Abstract
Members of the heat-shock protein (HSP)40 regulate the protein folding activity of HSP70 proteins and help the functional specialization of this molecular chaperone system in various types of cellular events. We have recently identified Hsp40 as a component of flagellar axoneme in the ascidian Ciona intestinalis, suggesting a correlation between Hsp40 related chaperone system and flagellar function. In this study, we have found that Ciona 37-kDa Hsp40 is extracted from KCl-treated axonemes with 0.5 M KI solution and comigrates with radial spoke protein (RSP)3 along with several proteins as a complex through gel filtration and ion exchange columns. Peptide mass fingerprinting with matrix-assisted laser desorption ionization/time of flight/mass spectrometry revealed that other proteins in the complex include a homolog of sea urchin spokehead protein (homolog of RSP4/6), a membrane occupation and recognition nexus repeat protein with sequence similarity with meichroacidin, and a functionally unknown 33-kDa protein. A spoke head protein, LRR37, is not included in the complex, suggesting that the complex constructs the stalk of radial spoke. Immunoelectron microscopy indicates that Hsp40 is localized in the distal portion of spoke stalk, possibly at the junction between spoke head and the stalk.
INTRODUCTION
The axonemes in eukaryotic cilia and flagella are highly organized structures consisting of ∼250 proteins (for review, see Mitchell, 2000; Porter and Sale, 2000; Inaba, 2003). Functionally diverse protein complexes are bound to the 9 + 2 microtubule structure, resulting in construction of sophisticated micromachinery that generates wave propagation. Recent molecular approaches have revealed several molecules that constitute these substructures of the axonemes, especially the outer and inner arms (King, 2000; Kamiya, 2002) and radial spokes (Yang et al., 2001).
For complete understanding of the molecular architecture of the axonemes, we have selectively isolated the cDNAs for sperm axonemal proteins in the ascidian Ciona intestinalis by using immunoscreening with anti-axoneme antisera (Padma et al., 2003). A leucine-rich repeat protein, LRR37, seems highest, with 11 occurrences in 76 positive clones. Immunoelectron microscopy revealed that LRR37 is localized at the tip of radial spoke, most likely at the spoke head. CiRSP3, a homolog of Chlamydomonas radial spoke protein 3 (RSP3) is localized at the base of radial spoke, coincident with the localization in Chlamydomonas flagella (Huang et al., 1981; Padma et al., 2003). Several lines of evidence support the idea that the central pair determines the plane of flagellar bending by sending signals to radial spokes (Smith and Lefebvre, 1997). Radial spokes regulate inner arm dynein by phosphorylation/dephosphorylation (Porter and Sale, 2000). In fact, among the 23 polypeptides identified as radial spoke components in Chlamydomonas, several components potentially participates in cAMP-dependent or Ca2+-dependent signaling pathway (Yang et al., 2001, 2004; Gaillard et al., 2001; Patel-King et al., 2004). The RSP3 has been shown to possess a domain of A-kinase anchoring protein (Gaillard et al., 2001).
Evidence suggests that molecular chaperone system is involved in the assembly or turnover of axonemal proteins. The localization of HSP70 in eukaryotic flagella was first reported in Chlamydomonas (Bloch and Johnson, 1995). The existence of HSP70 proteins in ciliary and flagellar axonemes was further shown in a variety of organs (Stephens, 1997; Williams and Nelsen, 1997; Stephens and Lemieux, 1999), suggesting that HSP70-related molecular chaperone system might be deeply involved in the assembly and/or protein turnover in 9 + 2 axonemes.
Members of the 40-kDa molecular chaperone (Hsp40) family are structurally more diverse than Hsp70s (Ohtsuka and Hata, 2000). Although Hsp40s exert autonomous, Hsp70-independent chaperone activity, they generally function as Hsp70-dependent “cochaperone.” They have a highly conserved, N-terminal J-domain, which is most strongly implicated in the regulation of protein folding through modulation of ATPase activity of Hsp70s (Lu and Cyr, 1998). The diversity of Hsp40s exerts the determination of each intracellular compartment and specific function of the each chaperone machinery (Rassow et al., 1995).
Immunoscreening with anti-axoneme antisera revealed that a heat-shock protein, Hsp40, is a component of the axoneme (Padma et al., 2003). It showed the second highest occurrence (6 of 76 positive clones), suggesting that it is an integral component of the axoneme, although its localization and function in the axonemes have not been clear. Recently, we have established a system for the proteomic analysis of testis-expressed proteins in C. intestinalis. This is composed of separation of proteins by two-dimensional gel electrophoresis and peptide mass fingerprinting by matrix-assisted laser desorption ionization/time of flight/mass spectrometry (MALDI-TOF/MS), followed by a local MS database (MSCITS) and the search program (PerMS) (Hozumi et al., 2004). This system enables quick and multiple identifications of testis or sperm proteins. In this study, we have carried out an analysis of Hsp40 and Hsp40-containing complex and showed that Hsp40 is a component of radial spoke of the axonemes. We also have identified some novel proteins as components of radial spokes in the complex.
MATERIALS AND METHODS
Experimental Materials
Adults of C. intestinalis were mainly collected near the Education & Research Center of Marine Bio-Resources, Tohoku University, and kept under constant light for several days until the accumulation of sperm in sperm duct could be observed. Sperm were collected from sperm duct by dissection into a sample tube and kept on ice until use.
Extraction and Purification of Radial Spoke
Sperm flagella and axonemes were prepared from C. intestinalis sperm pellet as described previously (Inaba et al., 1998; Padma et al., 2001). High salt extraction with a solution containing 0.6 M KCl, 20 mM Tris-HCl, pH 8.0, 1 mM MgSO4, 0.5 mM EGTA, 0.2 mM dithiothreitol (DTT) (Inaba et al., 1998) was repeated twice for careful removal of outer arm dyneins. Then, 0.5 M KI extraction was carried out against KCl-extracted axoneme as described previously (Yang et al., 2001; Padma et al., 2003). After centrifugation at 12,000 × g for 15 min at 4°C, the clear supernatant was collected and dialyzed against excess amount of buffer A (0.15M KCl, 20 mM Tris-HCl, pH 8.0, 1 mM MgSO4, 0.5 mM EGTA, 0.2 mM DTT) for 6 h with two changes of the buffer. The retentate in the dialysis bag was centrifuged at 100,000 × g for 40 min at 4°C, and the supernatant was separated on a Poros HQ anion exchange column (4.6 × 100 mm; Applied Biosystems, Foster City, CA) with a BioCAD HPLC Workstation (Global Medical Instrumentation, Albertville, MN) at a flow rate of 3.0 ml/min. Fractions of 0.3 ml each were collected. The main fractions showing immunoreactivity to anti-CiRSP3 antibody were pooled and subjected to gel filtration on a Superose 6 gel column (10 × 300 mm; Amersham Biosciences, Piscataway, NJ) at flow rate of 0.2 ml/min. Fractions (0.6 ml) were collected from the elution time 30 to 180 min.
