Abstract
The radial spoke is a stable structural complex in the 9 + 2 axoneme for the control of flagellar motility. However, the spokes in Chlamydomonas mutant pf24 are heterogeneous and unstable, whereas several spoke proteins are reduced differentially. To elucidate the defective mechanism, we clone RSP16, a prominent spoke protein diminished in pf24 axonemes. Unexpectedly, RSP16 is a novel HSP40 member of the DnaJ superfamily that assists chaperones in various protein-folding-related processes. Importantly, RSP16 is uniquely excluded from the 12S spoke precursor complex that is packaged in the cell body and transported toward the flagellar tip to be converted into mature 20S axonemal spokes. Rather, RSP16, transported separately, joins the precursor complex in flagella. Furthermore, RSP16 molecules in vitro and in flagella form homodimers, a characteristic required for the cochaperone activity of HSP40. We postulate that the spoke HSP40 operates as a cochaperone to assist chaperone machinery at the flagellar tip to actively convert the smaller spoke precursor and itself into the mature stable complex; failure of the interaction between the spoke HSP40 and its target polypeptide results in heterogeneous unstable radial spokes in pf24.
INTRODUCTION
The radial spoke is a conserved macromolecular complex required for regulation of bending in motile 9 + 2 cilia and flagella (Smith and Yang, 2004). The T-shaped structure anchors to the nine outer doublets with a thin stalk, while its bulbous head contacts central pair apparatus periodically during the oscillatory beating (Warner and Satir, 1974; Goodenough and Heuser, 1985). It is hypothesized that this intermittent interaction with central pair apparatus enables the radial spokes to distribute “signals,” originating from central pair apparatus, to subsets of outer doublets for localized control of dynein-driven microtubule sliding (Huang et al., 1982).
Consistent with this “distributor” model, the central apparatus is asymmetric in structure (Dutcher et al., 1984; Goodenough and Heuser, 1985; Mitchell and Sale, 1999; Mitchell, 2003; Wargo and Smith, 2003; reviewed in Smith and Yang, 2004) and in some organisms central pair rotates once per beat cycle (reviewed by Omoto et al., 1999). The radial spoke is thought to be relatively rigid to endure physical force arising from the interaction with central pair (Warner and Satir, 1974; Goodenough and Heuser, 1985; Lindemann, 2003; Smith and Yang, 2004; Yang et al., 2004) and is extracted as a stable 20S complex of 23 polypeptides that resists disassembly even in buffers containing 0.6 M KI (Yang et al., 2004; Patel-King et al., 2004). In addition to the mechanical properties, the radial spokes also contain proteins thought to be involved in chemical signaling through calcium or nucleotide binding (Patel-King et al., 2004; Yang et al., 2001, 2004). Based on these data and other studies, it has been postulated that the radial spoke operates as both mechano- and chemo-transducers (Yang et al., 2004; reviewed by Smith and Yang, 2004).
Among the most important issues is the assembly mechanism for the proposed mechanochemical signal transducers. It has been demonstrated that the proteins for radial spoke and other axonemal complexes are packaged into precursor complexes in the cell body (Fowkes and Mitchell, 1998; Qin et al., 2004). For example, the spoke precursors, found in both cell body and flagellar membrane matrix fraction (M+M) are 12S particles (Qin et al., 2004). The precursors become engaged and are transported by the intraflagellar transport (IFT) system to the flagellar tip for the final assembly (Rosenbaum and Child, 1967; Witman, 1975; Johnson and Rosenbaum, 1992; Piperno et al., 1996; Qin et al., 2004). Although much has been learned about the IFT system, anterograde and retrograde movement of axonemal protein complexes, turnover of axonemes, and the control of assembly and disassembly (Stephens, 1997; Marshall and Rosenbaum, 2001; Song and Dentler, 2001; Berman et al., 2003; Tam et al., 2003; Iomini et al., 2004; Pan et al., 2004; Qin et al., 2004; reviewed by Rosenbaum and Witman, 2002; Cole, 2003; Snell et al., 2004), little is known about how cargo precursors are assembled into the axoneme at the distal tip of flagella.
Notably, the 12S spoke precursors are much smaller than the 20S mature spokes in chaotropic solution (Yang et al., 2001) or from axoneme turnover (Qin et al., 2004). The difference in sizes indicates that the precursor complex is not the furbished product ready to be incorporated into the axoneme as a single module. Furthermore, the stability of mature spokes that do not disintegrate back to 12S or individual components suggests that the precursor is altered significantly during the final assembly process, possibly involving HSP70 chaperones enriched at the flagellar tips (Bloch and Johnson, 1995).
Several Chlamydomonas mutants are defective in the assembly of radial spokes. They either lack the entire spoke head (pf1 and pf17) or spoke (pf14), possibly because of failures in the assembly of precursors or anchoring the complex to the axonemes (Diener et al., 1993). In contrast, pf24 contains all of the spoke proteins but the spokes are heterogeneous, appearing either intact, headless, truncated, or completely absent (Huang et al., 1981). An ATA mutation at the initiation codon of RSP2 gene in pf24 impairs translation (Yang et al., 2004). As a consequence, pf24 axonemes contain diminished RSP2 and two other spoke stalk proteins, RSP23 (Patel-King et al., 2004) and RSP16, while spoke head proteins are reduced (Huang et al., 1981). This molecular phenotype suggests that the three stalk proteins, RSP2, 16, and 23, are closely associated underneath spoke head for connecting the stalk and head domains of the radial spoke (Huang et al., 1981; Piperno et al., 1981; Curry and Rosenbaum, 1993). However, earlier characterization of RSP2 and 23 as the calmodulin-binding proteins (Patel-King et al., 2004; Yang et al., 2004) has not yet provided insight into the assembly of the heterogeneous spoke structures.
Here we characterize the defective spokes of pf24 in detail and provide evidence suggesting that the assembly defect is also related to RSP16, the third spoke protein diminished in pf24 axonemes. RSP16, a novel HSP40, is transported separately from spoke precursors in flagella and may function as a cochaperone in converting the 12S spoke precursor into the 20S structural complex at the flagellar tip. This finding brings new light toward the assembly and configuration of radial spokes as well as the cargoes of the intraflagellar transport, the enriched HSP70 chaperone at the flagellar tip and the polarity of flagellar assembly.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas wild-type strain (cc125) and the defined radial spoke mutants including pf14, pf17, and pf24 (Huang et al., 1981) were provided by Dr. E. H. Harris (Duke University, Chlamydomonas Genetics Center). The pf28pf30 strain lacks both the 20S outer arm dynein and inner arm dynein I1 as described previously (Piperno et al., 1990) and was used for the experiments requiring radial spoke purification. All cells were grown in liquid modified medium I with aeration over a 14/10 light/dark cycle (Witman, 1986).
Fractionation of Axonemes and Flagellar Membrane Matrix
Preparation of flagella and axonemes, KI extraction of axonemes and velocity sedimentation of radial spoke complexes on sucrose gradients were carried out as described before (Yang et al., 2001). To purify RSP16 protein for mass spectrometry, the peak spoke fractions from sucrose gradient of pf28pf30 KI extract were concentrated using Nanosep 300 OMEGA centrifugal device (Pall Co., Glen Cove, NY).
