Abstract
Actin-depolymerizing factor (ADF)/cofilins are small actin-binding proteins found in all eukaryotes. In vitro, ADF/cofilins promote actin dynamics by depolymerizing and severing actin filaments. However, whether ADF/cofilins contribute to actin dynamics in cells by disassembling “old” actin filaments or by promoting actin filament assembly through their severing activity is a matter of controversy. Analysis of mammalian ADF/cofilins is further complicated by the presence of multiple isoforms, which may contribute to actin dynamics by different mechanisms. We show that two isoforms, ADF and cofilin-1, are expressed in mouse NIH 3T3, B16F1, and Neuro 2A cells. Depleting cofilin-1 and/or ADF by siRNA leads to an accumulation of F-actin and to an increase in cell size. Cofilin-1 and ADF seem to play overlapping roles in cells, because the knockdown phenotype of either protein could be rescued by overexpression of the other one. Cofilin-1 and ADF knockdown cells also had defects in cell motility and cytokinesis, and these defects were most pronounced when both ADF and cofilin-1 were depleted. Fluorescence recovery after photobleaching analysis and studies with an actin monomer-sequestering drug, latrunculin-A, demonstrated that these phenotypes arose from diminished actin filament depolymerization rates. These data suggest that mammalian ADF and cofilin-1 promote cytoskeletal dynamics by depolymerizing actin filaments and that this activity is critical for several processes such as cytokinesis and cell motility.
INTRODUCTION
Actin filaments in nonmuscle cells are highly dynamic and play a critical role in numerous cellular processes, including cell migration, cytokinesis, and polarized growth. These processes rely on the correct spatial and temporal organization of actin filaments that is regulated by numerous actin-binding proteins. The actin-depolymerizing factor (ADF)/cofilins are a family of small (Mr = 15-20) proteins that bind monomeric and filamentous actin. Unicellular organisms such as yeasts typically have only one ADF/cofilin, whereas multicellular organisms can have several isoforms (reviewed by Bamburg et al., 1999). In mammals, there are three different ADF/cofilins: cofilin-1, cofilin-2, and ADF. These proteins have distinct expression patterns: cofilin-1 is expressed in most embryonic and adult mouse cells, cofilin-2 is expressed in muscle, and ADF is mainly found in epithelial and neuronal cells (Vartiainen et al., 2002).
Based on in vitro studies, ADF/cofilins enhance the rate of actin filament turnover by depolymerizing filaments at their pointed ends, thereby providing a pool of actin monomers for filament assembly. ADF/cofilins also sever actin filaments and consequently increase the number of filaments ends (reviewed by Bamburg et al., 1999; Carlier et al., 1999). The mammalian ADF/cofilins are quantitatively different in their activities. ADF is the most efficient at turning over actin filaments and promotes a stronger pH-dependent actin filament disassembly than cofilin-1 or cofilin-2. The muscle cell-specific cofilin-2 has a weaker actin filament depolymerization activity than the other two and promotes filament assembly rather than disassembly in steady-state assays (Vartiainen et al., 2002; Yeoh et al., 2002).
Genetic studies on Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans demonstrated that ADF/cofilins are essential for viability (Moon et al., 1993; McKim et al., 1994; Gunsalus et al., 1995). However, whether ADF/cofilins contribute to cytoskeletal dynamics by depolymerizing actin filaments at their pointed ends, or by creating new filament barbed ends for F-actin assembly through their severing activity has remained unclear. Studies on the motility of Listeria and analysis of loss-of-function cofilin mutants in yeast indicated that ADF/cofilins enhance actin dynamics by depolymerizing actin filaments and provide actin monomers to the cytoplasmic pool (Carlier et al., 1997; Rosenblatt et al., 1997; Lappalainen and Drubin, 1997). Furthermore, cytoplasmic actin filaments accumulate when ADF/cofilins are mutated in Drosophila or Caenorhabditis elegans or when ADF/cofilins are inactivated by overexpressing LIM kinase (Gunsalus et al., 1995; Arber et al., 1998; Yang et al., 1998; Ono et al., 1999; Chen et al., 2001). In contrast, studies on epidermal growth factor (EGF)-stimulated rat mammary adenocarcinoma cells suggested that ADF/cofilin's biological role is to increase actin filament nucleation by severing actin filaments and thus create new filament barbed ends for actin assembly (Chan et al., 2000; Zebda et al., 2000; Ichetovkin et al., 2002; Ghosh et al., 2004).
It is important to note that the biological role(s) of ADF/cofilins in mammalian cells has been mainly examined by inactivating these proteins by LIM kinase. Recent studies revealed that LIM kinase also has other targets than cofilin (Roovers et al., 2003), and to accurately understand the role of ADF/cofilins in actin dynamics in mammalian cells, more direct and specific methods are required. Furthermore, ADF and cofilin-1 are coexpressed in many mammalian cells (Vartiainen et al., 2002), but whether these proteins are involved in same or different biological processes has not been examined. To elucidate the biological roles of mammalian ADF/cofilins, we depleted ADF and cofilin-1, either individually or in combination with each other, from various mouse cell-lines by small interfering RNA (siRNA)-induced gene silencing (Elbashir et al., 2001). Analyses of the ADF and cofilin-1 knockdown cells showed that these proteins promote rapid F-actin depolymerization and provide new monomers to the cytoplasmic actin pool. Our studies also demonstrated that the actin dynamics induced by ADF and cofilin-1 are important for normal actin organization, as well as for morphogenesis, motility, and cytokinesis in cultured mammalian cells.
MATERIALS AND METHODS
Proteins and Antibodies
Recombinant cofilin-1 and ADF were purified as described previously (Vartiainen et al., 2002). Rabbits were immunized with recombinant mouse cofilin-1 and hens with recombinant mouse ADF. Antibodies were collected after four (rabbits) or three (hens) immunizations and affinity purified as described previously (Vartiainen et al., 2000).
Cell Culture, Immunofluorescence, and Western Blotting
NIH 3T3, Neuro 2A, and B16F1 cells were maintained in DMEM supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) or 10% fetal calf serum (PAA Laboratories, Pasching, Austria), 2 mM l-glutamine, penicillin, and streptomycin (Sigma-Aldrich, St. Louis, MO). B16F1 cells stably expressing green fluorescent protein (GFP)-actin (Ballestrem et al., 1998) were maintained in medium supplemented with 1.5 mg/ml Geneticin (Invitrogen, Carlsbad, CA). For immunofluorescence, the NIH 3T3 cells were plated on coverslips. For the B16F1 cells, coverslips were precoated with laminin (25 μg/ml) or fibronectin (50 μg/ml). Immunofluorescence was performed as described previously (Vartiainen et al., 2000). As an exception, for the AC-15 and DNAseI stainings 3% paraformaldehyde with 0.2% glutaraldehyde was used as a fixative to achieve a better preservation of the G-actin. Free aldehyde groups were then blocked with 0.1 M glycine. Cofilin-1 was visualized with rabbit anti-cofilin-1, ADF with hen anti-ADF, myosin II with rabbit anti-nonmuscle-myosin (Biomedical Technologies, Cambridge, MA), myc-tagged fusion proteins with mouse anti-myc antibodies, and secondary antibodies conjugated to fluorescein, rhodamine, or cy5 (Molecular Probes, Eugene, OR). G-actin was visualized with Alexa-594-DNAseI (Molecular Probes), F-actin with Alexa-488-phalloidin, or rhodamine-phalloidin (Molecular Probes). β-Actin monoclonal antibody AC-15 (Sigma-Aldrich) was used for labeling the actin cytoskeleton, excluding stress fibers. DAPI was the nuclear counterstain. Images were acquired through a SenSys (Photometrics, Tucson, AZ) or DP70 (Olympus, Tokyo, Japan) charge-coupled device camera on an AX70 Provis microscope (Olympus). For Western blotting, cell lysates were prepared as described previously (Vartiainen et al., 2000), and the total protein concentrations were measured using Bradford reagent (Sigma-Aldrich) according to the manufacturer's instructions. Western blotting was performed according to instructions from AgriSera (Vannas, Sweden). Anti-ADF and anti-cofilin-1 antibodies were used at a 1:1000 dilution. Horseradish peroxidase-conjugated anti-hen, anti-mouse, and anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at a 1:20000 (hen) or 1:5000 (mouse and rabbit) dilutions. The intensities of immunoblot bands were quantified by TINA software.
