implicate in PTS2 import is necessary, but may not be sufficient, for the PEX5-PEX7 interaction driving PTS2 import in vivo.
Whereas both pex7-1 and pex5-1 single mutants are peroxisomally deficient in only the most sensitive assays, combining the two defects yields a severely affected double mutant with numerous developmental abnormalities, likely resulting from reduced PEX7 expression combined with inefficient PEX7-PEX5 interaction (Figure 7). PTS2-targeted thiolase import is severely affected in pex7-1 pex5-1, whereas PEX7 expression and PEX5 levels are similar to those in the respective single mutants (Figure 4); this result is consistent with the double mutant harboring two partial loss-of-function mutations in the same pathway. pex7-1 pex5-1 requires exogenous sucrose for seedling establishment (Figure 2E), but not for growth as an adult plant. The sucrose dependence of this mutant demonstrates the necessity of PTS2 protein import for utilization of seed-storage lipids by seedlings before photosynthesis is established, a process of specialized peroxisomes termed glyoxysomes (Beevers, 2002).
Figure 7.

Model for PTS2 protein import into plant peroxisomes. (A) PEX7 binds both PTS2 protein cargo and the PTS1 protein receptor PEX5. Delivery of PTS2 protein into the peroxisome is dependent on complex formation with PEX5. PTS2 protein cargo is necessary for peroxisomal processes of fatty acid β-oxidation and conversion of IBA into the active auxin IAA. (B) Functional PEX7 may be expressed at reduced levels in the pex7-1 mutant, resulting in reduced PTS2 protein import and IBA resistance. (C) PEX7 binding with pex5-1 is inefficient, resulting in decreased PTS2 protein import and IBA resistance. (D) Two partial defects are combined in pex7-1 pex5-1 to nearly eliminate PTS2 protein import, resulting in severe developmental defects.
Though the PTS2 signal sequence is less common than PTS1, it is more common in plants than other organisms (Johnson and Olsen, 2001; Mullen, 2002) and is greatly over-represented among β-oxidation enzymes highly expressed during the glyoxysome-dependent seedling establishment phase (Kamada et al., 2003). PTS2-bearing β-oxidation enzymes with peak expression in seedlings (Kamada et al., 2003) include the acyl-CoA synthases LACS6 and LACS7 (Fulda et al., 2002), the acyl-CoA oxidases ACX2 (Hooks et al., 1999) and ACX3 (Froman et al., 2000), and the thiolase PED1/KAT2 (Hayashi et al., 1998; Germain et al., 2001). Among these enzymes, ped1/kat2 mutants (Hayashi et al., 1998; Germain et al., 2001) and the lacs6 lacs7 double mutant require exogenous sucrose for seedling establishment (Fulda et al., 2004). Although both LACS6 and LACS7 have functional PTS2 signal sequences, LACS7 also possesses a functional PTS1 sequence, and the lacs6 single mutant is sucrose-independent (Fulda et al., 2002), making these proteins unlikely causes of sucrose dependence in the PTS2-import deficient pex7-1 pex5-1. However, sucrose dependence in pex7-1 pex5-1 could result from mislocalization of the PTS2-targeted PED1/KAT2 to the cytosol (Figure 4A). In contrast to these β-oxidation enzymes, all known enzymes directly involved in photorespiration in green leaf peroxisomes bear PTS1 signal sequences (Reumann, 2002); the PTS2-independence of photorespiration could account for the healthy green coloration of the pex7-1 pex5-1 mutant (Figure 5C), which is unlike the pale-colored pex14 and pex6 mutants (Hayashi et al., 2000; Zolman and Bartel, 2004).
pex7-1 pex5-1 plants germinated on sucrose and transferred to soil grow into stunted adults that produce few normal seeds, only a minority of which germinate, even when supplied with sucrose (Figure 5). Thus, PTS2 import is required not only for seedling establishment, but also for vegetative growth and normal seed development. Strikingly, the sse1 mutant, which is defective in PEX16, a peroxin implicated in early steps in peroxisome formation, was isolated because it bears ∼90% inviable shrunken seeds (Lin et al., 1999). Further, null mutations in PEX2 and PEX10 are lethal at stages before seed maturation (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003). Thus, the high frequency of shrunken and inviable seeds in the pex7-1 pex5-1 double mutant again implicates peroxisomes in proper seed development and reveals the necessity of PTS2 protein import for this process.