Antibodies
Sequence analysis and production of antibodies against bacterially expressed proteins were performed as described previously (Padma et al., 2003). Polymerase chain reaction (PCR) primers used for the amplification of open reading frame are 5′-CGCGGAATTCATGTCGGATTTAGGATCA-3′ (sense) and 5′-CGCGCTCGAGCGGTATACAGTGTACAAA-3′ (antisense) for membrane occupation and recognition nexus (MORN) repeat protein, 5′-CGCGGAATTCATGGATGCCAATGGACTT-3′ (sense) and 5′-CGCGCTCGAGACAAACAACAGTACATAC-3′ (antisense), for 33-kDa protein. The clone (CiAx12), including C. intestinalis axonemal heat-shock protein 40 (CiAxHsp40), was obtained by immunoscreening of testis-expressed λZAP II cDNA library with anti-axoneme antiserum (Padma et al., 2003). The insert was amplified by PCR with T3 and T7 primers, digested by EcoRI/XhoI, and subcloned into pET32c vector. Protein expression was induced by isopropyl β-d-thiogalactoside. The fusion protein contained two third C-terminal region of CiAxHsp40 (from Ile76 to C-terminus Ile240). Production of the antibody against thioredoxin-CiAxHsp40 fusion protein was performed as described previously (Padma et al., 2003).
MS Analysis by MALDI-TOF/MS
Proteins were separated by SDS-PAGE (Laemmli, 1970). In-gel digestion was performed according to Toda et al. (1998). Coomassie-stained protein band was cut out and trimmed into small pieces, followed by sequential treatment with 60% methanol/50 mM NH4HCO3 twice, 50 mM NH4HCO3/50% acetonitrile three times, and 100% acetonitrile once. The gel pieces were dried up completely. Reduction or alkylation of cysteines was performed with 10 mM DTT/100 mM NH4HCO3 or with 54 mM iodoacetoamide/100 mM NH4HCO3, respectively. The gel pieces were then washed with 60% methanol/50 mM NH4HCO3 twice, 50% methanol/50 mM NH4HCO3 once, and finally completely dehydrated by washing twice with 100% acetonitrile. The dried gels were incubated in 10 μg/ml trypsin (Promega, Madison, WI) in 30% acetonitrile/50 mM NH4HCO3 at 30°C overnight. The tryptic fragments eluted into supernatant were mixed with equal volume of matrix solution containing 10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 40% acetic acid, and 0.1% trifluoroacetic acid. Peptide mass spectra were obtained on an AXIMA-CFR (Shimadzu Biotech, Kyoto, Japan) MALDI-TOF/MS with reflectron mode and the measurement range of 500-4000 m/z. Each mass spectrum was then calibrated and edited using Kompact (Kratos Analytical, Manchester, United Kingdom).
Peptide Mass Fingerprinting
Peptide mass fingerprinting was performed by a system that we have recently developed (Hozumi et al., 2004). Briefly, the mass data processed by Kompact were searched against a mass database for Ciona testis proteins MSCITS by a perl-based search program PerMS. MSCITS were constructed from Ciona testis expressed sequence tag (EST) data (Inaba et al., 2002; http://ghost.zool.kyoto-u.ac.jp/indexr1.html) and the genome draft sequence (Dehal et al., 2002; http://genome.jgi-psf.org/Ciona4/Ciona4.home.html) by the help of ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and MS-digest (http://prospector.ucsf.edu/). PerMS search was performed with the molecular mass tolerance at 0.15 Da.
Immunoprecipitation
Antiserum was diluted to 1:50 with phosphate-buffered saline (PBS), and 1 ml of the solution was mixed with 50 μl of protein A-Sepharose beads (Amersham Biosciences). After gentle agitation for 2 h, the beads were washed with PBS repetitively. Antibody was then immobilized onto the beads with 2.5 mM disuccinimidyl suberate (Pierce Chemical, Rockford, IL) in PBS for 1 h. The beads were washed with 50 mM glycine, pH 2.5, four times to terminate the cross-linking reaction and to ablate nonspecific binding, followed by equilibration with buffer A. The KI extract (∼20 mg) was dialyzed against buffer A and centrifuged to remove the aggregates. Triton X-100 was added to both beads and KI extract to 1%. Both were gently mixed on a rotator at 4°C overnight. The beads were collected by brief centrifugation and washed with buffer A containing 1% Triton X-100 repetitively. Proteins were eluted with equal volume of 100 mM glycine (pH 2.5).
Immunohistochemistry
Immunofluorescence microscopy and immunogold electron microscopy were carried out according to the methods described previously (Padma et al., 2003). The immunofluorescent specimen also was counterstained with 1 μM 4,6-diamidino-phenylindole (DAPI).
Computational Analysis
Translation of DNA into amino acids, calculation of molecular mass, design of PCR primers, and estimation of isoelectric points were done by GENETYX-Mac software. BLASTP program was used to search for homologous proteins. Multiple sequences alignment and drawing of phylogenic trees were carried out by ClustalW. Output to a phylogenic tree was conducted by TreeView-PPC. ProfileScan was used for prediction of functional sites or domains in amino acid sequence. Search and analysis of Ciona genes and proteins were done using online databases at http://ghost.zool.kyoto-u.ac.jp/indexr1.html and http://genome.jgi-psf.org/Ciona4/Ciona4.home.html.
RESULTS
Characterization of Ciona Axonemal Hsp40
Immunoscreening of testis cDNA library with anti-axonemal proteins antibody resulted in the isolation of six clones showing sequence homology to Hsp40 (Padma et al., 2003). All of them showed identical cDNA sequences containing an open reading frame encoding 313 amino acids with the predicted molecular mass or the isoelectric point of 35 kDa or 8.9, respectively (accession no. BAB85846). The BLAST search of Ciona axonemal Hsp40 (CiAxHsp40) showed high similarity to human testis spermatogenesis apoptosis-related gene 6 (accession no. NP_705842) with 64% identity and 80% homology, or to mouse spermatogenesis apoptosis-related protein (accession no. NP_705755) with 65% identity and 80% homology. Among all the Hsp40 species in human genome database, CiAxHsp40 showed highest similarity to human Hsp40 homolog subfamily B member 4 (DjB4) (52% identity and 68% homology) (Ohtsuka and Hata, 2000). Motif search by ProfileScan revealed that N-terminal region from amino acid residues 4-68 or C-terminal region from the residues 189-311 showed significant match with DnaJ domain or DnaJ_C domain, respectively.
Phylogenic Analysis and Expression Pattern of Ciona Axonemal Hsp40
Hsp40 family shows much diversity that is suggested to render localization or functional variety for the molecular chaperone system (Rassow et al., 1995; Ohtsuka and Hata, 2000). Search of Hsp40s in Ciona testis EST database revealed eight distinct clones out of 4717 clones. Four clones encode the CiAxHsp40, two encode DnaJ homolog subfamily A member 1, and the other clones encode DnaJ homolog subfamily B member 12 and a putative DnaJ domain-containing protein. The Ciona CiAxHsp40 shows high similarity to DjB4, but its specific role in the assembly or folding of axonemal proteins has not been elucidated.