To obtain extract from flagellar membrane and matrix compartment (M+M), ∼20-mg flagella were treated with 0.7 ml 0.025% NP-40 in buffer A (10 mM HEPES, 5 mM MgSO4, 1 mM dithiothreitol, 0.5 mM EDTA, 30 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and 0.5 TIU/ml aprotinin, pH 7.4) followed by 12,500 × g centrifugation for 15 min. For fractionation of protein complexes, the M+M supernatant fractions, typically containing ∼5 mg/ml protein, were subjected to velocity sedimentation in 5-20% sucrose gradient at 36K rpm (SW41 rotor, Beckman Coulter, Fullerton, CA) for 16 h.
Blue-Native Polyacrylamide Gel Electrophoresis
Blue-native polyacrylamide gel electrophoresis (BN-PAGE), developed to fractionate mitochondrial ATPase complex, was carried out as described (Schagger and von Jagow, 1991) with the following modifications. Native gels of 5-10 and 13% were made using an acrylamide mixture of 30% polyacrylamide and 0.2% bis-acrylamide. For sample preparation, KI extract or M+M extract were mixed with 10× sample buffer (5% Coomassie Blue in 500 mM amino capronic acid). Electrophoresis was performed using “blue cathode buffer” for the first 50 min at 50 V with the mini-gel electrophoresis system (Bio-Rad Laboratories, Richmond, CA) and then the buffer was exchanged into cathode buffer for another 4-h electrophoresis at 100 V. For further separation in SDS-PAGE, the lane containing the sample separated in blue native gel was excised and then equilibrated in 5× SDS-PAGE sample buffer for 30 min at room temperature. Subsequently, the denatured gel strip was inserted into the sample well of a preparatory mini-gel, and standard SDS-PAGE was performed.
Two-dimensional Electrophoresis
The electrophoresis of first dimension nonequilibrium electrophoresis followed by SDS-PAGE was carried out as previously described (Yang et al., 2001). The proteins spots in the second dimension SDS-PAGE gel was visualized using Sliver Stain Plus kit (Bio-Rad Laboratories) and the results were recorded using a digital camera. The RSP16 spot, identified based on its pI and molecular weight, was excised and subjected for trypsin digestion and MALDI-TOF mass spectrometry (Dr. John Leszyk, Proteomic Mass Spectrometry Laboratory, University of Massachusetts) to obtain peptide sequences for cloning.
Fast-performance Liquid Chromatography Gel Filtration
An AKTA fast-performance liquid chromatography (FPLC) system equipped with a Superose 6 HR 10/30 chromatography column (Amersham Pharmacia Biotech) was used to fractionate M+M. The column with an optimal separation range of 5K-5000K Mr, was calibrated with marker proteins (Sigma-Aldrich, St. Louis, MO) including bovine serum albumin (BSA, 66 kDa); alcohol dehydrogenase (150 kDa), and thyroglobulin (669 kDa). An aliquot of 200 μl M+M (protein concentration 5-10 mg/ml) was loaded onto the column followed by the elution with buffer A at a flow rate of 0.1 ml/min. The eluent was collected into 0.25-ml fractions following 0.2× column volume (Vc) of flow through. The fractions were concentrated 10-fold by lyophilization and then treated with SDS-PAGE sample buffer for Western analysis.
Molecular Cloning and Protein Expression
The primers used for RT-PCR and cloning RSP16 included the following: 1: cggcgagctttgggcgattggta; 2: agctcaggctgacggcatagccg; 3: accatggcgcggggcttggactattacg; 4: gggaattcctctaccgcttgaacacggg; and 5: gggaattcctataccgcttgaacacggg.
The primer pair, 1 and 2, was used to amplify the entire RSP16 message by RT-PCR. The 1.6-kb product was cloned into pGEM-T Easy vector (Promega, Madison, WI). The construct, plasmid A, was sequenced and used as a template for PCR amplification of 1-kb coding sequence. The PCR products were used to create expression constructs for untagged and 6His-tagged RSP16. For expression of untagged RSP16, the primer pair, 3 and 4, with NcoI and EcoRI restriction sites for cloning purposes, was used and the PCR product was cloned into pGEM-T Easy first to generate plasmid B. To engineer a construct for the 3′ 6His-tagged protein, plasmid C was created similarly but 5 primer without the stop codon (compare the bold codons) in 4 was used to pair with 3 primer. The inserts of RSP16 coding sequence in plasmids B and C were purified and ligated into the respective restriction sites in the pET28(a) expression vector (Novagen, Madison, WI), which carry kanamycin-resistant gene. To engineer the 5′ GST-tagged construct, the insert was released from plasmid C by sequential treatment with NcoI digestion, a Klenow filling-in reaction and then with NotI digestion. The pGEX-4T-1 vector (Amersham Pharmacia Biotech) was prepared by SmaI and NotI digestion. The resulting construction, pGEX-RSP16 carries the Amp-resistant gene and expresses the N-terminal-tagged GST-RSP16.
For protein expression, the expression constructs were transformed into BL21(DE3) bacterial strain individually or in pair under double antibiotic selections. Recombinant proteins were induced with 1 mM IPTG at room temperature for 4 h. Bacteria were harvested and the extract was subjected to Ni-NTA (Qiagen, Chatsworth, CA) affinity purification under natural condition as described by the manufacturer.
Sequence analyses were carried out using the programs accessible through the Internet, including Blastn, Blastp, Multiple Sequence Alignment, ClustalW and MEGA2.
Chemical Cross-linking
Chemical cross-linking of axoneme suspension, axonemal KI extract and M+M with EDC (1-ethyl-3-(3-dimerthylaminoporopyl) carbodiimide HCl, Pierce Chemical, Rockford, IL) was carried out as previously described (Yang et al., 2001). The cross-linked samples were fixed by the appropriate sample buffer for either two-dimensional (2D) gel or SDS-PAGE.
Antibodies
The rabbit anti-IC140 and anti-RSP1, anti-RSP2 and anti-RSP3 have been described previously (Yang and Sale, 1998; Yang et al., 2001; Qin et al., 2004). Ni-affinity-purified 6His-tagged recombinant proteins expressed in bacterial strain BL21 (DE3) were used as the immunogen for raising anti-RSP16 and anti-RSP11 serum.
To raise spoke complex antibodies, the entire spoke complex in the 20S spoke fractions that was dropped frozen in liquid nitrogen was used for immunogen preparation. Mature Leghorn laying hens were initially immunized with 400 μg thawed 20S spokes that was solubilized in 500 μl Freund's complete adjuvant. This mixture was administered to the laying hens via an intramuscular injection into the breast musculature. Two and four weeks after the initial immunization, similar booster immunizations were performed with the exception that Freund's incomplete adjuvant was used. The IgY fraction from egg yolks of pre- and postimmunized hens was isolated using the polyethylene glycol (PEG) precipitation method (Polson et al., 1980). Briefly, egg yolks were diluted 4:1 with a Tris/NaCl buffer (10 mM Tris, pH 7.4, 100 mM NaCl), and nonimmunoglobulin yolk proteins were precipitated by the addition of 3.5% PEG 8000 and centrifugation at 14,000 × g for 10 min at 4°C. The supernatant fraction was collected and the immunoglobulin (Ig) fraction was precipitated by the addition of 9% PEG 8000. The Ig containing pellet was solublized in Tris/NaCl buffer and the 3.5 and 9% PEG 8000 precipitation procedures were repeated. This procedure typically yields ∼50 mg of IgY per egg yolk that is 80-90% pure via SDS-PAGE analysis.
The monoclonal antibody, 3a3, for HSP70 (Affinity BioReagents, Golden, CO), was used as described (Bloch and Johnson, 1995). Tubulin was identified by monoclonal anti-α-tubulin B-5-1-2 (Sigma-Aldrich).