siRNA Treatment
For the siRNA experiments, 20-40 pmol of preannealed fluorescein (5F1)-labeled or unlabeled Cof1-siRNA [r(GGAGGACCUGGUGUUCAUC)d(TT), r(GAUGAACACCAGGUCCUCC)d(TT)] (Xeragon Qiagen, Valencia, CA), fluorescein-labeled (5F1), TAMRA (rhodamine, 5Rh)-labeled or -unlabeled ADF-siRNA [r(GUGAUUGCAAUCCGUGUAU)d(TT), r(AUACACGGAUUGCAAUCAC)d(TT)] (Xeragon), or unlabeled control siRNA [r(AGCUUCAUAAGGCGCAUGC)d(UU), r(GCAUGCGCCUUAUGAAGCU)d(UU)] (made with Ambion Silencer siRNA construction kit) duplexes were transfected into cells on 24-well plates by using GeneSilencer's siRNA transfection reagent (Gene Therapy Systems, San Diego, CA) as described previously (Elbashir et al., 2001). After 48 h, the cells were detached with trypsin-EDTA, diluted, and plated on coverslips for immunofluorescence. The assays with NIH 3T3 cells were carried out 20 h after replating and with B16F1 cells 5-20 h after replating.
siRNA Rescue Experiments
Construction of the ADF-myc expression plasmid (pPL112) is described (Vartiainen et al., 2002). Because the ADF-siRNA oligonucleotide was targeted to the 3′ untranslated region of ADF mRNA, the ADF-myc construct is refractory to this siRNA duplex. The cofilin-1-myc (pPL108) construct (Vartiainen et al., 2002) was mutated by inverse PCR by using primers (5′-TTGGTATTCATCTTCTGGGCCCCCGAG and 5′-ATCTTCCTTCTTGCTCTCCTTGGTCTC). This generated four nucleotide changes to the cofilin-1 siRNA-target sequence without altering the amino acid sequence of the protein [GGAGGACCTGGTGTTCATCTT sequence was mutated to GGAAGATTTGGTATTCATCTT, resulting in a cofilin-1-myc rescue construct (pPL256)]. The cells were treated with siRNA oligonucleotide duplexes as described above and transiently transfected with one of the rescue constructs 24 h before fixation. Transfection of B16F1 cells was performed with Superfect (QIAGEN, Valencia, CA) and for NIH3T3 cells with FuGENE6 (Roche Diagnostics, Mannheim, Germany) according to manufacturers' recommendations.
Cell Motility Assays
Cell motility assay with NIH 3T3 cells was performed with the Cell Motility HitKit (Cellomics, Pittsburgh, PA). NIH 3T3 cells were treated for 54 h with ADF or cofilin-1 siRNA, replated at a density of 3000-5000 cells/ml on coverslips coated with fibronectin (10 μg/ml) and blue fluorescent beads, grown for 20 h, and fixed with 3.7% formaldehyde for immunofluorescence. The minimum distance of migration was quantified by measuring the shortest path from the cell body to the most distant area cleared from the beads. For the live-imaging assay, the B16F1 cells or B16F1 cells stably expressing GFP-actin (Ballestrem et al., 1998) were plated on laminin (25 μg/ml)-coated glass bottom dishes (MatTek). The time-lapse images of wild-type and cofilin-1 knockdown cells were acquired with an IX70 inverted microscope (Olympus) equipped with a Polychrome IV monochromator (TILL Photonics, Martinsried, Germany) with the appropriate filters and heated sample environment. To enhance motility of B16F1 cells, final concentration of 50 μM AlCl3 and 30 mM NaF were added to medium. After the experiment, cells were fixed and cofilin-1 knockdowns were confirmed by immunofluorescence as described above. For statistical analysis, migration of 35 wild-type and 25 cofilin-1 knockdown cells (that do not express GFP-actin) were examined by tracking the position of the nucleus every 20 min. To follow the cytokinesis in living cells, the NIH 3T3 cells were plated on glass bottom dishes (MatTek, Ashland, MA), grown for further 20 h, and then monitored by acquiring differential interference contrast time-lapse images for 200 min with the setup described above.
Actin Filament Turnover Assays
The actin filament depolymerization rates in B16F1 cells were determined by using the actin monomer-sequestering drug, latrunculin-A. Wild-type and siRNA-treated cells were replated on fibronectin-coated coverslips after 48 h of transfection, and grown for further 20 h. Latrunculin-A (Sigma-Aldrich) was added to the plates at a final concentration of 2 μM, and the cells were fixed with 4% paraformaldehyde at 5, 10, or 30 min after addition of latrunculin-A. Control cells were treated with medium supplemented with dimethyl sulfoxide (DMSO), which was used as a vehicle for latrunculin-A. Actin filaments, ADF, and cofilin-1 were visualized as described above. Fluorescence recovery after photobleaching (FRAP) was applied to measure the actin treadmilling rates in B16F1 cells. Confocal imaging for examining the dynamics of stress fibers was carried out on a Zeiss LSM 510 confocal microscope equipped with an argon-ion laser (Carl Zeiss, Jena, Germany) and LSM 3.0 software as described previously (Bertling et al., 2004). GFP-actin-expressing B16F1 cells (wild type and ADF or cofilin-1 siRNA treated) were grown for 5-12 h on fibronectin-coated glass bottom dishes (MatTek). Wild-type, cofilin-1 knockdown and ADF knockdown cells with strong stress fibers were selected. After one prebleach scan of an entire image, 100 scan iterations of a rectangular region of interest (ROI) were scanned with 100% intensity of 30-mW argon-ion 488-nm laser (transmission intensity). Directly after bleaching the fluorescence recovery was measured automatically after every 10 s (20 times, entire image). After the experiment, cells were fixed with 4% paraformaldehyde and cofilin-1 was visualized by immunofluorescence. ADF-transfection was confirmed by detecting the rhodamine-labeled siRNA-duplexes. The recovery of the GFP-actin intensity was measured by TINA software. The intensity of the bleached area was normalized to neighboring nonbleached area to diminish the error caused by normal photobleaching during the monitoring period. Confocal imaging for examining the dynamics of cortical actin structures was carried out on a Leica TCS SP2 AOBS confocal microscope (Leica Microsystems, Wetzlar, Germany). For GFP-imaging, 488-nm line and a 63× numerical aperture 1.2 water immersion objective was used. GFP-actin-expressing B16F1 cells (wild type and ADF or cofilin-1 siRNA treated) were grown for 5 h on laminin-coated glass bottom dishes (MatTek). After two prebleach scans of an entire image, 10 scan iterations of a rectangular ROI were scanned with 100% intensity of 30 mW argon-ion 488 nm laser (transmission intensity). After bleaching, the fluorescence recovery was measured automatically after every 3 to 4 s. After the experiment, cells were fixed with 4% paraformaldehyde and cofilin-1 was visualized by immunofluorescence.
RESULTS
Cofilin-1 Is the Predominant Isoform in Most Mammalian Nonmuscle Cells
Two ADF/cofilin isoforms, cofilin-1 and ADF, are expressed in mammalian nonmuscle cells. Based on in situ hybridizations, cofilin-1 is found in most embryonic and adult mouse cells, whereas ADF is most strongly expressed in epithelial cells and neurons (Vartiainen et al., 2002). To examine the levels of cofilin-1 and ADF proteins in various mouse nonmuscle cell-lines, we carried out a Western blot assay by using two affinity-purified polyclonal antibodies, one of which is specific to ADF, and the other one recognizes both cofilin-1 and cofilin-2 (Figure 1A). We compared the amounts of ADF and cofilin-1 relative to known concentrations of purified recombinant mouse cofilin-1 and ADF (Figure 1B). We found that cofilin-1 is the major ADF/cofilin isoform in NIH 3T3, B16F1, and Neuro 2A cells (Figure 1B, lanes 3-5, respectively). It is present at approximately sixfold (NIH 3T3 cells) to 11-fold (B16F1 cells) higher molar amounts than ADF (Figure 1B). Because cofilin-2 expression in mice is almost exclusively restricted to striated muscle cells (Ono et al., 1994; Vartiainen et al., 2002), and because siRNA treatment with a cofilin-1-specific duplex oligonucleotide resulted in an almost complete loss of cofilin staining in these cells (Figure 2, B and C), it seems that cofilin-2 is not expressed in significant amounts in NIH 3T3, B16F1, or Neuro 2A cells. Immunofluorescence microscopy studies with cofilin-1- and ADF-specific antibodies showed that in NIH 3T3 (Figure 1C) and B16F1 cells (our unpublished data) ADF and cofilin-1 have similar subcellular localizations. Both proteins show diffuse cytoplasmic and perinuclear stainings but are also concentrated to the cortical actin cytoskeleton.
Figure 1.