There are 23 Hsp40s in mammals classified based on PSORT analysis (Ohtsuka and Hata, 2000). We have searched the members of Hsp40s in Ciona genome by first BLAST search of CiAxHsp40 in Ciona genome with the threshold of probability of e-10. The BLAST search result with the threshold of e-5 resulted in no difference from that with e-10. The data were supplemented by BLAST search of each member of mouse and human Hsp40s in Ciona database. These lines of search in Ciona genome database found 15 members of Hsp40 in C. intestinalis (Figure 1). Multiple alignment of mouse, human, and Ciona Hsp40s by ClustalW, followed by phylogenic analysis confirmed that CiAxHsp40 belongs to mammalian DjB1/B4 class (Figure 1).
Figure 1.
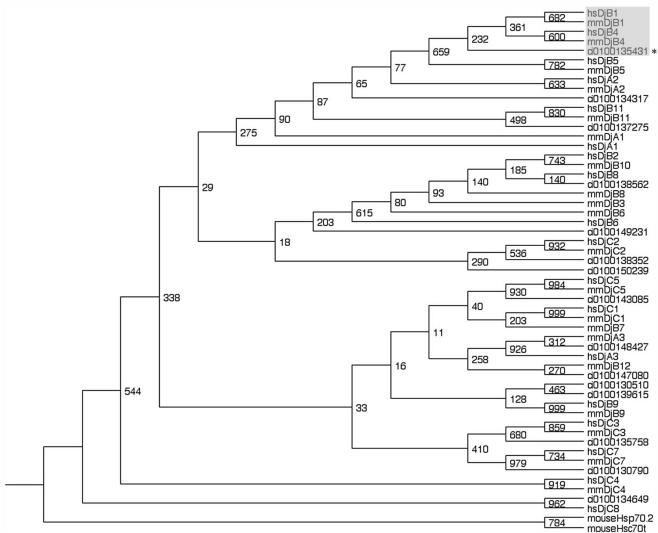
Phylogenic tree of the members of Hsp40 family in human, mouse, and Ciona. Note that CiAxHsp40 (asterisk) belongs to DjB1/B4 class (shaded). Two Hsp70s, mouse Hsp70-2 (accession no. I49761) and mouse Hsp70t (accession no. BC004714) were used as outgroups. Ciona Hsp40s were searched at http://genome.jgi-psf.org/Ciona4/Ciona4.home.html. The names of the gene number are indicated. The number on the branch represents the percentage of times that a node was supported in 1000 bootstrap pseudoreplications. The accession numbers of human mouse Hsp40s are hsDjA1, D13388; hsDjA2, AJ001309; hsDjA3, AF061749; hsDjB1, X62421; hsDjB2, X63368; hsDjB4, U40992; hsDjB5, AF088982; hsDjB6, AB014888; hsDjB8, BC029521; hsDjB9, AF083247; hsDjB11, AJ250137; hsDjC1, AK027263; hsDjC2, X98260; hsDjC3, U28424; hsDjC4, AF036874; hsDjC5, AL118506; hsDjC7, U46571; hsDjC8, AF083190; mmDjA1, AF055664; mmDjA2, AB028853; mmDjA3, AY009320; mmDjB1, AB028272; mmDjB3, U95607; mmDjB4, AK008537; mmDjB5, AF092536; mmDjB6, AF035962; mmDjB7, AB028855; mmDjB8, AB028856; mmDjB9, AB028857; mmDjB1, 0AB028858; mmDjB1, 1BC003999; mmDjB1, 2AB028860; mmDjC1, L16953; mmDjC2, U53208; mmDjC3, U28423; mmDjC4, AF036875; mmDjC5, U39320; and mmDjC7, BC023681.
We compared the gene expression of Ciona Hsp40s among several adult tissues to know the tissue distribution of CiAxHsp40s, by the use of Ciona cDNA database (http://ghost.zool.kyoto-u.ac.jp/indexr1.html). A DjC3 class of Hsp40 (ci0100130790), which regulates the activity of the inhibitor for RNA-activated protein kinase (Melville et al., 1997), shows the highest expression among all classes of Hsp40s in testis, but it is abundantly expressed in all tissues. The CiAxHsp40 (gene name ci0100135431; DjB1/B4 class) as well as ci0100147080 (DjB12 class) are shown to be testis specific (Figure 2).
Figure 2.
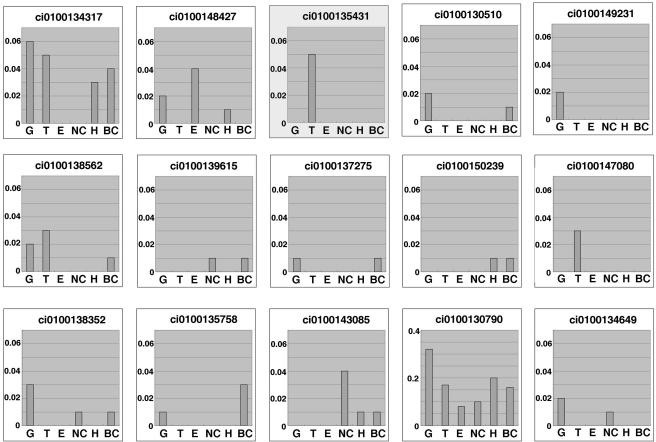
Expression patterns of several members of Hsp40 in adult tissues of C. intestinalis. Data are based on >480,000 EST data collected from several Ciona tissues (http://ghost.zool.kyoto-u.ac.jp/indexr1.html). The vertical values in the histogram reflect the occurrence of the gene (number of occurrence/number of total ESTs collected). The DjB1/B4 class of Hsp40 is highlighted. BC, blood cells; E, endostyle; G, gonad; H, heart; NC, neural complex; T, testis.
Distribution of CiAxHsp40 in the Fractions from Chemically Dissected Axonemes
Localization of CiAxHsp40 in sperm cell was determined using an antibody against thioredoxin-CiAxHsp40 fusion protein. The antibody recognized a single band with molecular mass of 37 kDa (Figure 4). Immunofluorescent microscopy showed that it is localized along sperm flagellar axonemes (Figure 3).
Figure 4.