RESULTS
Altered Radial Spoke Assembly and Stability in pf24 Axonemes
Electron microscopy and 35S labeling revealed heterogeneous radial spokes and defective spoke proteins in pf24 (Huang et al., 1981), suggesting that RSP2, encoded by PF24, attach spoke head to stalk (Huang et al., 1981; Curry and Rosenbaum, 1993). To further evaluate the extent of defect, Western analysis with the available antibodies (Yang et al., 2001; Qin et al., 2004) was first carried out to compare radial spoke head protein RSP1 and stalk proteins, RSP2 and 3 in the axonemes of pf24, the wild-type cells, dynein mutant pf28pf30, headless spoke mutant pf17, and spokeless pf14. Compared with the axonemes containing wild-type spoke stalks (first three lanes in Figure 1), RSP1 and 2 in pf24 was reduced (Figure 1, arrowhead) significantly, but interestingly the reduction of RSP1 was less dramatic. This result clarified the “diminished” or “reduced” spoke proteins in pf24 (Huang et al., 1981) and indicated more head proteins than the stalk connector proteins in pf24 axonemes. Contrarily, RSP3, which mediated the binding of radial spokes to outer doublets (Diener et al., 1993), appeared normal in pf24.
Figure 1.

Western blot revealed that only the proteins located at the head end of pf24 spokes were defective. Axonemes were prepared from wild-type cell, dynein mutant pf28pf30, and spoke mutants, pf17, pf24, and pf14. RSP1, 2, and 3 were barely detectable in the spokeless mutant, pf14. RSP1, a head protein, was absent in headless pf17. RSP2, a stalk protein underneath spoke head, was normal in pf17 but was dramatically reduced in pf24, which contained less RSP1 but the wild-type level of RSP3.
To evaluate the partially assembled spokes in pf24 and their stability, Western analysis was carried out to compare spoke particles extracted from the axonemes of pf24, WT and headless pf17, with KI buffer and fractionated in 10-25% sucrose gradients (Figure 2). A chicken antibody was raised against the entire spoke particles in 20S fractions. The IgY recognized several larger spoke proteins, including RSP1, 2, 3, 5, and 7, and tubulin (T), cosedimenting with RSP11, recognized by specific rabbit antibody, in the monodispersed 20S wild-type spokes and the 15S spoke stalk from the headless pf17 (Figure 2A, top 2 panels; Yang et al., 2001). The result was supported by Western blot of 2D gel and using the rabbit antibodies specific to RSP1, 2, or 3 (unpublished data). However, in pf24 extract, spoke proteins no longer cosediment. RSP3, 7, and 11 sediment into two poorly resolved ∼12-14S peaks (Figure 2A, bottom, arrowheads) and the reduced RSP1, 2, and 5 primarily sediment into a 16S peak and a ∼6S peak (Figure 2A, bottom, arrows). The prominent 6S fractions supported the prediction that the stalk proteins, RSP2 and 5, were located toward the head end of stalk (Piperno et al., 1981; Curry and Rosenbaum, 1993) and indicated the head end of pf24 spokes dissociated significantly upon KI extraction.
Figure 2.
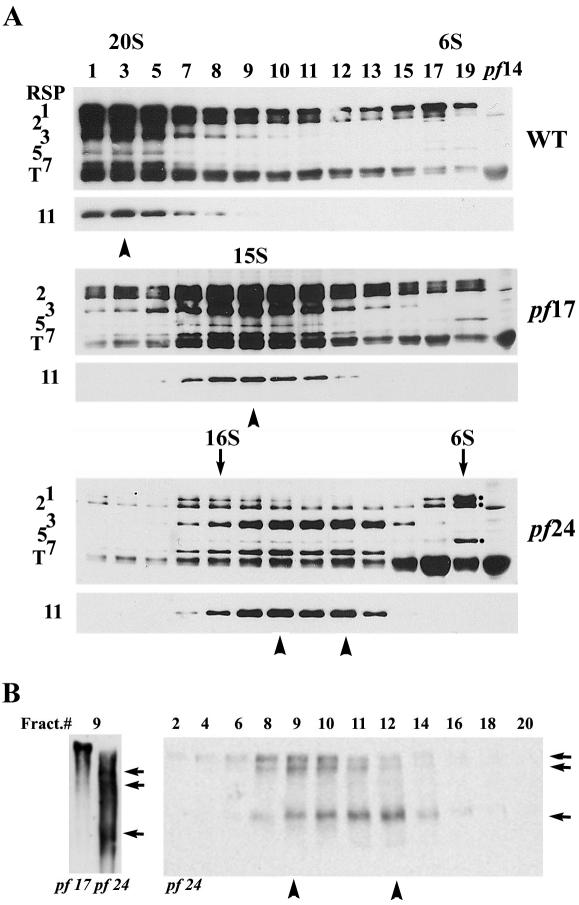
The head end of the defective pf24 spokes further dissociated during KI extraction and the extracted spoke particles were different in sizes. (A) Western analyses of 10-25% sucrose gradient fractionations of axonemal KI extract were probed with anti-RSP11 and antispoke IgY that recognized RSP1, 2, 3, 5, and 7 and tubulin (T). Extracted spokes from wild-type and pf17 sediment into single 20 and 15S peaks, respectively. Note that RSP3, 7, and 11 from pf24 (arrowheads) sediment into two poorly resolved ∼12-14S peaks, whereas RSP1, 2, and 5 were found in 16S and 6S peaks (arrows). The 6S fraction is much more prominent. RSP1 and 2 in pf24 are much reduced compared with RSP3. (B) Electrophoresis of 5-10% blue native gel and RSP3 Western analysis were used to reveal the three smaller spoke particles in 9 fractions of pf24 (arrows, left) compared with the single and larger stalk in the equivalent fraction from pf17. The pf24 spoke particles of at least three sizes sediment differentially in the entire sucrose gradient (right).
The different sedimentation pattern among spoke proteins (Figure 2A, bottom, compare arrow and arrowhead) suggested that pf24 spokes were extracted as particles with various degrees of defects. To test this independently, sucrose gradients fractions were resolved in 5-10% native gels, and spoke particles were revealed by Western analysis using the RSP3 antibody (Figure 2B). Compared with the monodispersed particles from wild-type (unpublished data, too large to enter the gel) and pf17, pf24 spoke particles in the same fraction number (9) appeared as three smaller particles (Figure 2B, left, arrows) even in the presence of spoke head proteins (Figure 2A, bottom, left arrow). Native gel analysis of the entire gradient further confirmed that at least three distinct pf24 spoke particles (Figure 2B, right panel, arrowheads) sediment differentially in sucrose gradient.
Together, the evidence showed that 1) the radial spokes in pf24 were heterogeneous (Huang et al., 1981); 2) the proteins in the head end of pf24 spokes were assembled differentially and underwent dissociation upon extraction, whereas the base of pf24 spoke stalk was not obviously affected; 3) RSP7 and 11 were located near RSP3, the basal end of the stalk, whereas RSP5 was located toward the head end of the stalk (Piperno et al., 1981); and 4) the higher ratio of head proteins to the stalk proteins, RSP2, 23, and 16 (Huang et al., 1981; Patel-King et al., 2004; Yang et al., 2004) suggested that these stalk proteins are not required to link head proteins to the base of stalk in a strict stoichiometric manner and may play additional roles other than as connectors of head and stalk of the radial spoke.