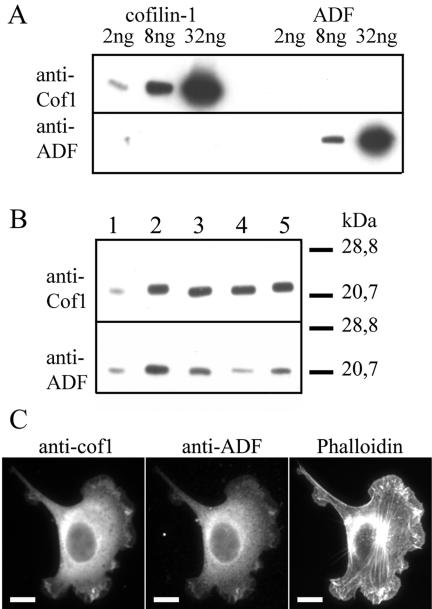
Expression levels and subcellular localizations of cofilin-1 and ADF. (A) Western blot assay demonstrating the specificities of the anti-ADF and anti-cofilin-1 antibodies. Purified recombinant cofilin-1 (lane 1, 2 ng; lane 2, 8 ng; lane 3, 32 ng) and ADF (lane 4, 2 ng; lane 5, 8 ng; lane 6, 32 ng) were visualized on Western blots by using anti-cofilin-1 (top) and anti-ADF (bottom) antibodies. (B) The levels of cofilin-1 and ADF in NIH 3T3, B16F1, and Neuro 2A cells were compared with known concentrations of cofilin-1 (top) and ADF (bottom). Lane 1, 30 ng of cofilin-1 (top) and 7.5 ng of ADF (bottom); lane 2, 80 ng of cofilin-1 (top) and 20 ng of ADF (bottom); lane 3, 10 μg of NIH 3T3 extract; lane 4, 10 μg of B16F1 extract; lane 5, 10 μg of Neuro 2A extract. Cofilin-1 is expressed approximately in sixfold (NIH 3T3), 11-fold (B16F1), and sevenfold (Neuro 2A) higher molar amounts than ADF. (C) Cofilin-1, ADF, and F-actin were visualized by immunofluorescence in NIH 3T3 cells. Cofilin-1 and ADF show similar subcellular localizations and are concentrated in F-actin-rich ruffles. Bar, 10 μm.
Figure 2.
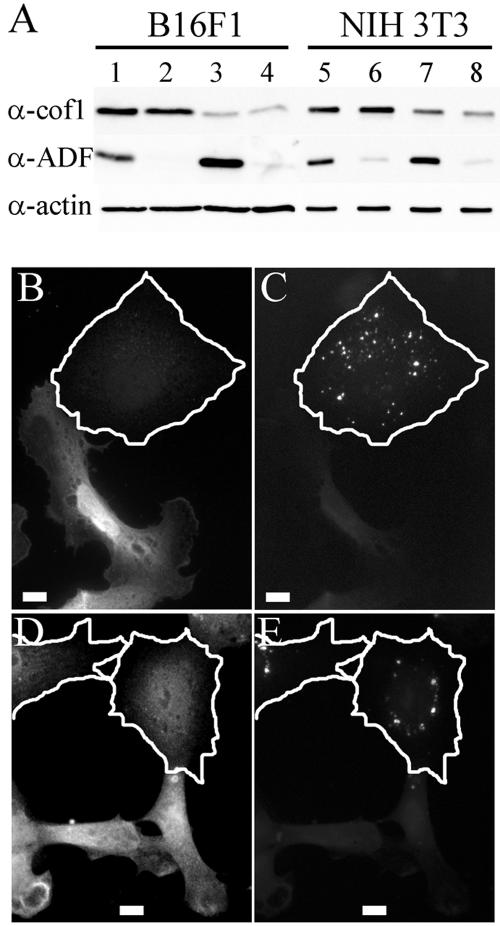
siRNA induced gene silencing of ADF or cofilin-1. (A) Western blot analysis demonstrating the ADF and/or cofilin-1 protein levels in B16F1 (lanes 1-4) and NIH 3T3 cells (lanes 5-8) transfected with control (lanes 1 and 5), ADF-specific (lanes 2 and 6), cofilin-1-specific (lanes 3 and 7), and with both ADF- and cofilin-1-specific (lanes 4 and 8) siRNA oligonucleotide duplexes. Equal amounts of cell lysates were run on polyacrylamide gels, and cofilin-1, ADF, and β-actin were visualized by Western blotting. (B-E) Cofilin-1 and ADF antibody stainings are decreased in siRNA-transfected cells. (B and C) B16F1 cells transfected with FITC-labeled cofilin-1-specific siRNA. (B) Anti-cofilin-1 antibody staining. (C) FITC-siRNA. (D and E) NIH 3T3 cells transfected with FITC-labeled ADF-specific siRNA. (D) Anti-ADF antibody staining. (E) FITC-siRNA. The borders of the transfected cells are indicated by white lines. Bars, 10 μm.
siRNA-induced Gene Silencing of Cofilin-1 or ADF Results in Formation of Abnormal Stress Fibers and Increase in Cell Size
To reveal the role of ADF/cofilins in actin filament turnover (depolymerization of “old” filaments versus promotion of filament assembly through the severing activity) and to elucidate how these proteins contribute to various processes in mammalian cells, we depleted ADF and cofilin-1 proteins, either alone or together, from NIH 3T3 and B16F1 cells by siRNA. After 72 h of transfection with a duplex cofilin-1-specific oligonucleotide, cofilin-1 was substantially depleted as determined by Western blotting (Figure 2A, lanes 3 and 7). Quantification of the Western blots demonstrated that in B16F1 and NIH3T3 cell populations the amount of cofilin-1 was reduced to ∼20 and ∼60% levels of the ones in wild-type cells (Supplemental Figure 1). Cofilin-1 siRNA treatment did not decrease the cellular ADF level. However, the ADF levels in cofilin-1 knockdown B16F1 and NIH3T3 cells were increased by ∼2- and 1.5-fold, respectively. Similarly, a duplex oligonucleotide specific for ADF efficiently depleted ADF protein (to <5% in B16F1 and ∼10% in NIH3T3 compared with wild-type cells) (Figure 2A, lanes 2 and 6; Supplemental Figure 1). In ADF knockdown B16F1 cells, the cofilin-1 levels were unaffected, but in NIH 3T3 cells ADF knockdown resulted in a small (∼40%) increase in cofilin-1 levels (Supplemental Figure 1). Treatment of these cells at the same time with ADF- and cofilin-1-specific oligonucleotides depleted both proteins simultaneously (Figure 2A, lanes 4 and 8; Supplemental Figure 1).
To confirm that the depletion of cofilin-1 or ADF is indeed a result of siRNA transfection, we used fluorescein- and rhodamine-labeled siRNA oligonucleotides and compared the antibody staining of transfected cells with the appearance of fluorescent oligonucleotides (Figure 2, B-E). The cells with significantly decreased cofilin-1 or ADF antibody staining displayed punctate oligonucleotide labeling, whereas the cells with normal cofilin-1 or ADF levels did not contain detectable amounts of fluorescent oligonucleotides (Figure 2, B-E). The cells with significantly reduced cofilin-1 or ADF antibody staining will be referred as cofilin-1 or ADF knockdown cells, respectively.
Immunofluorescence microscopy demonstrated that 72 h after cofilin-1 or ADF siRNA transfection, ∼60-90% of the cells contained fluorescent oligonucleotides and did not show detectable cofilin-1 or ADF staining (Figure 3). The depletion of cofilin-1 from B16F1 cells induced a formation of very long and thick stress fibers (Figure 3, D-F). The stress fibers often ended to large F-actin clusters that were defined to be enlarged focal contacts by vinculin staining (our unpublished data). Some cells exhibited also increased F-actin levels in lamellipodia. Most cofilin-1 knockdown cells were also significantly larger than the wild-type cells (Figure 3). The increase in cell size may partly result from a flattened cell shape, but it is also possible that the total cell volume is increased. The depletion of ADF from B16F1 cells induced a small increase in the amount of filamentous actin, but the phenotype was much milder than the one in cofilin-1 knockdown cells. The ADF knockdown cells were often elongated or spindle-shaped and contained abnormally long stress fibers. However, in comparison with cofilin-1 depletion-induced actin stress fibers, the ADF depletion-induced stress fibers were much thinner (Figure 3, G-I). The double-knockdown cells in which both cofilin-1 and ADF were depleted displayed a very similar phenotype than cofilin-1 depletion alone. However, these cells contained more abnormal stress fibers than the individual knockdown cells (Figures 3, J-L).
Figure 3.