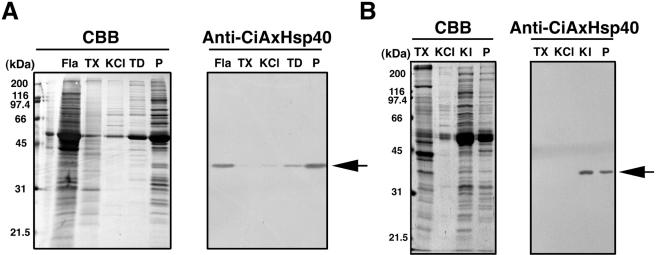
Western blot of several fractions from chemical dissection of sperm axonemes. (A) Sperm flagella (Fla) were fractionated into Triton X-100 extract (TX), 0.6 M KCl extract of the axonemes (KCl), the supernatant (TD), and the pellet (P) after Tris-EDTA treatment of the KCl-extracted axonemes. (B) Sperm flagella (Fla) were fractionated into Triton X-100 extract (TX), 0.6 M KCl extract of the axonemes (KCl), the supernatant (KI), and the pellet (P) after 0.5 M KI treatment of the KCl-extracted axonemes. Both data show the Coomassie-stained protein pattern (left) and corresponding Western blot with anti-CiAxHsp40 (right). The arrows show the position of CiAxHsp40.
Figure 3.
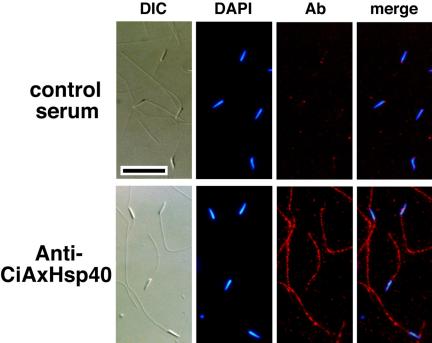
Immunofluorescence microscopy showing the localization of CiAxHsp40 along sperm flagella. Ciona sperm were fixed, stained with anti-CiAxHsp40 antibody (red), and counterstained with DAPI (blue). Differential interference contrast and merged fluorescent images also are shown. Normal mouse serum was used in the control experiment. Anti-CiAxHsp40 antibody clearly stained the flagellum, although slight nonspecific staining on the head was observed, as also with control serum. Bar, 10 μm.
Chemical dissection of the axonemes, followed by immunoblotting by a specific antibody, gives an important clue for the localization of a protein in the axonemes (Gibbons, 1981; Piperno et al., 1981;Padma et al., 2003). Sperm flagella were treated with Triton X-100 solution, resulting in the extraction of membrane and matrix fraction. The axonemal pellet was sequentially extracted with 0.6 M KCl solution and Tris-EDTA solution. Western blotting of each extract with anti-CiAxHsp40 showed that CiAxHsp40 is mostly detected in the pellet after Tris-EDTA treatment (Figure 4A). Previous study showed that CiRSP3 or LRR37 is extracted into the pellet or the supernatant, respectively, after Tris-EDTA treatment (Padma et al., 2003). The extraction pattern of CiAxHsp40 was similar to that of CiRSP3, although a part of CiAxHsp40 was extracted by Tris-EDTA treatment. Furthermore, the extraction by 0.5 M KI solution, which was used for selective extraction of radial spokes in Chlamydomonas flagella (Yang et al., 2001), efficiently solubilized the CiAxHsp40 from KCl-extracted axonemes (Figure 4B). These results raised a possibility that CiAxHsp40 is a component of radial spokes.
Immunoelectron microscopy was carried out to elucidate the possibility that CiAxHsp40 is localized in radial spokes. We first tried to perform with intact axonemes by preembedding procedure, almost no gold signal was observed, probably due to antibody accessibility. Because most part of CiAxHsp40 remained in the pellet after 0.6 M KCl treatment (Figure 4), the preembedding immunogold electron microscopy was applied to the pellet after KCl treatment. This treatment extracted the outer arm dyneins. We observed that the central pair apparatus was often lost from the axonemes. The gold particles were clearly found on the radial spokes, most likely at the distal region of radial spoke stalk (Figure 5).
Figure 5.
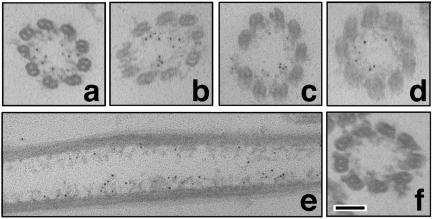
Immunoelectron microscopy showing the localization of CiAxHsp40 in the Ciona axonemes after 0.6 M KCl treatment. Images a-e show immunogold labeling with anti-CiAxHsp40 antibody. Image f shows control (secondary antibody alone). The gold particles are observed at the distal part of the radial spokes. Bar, 100 nm.
The distance from the base of radial spoke to the gold particle was 27.6 ± 7.0 nm (n = 42). We previously estimated the full length of radial spoke (33.6 ± 2.6 nm), the distance from the base of radial spoke to the location of a spoke stalk protein CiRSP3 (14.9 ± 4.8 nm) and that to the location of a spoke head protein LRR37 (25.9 ± 4.3 nm) (Padma et al., 2003). The CiRSP3 or LRR37 is suggested to be located at the base of spoke stalk or the spoke head, respectively. Thus, CiAxHsp40 seems to be located in spoke head or at the vicinity of spoke head.
Isolation of a complex containing both RSP3 and CiAxHsp40
We have biochemically isolated a complex containing CiRSP3 by selective extraction of the axoneme, followed by column chromatographies. The CiAxHsp40 was solubilized by 0.5 M KI treatment from the KCl-treated axonemes (Figure 4B). Anion-exchange column chromatography of the KI extract by using Poros HQ column yielded a fraction with a sharp peak of the immunoreactivity with anti-CiRSP3 (Figure 6). Subsequent Superose 6 gel filtration of the fraction resulted in the isolation of a complex containing CiRSP3 (Figure 7). The complex contained at least 10 distinctive proteins with the molecular mass from 97 to 28 kDa. Anti-CiAxHsp40 antibody clearly recognized a 37-kDa protein band with similar distribution to that of CiRSP3 (Figure 7), indicating that CiAxHsp40 is included in the same subcomplex of radial spokes.
Figure 6.
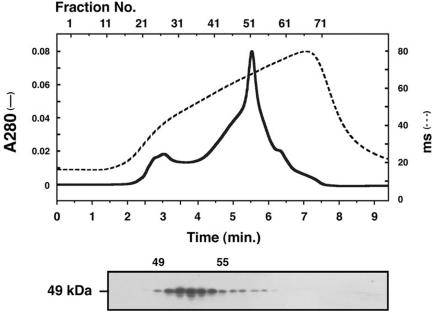
Poros HQ ion exchange column chromatography of the KI extract from KCl-treated axonemes. Proteins were eluted by a programmed NaCl gradient as shown by dotted line. The bottom blot shows the elution profile of CiRSP3, obtained by Western blotting with anti-CiRSP3 antibody.
Figure 7.