RSP16 Is a Type II DnaJ HSP40 Homologue
To assess the role of RSP16, the third stalk protein that was reduced in pf24 axonemes, we cloned RSP16 gene by first purifying the polypeptides. The 20S intact radial spokes were isolated as described (Yang et al., 2001). The spoke proteins in the 20S particles were then separated by 2D SDS-PAGE. RSP16 (Figure 3A, arrowhead) is abundant and particularly distinctive because it is the most basic spoke protein (pI ∼ 6.4; Piperno et al., 1981; Yang et al., 2001, see Figures 2A and 5A). The RSP16 spot was excised for MALDI-TOF mass spectrometry to obtain the MS/MS data for Mascot analysis.
Figure 3.

Cloning of RSP16. (A) Purification of RSP16 polypeptide. The 20S fractions of pf28pf30 KI extract were separated by 10% 2D gel. The acidic end is at the right. Individual radial spoke proteins, revealed by silver staining were designated in numbers. RSP16 (arrowhead), the most basic spoke protein, was excised for mass spectrometry. (B) Cloning Strategy. Mass spectrometry of the purified RSP16 resulted in a single peptide sequence (black bar). The EST that encoded the peptide and contained the adjacent cDNA sequences (gray line) were identified by Mascot analysis and Blastn search. A primer pair (arrows) was used to recover the cDNA including the entire coding sequence and UTR by RT-PCR. Further Blastn search the Chlamydomonas genome database revealed the coding sequences (gray bars) disrupted by six introns and untranslated regions (open bar) of RSP16 gene. (C) Blastp search with the predicted RSP16 polypeptide revealed molecular domains, including DnaJ-J and DnaJ-C domains unique for HSP40.
Figure 5.

The antibody raised against the recombinant HSP40 recognized a 40-kDa spoke protein. (A) Coomassie-stained SDS-PAGE showed the purification of the 6His-tagged novel HSP40 by Ni-NTA chromatography. Comparing with control (-), a 40-kDa protein was in the IPTG induced supernatant (Pre), depleted after the Ni-NTA chromatography (Post), and was eluted by imidazole buffer (Elute). HSP40 Western analysis revealed a 40-kDa proteins in axonemes (B), cell body extract (C), and sucrose gradients of wild-type extract (D). The 40-kDa spoke protein, present in the axonemes of wild-type cells, dynein mutant pf28pf30, and headless spoke mutant pf17, was absent and diminished, respectively, in the axonemes of spoke-less pf14 and pf24 (empty and gray arrowheads), although the 40-kDa protein was synthesized in cell body of pf24, which was defective in RSP2 gene and had little RSP2 (C). IC140 and an unknown cell body protein (CBP) demonstrated equal loading of samples. Furthermore, the novel HSP40 cosediment with RSP2, 3, and 11 in the 20S spoke particles (D).
Sequence analysis revealed that several Chlamydomonas EST clones (e.g., BI721962) matched one tryptic peptide (GLDYYEVMGLTR, 1416.60 Da, Figure 3B, black bar) and contained the 5′ untranslated region (UTR). Blast search (Blastn) with the 5′-end EST clones identified the overlapping 3′ EST clones. All of the sequences were subsequently localized in the predicted gene designated as C_490039 listed in the Chlamydomonas genome database adjacent to CNC53 molecular marker and GSK3 gene in linkage group XII/XIII (http://www.biology.duke.edu/chlamy_genome/BAC/LG12_13.html). The result was confirmed by screening a BAC library with a PCR product (unpublished result by N. Haas and Dr. C. D. Silflow, University of Minnesota). No motility mutants were mapped near this locus. The compiled EST sequences included both 5′ and 3′ UTR and an open reading frame (ORF) encoding a 39-kDa protein of 346 a.a. and a calculated pI 6.5, as anticipated for RSP16 (Figure 3A). RT-PCR resulted in the predicted 1.6-kb product using the primer pair, 1 and 2, located, respectively, near the 5′ and 3′ ends of the predicted cDNA (Figure 3B). Comparison of cDNA and genomic sequence from database revealed that the predicted gene consisted of seven exons and six introns (Accession No. AY805324).
Protein Blast search (Blastp) showed that the predicted protein contained two domains, DnaJ-J and DnaJ-C (Figure 3C), characteristics of HSP40 molecules within the DnaJ superfamily. Furthermore, the protein was homologous to numerous HSP40 molecules (E value 10-40 and less), many of which are from the same species. Specifically, RSP16 belongs to type II DnaJ, which contains a G/F-rich region between DnaJ and DnaJ-C but lacked the 4 zinc-finger repeats (Cheetham and Caplan, 1998). The sequences and molecular domains of RSP16 were listed in separate report describing the proteomic studies of spoke complex (unpublished results).
The DnaJ molecules from Ciona (BAB85846), mouse (AAH48501), and human (AAN15925 and AAH19827) share the highest similarity with RSP16. Multiple Sequence Alignment (Figure 4A) was carried out to compare these homologues as well as a DnaJ homologue found in the human cilia proteomic project, MRJ (O75190; Ostrowske et al., 2002) and the DnaJ from Ciona sperm proteomic project (BAB85846, Padma et al., 2003). The most homologous region among these DnaJ molecules was the N-terminal 75-a.a. DnaJ-J domain, which stimulated the ATPase activity of DnaK HSP70 (Wall et al., 1994; reviewed by Fink, 1999). Notably, mouse AAH48501 and human AAN15925 were annotated as testis spermatogenesis apoptosis-related proteins, but the cDNA sequences were present in the EST database for ciliated cells (NCBI database). A dendrogram (Figure 4B) revealed that these two genes (AAH48501 and AAN15925) as well as the homologue in Ciona sperm flagella were most closely related to RSP16 and, hence, are likely to be the orthologues of RSP16. On the other hand, the EST of human AAH19827 and MRJ were expressed in cell types lacking 9 + 2 cilia and flagella as well. Thus, the latter two homologues may have broader roles than spoke HSP40. These findings suggested that radial spoke HSP40 is not unique to Chlamydomonas flagella and is not the sole DnaJ molecule in these organelles.
Figure 4.

Multiple sequence alignment (A) demonstrated the homology between Chlamydomonas (C.R.) RSP16 and its most homologous HSP40 molecules in human (H.S.), mouse (M.M.), and Ciona intestinalis (C.I.) as well as a DnaJ (MRJ, O75190) found in human cilia proteomic project. The identical residues were shaded in black. Those sharing similar property were shaded in gray. The alignment was generated by ClustalW and ShadyBox programs. The similarity among the DnaJ molecules were demonstrated by a dendrogram (B), plotted by MEGA2.
Defective HSP40 in Spoke Mutants
The ultimate proof that the novel HSP40 was the constitutive RSP16 in the spoke complex was to raise antibodies against the recombinant HSP40. This antibody should recognize the 40-kDa RSP16 in the wild-type axonemes and 20S isolated radial spokes but was defective in the spoke stalk mutants.
To prepare the HSP40 immunogen, PCR of the entire coding sequence of the novel HSP40 was carried out using the RT-PCR clone (see Figure 3B, RT-PCR primers) as a template. The resulting 1-kb PCR product was cloned into the 6His-tag expression vector, pET28(a) and the construct was transformed into BL21(DE3) cells to express a 3′ 6His-tagged recombinant HSP40. The expression of a 40-kDa recombinant protein was induced by IPTG (compare first two lanes in Figure 5A). The soluble protein was subsequently purified by Ni-NTA chromatography and was used to raise rabbit polyclonal antibodies and for further studies described below.