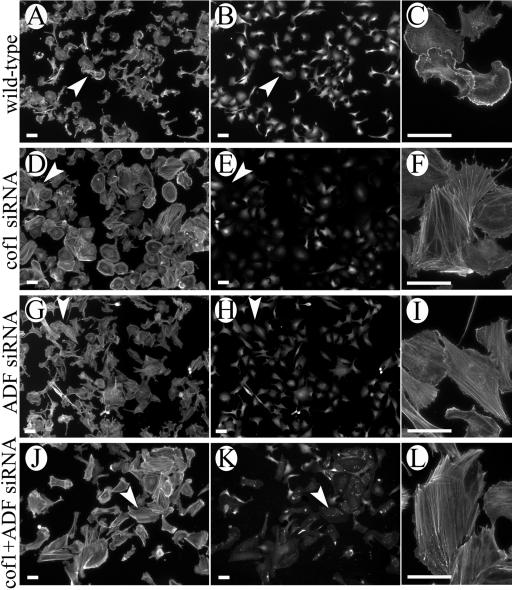
Depletion of ADF or cofilin-1 resulted in an accumulation of thick actin stress fibers and an increase in cell size. B16F1 cells were treated with cofilin-1-(D-F) or ADF (G-I)-specific duplex oligonucleotides, or simultaneously with both oligonucleotides (J-L). Cofilin-1 (B, E, and K) and ADF (H) were visualized by isoform-specific antibodies and F-actin (A, C, D, F, G, I, J, and L) with rhodamine-phalloidin. (K) Both the FITC-labeled siRNA and cofilin-1 antibody staining. Representative cells from each population are indicated by white arrowheads and shown with larger magnifications (C, F, I, and L) (L is rotated 90° counterclockwise). Bars, 50 μm.
It is important to note that ADF is expressed in B16F1 cells only in very small amounts (the molar ratio of ADF:cofilin-1 in these cells is ∼1:11; Figure 1). Therefore, we examined the effects of ADF and cofilin-1 depletion also in NIH 3T3 cells, in which the ADF:cofilin-1 M ratio is ∼1:6 (Figure 1). Depletion of cofilin-1 and/or ADF from NIH 3T3 cells induced formation of stress fibers similarly to B16F1 cells. Typical knockdown cells also exhibited large, smooth lamellipodia instead of small, intensively ruffling lamellipodia characteristic to the wild-type NIH 3T3 cells (Figure 8, G-L). However, in NIH 3T3 cells ADF depletion induced a significantly stronger phenotype than in B16F1 cells, suggesting that the relatively mild effects of ADF depletion in B16F1 cells are a consequence of very low levels of ADF in this cell line.
Figure 8.
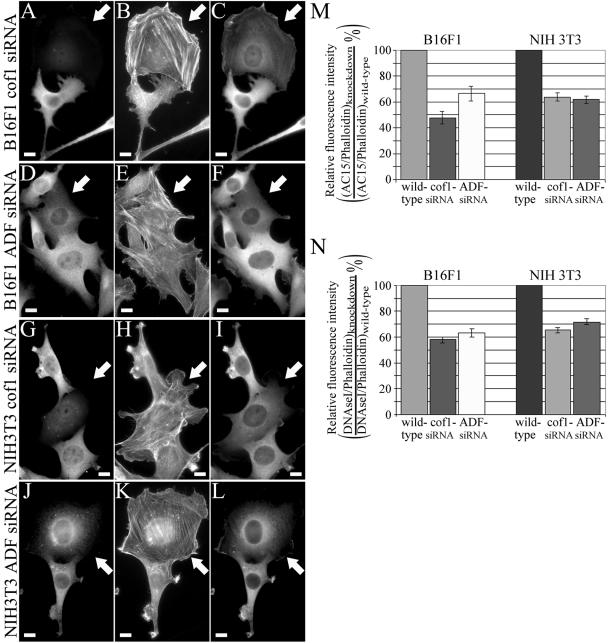
G-actin/F-actin ratio is decreased in cofilin-1/ADF knockdown cells. B16F1 (A-F) and NIH 3T3 (G-L) cells were treated with cofilin-1-(A-C and G-I) or ADF (D-F and J-L)-specific duplex oligonucleotides. Cofilin-1 (A and G) and ADF (D and J) were visualized by isoform-specific antibodies, F-actin with Alexa488-phalloidin (B, E, H, and K), and G+F-actin with β-actin AC-15 antibody (C, F, I, and L). White arrows indicate the siRNA-treated cells with dramatically reduced cofilin-1/ADF levels. Bars, 10 μm. (M and N) The intensities of phalloidin and AC-15 (M) or phalloidin and DNAseI (N) stainings were analyzed from 20 ADF and cofilin-1 knockdown B16F1 and NIH 3T3 cells by TINA software, and the relative AC-15/phalloidin or DNAseI/phalloidin stainings were compared with the ones from neighboring wild-type cells to yield the ratio of (AC-15/phalloidin)knockdown/(AC-15/phalloidin)wild-type (M) or (DNAseI/phalloidin)knockdown/(DNAseI/phalloidin)wild-type (N). The knockdown cells showed a decrease in the amount of G-actin compared with nontransfected wild-type cells. SEMs are indicated in the graphs.
Rescue of the Knockdown Phenotypes
To confirm that the phenotypes of ADF and cofilin-1 knockdown cells indeed result from a decrease in ADF and cofilin-1 protein levels, we attempted to rescue ADF and cofilin-1 knockdown cells by expressing myc-tagged ADF and cofilin-1 that are refractory to the siRNA oligonucleotide duplexes. Cells were first transfected with fluorescein isothiocyanate (FITC)-siRNA and on day 3 further transfected with one of the rescue constructs. siRNA transfection was detected by the appearance of fluorescent oligonucleotides in these cells, and the rescue-construct transfection was detected by anti-myc antibody. B16F1 and NIH 3T3 cells transfected with cofilin-1-siRNA and cofilin-1-myc rescue construct exhibited similar cell morphology and F-actin phenotype to wild-type cells (Figure 4, top; our unpublished data). Similarly, B16F1 and NIH 3T3 cells transfected with ADF-siRNA and ADF-myc rescue construct exhibited a wild-type phenotype (Figure 4, middle; our unpublished data).
Figure 4.
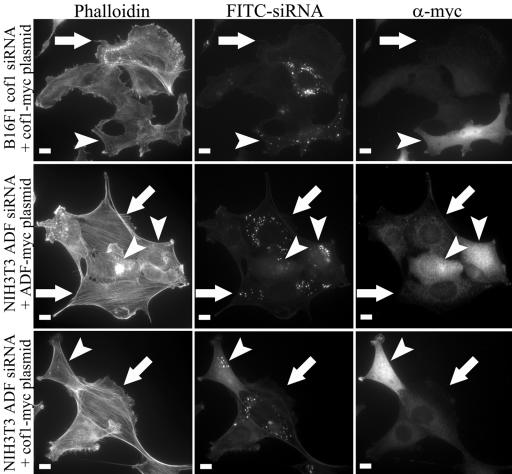
Rescue of ADF or cofilin-1 knockdown phenotype. B16F1 and NIH 3T3 cells were treated with cofilin-1- or ADF-specific FITC-siRNA oligonucleotides followed by a transfection with myc-tagged rescue constructs refractory to siRNA. The cells transfected only with siRNA oligonucleotides (white arrows) were identified by presence of FITC-oligonucleotides (middle row) and by lack of myc-tag staining (right). These cells showed a typical knockdown phenotype with an accumulation of abnormal F-actin structures. The cells transfected with both siRNA and rescue construct (arrowheads) were identified by the simultaneous presence of FITC-oligonucleotides and anti-myc staining. These cells displayed similar actin phenotype to the nontransfected wild-type cells. It is also important to note that the ADF knockdown phenotype in NIH3T3 cells could be rescued by overexpression of cofilin-1 (bottom row). Bars, 10 μm.
Because the cofilin-1 and ADF knockdown phenotypes were very similar to each other (Figures 3 and 8) and the depletion of cofilin-1 or ADF increased the expression level of the other isoform (Figure 2 and Supplemental Figure 1), we also examined whether cofilin-1 knockdown phenotype could be rescued by overexpression of ADF and whether ADF-depletion could be rescued by cofilin-1 overexpression. NIH 3T3 cells transfected with ADF-siRNA and cofilin-1 rescue construct exhibited phenotype indistinguishable from wild-type cells, indicating that overexpression of cofilin-1 can rescue the depletion of ADF (Figure 4, lowest panel). Similarly, NIH 3T3 cells transfected with cofilin-1-siRNA could be rescued by ADF-myc overexpression (our unpublished data), suggesting that these proteins display overlapping functions in cells.