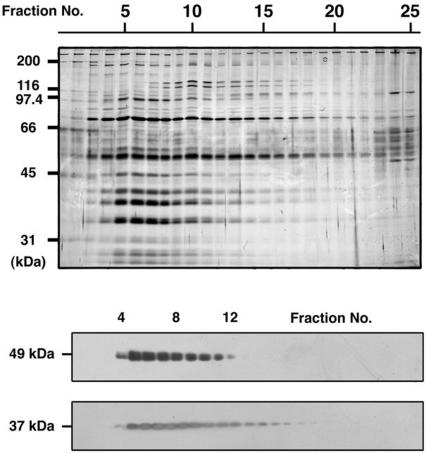
Elution profiles of CiRSP3 and CiAxHsp40 through Superose 6 gel filtration. Top, protein pattern in each fraction, visualized by silver staining. Bottom, elution patterns of CiRSP3 (49 kDa) and CiAxHsp40 (37 kDa), examined by Western blotting.
Identification of the Major Components in the Subcomplex of Radial Spoke
To identify the proteins copurified with CiAxHsp40 by using peptide mass fingerprinting, the fractions (numbers 5-8) from Superose 6 column were pooled, concentrated, and separated by SDS-PAGE. Proteins with molecular masses of 76, 53, 52, 49, 40, 37, and 33 kDa were obviously stained with Coomassie Brilliant Blue (Figure 8). As judged by their wide distribution in most fractions, the 53- and 52-kDa proteins seemed to be α- and β-tubulins. This was confirmed by Western blotting and by peptide mass fingerprinting (our unpublished data). The 49- or 37-kDa protein was clearly recognized by anti-CiRSP3 or anti-CiAxHsp40 antibody, respectively, as shown in Figure 7. The other 76-, 40-, and 33-kDa proteins, along with 37-kDa CiAxHsp40, were subjected to MS analysis for peptide mass fingerprinting.
Figure 8.
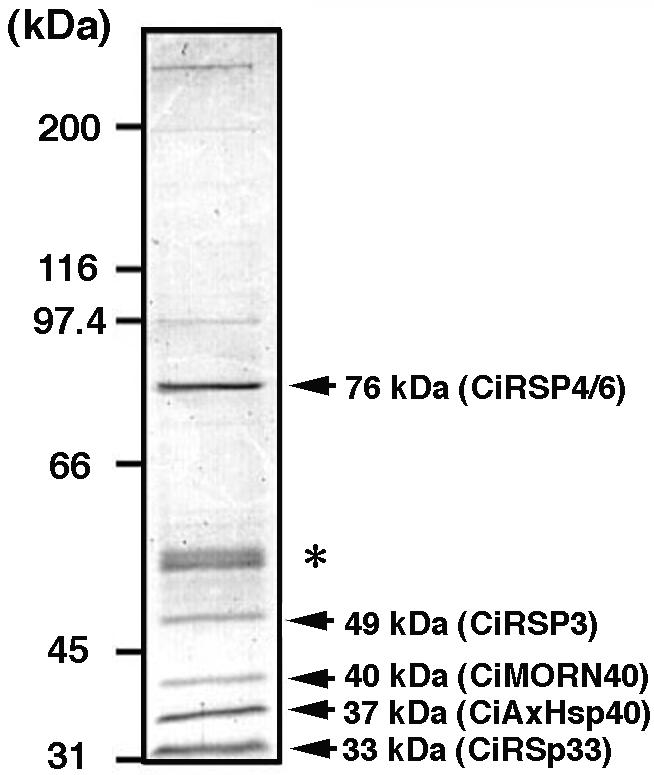
Protein composition of RSP3-containing fraction obtained after Superose 6 gel filtration. The gel was visualized by Coomassie Brilliant Blue. Proteins subjected to peptide mass fingerprinting are shown by arrows with their molecular masses on the right. The asterisk shows the 53- and 52-kDa bands of α- and β-tubulins.
Tryptic fragments derived from each protein was harvested and analyzed by MALDI-TOF/MS. The local MS database of adult testis proteins in C. intestinalis (MSCITS) was mined for each data set, and the clusters showing high hit rates (numbers of matched fragments/submitted fragments) were listed in order. Analysis of all four proteins results in selection one cluster with prominently high hit rates, which could be distinguished from the other listed clusters (Table 1, bold lines). Homology search of the amino acid sequence of the cluster by BLASTP could identify each protein (Table 2). The 76-kDa proteins showed sequence similarity to a spoke head protein in sea urchin (accession no. U73123; Chlamydomonas RSP4/6 homolog) with hit rate of 23/27 (termed CiRSP4/6). The 40-kDa protein turned out to be to a protein with seven MORN repeats (accession no. AL136131) (termed CiMORN40). The 33-kDa proteins showed high homology to human chromosome 6 open reading frame 205 protein (accession no. BC029519), with the hit rate of 15/23 (termed CiRSp33, for 33-kDa radial spoke protein of C. intestinalis). As expected from Western blotting (Figure 7), the 37-kDa proteins showed complete match with CiAxHsp40.
Table 1.
MS search result of radial spoke proteins by using MSCITS/PerMS
| Protein (input) | CLSTR | Match | Score | Score-M | Score/size | pI | Size (Da) |
|---|---|---|---|---|---|---|---|
| 76 kDa (27) | 09453.3.1.0 | 23 | 167595.09 | 7286 | 4.95 | 4.3 | 59708 |
| 09453.1.1.0 | 23 | 167595.09 | 7286 | 4.95 | 4.3 | 59708 | |
| 11200.0.1.0 | 12 | 63809.28 | 5317 | 1.52 | 6.3 | 135039 | |
| 00123.0.1.1 | 11 | 114702.93 | 10427 | 1.05 | 9 | 98445 | |
| 01828.0.1.0 | 11 | 540393.89 | 49126 | 1.32 | 9.1 | 102480 | |
| 40 kDa (13) | 02632.0.1.0 | 13 | 35048.82 | 2696 | 5.32 | 4.4 | 32868 |
| 03032.0.1.0 | 9 | 27575.85 | 3063 | 0.53 | 6.5 | 122090 | |
| 03619.0.1.0 | 8 | 132916.22 | 16614 | 1.02 | 9.7 | 90654 | |
| 08480.0.1.0 | 6 | 7208.75 | 1201 | 1.55 | 9.3 | 64525 | |
| 16217.0.1.0 | 6 | 7994.91 | 1332 | 0.79 | 7.7 | 56838 | |
| 37 kDa (46) | 10996.0.1.0 | 18 | 665626.10 | 36979 | 11.61 | 8.8 | 34864 |
| 12449.0.1.0 | 13 | 26107.69 | 2008 | 1.99 | 9.5 | 51446 | |
| 00555.2.1.2 | 11 | 1016084.84 | 92371 | 2.511 | 8.8 | 51192 | |
| 02454.2.1.2 | 11 | 1016084.84 | 92371 | 2.511 | 8.8 | 51192 | |
| 02454.1.1.2 | 11 | 1016084.84 | 92371 | 2.511 | 8.8 | 51192 | |
| 33 kDa (23) | 08605.0.1.0 | 15 | 153004.70 | 10200 | 3.846 | 5.5 | 31055 |
| 12167.0.1.0 | 12 | 45401.06 | 3783 | 1.009 | 8.4 | 143510 | |
| 00089.3.1.0 | 11 | 28955.05 | 2632 | 0.941 | 8.3 | 143971 | |
| 00089.2.1.0 | 11 | 28955.05 | 2632 | 0.941 | 8.3 | 143971 | |
| 00089.1.1.0 | 11 | 28955.05 | 2632 | 0.941 | 8.3 | 143971 |
Bold letters indicate the clusters (CLSTR) with the significant values of Match, pI, and Size.