Western analysis using the anti-HSP40 antibody (Figure 5B) was carried out to compare axonemes from spoke mutants with wild-type and dynein mutant, pf28pf30. Antibodies to inner arm IC140 and RSP3 demonstrated normalized sample loading and revealed spoke particles, respectively (Figure 5B). A 40-kDa protein recognized by the HSP40 antibody was present in wild-type, pf28pf30, and spoke mutant pf17 (Figure 5B). All of these strains had spoke stalk and RSP16. In contrast, this 40-kDa protein was diminished in pf24 and was not detectable in the spokeless pf14 (gray and empty arrowheads, respectively, Figure 5B). Importantly, Western analysis of cell body extract showed that RSP2 was undetectable in pf24 but RSP16 in pf24 and wild-type cells (Figure 5C) was identical. The result indicated that RSP16 failed to assemble into pf24 radial spokes because of the diminished RSP2. To further confirm that HSP40 is an integral spoke component, Western analysis of extracted wild-type spokes sedimenting on a 10-25% sucrose gradient was carried out. Again, the 40-kDa molecule cosediment with 20S radial spoke, represented by RSP2, 3, and 11 (Figure 5D). Together, the evidence conclusively indicated that RSP16 is a novel HSP40.
The Spoke HSP40 Was Excluded from the Radial Spoke Precursors
On the basis of the molecular domains, assembly defect of spokes and reduction of RSP16 in pf24 axonemes, we postulated that RSP16 indeed operated as a cochaperone for the assembly of radial spokes. However, RSP16 was a constitutive component in spokes, whereas many DnaJ or HSP40 cochaperones interacted dynamically with their polypeptide substrates for refolding, disassembly, or assembly (reviewed by Bukau and Horwich, 1998). We predicted that the recently discovered 12S radial spoke precursors (Qin et al., 2004) in soluble flagellar membrane matrix (M+M) was the substrate of RSP16 and thus RSP16 was not present in the 12S particle.
To test this, M+M of pf28pf30 was fractionated with a 5-20% sucrose gradient, and the spoke proteins were revealed by Western blots. A sample of purified 20S axonemal spokes was included as a control. RSP2 and 3 (Qin et al., 2004) and RSP11 (Figure 6A, large and small arrows) cosediment in the 20S mature spokes from axoneme turnover as well as the 12S spoke precursors. In contrast, while RSP16 was present in the mature 20S spokes, it was absent in the 12S precursors. Importantly, a peak of RSP16 was found in ∼6S (arrowhead, Figure 6A), suggesting that RSP16 at preassembled state was transported separately as a smaller particle or dissociated from 12S precursors. If the latter were true, the 6S peak in pf24 M+M would be reduced as RSP2 because of inefficient translation of RSP2 (Yang et al., 2004). However, if RSP16 were transported independently, the 6S fraction of RSP16 from pf24 M+M would not be affected.
Figure 6.

Western analysis showed that RSP16 in M+M was excluded from the major radial spoke precursors. (A) In sucrose gradient sedimentation of pf28pf30 M+M, RSP2, 3, and 11 were present in 20S mature spokes from axonemal breakdown (large arrow), 12S spoke precursor (small arrow), and control, and the 20S extracted axonemal spokes (last lane). RSP16 was in 20S but absent in the 12S precursor. Notably, a prominent fraction of RSP16 was present in 6S fractions (arrowhead). (B) The smaller defective breakdown product (large arrow) and precursor (small arrow) in the gradient of pf24 M+M were revealed by RSP3 and RSP11 Western analyses. No RSP2 and 16 were detected in either peak. Importantly, the ∼6S fraction of RSP16 remain abundant (arrowhead) compared with the less abundant and smaller particle of RSP2.
To differentiate these two possibilities, pf24 M+M was fractionated by sucrose gradient as well (Figure 6B). As anticipated, the pf24 breakdown spokes and precursors, represented by RSP3 and 11, were smaller (compare large and small arrows in Figure 6A and 6B) and did not contain detectable RSP2 and 16. Importantly, the 6S fractions of RSP16 was still as abundant in pf24 M+M as in pf28pf30 (compare arrowhead Figure 6A and 6B and RSP16 in 20S spoke in Figure 6A), even though RSP2 was reduced significantly (compare RSP2 fractions in Figure 6A and 6B) and was present in particles slightly smaller than 6S RSP16 particles. This result supported the independent transport of RSP16 and disagreed with the dissociation.
To independently evaluate spoke precursors and the size of the small RSP16 particles, pf28pf30 M+M was fractionated by gel-filtration chromatography and native gel electrophoresis and the fractions were evaluated by Western analysis (Figure 7, A and B). Fractionation was carried out using a FPLC system equipped with a Superose 6 HR sizing column that has an optimal separation range of 5K-5000K Mr and the fractions were collected after void volume. Spoke particles represented by RSP3 were eluted into two peaks, 1-4 and 8-12 (Figure 7A, bottom). A majority of proteins in M+M revealed by UV absorption was coeluted in the first peak and the high concentration of proteins distorted the SDS-PAGE (Figure 7A, fractions 1-4). RSP16, was present in the 1-4 as well but was not detectable in fraction 8-12. Importantly, a large fraction of RSP16 migrated similarly as BSA at 33-36 (Figure 7A) and smaller than 150-kDa alcohol dehydrogenase.
Figure 7.

Western analysis of gel filtration fractionation (A) and blue native gel (B) showed that the RSP16 particle in pf28pf30 M+M is ∼100 kDa. (A) Spoke particles in pf28pf30 M+M, represented by RSP3, were fractionated into two peaks (1-4 and 8-11) ahead of the 669-kDa thyroglobulin marker (TG). Notably, a prominent RSP16 peak was fractionated at the range of 66-kDa BSA and 150-kDa alcohol dehydrogenase (AD). (B) Independently, M+M was separated by 1D 13% Blue native gel and the strip (schematics) was inserted into a preparatory gel for 2D SDS-PAGE and Western analysis. A fraction of RSP16 in M+M migrated as a particle smaller than the smallest tubulin particles (T). HSP70 migrated as a distinct larger particle of ∼150 kDa.
Electrophoresis of 13% blue native gel was also carried out to fractionate M+M. To reveal RSP16 particles by Western blots, the M+M lane was sliced and then further subjected to a 2D SDS-PAGE (Figure 7B, schematic drawing), because anti-RSP16 cannot recognize native RSP16 in either native gel or immunofluorescence. Smaller particles migrating faster are on the right side of the gel. The smear of abundant tubulin oligomers revealed by anti-α-tubulin serves as an molecular marker. As shown by RSP16 Western analysis, RSP16 particles were smaller than the smallest tubulin particles, presumably 110-kDa dimers (T, in Figure 7B). In addition, Western analysis of the same membrane showed that HSP70 migrated as a separate particle larger than the tubulin dimer and RSP16 (Figure 7B, top). The 12-20S spoke particles, which were separated in a 5-10% gel (Figure 2B), were too large to be fractionated in the 13% gel.
Taken together, these results from two strains and three independent methods strongly suggested that 1) RSP16 was transported in a ∼100-kDa complex separately from the major spoke precursors; and 2) RSP16 was transported into pf24 flagella but failed to incorporate into spokes because of insufficient interacting polypeptides such as RSP2 or 23.