Those pex7-1 pex5-1 seeds that do germinate often display fused cotyledons (Figure 6). Several Arabidopsis mutants with defective cotyledon separation have been described, and these link cotyledon development with auxin response. The pin-formed1 mutant has frequent cotyledon fusion and is defective in an auxin efflux facilitator that is necessary to establish auxin gradients in developing embryos (Gälweiler et al., 1998). The monopteros mutant likewise has variably fused cotyledons and is defective in an auxin response factor that interprets auxin gradients (Hardtke and Berleth, 1998). The cup-shaped cotyledon double mutant cuc1 cuc2 was isolated on the basis of cotyledon fusion and is defective in functionally redundant transcription factor genes (Aida et al., 1997; Takada et al., 2001) that are misregulated in monopteros and pin-formed1 (Aida et al., 2002). Thus, auxin signaling is critical for proper cotyledon development.
The requirement of auxin response for proper embryonic symmetry, the requirement for peroxisome function to allow IBA conversion into active IAA, and the reduced auxin response phenotype of pex7-1 pex5-1 implicate IBA as a critical auxin reservoir during embryogenesis. Though IBA levels have not been quantified in seeds, IBA is present in seedling tissue at nearly the levels of free IAA (Ludwig-Müller et al., 1993).
PEX7 is necessary for peroxisome function in mammals and yeast (Rehling et al., 1996; Mukai et al., 2002), and characterization of the first plant pex7 mutant reveals a reduction in PTS2 protein import into peroxisomes. We also show that the sole described plant pex5 mutant is defective in PTS2-, but not PTS1-protein import. pex7-1 pex5-1 double mutants have severe PTS2 import defects, several developmental abnormalities, and a high frequency of embryonic death and deformities, some of which may result from defective IBA metabolism. It will be interesting to observe the phenotypes of hypothetical pex5 mutants specifically defective in PTS1 rather than PTS2 import, which will reveal the roles for PTS1 import in plant development. In addition, the functional necessity of PEX7 interaction with PEX5 observed previously only in mammals further establishes Arabidopsis as an excellent model for human peroxisome biogenesis disorders.
Acknowledgments
We are grateful to Makoto Hayashi for the anti-thiolase antibody, Bethany Zolman for pex5-1 PTS1-GFP seeds, Mary Ellen Lane for use of microscopy facilities, and Raquel Adham, Diana Dugas, Melanie Monroe-Augustus, Rebekah Rampey, and Bethany Zolman for critical comments on the manuscript. We thank the Salk Institute and Arabidopsis Biological Resource Center for T-DNA insertion lines and a thiolase cDNA, and the Kazusa DNA Research Institute for a PEX7 cDNA. This research was supported by the National Science Foundation (IBN-0315596), the Robert A. Welch Foundation (C-1309), and the Houston Livestock Show and Rodeo (fellowship to A.W.W.).
Article published online ahead of print in MBC in Press on November 17, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-05-0422).
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Vernoux, T., Furutani, M., Traas, J., and Tasaka, M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965-3974. [DOI] [PubMed] [Google Scholar]
- Albertini, M., Rehling, P., Erdmann, R., Girzalsky, W., Kiel, J.A.K.W., Veenhuis, M., and Kunau, W. H. (1997). Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83-92. [DOI] [PubMed] [Google Scholar]
- Alonso, J. M. et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7, 175-180. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1995). Short Protocols in Molecular Biology, New York: John Wiley and Sons, Inc.
- Bartel, B., LeClere, S., Magidin, M., and Zolman, B. K. (2001). Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J. Plant Growth Regul. 20, 198-216. [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M., and Sonnhammer, E. L. (2002). The Pfam protein families database. Nucleic Acid Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers, H. (2002). Early research on peroxisomes in plants. In: Plant Peroxisomes: Biochemistry, Cell Biology, and Biotechnological Applications, ed. A. Baker and I. A. Graham, Dordrecht, The Netherlands: Kluwer, 1-17.