Table 2.
Gene descriptions of the clusters (CLSTRs) derived from MS search
| Protein | CLSTR | Best hit gene in BLAST search (DDBJ/GenBank/EMBL) | Accession No. | Probability |
|---|---|---|---|---|
| 76 kDa | 09453 | Strongylocentrotus purpuratus radial spokehead protein | U73123 | 0.0 |
| 40 kDa | 02632 | Mus musculus testis-specific gene A2. | BC049584 | 6e-92 |
| 37 kDa | 10996 | C. intestinalis heat-shock protein 40. | AB079075 | 0.0 |
| 33 kDa | 08605 | Homo sapiens dJ261G23.2.2 (novel protein, isoform 2) protein. | AL136131 | 3e-93 |
DDBJ, DNA Data Bank of Japan; EMBL, European Molecular Biology Laboratory.
Immunoprecipitation Confirmed the Protein MS Analysis and Identified New Components
The Ci-RSP3 was immunoprecipited from 0.5 M KI extract and analyzed by SDS-PAGE, followed by Western blotting with several antibodies (Figure 9). All the proteins with molecular masses of 76, 53, 52, 49, 40, 37, and 33 kDa were identified in the precipitate (Figure 9, left). The 49-kDa CiRSP3 was detected as a faint band relative to the other bands. The α- and β-tubulins also were found in a trace amounts, suggesting that tubulins are associated with this subcomplex in less amounts. The 37- (CiAxHsp40) and 33-kDa (CiRSp33) proteins seemed to be present in high amounts in the subcomplex. A protein with molecular mass of 28 kDa also was immunoprecipitated by anti-CiRSP3 antibody. This protein also comigrated in HQ anion exchange column and seems to be components of the subcomplex as well (Figures 7 and 8).
Figure 9.
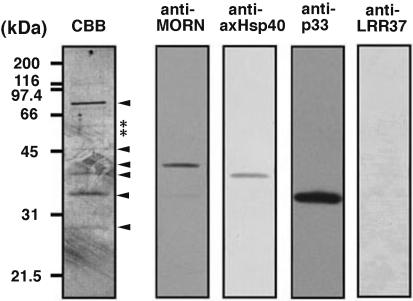
Immunoprecipitation of CiRSP3-containing subcomplex in the KI extract of KCl-treated axonemes. The Coomassie-stained blot on the left shows the protein composition of the precipitate, with the major components indicated by arrowheads. Asterisks show the position of tubulins. Other four blots show the immunoblots of the precipitate with antibodies against CiMORN40, CiAxHsp40, CiRSp33, and LRR37.
The cDNA clone cits044n24 or cits039k01 from cluster 2632 or 8605 was picked up as the representative clone for 40- or 33-kDa protein, respectively. The protein-coding region was amplified by PCR and subcloned into pET32a expression vector. Antibodies were raised against thioredoxin fusion proteins (Figure 9). The anti-MORN or anti-CiRSp33 antibody specifically recognized 40- or 33-kDa axonemal protein, respectively, in purified radial spoke subcomplex (Figure 9), confirming the result obtained from MS analysis (Tables 1 and 2). An antibody to a potential radial spoke head protein LRR37 (Padma et al., 2003) gave no signal in the complex, indicating that LRR37 is not included in the subcomplex.
DISCUSSION
In this study, we have isolated a subcomplex containing a subset of radial spoke components and demonstrated the localization of a member of the HSP40 family, CiAxHsp40, at flagellar radial spokes. Numerous members of Hsp40s have been so far reported and classified into 23 classes in mammals (Ohtsuka and Hata, 2000). Phylogenic tree analysis indicated that it belongs to DjB1/B4 class of Hsp40 (Figure 2). Some classes of Ciona Hsp40 member, for example, ci0100134317 (DjB5/A2), belongs to a class covering multiple classes of mammalian Hsp40 (Figure 1). Large-scale of genomic duplications are suggested to have occurred twice during an evolutionary process to vertebrate. Ascidians, however, have a nonduplicated, basic set of the chordate-type genome (Holland et al., 1994; Sidow, 1996). Ciona genome seems to contain basic complement of vertebrate genome, and the gene duplications during evolution from basic chordates to vertebrates are suggested in many genes (Dehal et al., 2002). In this point of view, some classes of mammalian Hsp40s may be evolved through gene duplication from the ancient Hsp40s.
The expression pattern deduced from Ciona cDNA database showed that CiAxHsp40s are highly testis specific, as well as DjB12 class of Ciona Hsp40 (Figure 2). In mouse, a testis-specific Hsp40, MSJ-1, has been identified and classified into DjB3 (Ohtsuka and Hata, 2000; Berruti and Martegani, 2001). MSJ-1 is highly expressed in elongating spermatid and localized at developing acrosome and postnuclear region (Berruti and Martegani, 2001), which is the feature different from that of CiAxHsp40 (Figure 3). The mouse homolog of Hlj1, a human DjB4 class of Hsp40, is highly expressed in testis, although a certain level of the message is detected in all tissues (Hasegawa et al., 2000). A different size of Hlj1 transcript is specifically found in testis at high level (Hasegawa et al., 2000). Although it is unclear which type of Hlj1 corresponds to CiAxHsp40, it is possible that a specific type of Hsp40 functions in axonemal architecture or movement as a component of radial spoke in human.
The function of CiAxHsp40 has not been elucidated in the present study. Hsp40 generally functions as the cochaperone of Hsp70. Hsp70, as well as Hsp90, have been shown to be widely present in eukaryotic cilia and flagella (Bloch and Johnson, 1995; Stephens and Lemieux, 1995; Stephens, 1997; Williams and Nelsen, 1997). In Chlamydomonas, Hsp70 is concentrated at the tip of flagella, suggesting that it is involved in the assembly of the axonemal components at the tip (Bloch and Johnson, 1995), through intraflagellar transport (for review, see Rosenbaum and Witman, 2002). In mouse sperm, testis-specific Hsp40, Msj1, interacts with testis-specific Hsp70-2 and is colocalized in acrosomal vesicle and postnuclear region (Berruti and Martegani, 2001), suggesting that Msj1 functions as cochaperone of Hsp70-2 in mature sperm. In sea urchin embryo, the level of Hsp40 increases at the regeneration of embryonic cilia (Casano et al., 2003). In light of the localization in the axoneme, the CiAxHsp40 might work as the cochaperone of flagellar Hsp70 in the intraflagellar transport. However, the mechanism for the achievement of two different functions of CiAxHsp40, i.e., cochaperone of Hsp70 and construction of radial spoke, has not been elucidated in this study.