Homodimerization of RSP16
It was demonstrated previously that HSP40 molecules formed homodimers (for example, Zylicz et al., 1985; Harrison et al., 1997). Furthermore, Sis1, a yeast HSP40 homologue, homodimerized into an U-shaped particle via its C-terminus and the dimerization was required for recruiting substrates for HSP70 (Sha et al., 2000). To test homodimerization of RSP16 molecules, two independent approaches, coexpression/copurification and chemical cross-linking, were taken.
For the first approach, the plasmids for expressing RSP16-His and GST-RSP16 were cotransformed into bacteria. The extract from coexpressing cells was subjected to Ni-affinity purification. As predicted, Ni-matrix pulled down not only 6His-tagged RSP16 but also GST-RSP16 (Figure 8A, left). In contrast, in the control, the coexpressed untagged RSP16 and GST (Figure 8A, right) failed to bind to Ni-NTA beads. The result indicated that recombinant RSP16 homodimerized or oligomerized in vitro.
Figure 8.

Homodimerization of RSP16 both in vitro (A) and in flagella (B and C). (A) Coomassie protein gel showed that GST-tagged RSP16 was copurified with 6His-tagged RSP16 by Ni-NTA affinity chromatography. Expression constructs for each recombinant proteins were cotransformed into bacteria. The supernatant (Pre) of IPTG-induced bacterial pellet was applied to a Ni-NTA column. Both recombinant proteins were reduced in the flow through (Post) and recovered in the Elute (left). In contrast, coexpressed untagged RSP16 and GST in the control supernatant (right) did not show visible affinity toward the Ni-NTA matrix. (B) Western analysis showed that after treatment with the zero-length cross-linker, EDC, ∼80-120-kDa bands containing RSP16 appeared in the samples of purified recombinant protein (arrowheads, left) and M+M and pf17 KI axonemal extract (right). Compared with the recombinant sample (first lane, right panel), the cross-linked products from flagella migrated slightly slower. (C) Western analysis of 2D SDS-PAGE revealed that ∼80-120-kDa spots of RSP16 in the EDC-treated pf17 KI extract migrated at the same pI in 2D Western analysis as the monomer, indicating the larger cross-linked product was RSP16 homodimer. Acidic end was indicated as A at the right side of the blot.
For the second approach, affinity-purified RSP16-His (Figure 5A, last lane) was treated with a stringent zero-length cross-linker, EDC, which has been used to reveal molecular interactions in axonemal complexes without spurious cross-linked products (for example, Diener et al., 1993; Yang and Sale, 1998). Significant amounts of purified recombinant RSP16, revealed by RSP16 Western analysis, became ∼80-kDa and slightly larger particles possibly due to variation in cross-linked residues (arrowheads, Figure 8B, left). This result indicated that EDC could be used to reveal the homodimerization. This cross-linked sample was used as a standard to test whether similar cross-linked RSP16 products were present in the EDC-treated flagellar M+M and in extracted pf17 spoke stalk (to rule out the cross-linking with spoke head proteins). EDC treatment resulted in a ∼100-kDa RSP16 product in both samples (Figure 8B, right) and in cross-linked wild-type axonemes. However, the cross-linked flagellar RSP16 migrated similar but not identical to the EDC-treated recombinant RSP16. The difference in size could be due to excess of tubulins in M+M (Qin et al., 2004) and KI extract or due to heterodimerization.
To differentiate the two possibilities, EDC-treated pf17 KI extract was evaluated by 2D gel Western analysis. The prediction was that homodimeric and monomeric RSP16 will have the same pI in the 2D gel. On the other hand, heterodimer will migrate toward a more acidic position than RSP16, because RSP16 is the most basic spoke protein (Figure 3A; Piperno et al., 1981; Yang et al., 2001). Compared with no-EDC control, a cluster of ∼80-kDa and larger spots containing RSP16 appeared in the EDC-treated KI extract as seen in 1D Western analysis (Figure 8C, compared with 8B). Importantly, these spots migrated approximately at the same pI as the 40-kDa RSP16 monomer. No other RSP16 spots of this size were found at different pI in 2D Western analysis. The cross-linking result along with the copurification of recombinant RSP16 strongly indicated that the RSP16 formed homodimer as HSP40 cochaperone in vitro as well as in flagellar matrix and radial spokes.
DISCUSSION
The Constitutive HSP40 in the Radial Spoke
This study demonstrates that RSP16 is a novel homologue of HSP40, a subtype of DnaJ cochaperones. The evidence includes the following: 1) the protein sequence resulting from purified RSP16 in 2D gel of the isolated 20S radial spokes is highly homologous to the HSP40 molecules, a subtype of the DnaJ superfamily; and 2) Western blots of 1D and 2D gels showing that the antibody against the novel HSP40 recognizes a single 40-kDa protein that is absent in spokeless mutant, diminished in pf24 axoneme, and importantly, cosediment with 20S radial spokes in KI extract.
Although numerous DnaJ and HSP40 molecules have been found in eukaryotic cells (Ohtsuka and Hata, 2000), the constitutive HSP40 in the radial spoke is intriguing in two aspects. First of all, DnaJ molecules are often found in dynamic chaperone-like systems mediating protein folding or protein transport. In contrast, the radial spoke is known as a rigid and stable structural complex postulated as a mechanochemical transducer for the control of flagellar motility. Second, analysis of spokeless mutant (Piperno et al., 1981) clearly indicates that RSP16 is a structural component only present in radial spoke, one of the many macromolecular complexes in flagella. The unorthodox presence invites a series of intriguing questions. Do other axonemal complexes contain DnaJ molecules as well? Why would the radial spoke need a DnaJ? Does this spoke HSP40 function also like a cochaperone or simply moonlight for Chlamydomonas radial spokes? Is its function restricted to the radial spoke?
The potential RSP16 orthologues present in organisms as diverse as human and Ciona suggest that spoke HSP40 is conserved to play a central role for the conserved structure in the 9 + 2 axonemes. The role appears to be particularly unique compared with other spoke proteins, which are likely either structural or regulatory components for the proposed mechanochemical transducers (Smith and Yang, 2004). Rather, several independent lines of evidence, despite indirect, suggest that RSP16 may involve in the assembly of the macromolecular complex as a cochaperone. The evidence includes 1) its highly conserved DnaJ-J domain, including the conserved His-ProAsp residues, crucial for activating ATPase of HSP70 (Wall et al., 1994; Laufen et al., 1999; reviewed by Fink, 1999); 2) the exclusion of the ∼100-kDa RSP16 particle from the 12S spoke precursors that contain all of the remaining known spoke proteins (Qin et al., 2004); 3) homodimerization of RSP16, a characteristic of HSP40 molecules and a requisite for substrate recruitment (Sha et al., 2000); 4) correlation of the inconsistently assembled spokes and diminished RSP16 in pf24 axonemes; and 5) the nature of chaperone machinery, as discussed below.
Despite the homologous J-domain, we have not yet succeeded in using recombinant RSP16 to activate the ATPase activity of recombinant human HSP70 or facilitate the refolding of denatured luciferase. This is not unexpected since each organism expresses multiple types of HSP70 and DnaJ molecules. Specific pairing of certain DnaJ, HSP70 as well as substrates has been demonstrated (reviewed by Kelly, 1999; Fink, 1999). The definitive experiment includes direct test of the interaction of spoke HSP40 and flagellar HSP70, a protein that remains to be cloned and characterized.