Cofilin-1 and ADF Play Overlapping Roles in Cytokinesis and Cell Motility
We next examined the roles of ADF and cofilin-1 in various cellular processes. To elucidate the roles of ADF and cofilin-1 in cytokinesis of cultured mouse cells, we stained wild-type and ADF/cofilin knockdown cells with DAPI 72 h after siRNA treatment (Figure 5A). Approximately 11 and 7% of cofilin-1 and ADF knockdown NIH 3T3 cells, respectively, contained multiple nuclei, whereas ∼2% of wild-type cells had two nuclei (Figure 5B). Interestingly, almost 30% of the cells depleted of both cofilin-1 and ADF had multiple nuclei, indicating that ADF and cofilin-1 have synergistic effects on division of NIH 3T3 cells (Figure 5B). Most multinucleated knockdown cells contained two nuclei, although ∼5% of the double knockdown cells had three or four nuclei. These results suggest that ADF and cofilin-1 do not affect chromosome replication, but they play an important role in cytokinesis.
Figure 5.
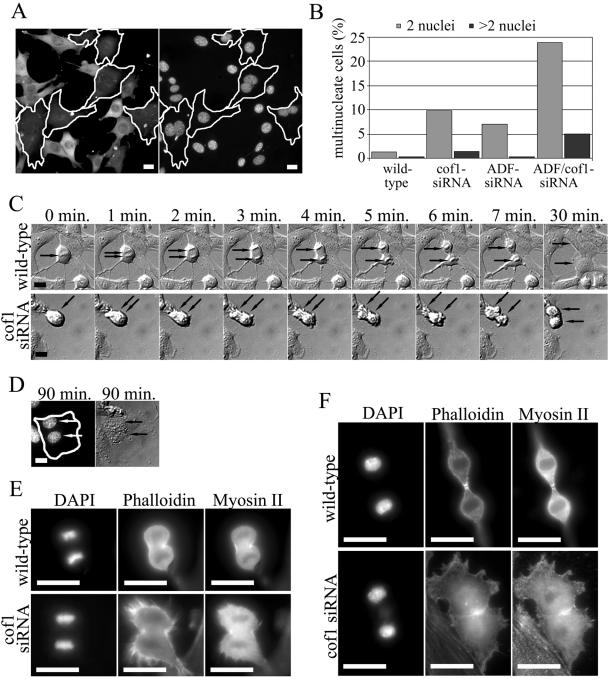
Cofilin-1 and ADF play overlapping roles in cytokinesis. (A and B) Wild-type and cofilin-1 or ADF knockdown NIH 3T3 cells were fixed, and DNA was visualized by DAPI staining. (A) Representative examples of a cofilin-1 knockdown cells (cell borders are indicated by white lines). Cofilin-1 was visualized with an anti-cofilin-1 antibody (left), and DNA with DAPI staining (right). (B) The number of multinucleated cells was counted from at least 700 wild-type and knockdown cells from four independent experiments. The depletion of ADF or cofilin-1 resulted in a small increase in the number of multinucleated cells, whereas the silencing of both genes resulted in a synergistic increase in the amount of multinucleated cells. (C) Time-lapse analysis of cytokinesis of wild-type (top and Supplementary Video 1) and cofilin-1 knockdown (bottom and Supplementary Video 2) NIH 3T3 cells. Frames “0 min.” represent metaphase. Black arrows indicate the positions of chromosomes. Wild-type cells undergo cell division and spreading within 30 min after the metaphase (last frame in C), whereas the process in cofilin-1 knockdown cell is significantly slower and the cell spreading is complete after ∼90 min of metaphase. Supplementary videos display the entire division processes. (D) The same cofilin-1 knockdown cell as shown in C was fixed and stained with cofilin-1 antibodies and with DAPI (first panel) to visualize the two nuclei present in the cofilin-1 knockdown cell. (E and F) Visualization of F-actin and myosin II during cytokinesis in wild-type and cofilin-1 knockdown cells. Wild-type and cofilin-1 knockdown B16F1 cells were fixed, DNA was visualized by DAPI, F-actin by phalloidin, and myosin II by an anti-myosin II antibody. Representative cells undergoing telophase (E) and late telophase (F) are shown. Bars, 20 μm.
To further elucidate the role of ADF/cofilins in cytokinesis, we monitored dividing wild-type, cofilin-1, and ADF knockdown NIH3T3 cells by acquiring time-lapse images every 60 s during the cell division process. Cofilin-1 knockdown cells displayed severe problems after metaphase. These knockdown cells typically succeeded in forming a primitive cleavage furrow (Figure 5C, time-lapse images 3-6 min), but they were defective in the final stage of the cytokinesis when the two cells are supposed to separate from each other. A very similar phenotype was seen in ADF knockdown cells (our unpublished data). Typically, the knockdown cells tried unsuccessfully to separate from each other for a relatively long time, after which they either managed to divide by struggling or failed to separate and fused back with each other to form a cell with two nuclei (Figure 5, C and D, and Supplementary Video 2). The complete cytokinesis process was typically much slower in cofilin-1 knockdown cells (∼90 min) than in wild-type cells (∼30 min). After the time-lapse monitoring, the cells were stained with DAPI to visualize the nuclei (Figure 5D) and with ADF or cofilin-1 antibody to distinguish knockdown cells from wild-type cells.
To elucidate the mechanism by which ADF/cofilins participate in cytokinesis, we compared the localization of F-actin and myosin II in wild-type and cofilin-1 knockdown cells during various mitotic stages. In B16F1 cells, both F-actin and myosin II localized to cleavage furrow, whereas in NIH 3T3 cells clear myosin II localization to cleavage furrow was not observed. Thus, B16F1 cells were chosen for further analysis. Representative wild-type and cofilin-1 knockdown cells during telophase (Figure 5E) and late telophase (Figure 5F) were selected based on morphological structures of the chromosomes. The cells also were stained with an anti-tubulin antibody to further confirm the stage of cell division (our unpublished data). In wild-type B16F1 cells, both myosin II and F-actin were aligned around the cleavage furrow during early telophase (Figure 5E, top). As cytokinesis proceeded, the diameter of the contractile ring decreased (Figure 5F, top), resulting in a separation of the two daughter cells. In cofilin-1 knockdown cells, the alignment of myosin II and F-actin occurred similarly to wild-type cells during telophase. However, cofilin-1 knockdown cells exhibited relatively strong F-actin staining throughout the cytoplasm, whereas in wild-type cells the F-actin staining was mainly concentrated to the cleavage furrow (Figure 5E). During late telophase, an aberrant accumulation of myosin II and F-actin was detected in the contractile rings of cofilin-1 knockdown cells, and the contraction of this actin structure was defective (Figure 5F, lower panel).
Actin dynamics is central to cell motility (reviewed in Pollard et al., 2000; Pantaloni et al., 2001) and thus we monitored the migration of ADF, cofilin-1, and ADF/cofilin-1 siRNA-treated NIH 3T3 cells on fibronectin during a 20-h period. The degree of cell migration was assayed by analyzing the phagokinetic tracks of these cells on coverslips coated with blue fluorescent beads. After 20 h, the cells were fixed with 3.7% formaldehyde and stained with anti-ADF, anti-cofilin-1, or a mixture of the two antibodies to distinguish knockdown cells from wild-type cells. The cells with normal ADF/cofilin levels migrated over relatively long distances during the 20-h period and displayed clear directional motility (Figure 6A). In contrast, the cells lacking ADF or cofilin-1 cleared beads only in the area immediately around the cells or displayed very short directional tracks (Figure 6, B-D). We quantified the minimum distance of migration for >24 wild-type and knockdown cells and showed that depleting ADF or cofilin-1 resulted in a 2- or 2.5-fold decrease in track length (Figure 6E). The cells in which both ADF and cofilin-1 were depleted showed a threefold decrease in their track lengths compared with wild-type cells (Figure 6E). Because knockdown cells are generally much larger than wild-type cells, their true defects in directional migration are likely to be even larger. The observation that knockdown cells cleared fluorescent beads in their vicinities suggests that they were able to extend and retract their lamellipodia despite a severe defect in motility. Therefore, the lack of motility of ADF and cofilin-1 knockdown cells may result from defects in cell polarization (Figure 3).
Figure 6.