The distribution of CiAxHsp40 in the fractions from chemical dissection of the axonemes is similar to that of stalk protein CiRSP3 (Padma et al., 2003). The potential radial spoke head protein LRR37 is not included in the complex containing both CiRSP3 and CiAxHsp40. Thus, CiAxHsp40 seems to construct a part of radial spoke stalk. However, a part of CiAxHsp40 could be extracted by Tris-EDTA treatment, which solubilizes the radial spoke heads. Furthermore, Western blotting with anti-CiaxHsp40 showed broader distribution to lower molecular range in the gel filtration column chromatography than that of RSP3 (Figure 7). Immunoelectron microscopy suggests that CiAxHsp40 is localized near the spoke head (Figure 5). A single mutation in each component of the radial spoke head results in loss of the whole structure in Chlamydomonas, suggesting that the components of radial spokes are tightly associated each other (Piperno et al., 1981). Together, it is possible that CiAxHsp40 is located at spoke stalk but may be associated near the binding site of spoke head. This idea well agrees with recent hypothesis obtained in Chlamydomonas that Hsp40 (RSP16) forms a dimer and may sit at the junction between spokehead and the thin stalk, along with RSP2 calmodulin-binding protein (Yang, Compton, and Yang, unpublished data).
The Ciona 76-kDa protein (CiRSP4/6) showing sequence similarity to Chlamydomonas RSP4/6 was isolated with CiRSP3 as a potential stalk component. However, both RSP4 and RSP6 are the components of spoke head in Chlamydomonas. The reason for the difference in the localization between homologs in two organisms is unknown, but a different feature in the sequence of 90-kDa RSP4/6 homolog of sea urchin has been demonstrated: the presence of Glu-rich region throughout its sequence instead of more basic regions containing proline-rich sequences in Chlamydomonas RSP4/6. These repeats showed a strong probability of forming α-helical coiled-coil structures that may play important roles in protein-protein interaction (Gingras et al., 1998). Such Glu-rich regions are also present in CiRSP4/6 (amino acid residues 179-213 and 317-413). Search in Ciona genome revealed no other homolog of RSP4/6 in the genome sequence, suggesting that only a single copy of RSP4/6 is present in Ciona, although two relative genes (RSP4 and RSP6) are present in Chlamydomonas. Therefore the homologs of Chlamydomonas RSP4/6 in metazoan sperm axonemes might interact with the other spoke components in a different manner and hence they could be localized in spoke stalks.
Similar discrepancy is present in the sublocalization of LRR37, which has been identified as a radial spoke head component in Ciona (Padma et al., 2003), whereas recent proteomic study implies that it is a radial spoke stalk components in Chlamydomonas (Yang, personal communication). Many of Chlamydomonas radial spoke proteins are known to be highly acidic proteins (Piperno et al., 1981; Yang et al., 2001). This is also the case in those in Ciona sperm: the isoelectric point of CiRSP3, LRR37, CiRSP4/6, CiMORN40, or CiRSp33 is deduced as 5.01, 4.65, 4.10, 4.22, and 5.36, respectively. However, the predicted isoelectric point of CiAxHsp40 is rather basic (pI = 8.91), which was confirmed by two-dimensional gel electrophoresis showing single Hsp40 spot with the isoelectric point at around 9.0 (our unpublished data). The basic property of Hsp40 seems to be unique in Metazoa, because the isoelectric points of DjB1 and DjB4 species in mouse and human are calculated as ranging from 8.7 to 8.9. On the other hand, the isoelectric point of Hsp40 in Chlamydomonas flagella (RSP16) shows acidic (pI = 6.5) (Yang, Compton, and Yang, unpublished data). The electrostatic difference between Hsp40s of Chlamydomonas and metazoan flagella may influence molecular interactions within radial spokes, which may result in the subtle differences in sublocalization of LRR37 and RSP4/6 in radial spokes.
Peptide mass fingerprinting have identified two novel proteins as components of radial spokes, CiMORN40 and CiRSp33. The exact roles of these proteins in the function of radial spokes are unknown. The MORN repeat is found in multiple copies in several proteins, including junctophilins (Takeshima et al., 2000), but the function of this motif is unknown. Considering the protein-membrane interaction, it is possible that CiMORN40 also is involved in the process of intraflagellar transport as discussed above for the function of CiAxHsp40, but the finding of the protein as a component of radial spoke may imply another functions of MORN repeat in protein-protein interaction. CiMORN40 shows sequence similarity to mouse meichroacidin (Tsuchida et al., 1998). Meichroacidin is expressed almost specifically in testis, although trace amount of the protein is detected in ovary in mouse. The protein was localized at the metaphase chromosomes and spindles at both the first and second meiotic divisions, suggesting that it plays important roles in male meiosis (Tsuchida et al., 1998). Considering the common aspects in both male meiosis and the structure of axonemes, the CiMORN40/meichroacidin may play roles in mediating the binding of other proteins to microtubules.
In conclusion, we have isolated a subcomplex of radial spokes, most likely that of spoke stalk, containing RSP3 and Hsp40. The complex contains at least another four proteins, including RSP4/6, MORN repeat protein, and a protein with unknown functions. The analysis of molecular interaction of the radial spoke components in Ciona axonemes, along with a comparative study with Chlamydomonas axonemes, would shed light not only on the function of the Hsp40-mediated molecular chaperone system in the assembly and motility of axonemes but also on the mechanism of signal transduction for the modulation of dynein activity in the radial spokes.
Acknowledgments
We thank the members of Education and Research Center of Marine Bio-Resources, Tohoku University, and Otsuchi Marine Research Center, University of Tokyo, for supplying C. intestinalis and the staff of Electron Microscopy Facility in National Institute for Basic Biology for the support in electron microscopy (National Institute of Basic Biology Cooperative Research Program No. 3-133). Y. S. is a research fellow of the Japan Society for the Promotion of Science. This work was supported in part by grants from Intelligent Cosmos Academic Foundation and NOVARTIS Foundation Japan for the promotion of Science and by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan to K. I. This work also was part of a research project “Development, Differentiation and Regeneration”, Core Research for Evolutional Science and Technology, Japan Science and Technology, to N. S.
Article published online ahead of print in MBC in Press on November 24, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0784).