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman, N., Dodt, G., Gould, S. J., and Valle, D. (1998). An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7, 1195-1205. [DOI] [PubMed] [Google Scholar]
- Braverman, N., Steel, G., Obie, C., Moser, A., Moser, H., Gould, S. J., and Valle, D. (1997). Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat. Genet. 15, 369-376. [DOI] [PubMed] [Google Scholar]
- Brickner, D. G., Brickner, J. H., and Olsen, L. J. (1998). Sequence analysis of a cDNA encoding Pex5p, a peroxisomal targeting signal type 1 receptor from Arabidopsis. Plant Physiol. 118, 330. [Google Scholar]
- Chang, C. C., Warren, D. S., Sacksteder, K. A., and Gould, S. J. (1999). PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J. Cell Biol. 147, 761-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton, W. A., and López-Huertas, E. (2002). PEX genes in plants and other organisms. In: Plant Peroxisomes: Biochemistry, Cell Biology, and Biotechnological Applications, ed. A. Baker and I. A. Graham, Dordrecht, The Netherlands: Kluwer, 385-426.
- Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- Collins, C. S., Kalish, J. E., Morrell, J. C., McCaffery, J. M., and Gould, S. J. (2000). The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol. Cell Biol. 20, 7516-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammai, V., and Subramani, S. (2001). The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisome matrix and recycled to the cytosol. Cell 105, 187-196. [DOI] [PubMed] [Google Scholar]
- Davis, S. J., and Vierstra, R. D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521-528. [DOI] [PubMed] [Google Scholar]
- Dodt, G., Braverman, N., Wong, C., Moser, A., Moser, H. W., Watkins, P., Valle, D., and Gould, S. J. (1995). Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9, 115-125. [DOI] [PubMed] [Google Scholar]
- Dodt, G., and Gould, S. J. (1996). Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135, 1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt, G., Warren, D., Becker, E., Rehling, P., and Gould, S. J. (2001). Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J. Biol. Chem. 276, 41769-41781. [DOI] [PubMed] [Google Scholar]
- Einwächter, H., Sowinski, S., Kunau, W. H., and Schliebs, W. (2001). Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfil a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2, 1035-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, C. R., Mullen, R. T., and Trelease, R. N. (1998). Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. Plant J. 16, 709-720. [DOI] [PubMed] [Google Scholar]
- Footitt, S., Slocombe, S. P., Larner, V., Kurup, S., Wu, Y., Larson, T., Graham, I., Baker, A., and Holdsworth, M. (2002). Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 21, 2912-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman, B. E., Edwards, P. C., Bursch, A. G., and Dehesh, K. (2000). ACX3, a novel medium-chain acyl-Coenzyme A oxidase from Arabidopsis. Plant Physiol. 123, 733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda, M., Schnurr, J., Abbadi, A., Heinz, E., and Browse, J. (2004). Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16, 394-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda, M., Shockey, J., Werber, M., Wolter, F. P., and Heinz, E. (2002). Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant J. 32, 93-103. [DOI] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226-2230. [DOI] [PubMed] [Google Scholar]
- Gatto, G. J., Jr., Geisbrecht, B. V., Gould, S. J., and Berg, J. M. (2000). Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091-1095. [DOI] [PubMed] [Google Scholar]
- Gerhardt, B. (1992). Fatty acid degradation in plants. Prog. Lipid Res. 31, 417-446. [DOI] [PubMed] [Google Scholar]
- Germain, V., Rylott, E. L., Larson, T. R., Sherson, S. M., Bechntold, N., Carde, J. P., Bryce, J. H., Graham, I. A., and Smith, S. M. (2001). Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J. 28, 1-12. [DOI] [PubMed] [Google Scholar]