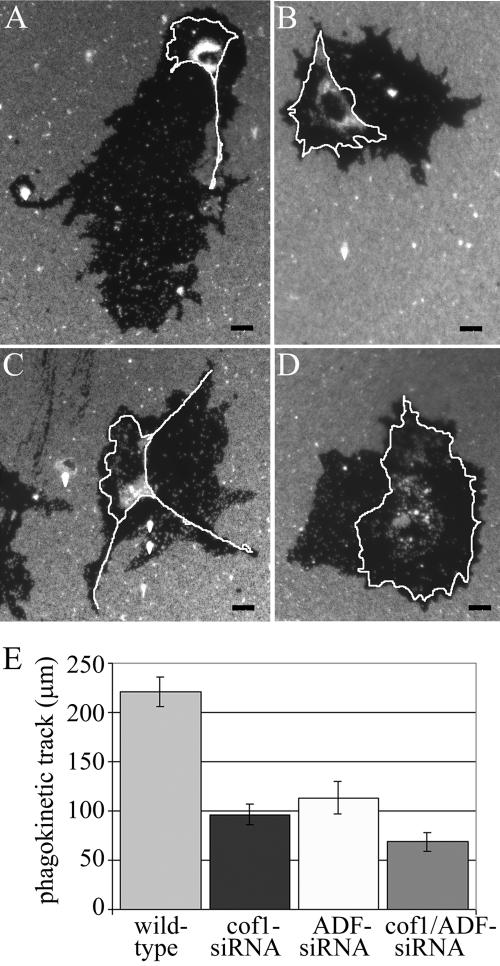
Cofilin-1 and ADF are required for cell migration. Wild-type and cofilin-1/ADF knockdown NIH 3T3 cells were plated on coverslips coated with fibronectin and blue fluorescent beads, grown for 20 h, and fixed for immunofluorescence. The wild-type cells (A) exhibited relatively long and thin phagokinetic motility tracks. In contrast, the cofilin-1 (B), ADF (C) and cofilin-1/ADF (D) knockdown cells showed clearance of beads only in their immediate vicinity and seldom displayed directional motility tracks. The borders of the cells are indicated with white lines. Bars, 20 μm. (E) The minimum motility distances were quantified from wild-type, cofilin-1, ADF, and cofilin-1/ADF knockdown cells (n ≥ 24). The silencing of cofilin-1 or ADF results in 2- to 2.5-fold decreases, and cofilin-1/ADF results in threefold decrease in the length of the phagokinetic motility tracks. SEMs are indicated in the graph.
The role of ADF/cofilins in the cell migration was also investigated by videomicroscopy. In these assays, we used highly motile B16F1 cells, and the migration of the cells was further induced by addition of AlF4- to the medium (Hahne et al., 2001). After AlF4- treatment, wild-type cells were highly polar and displayed directional migration. Although these cells occasionally changed the direction during migration, they displayed clear protruding leading edge and retracting tail. In contrast, cofilin-1 knockdown cells were typically nonpolarized and often projected multiple lamellipodia to different directions. Quantification of velocities of 35 wild-type and 25 cofilin-1 knockdown cells demonstrated that cofilin-1 depletion results in ∼twofold decrease in cell migration (Figure 7C).
Figure 7.
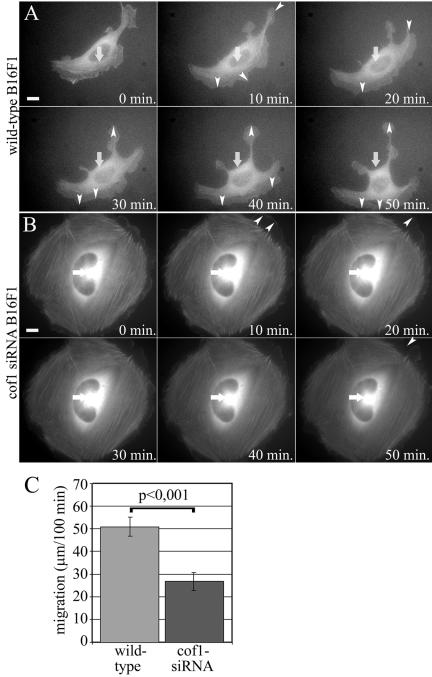
Live cell analysis of wild-type and cofilin-1 knockdown B16F1 cell migration. Wild-type B16F1 cells expressing GFP-actin (A and Supplementary Video 3) displayed fast actin dynamics in the lamellipodia and directional cell motility. Cofilin-1 knockdown cells (B and Supplementary Video 4) were unable to migrate but were still capable of slowly extending and retracting their lamellipodia. White arrows indicate the locations of the nuclei in the first frame. White arrowheads indicate largest protrusions and retractions. Bars, 10 μm. (C) Migration of 35 wild-type and 25 cofilin-1 knockdown B16F1 cells were monitored for 100 min, and the positions of the nuclei was tracked every 20 min. The average motility distances of wild-type cells are 51.0 μm and cofilin-1 knockdown cells 26.9 μm. SEMs and statistical significance of the data are indicated in the graph.
To visualize cytoskeletal defects in ADF- or cofilin-1 knockdown cells during migration, we examined GFP-actin-expressing wild-type and knockdown cells. Similarly to wild-type B16F1 cells described above, also B16F1 cells expressing GFP-actin were highly motile. The actin cytoskeletons in these cells were constantly and rapidly reorganizing (Figure 7A and Supplementary Video 3). ADF knockdown cells did not significantly differ from wild-type cells (our unpublished data), whereas a dramatic accumulation of stable stress fibers was observed in cofilin-1 knockdown cells (Figure 7B and Supplementary Video 4). Similarly to the cytokinesis studies shown in Figure 5, the cells were fixed after the experiment and stained with cofilin-1 or ADF antibody to distinguish cofilin-1/ADF knockdown cells from wild-type cells. Together with the data shown in Figure 6, these experiments demonstrate that ADF/cofilins are essential for motility of mammalian cells.
Cofilin-1 and ADF Contribute to Fast Actin Treadmilling by Depolymerizing Actin Filaments
Phalloidin staining of cofilin-1 and ADF knockdown cells (Figure 3) suggests that they contain more F-actin than wild-type cells. Also the directional motility was abolished in cofilin-1/ADF knockdown cells (Figures 6 and 7), suggesting that the cytoplasmic G-actin pool may be diminished. We thus next investigated the relative ratios of G-actin versus F-actin in wild-type B16F1 and NIH 3T3 cells and in ADF/cofilin-depleted cells. In this experiment, F-actin was visualized by Alexa488-phalloidin and the “total” F+G-actin was visualized by an AC-15 antibody. However, it is important to note that the AC-15 antibody does not stain stress fibers presumably due to the presence of certain actin-binding proteins, which block the access to the epitope in vivo (Mies et al., 1998). Therefore, this antibody is expected to visualize the total cellular actin excluding the stress fibers. Quantification of the intensities of phalloidin and AC-15 antibody stainings in knockdown cells and in neighboring wild-type cells showed that the ratio of AC-15 versus phalloidin intensity was decreased by 40-50% in ADF or cofilin-1-depleted cells compared with the wild-type cells, indicating that there is more F-actin and less G-actin in knockdown cells than in wild-type cells (Figure 8M). Similar results also were obtained when G-actin was visualized by DNAseI and compared with phalloidin staining similarly to AC-15/phalloidin experiment described above (Figure 8N).
To investigate actin filament treadmilling rates in wild-type and cofilin-1 or ADF knockdown cells, we used FRAP. In these experiments, we bleached a region of B16F1 cells expressing GFP-actin (Ballestrem et al., 1998) by intense laser irradiation and then monitored the exchange between the bleached and unbleached populations of GFP-actin. The experiment was first carried out to compare the dynamics of stress fibers of eight wild-type, nine cofilin-1, and nine ADF knockdown cells. In each case, the recovery of fluorescence at the bleached region was monitored for 255 s after laser irradiation. After the FRAP experiment, the transfection of the knockdown cells was confirmed. Representative examples of wild-type, cofilin-1, and ADF knockdown cells are shown in Figure 9. In wild-type cells, the fluorescence recovery at the bleached region was relatively rapid with a nearly complete exchange at stress fibers within 140 s (Figure 9, A and B). In contrast, the exchange between the bleached and unbleached regions in cofilin-1 knockdown cells was significantly slower and was not completed during the 255-s monitoring period (Figure 9, A and B). The rate of fluorescence recovery of the ADF knockdown cells seemed to be slightly slower than in wild-type cells, although the initial (up to 70 s after the bleaching) fluorescence recovery in ADF knockdown cells was close to the one in wild-type cells.
Figure 9.