References
- Berruti, G., and Martegani, E. (2001). MSJ-1, a mouse testis-specific DnaJ protein, is highly expressed in haploid male germ cells and interacts with the testis-specific heatshock protein Hsp70-2. Biol. Reprod. 65, 488-495. [DOI] [PubMed] [Google Scholar]
- Bloch, M. A., and Johnson, K. A. (1995). Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 108, 3541-3545. [DOI] [PubMed] [Google Scholar]
- Casano, C., Gianguzza, F., Roccheri, M. C., Di Giorgi, R., Maenza, L., and Ragusa, M. A. (2003). Hsp40 is involved in cilia regeneration in sea urchin embryos. J. Histochem. Cytochem. 51, 1581-1587. [DOI] [PubMed] [Google Scholar]
- Dehal, P., et al., (2002). The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- Gaillard, A. R., Diener, D. R., Rosenbaum, J. L., and Sale, W. S. (2001). Flagellar radial spoke protein 3 is an A-Kinase anchoring protein (AKAP) J. Cell Biol. 153, 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, I. R. (1981). Cilia and flagella of eukaryotes. J. Cell Biol. 91, 107s-124s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, D., White, D., Garin, J., Cosson, J., Huitorel, P., Cibert, H.Z.C., and Gagnon, C. (1998). Molecular cloning and characterization of a radial spoke head protein of sea urchin sperm axonemes: involvement of the protein in the regulation of sperm motility. Mol. Biol. Cell 9, 513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, T., Xiao, H., Hamajima, F., and Isobe, K. (2000). Interaction between DNA damage protein GADD34 and a new member of the Hsp40 family of heat shock proteins that is induced by a DNA damaging reagent. Biochem. J. 352, 795-800. [PMC free article] [PubMed] [Google Scholar]
- Holland, P.W.H., Garcia-Fernandes, J., William, N. A., and Sidow, A. (1994). Gene duplications and the origins of vertebrate development. Dev. Suppl. 125-133. [PubMed]
- Hozumi, A., Satouh, Y., Ishibe, D., Kaizu, M., Konno, A., Ushimaru, Y., Toda, T., and Inaba, K. (2004). Local database and the search program for proteomic analysis of sperm proteins in the ascidian Ciona intestinalis. Biochem. Biophys. Res. Commun. 319, 1241-1246. [DOI] [PubMed] [Google Scholar]
- Huang, B., Piperno, G., Ramanis, Z., and Luck, D.J.L. (1981). Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88, 80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, K. (2003). Molecular architecture of the sperm flagella: molecules for motility and signalings. Zool. Sci. 20, 1043-1056. [DOI] [PubMed] [Google Scholar]
- Inaba, K., Morisawa, S., and Morisawa, M. (1998). Proteasomes regulate the motility of salmonid fish sperm through modulation of cAMP-dependent phosphorylation of an outer arm dynein light chain. J. Cell Sci. 111, 1105-1115. [DOI] [PubMed] [Google Scholar]
- Inaba, K., Padma, P., Satouh, Y., Shin-i, T., Kohara, Y., Satoh, N., and Satou, Y. (2002). EST analysis of gene expression in testis of the ascidian, Ciona intestinalis. Mol. Reprod. Dev. 62, 431-445. [DOI] [PubMed] [Google Scholar]
- Kamiya, R. (2002). Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219, 115-155. [DOI] [PubMed] [Google Scholar]
- King, S. M. (2000). The dynein microtubule motor. Biochem. Biophys. Acta 1496, 60-75. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 22, 680-685. [DOI] [PubMed] [Google Scholar]
- Lu, Z., and Cyr, D. M. (1998). The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273, 27824-27830. [DOI] [PubMed] [Google Scholar]
- Melville, M. W., Hansen, W. J., Freeman, B. C., Welch, W. J., and Katze, M. G. (1997). Molecular chaperone hsp40 regulates the activity of p58IPK, the cellular inhibitor of PKR. Proc. Natl. Acad. Sci. USA 94, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D. R. (2000). Chlamydomonas flagella. J. Physiol. 36, 261-273. [Google Scholar]
- Ohtsuka, K., and Hata, M. (2000). Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5, 98-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma, P., Hozumi, A., Ogawa, K., and Inaba, K. (2001). Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275, 177-183. [DOI] [PubMed] [Google Scholar]
- Padma, P., Satouh, Y., Wakabayashi, K., Hozumi, A., Ushimaru, Y., Kamiya, R., and Inaba, K. (2003). Identification of a novel leucine-rich repeat protein as a component of flagellar radial spoke in the ascidian Ciona intestinalis Mol. Biol. Cell 14, 774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King, R. S., Gorbatyuk, O., Takebe, S., and King, S. M. (2004). Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell 15, 3891-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Huang, B., Ramanis, Z., and Luck, D. J. (1981). Radial spokes Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88, 73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M. E., and Sale, W. S. (2000). The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151, F37-F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow, J., Voos, W., and Pfanner, N. (1995). Partner proteins determine multiple functions of Hsp70. Trends Cell Biol. 5, 207-212. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, J. L., and Witman, G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3, 813-825. [DOI] [PubMed] [Google Scholar]
- Sidow, A. (1996). Gen(om)e duplications in the evolution of early vertebrates. Curr. Opin. Genet. Dev. 6, 715-722. [DOI] [PubMed] [Google Scholar]
- Smith, E. F., and Lefebvre, P. A. (1997). The role of central apparatus components in flagellar motility and microtubule assembly. Cell Motil. Cytoskeleton 38, 1-8. [DOI] [PubMed] [Google Scholar]
- Stephens, R. E. (1997). Synthesis and turnover of embryonic sea urchin ciliary proteins during selective inhibition of tubulin synthesis and assembly. Mol. Biol. Cell 8, 2187-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, R. E., and Lemieux, N. A. (1999). Molecular chaperones in cilia and flagella: implications for protein turnover. Cell Motil. Cytoskeleton 44, 274-283. [DOI] [PubMed] [Google Scholar]
- Takeshima, H., Komazaki, S., Nishi, M., Iino, M., and Kangawa, K. (2000). Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell 6, 11-22. [DOI] [PubMed] [Google Scholar]
- Toda, T., Kaji, K., and Kimura, N. (1998). TMIG-2DPAGE: a new concept of two-dimensional gel protein database for research on aging. Electrophoresis 19, 344-348. [DOI] [PubMed] [Google Scholar]
- Tsuchida, J., Yukio Nishina, Y., Wakabayashi, N., Nozaki, M., Sakai, Y., and Nishimune, Y. (1998). Molecular cloning and characterization of meichroacidin (male meiotic metaphase chromosome-associated acidic protein). Dev. Biol. 197, 67-76. [DOI] [PubMed] [Google Scholar]
- Yang, P., Diener, D. R., Rosenbaum, J. L., and Sale, W. S. (2001). Localization of calmodulin and dynein light chain LC8 in flagellar radial spoke J. Cell Biol. 153, 1315-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Yang, C., and Sale, W. S. (2004). Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot Cell 3, 72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, N. E., and Nelsen, E. M. (1997). HSP70 and HSP90 homologs are associated with tubulin in hetero-oligomeric complexes, cilia and the cortex of Tetrahymena. J. Cell Sci. 110, 1665-1672. [DOI] [PubMed] [Google Scholar]