- Gould, S. J., Keller, G. A., Hosken, N., Wilkinson, J., and Subramani, S. (1989). A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I. A., and Eastmond, P. J. (2002). Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 41, 156-181. [DOI] [PubMed] [Google Scholar]
- Gurvitz, A., Langer, S., Piskacek, M., Hamilton, B., Ruis, H., and Hartig, A. (2000). Predicting the function and subcellular location of Caenorhabditis elegans proteins similar to Saccharomyces cerevisiae β-oxidation enzymes. Yeast 17, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C. S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn, G.W., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430-434. [Google Scholar]
- Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T., and Nishimura, M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19, 5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10, 183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks, M.A., Kellas, F., and Graham, I.A. (1999). Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 20, 1-13. [DOI] [PubMed] [Google Scholar]
- Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J., and Chory, J. (2002). A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297, 405-409. [DOI] [PubMed] [Google Scholar]
- Johnson, T. L., and Olsen, L. J. (2001). Building new models for peroxisome biogenesis. Plant Physiol. 127, 731-739. [PMC free article] [PubMed] [Google Scholar]
- Johnson, T. L., and Olsen, L. J. (2003). Import of the peroxisomal targeting signal type 2 protein 3-ketoacyl-Coenzyme A thiolase into glyoxysomes. Plant Physiol. 133, 1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, T., Nito, K., Hayashi, H., Mano, S., Hayashi, M., and Nishimura, M. (2003). Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 44, 1275-1289. [DOI] [PubMed] [Google Scholar]
- Kato, A., Hayashi, M., Takeuchi, Y., and Nishimura, M. (1996). cDNA cloning and expression of a gene for 3-ketoacyl-CoA thiolase in pumpkin cotyledons. Plant Mol. Biol. 31, 843-852. [DOI] [PubMed] [Google Scholar]
- Kindl, H. (1993). Fatty acid degradation in plant peroxisomes: function and biosynthesis of the enzymes involved. Biochimie 75, 225-230. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383-396. [Google Scholar]
- LeClere, S., and Bartel, B. (2001). A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol. Biol. 46, 695-703. [DOI] [PubMed] [Google Scholar]
- Liepman, A. H., and Olsen, L. J. (2001). Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 25, 487-498. [DOI] [PubMed] [Google Scholar]
- Lin, Y., Sun, L., Nguyen, L. V., Rachubinski, R. A., and Goodman, H. M. (1999). The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284, 328-330. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller, J., Sass, S., Sutter, E. G., Wodner, M., and Epstein, E. (1993). Indole-3-butyric acid in Arabidopsis thaliana. I. Identification and quantification. Plant Growth Regul. 13, 179-187. [Google Scholar]
- Matsumura, T., Otera, H., and Fujiki, Y. (2000). Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel PEX5-impaired CHO cell mutant. J. Biol. Chem. 275, 21715-21721. [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus, M. (2004). Genetic approaches to elucidating the mechanisms of indole-3-acetic acid and indole-3-butyric acid function in Arabidopsis thaliana. PhD thesis, Rice University, Houston.
- Motley, A. M. et al. (1997). Rhizomelic chondrodysplasia punctata is a peroxisomal protein targeting disease caused by a non-functional PTS2 receptor. Nat. Genet. 15, 377-380. [DOI] [PubMed] [Google Scholar]
- Motley, A. M., Hettema, E. H., Ketting, R., Plasterk, R., and Tabak, H. F. (2000). Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 1, 40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai, S., Ghaedi, K., and Fujiki, Y. (2002). Intracellular localization, function, and dysfunction of the peroxisome-targeting signal type 2 receptor, Pex7p, in mammalian cells. J. Biol. Chem. 277, 9548-9561. [DOI] [PubMed] [Google Scholar]
- Mullen, R. T. (2002). Targeting and import of matrix proteins into peroxisomes. In: Plant Peroxisomes: Biochemistry, Cell Biology, and Biotechnological Applications, ed. A. Baker and I. A. Graham, Dordrecht, The Netherlands: Kluwer, 339-383.