The separate spoke HSP40 particle is consistent to the role of a cochaperone that recruits the substrate, the 12S spoke, to HSP70. Alternatively, the HSP40 is assembled into the 12S particles like the other major spoke proteins (Qin et al., 2004) but becomes completely dissociated during the experimentation. Dissociation appears unlikely because a low concentration of detergent (0.025% NP-40) is used in preparing M+M. By contrast, RSP16 remained tightly associated with axonemes and extracted radial spokes after demembranation with 0.5% NP-40 and after KI extraction. Furthermore, three independent measures, including velocity sedimentation, gel filtration, and native gel electrophoresis, consistently reveals the ∼100-kDa RSP16 particles. Moreover, even though RSP2 is reduced in the M+M and axonemes of pf24 and RSP16 is reduced in pf24 axonemes, RSP16 is synthesized normally and there is no obvious change in the fractions of small RSP16 particles in the M+M of pf24. Thus, the simplest interpretation is that the particles of RSP16 are transported separately and joint the remaining spoke proteins in the precursors by interacting with RSP2 during the final assembly processes in flagella.
The RSP16 particles in M+M are consistent to the function of cochaperones that recruits polypeptide substrates to the chaperone system. In addition, the finding revealed the un-finished precursors and possible small cargoes, suggesting that the cargoes for IFT are not limited to large complexes or particular requirement.
Dimeric RSP16 and the Configuration of Mature Radial Spoke
Both in vivo and in vitro approaches demonstrate that RSP16 molecules form homodimers. This finding is useful in establishing the stoichiometry of spoke proteins in the T-shaped complex. The accumulated molecular mass of the 23 spoke polypeptides in 20S spokes is ∼700 kDa. The predicted total mass is significantly less than the 20S dyneins and the ∼1200-kDa Ciona sperm spoke particles (Padma et al., 2003). Furthermore, 35S autoradiography and silver staining (Piperno et al., 1981; Yang et al., 2001; Figure 3A) show that a group of radial spoke proteins, including RSP16, are obviously more abundant than the others. These observations together with the discovery of dimeric RSP16 strongly suggest that major spoke proteins of similar stoichiometry contribute at least two copies to each spoke particle. We envision two models that are not mutually exclusive. First, the RSP16 dimer along with two 12S precursors are assembled into a 20S particle. Alternatively, 20S spokes could be the combination of a RSP16 dimer, additional unknown spoke proteins and a 12S spoke precursor that at least contains two copies of each major spoke proteins. These two models may be differentiated by further characterization of 12S particles and unknown spoke proteins.
The Roles of Spoke HSP40 and Chaperones in Cilia and Flagella
In addition to the separate particles of dimeric spoke HSP40, independent lines of evidence implicate that a chaperone system is involved in the assembly of radial spokes as well as axonemes. It is well established that newly synthesized proteins are predominantly incorporated into the distal tip of flagella (Rosenbaum and Child, 1967; Witman, 1975; Marshall and Rosenbaum, 2001) where a unique capping structure is present (Dentler and Rosenbaum, 1977) during flagellar elongation or maintenance. Repair of the defective axoneme also preferentially occurs in a tip-to-base direction (Johnson and Rosenbaum, 1992; Piperno et al., 1996). These observations indicate that the majority of axonemal components, including radial spoke components, have to be first transported toward the tip to be converted into part of the axonemes. The different sizes between precursors and mature spoke particles and the stability of the latter suggest that the final assembly at the distal tip involve changes in protein folding possibly catalyzed by chaperone machines. Chaperones, HSP70 and HSP90, are present in cilia and flagella (Bloch and Johnson, 1995; Stephens and Lemieux, 1999; Ostrowske et al., 2002). In particular, HSP70 is enriched at the flagellar tip.
Importantly, based on the well-established mechanism of chaperone machinery, DnaJ molecules do not function independently and contain no enzymatic activity, except several controversial examples (reviewed by Cheetham and Caplan, 1998; Fink, 1999; Hartl and Hayer-Hartl, 2002). Rather, they work with chaperones. They primarily recruit substrates for HSP70 and activate its ATPase activity with the J-domain or cooperate with HSP90 in a few instances. On the other hand, HSP70 requires DnaJ to display sufficient ATPase activity in vivo. The dynamic interaction of HSP70 accumulated at the tip and the dimeric RSP16 particles in M+M are ideal partners to transform the 12S precursors into the stable axonemal spokes. It is baffling that cells do not make spoke precursors as the final 20S product once for all. One possibility is that the 12S precursor may be a more suitable format in the crowded cytosolic compartment and for anterograde intraflagellar transport.
Despite the pf24 restricted defect to the head ends of spokes, it remains to be tested whether RSP16 is involved in the assembly of the base of stalk or other axonemal complexes. It is important to point out that the phenotype of pf24 is not directly due to lacking RSP16 but RSP2, the potential binding partners of RSP16, because RSP16 is normal in pf24 cell body and 6S RSP16 fraction in pf24 and wild type appears similar (Figure 6, B and A, respectively). A null mutant or knockdown cell lines may be useful in differentiating the scope of the role of RSP16. If the sole function of RSP16 is limited to assist spoke assembly, it might be more economical that RSP16 dissociates from the mature spokes after assembly to carry out additional reactions like many other cochaperones. We speculate that this constitutive cochaperone may confer the structural stability of radial spokes that endure tension, distortion, or transverse force (Warner and Satir, 1978; Goodenough and Heuser, 1985; Lindemann, 2003) during the oscillatory beating due to the transient interaction between the free spoke heads and central pair while the stalk is immobilized to the sliding outer doublet microtubules.
If the prediction is correct, RSP16 will join the emerging group of DnaJ cochaperones that are localized in specific cytosolic compartments to carry out diversified activities related to protein folding (reviewed by Young et al., 2003). In this case, the role is the assembly of structural complexes. Despite the focus and insight on radial spoke, this study sheds light on the cargoes of intraflagellar transport, the polarized assembly of axonemes and the roles of molecular chaperones in cilia and flagella.
Acknowledgments
We are grateful to N. Hass and Dr. C. D. Silflow (University of Minnesota) for mapping the RSP16 gene and Drs. D. R. Diener and J. L. Rosenbaum (Yale University) for the anti-RSP1, 2, and 3. We also thank Dr. H. G. Tomasiewicz (Great Lakes Water Institute) for preparing preimmune IgY and thank Dr. W. S. Sale (Emory University) and Dr. D. R. Diener for helpful comment on manuscript. This work was supported by Marquette Faculty Summer Research Fellowship and a grant from the National Institute of General Medical Sciences (GM68101).
Article published online ahead of print in MBC in Press on November 24, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0787).