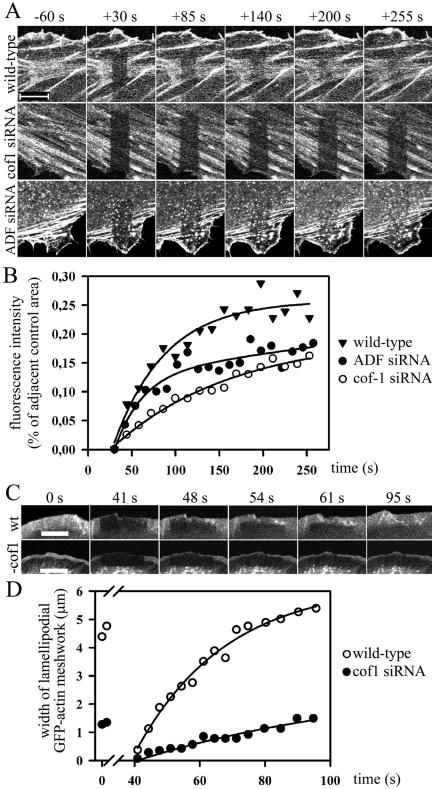
FRAP analysis of actin treadmilling rates in stress fibers and lammelipodia. B16F1 cells expressing GFP-actin were treated with a cofilin-1- or ADF-specific duplex oligonucleotides and incubated for 5-12 h before analysis. The selected cell region was bleached with an intense laser beam and the fluorescence recovery was monitored by taking time-lapse images. (A and B) Actin dynamics at stress fibers. Time-lapse images were acquired every 10 s starting at 30 s after bleaching. (A) Representative examples of wild-type (top), cofilin-1 (middle), and ADF (bottom) knockdown cells before and after bleaching. Bars, 10 μm. (B) The rate of fluorescence recovery of the bleached region was analyzed with TINA software from representative wild-type (black triangles) and ADF (black squares) or cofilin-1 knockdown (open squares) cells. In each frame, the fluorescence intensity of the bleached region was compared with the fluorescence of the control region (in same picture next to bleached region) to diminish the error caused by normal photobleaching during the monitoring period. The equilibration of fluorescence between bleached and un-bleached regions in cofilin-1 knockdown cells is significantly slower than in wild-type cells. (C and D) Actin dynamics at lamellipodial regions. Time-lapse images were acquired every 3 to 4 s immediately after bleaching. (C) Representative examples of wild-type (top) and cofilin-1 knockdown cells (bottom). It is important to note that the photobleaching was carried out during the time period 3.5-37.5 s and thus the 41-s time-lapse image represents the situation at 3.5 s after the end of bleaching. Bars, 10 μm. (D) The rate of lamellipodial actin meshwork growth was quantified from wild-type and knockdown cells. Time points 0 and 3.36 show the widths of lamellipodial actin meshworks before bleaching, and the time points at 41-95 s show the widths of the lamellipodial GFP-actin meshworks after the bleaching period.
Actin filament turnover rates in stress fibers are believed to be slower than the ones at the cortical actin cytoskeleton. This is probably due to the presence of tropomyosins in stress fibers (Des Marais et al., 2002). Tropomyosins compete with ADF/cofilins in actin-binding and inhibit their actin filament depolymerization/severing activities (Bernstein and Bamburg, 1982; Ono and Ono, 2002). Thus, we also examined the dynamics of lamellipodial actin meshwork in B16F1 cells by a FRAP experiment. A 4-μm-wide region at the leading edge of lamellipodia of wild-type and cofilin-1 knockdown cells was bleached by intense laser irradiation and the rate of GFP-actin accumulation to the leading edge of lamellipodia was followed every 3 to 4 s for a 35-s period and then every 5 s for a 25-s period. Data from a representative wild-type and cofilin-1 knockdown cell is shown (Figure 9, C and D). The width of the lamellipodial actin meshwork in wild-type cells grew significantly faster than in cofilin-1 knockdown cells. Quantification of the data from four wild-type and four cofilin-1 knockdown cells showed that the rate of lamellipodial actin meshwork growth in wild-type cells is ∼4.9 μm/min, whereas the corresponding growth rate in cofilin-1 knockdown cells is ∼2.1 μm/min. Very similar results were also obtained when the recovery of fluorescence intensity versus time was compared from the lamellipodia of wild-type and cofilin-1 knockdown cells (our unpublished data). Together, these data show that cofilin-1 depletion severely diminishes actin filament treadmilling rates in both stress fibers and lamellipodia of B16F1 cells.
Because the decrease in actin filament turnover rates in cofilin-1 knockdown cells can result either from decreased actin filament polymerization or depolymerization rates, we next compared the actin filament depolymerization rates of wild-type, ADF, cofilin-1, and ADF/cofilin-1 knockdown cells by using latrunculin-A, an actin monomer-sequestering drug. In cells, latrunculin-A causes a rapid and specific disruption of the actin cytoskeleton. Because latrunculin-A functions by sequestering actin monomers, the rate of disappearance of actin structures is expected to reflect the rate of actin monomer dissociation from filament ends (Coue et al., 1987; Ayscough et al., 1997). The actin filament depolymerization assay was carried out for B16F1 cells by using 2 μM latrunculin-A. The majority (80%) of wild-type cells lost all their stress fibers within 5 min after addition of latrunculin-A, and the remaining F-actin was concentrated to small aggregates (Figure 10). This was accompanied by significant changes in cell morphology. Similarly to wild-type cells, the majority of ADF knockdown cells lost their stress fibers and retracted already after 5 min. In contrast, the disappearance of stress fibers was much slower in cofilin-1 and cofilin-1/ADF knockdown cells. After 5 min of latrunculin-A addition, only ∼10% of the cells had lost their stress fibers (Figure 10, C, D, G, and H), and even after 30-min latrunculin-A treatment a significant proportion of these cells (80%) were morphologically nearly normal (Figure 10I). Although the depletion of ADF from B16F1 cells did not have significant effect on the stability of F-actin structures, an identical assay carried out for NIH 3T3 cells demonstrated that in this cell line ADF depletion resulted in comparable effects on actin filament depolymerization rates than cofilin-1 depletion does (our unpublished data). These data suggest that both cofilin-1 and ADF contribute to rapid actin dynamics by depolymerizing actin filaments. The low levels of ADF in B16F1 cells (Figure 1B) provide an explanation for the relatively small defects of ADF depletion for actin dynamics in these cells.
Figure 10.
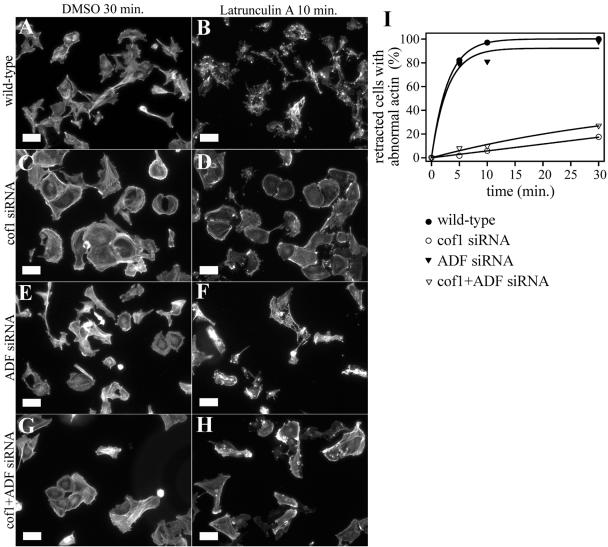
Actin filament depolymerization rates in wild-type and ADF/cofilin-1 knockdown cells. Filamentous actin was visualized by phalloidin staining in wild-type (A and B), cofilin-1 knockdown (C and D), ADF knockdown (E and F), and cofilin-1/ADF knockdown (G and H) B16F1 cells after the addition 2 μM latrunculin-A. Time points 30 min after DMSO addition (control) (A, C, E, and G) and 10 min after addition of 2 μM latrunculin-A (B, D, F, and H) are shown. (I) Percentage amount of retracted cells without clear stress fibers and with abnormal F-actin aggregates were counted (n = 50 cells) at time points 5, 10, and 30 min after latrunculin-A addition. The actin filament structures were rapidly disrupted in wild-type cells (black squares). In contrast, stress fibers disappeared much more slowly in cofilin-1 (open squares) and cofilin-1/ADF (open triangles) knockdown cells. The ADF knockdown cells (black triangles) lost their actin filament structures almost as quickly as the wild-type cells. Bars, 50 μm.
DISCUSSION
ADF/cofilins are small proteins that can sever and depolymerize actin filaments in vitro. However, the mechanism by which these ubiquitous proteins contribute to actin dynamics in cells has been a matter of controversy. Here, we analyzed the cellular roles of the two mammalian nonmuscle isoforms, ADF and cofilin-1, by using isoform-specific antibodies and siRNA knockdown methods. Our data provide direct evidence that 1) ADF/cofilins contribute to cytoskeletal dynamics in most mammalian cell types by promoting actin filament depolymerization; 2) ADF/cofilins play an important role in cytokinesis, cell motility, and morphogenesis in mammals; and 3) ADF and cofilin-1 are coexpressed in many cell-types where they play overlapping roles in actin filament depolymerization and above-mentioned cellular processes.