- Mullen, R. T., Flynn, C. R., and Trelease, R. N. (2001). How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci. 6, 256-261. [DOI] [PubMed] [Google Scholar]
- Neuberger, G., Maurer-Stroh, S., Eisenhaber, B., Hartig, A., and Eisenhaber, F. (2003). Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J. Mol. Biol. 328, 567-579. [DOI] [PubMed] [Google Scholar]
- Nito, K., Hayashi, M., and Nishimura, M. (2002). Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 43, 355-366. [DOI] [PubMed] [Google Scholar]
- Osumi, T., Tsukamoto, T., Hata, S., Yokota, S., Miura, S., Fujiki, Y., Hajikata, M., Miyazawa, S., and Hashimoto, T. (1991). Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem. Biophys. Res. Commun. 181, 947-954. [DOI] [PubMed] [Google Scholar]
- Purdue, P. E., Yang, X., and Lazarow, P. B. (1998). Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J. Cell Biol. 143, 1859-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue, P. E., Zhang, J. W., Skoneczny, M., and Lazarow, P. B. (1997). Rhizomelic chondrodysplasia punctata is caused by deficiency of human PEX7, a homologue of the yeast PTS2 receptor. Nat. Genet. 15, 381-384. [DOI] [PubMed] [Google Scholar]
- Rehling, P., Marzioch, M., Niesen, F., Wittke, E., Veenhuis, M., and Kunau, W. H. (1996). The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 15, 2901-2913. [PMC free article] [PubMed] [Google Scholar]
- Reumann, S. (2002). The photorespiratory pathway of leaf peroxisomes. In: Plant Peroxisomes: Biochemistry, Cell biology, and Biotechnological Applications, ed. A. Baker and I. A. Graham, Dordrecht, The Netherlands: Kluwer Academic Publishers, 141-189.
- Reumann, S. (2004). Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135, 783-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott, E. L., Rogers, C. A., Gilday, A. D., Edgell, T., Larson, T. R., and Graham, I. A. (2003). Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J. Biol. Chem. 278, 21370-21377. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., Sörensen, T. R., Stracke, R., Törjék, O., Altmann, T., Mitchell-Olds, T., and Weisshaar, B. (2003). Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13, 1250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, U., Gietl, C., and Schmid, M. (1999). Sequence analysis of a cDNA encoding Pex7p, a peroxisomal targeting signal 2 receptor from Arabidopsis thaliana. Plant Physiol. 120, 33910409053 [Google Scholar]
- Schumann, U., Wanner, G., Veenhuis, M., Schmid, M., and Gietl, C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 100, 9626-9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichting, M., Schell-Steven, A., Prokisch, H., Erdmann, R., and Rottensteiner, H. (2003). Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol. Biol. Cell 14, 810-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I. A., Brandizzi, F., Slocombe, S. P., El-Shami, M., Hawes, C., and Baker, A. (2003). An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 133, 1809-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I. A., and Baker, A. (2002). Peroxisome biogenesis and protein import in plants, animals and yeasts: enigma and variations? (Review). Mol. Membr. Biol. 19, 171-185. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos, T. C., and Hangarter, R. P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93, 1365-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127-1135. [DOI] [PubMed] [Google Scholar]
- van den Brink, D. M., Brites, P., Haasjes, J., Wierzbicki, A. S., Mitchell, J., Lambert-Hamill, M., de Belleroche, J., Jansen, G. A., Waterham, H. R., and Wanders, R.J.A. (2003). Identification of PEX7 as the second gene involved in Refsum disease. Am. J. Hum. Genet. 72, 471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C., and Hause, B. (2002). Jamonates and octadecanoids: signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 72, 165-221. [DOI] [PubMed] [Google Scholar]
- Yamada, K. et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842-846. [DOI] [PubMed] [Google Scholar]
- Zolman, B. K., and Bartel, B. (2004). An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. USA 101, 1786-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B. K., Monroe-Augustus, M., Thompson, B., Hawes, J. W., Krukenberg, K. A., Matsuda, S.P.T., and Bartel, B. (2001a). chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J. Biol. Chem. 276, 31037-31046. [DOI] [PubMed] [Google Scholar]
- Zolman, B. K., Silva, I. D., and Bartel, B. (2001b). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127, 1266-1278. [PMC free article] [PubMed] [Google Scholar]
- Zolman, B. K., Yoder, A., and Bartel, B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156, 1323-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]