References
- Berman, S. A., Wilson, N. F., Haas, N, A. and Lefebvre, P. A. (2003). A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13(13), 1145-1149. [DOI] [PubMed] [Google Scholar]
- Bloch, M. A., and Johnson, K. A. (1995). Identification of a molecular chaperone in the eukaryotic flagellum and its localization to the site of microtubule assembly. J. Cell Sci. 108, 3541-3545. [DOI] [PubMed] [Google Scholar]
- Bukau, B., and Horwich, A. L. (1998). The HSP70 and HSP60 chaperone machines. Cell 92, 351-366. [DOI] [PubMed] [Google Scholar]
- Cheetham, M. E., and Caplan, A. J. (1998). Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D. G. (2003). (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4(7), 435-442. [DOI] [PubMed] [Google Scholar]
- Curry, A. M., and Rosenbaum, J. L. (1993). Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil. Cytoskelet. 24, 224-232. [DOI] [PubMed] [Google Scholar]
- Dentler, W. L., and Rosenbaum, J. L. (1977). Flagellar elongation and shortening in Chlamydomonas: III. Structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 74, 747-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, D. R., Ang, L. H., and Rosenbaum, J. L. (1993). Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J. Cell Biol. 123, 183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S. K., Huang, B., and Luck, D.J.L. (1984). Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98, 229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, A. L. (1999). Chaperone-mediated protein folding. Physiol. Rev. 79, 425-449. [DOI] [PubMed] [Google Scholar]
- Fowkes, M. E., and Mitchell, D. R. (1998). The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell 9(9), 2337-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U. W., and Heuser, J. E. (1985). Substructure of inner dynein arms, radial spokes, and the central pair projection complex. J. Cell Biol. 100, 2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, C. J., Hayer-Hartl, M., Liberto, D. I., Hartl, F. U., and Kuriyan, J. (1997). Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276, 431-435. [DOI] [PubMed] [Google Scholar]
- Hartl, F. U., and Hayer-Hartl, M. (2002). Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852-1858. [DOI] [PubMed] [Google Scholar]
- Huang. B., Piperno, G., Ramanis, Z., and Luck, D.J.L. (1981). Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88, 80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Ramanis, Z., and Luck, D.J.L. (1982). Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28, 115-124. [DOI] [PubMed] [Google Scholar]
- Iomini, C., Tejada, K., Mo, W., Vaananen, H., and Piperno, G. (2004). Primary cilia of human endothelial cells disassemble under laminar shear stress. J. Cell Biol. 164(6), 811-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K. A., and Rosenbaum, J. L. (1992). Polarity of flagellar assembly in Chlamydomonas. J. Cell Biol. 119, 1605-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W. L. (1999). Molecular chaperones: how J domains turn on HSP70s. Curr. Biol. 9, R305-R309. [DOI] [PubMed] [Google Scholar]
- Laufen, T., Mayer, M. P., Beisel, C., Klostermeier, D., Reinstein, J., and Bukau, B. (1999). Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Nat. Acad. Sci. USA 96, 5452-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann, C. B. (2003). Structural-functional relationships of the dynein, spokes, and central-pair projections predicted from an analysis of the forces acting within a flagellum. Biophys. J. 84, 4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F., and Rosenbaum, J. L. (2001). Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155, 405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D. R. (2003). Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil. Cytoskelet. 56, 120-129. [DOI] [PubMed] [Google Scholar]
- Mitchell, D. R., and Sale, W. S. (1999). Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144, 293-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto, C. K., Gibbons, I. R., Kamiya, R., Shingyoji, C., Takahashi, K., and Witman, G. B. (1999). Rotation of the central pair microtubules in eukaryotic flagella. Mol. Biol. Cell 10, 1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowske, L. E., Blackburn, K., Radde, K. M., Moyer, M. B., Schlatzer, D. M., Moseley, A., and Boucher, R. C. (2002). A proteomic analysis of human cilia. Mol. Cell. Proteomics 1, 451-466. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, K., and Hata, M. (2000). Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones 5, 98-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma, P., Satouh, Y., Wakabayashi, K., Hozumi, A., Ushimara, Y., Kamiya, R., and Inaba, K. (2003). Identification of a novel leucine rich repeat protein as a component of the flagellar radial spoke in Ascidian Ciona intestinalis. Mol. Biol. Cell 14, 774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., Wang, Q., and Snell, W. J. (2004). An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell 6(3), 445-451. [DOI] [PubMed] [Google Scholar]
- Patel-King, R. S., Gorbatyuk, O., Takebe, S., and King, S. M., (2004). Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell 15: 3891-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Mead, K., and Henderson, S. (1996). Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1(FLA10) to reach the distal part of flagella in Chlamydomonas. J. Cell Biol. 133, 371-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Ramanis, Z., Smith, E. F., and Sale, W. S. (1990). Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J. Cell Biol. 110, 379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Huang, B., Ramanis, Z., and Luck, D. J. (1981). Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88, 73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Huang, B., and Luck, D. J. (1977). Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 74, 1600-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A., von Wechmar, M. M., and van Regenmortel, M.H.V. (1980). Isolation of viral IgY antibodies from yolks of immunized hens. Immunol. Commun. 9, 475-493. [DOI] [PubMed] [Google Scholar]
- Qin, H., Diener, D. R., Geimer, S., Cole, D. G., and Rosenbaum, J. L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J. L., and Child, F. M. (1967). Flagellar regeneration in protozoan flagellates. J. Cell Biol. 34, 345-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J. L., and Witman, G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3(11), 813-825. [DOI] [PubMed] [Google Scholar]
- Schagger, H., and von Jagow, G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatic ally active form. Anal. Biochem. 199, 223-231. [DOI] [PubMed] [Google Scholar]
- Sha, B., Lee, S., and Cyr, D. M. (2000). The crystal structure of the peptide-binding fragment from the yeast HSP40 protein Sis1. Structure 8, 799-807. [DOI] [PubMed] [Google Scholar]
- Smith, E. F., and Yang, P. (2004). The radial spokes and central apparatus: mechano-chemical sensors that regulate flagellar motility. Cell Motil. Cytoskelet. 57, 8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, W. J., Pan, J., and Wang, Q. (2004). Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell 117(6), 693-697. [DOI] [PubMed] [Google Scholar]
- Song, L., and Dentler, W. L. (2001). Flagellar protein dynamics in Chlamydomonas. J. Biol. Chem. 276, 29754-29763. [DOI] [PubMed] [Google Scholar]
- Stephens, R. E. (1997). Synthesis and turnover of embryonic sea urchin ciliary proteins during selective inhibition of tubulin synthesis and assembly. Mol. Biol. Cell 8, 2187-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, R. E., and Lemieux, N. A. (1999). Molecular chaperones in cilia and flagella: implications for protein turnover. Cell Motil. Cytoskelet. 44, 274-283. [DOI] [PubMed] [Google Scholar]
- Tam, L. W., Dentler, W. L., and Lefebvre, P. A. (2003). Defective flagellar assembly and length regulation in LF3 null mutants in Chlamydomonas. J. Cell Biol. 163, 597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, D., Zylicz, M., and Georgopoulos, C. (1994). The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269, 5446-5451. [PubMed] [Google Scholar]
- Wargo, M. J., and Smith, E. F. (2003). Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 100, 137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G. B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280-290. [DOI] [PubMed] [Google Scholar]
- Witman, G. B. (1975). The site of in vivo assembly of flagellar microtubules. Ann. NY Acad. Sci. 253, 178-191. [DOI] [PubMed] [Google Scholar]
- Yang, P., Yang, C., and Sale, W. S. (2004). Flagellar radial spoke protein 2 is a calmodulin binging protein required for motility in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Diener, D. R., Rosenbaum, J. L., and Sale, W. S. (2001). Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153, 1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., and Sale, W. S. (1998). The 140,000 Mr intermediate chain of Chlamydomonas inner arm dynein is a WD-repeat protein implicated in dynein anchoring. Mol. Biol. Cell 9, 3335-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. C., Barral, J. M., and Hartl, F. U. (2003). More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28, 541-547. [DOI] [PubMed] [Google Scholar]
- Zylicz, M., Yamamoto, T., McKittrick, N., Sell, S., and Georgopoulos, C. (1985). Purification and properties of the DnaJ replication protein of Escherichia coli. J. Biol. Chem. 260, 7591-7598. [PubMed] [Google Scholar]