ADF/Cofilins Promote Rapid Actin Filament Depolymerization in Cells
In this study, we used siRNA-induced gene silencing to deplete cofilin-1 or ADF, either individually or simultaneously, from various mouse cell lines. The analysis of NIH 3T3 and B16F1 cells demonstrated that depletion of ADF and/or cofilin-1 led to an accumulation of abnormal F-actin structures and to a simultaneous decrease in the cellular G-actin concentration. An increased number and thickness of stress fibers was observed by phalloidin staining as well as in knockdown cells expressing GFP-actin. Very similar actin phenotypes also were observed in ADF/cofilin-depleted Neuro 2A and N18 cells (our unpublished data). FRAP analysis of GFP-actin expressing B16F1 cells provided direct evidence that actin filament treadmilling rates in both stress fibers and lamellipodia are severely diminished in cofilin-1 knockdown cells. Furthermore, studies with actin-sequestering drug latrunculin-A showed that the loss of actin filament structures are much slower in ADF/cofilin-1 knockdown cells than in wild-type cells, indicating that actin filament depolymerization rates are severely diminished in cells lacking ADF/cofilin. It is important to note that actin filament turnover is too rapid at the cortical actin structures (such as the leading edge of a motile cell) to be examined with the time resolution of our latrunculin-A assay. Thus, in this assay we only concentrated on analyzing the depolymerization rates of stress fibers in wild-type and ADF/cofilin knockdown cells. Nevertheless, these studies provide direct evidence that ADF/cofilins contribute to cytoskeletal dynamics, at least in NIH 3T3, B16F1, Neuro 2A, and N18 cells, by depolymerizing actin filaments and thus providing new actin monomers to the cytoplasmic pool.
These results are in accordance with previous data obtained with yeast, Drosophila, and C. elegans strains carrying mutations in ADF/cofilins as well as with studies in which ADF/cofilins were inactivated in cells by overexpression of LIM-kinase or by depletion of cyclase-associated protein, which is an important ADF/cofilin recycling protein in cells (Gunsalus et al., 1995; Lappalainen and Drubin, 1997; Arber et al., 1998; Yang et al., 1998; Ono et al., 1999; Chen et al., 2001; Dong et al., 2001; Niwa et al., 2002; Bertling et al., 2004). In contrast to these data, recent studies on EGF-stimulated rat adenocarcinoma cells indicated that instead of promoting filament depolymerization, ADF/cofilins would contribute to cytoskeletal dynamics by increasing the number of assembly competent barbed ends through their filament-severing activity. Thus, ADF/cofilins would promote actin filament assembly rather than disassembly at the leading edge of the cell (Chan et al., 2000; Zebda et al., 2000; Ghosh et al., 2004). In a similar barbed-end assembly assay to the one used in the studies with rat adenocarcinoma cells, we could not detect significant effect of ADF/cofilin depletion to the number of barbed ends in B16F1 cells (our unpublished data). This suggests that in B16F1 cells, ADF/cofilins contribute to actin dynamics primarily by depolymerizing old filaments rather than by creating new barbed ends for actin assembly through the filament-severing activity. However, in certain cell types or after specific stimuli, ADF/cofilins also contribute to cytoskeletal dynamics by increasing actin filament assembly through their severing activity. We speculate that in cells having a very large G-actin pool, e.g., due to high expression levels of β-thymosins, the weak actin filament-severing activity of ADF/cofilins initially results in actin filament assembly rather than disassembly and that this would continue until the concentration of the cytoplasmic G-actin becomes limiting.
ADF and Cofilin-1 Play Overlapping Roles in Cells
Two ADF/cofilin isoforms, ADF and cofilin-1, are present in mammalian nonmuscle cells and are coexpressed in many cell types. ADF and cofilin-1 have quantitatively different effects on actin dynamics in vitro, but whether these proteins contribute to actin dynamics and various processes with similar or different mechanisms in cells was not previously examined (Vartiainen et al., 2002; Yeoh et al., 2002). The former methods for studying the cellular function of mammalian ADF/cofilins, such as overexpression of LIM-kinase and depletion of cyclase-associated protein, are not specific toward certain ADF/cofilin isoforms (Amano et al., 2001, 2002; Bertling et al., 2004). Thus, these studies could not reveal whether ADF and cofilin-1 have similar or different effects on actin dynamics in vivo.
By using isoform-specific antibodies and siRNA oligonucleotides, we provide evidence that ADF and cofilin-1 play overlapping roles in actin filament turnover and various cell processes. Depletion of either ADF or cofilin-1 resulted in similar effects to actin filament turnover, cell morphogenesis, motility, and cytokinesis in our assays. More importantly, in all cases a simultaneous depletion of ADF and cofilin-1 resulted in more pronounced phenotype than either one of the individual depletions. The overlapping roles of ADF and cofilin-1 are further supported by the fact that the ADF knockdown phenotype could be rescued by overexpression of cofilin-1 and cofilin-1 knockdown phenotype by ADF overexpression in NIH 3T3 cells. Similar results have been recently obtained from a study on mice carrying mutations in the ADF gene. The only observed phenotype in these mice was bilateral blindness, but the authors noticed that in ADF mutant mice cofilin-1 was strongly overexpressed in certain tissues where ADF was normally present (Ikeda et al., 2003).
Western blot analysis showed that cofilin-1 is the major isoform in NIH 3T3, B16F1, and Neuro 2A cells. ADF is expressed at approximately sixfold lower molar ratios in NIH 3T3 and Neuro 2A and at 11-fold lower molar ratio in B16F1 cells. Depletion of cofilin-1 or ADF induced comparable phenotypes in NIH 3T3 cells, whereas in B16F1 cells ADF depletion resulted in significantly weaker effect than cofilin-1 depletion. We suggest that the differences in the phenotypes of ADF depletion between NIH 3T3 and B16F1 cells result from the lower expression levels of ADF in B16F1 cells. It is also interesting to note that although the molar ratio of ADF:cofilin-1 in NIH 3T3 cells is ∼1:6, the depletion of ADF alone from this cell type resulted in a significant phenotype. However, previous studies have demonstrated that ADF has much stronger actin filament depolymerization activity than cofilin-1, suggesting that the depletion of ADF from cells should indeed result in significantly stronger effect on actin dynamics than what would be expected from its molar ratio to cofilin-1 (Vartiainen et al., 2002; Yeoh et al., 2002).
ADF and Cofilin-1 Are Necessary for the Proper Cytokinesis and Cell Motility
Our work provides direct evidence that ADF and cofilin-1 play overlapping roles in cytokinesis in mammalian cells. Significant proportions of ADF and cofilin-1 knockdown cells were multinuclear, and this phenotype was even more pronounced in ADF/cofilin-1 double knockdown cells. The involvement of ADF/cofilins in cytokinesis is supported by previous studies showing that inactivation of these proteins in other model organisms cause failures in cytokinesis (Gunsalus et al., 1995; Abe et al., 1996; Lappalainen and Drubin, 1997). Furthermore, ADF/cofilins have been shown to localize to the cleavage furrow and the midbody during the cytokinesis of mammalian cells and fertilized Xenopus eggs (Nagaoka et al., 1995; Abe et al., 1996).
Our time-lapse analysis demonstrated that the ADF and cofilin-1 knockdown cells could form a primitive cleavage furrow, but the proper contraction and final abscission seemed to be severely defective in these cells. Correspondingly, myosin II and phalloidin stainings demonstrated that contractile ring and primitive cleavage furrow were properly aligned in cofilin-1 knockdown cells, but the contraction and dynamics of the contractile ring were defective. During late telophase, an aberrant accumulation of F-actin and myosin II occurred in the contractile ring. Thus, these data indicate that ADF/cofilins play an important role in cytokinesis by regulating the turnover and disassembly of the contractile ring. However, the assembly of the contractile ring seems to proceed properly in cofilin-1 knockdown cells. These findings are supported by previous studies in which ADF/cofilins were depleted from Drosophila S2 tissue culture cells by RNA interference (Somma et al., 2002) and by Slingshot mutation, which increased the proportion of phosphorylated ADF/cofilin in cells (Kaji et al., 2003). Both studies showed that ADF/cofilins are important for cell division and that inactivation of these proteins leads to an abnormal F-actin accumulation near the midbody during the final stage of cytokinesis.
Analysis of knockdown cells also demonstrated that ADF and cofilin-1 play overlapping roles in cell motility. Together with FRAP and latrunculin-A assays and with quantification of G- versus F-actin ratios in wild-type versus knockdown cells, these data suggest that ADF and cofilin-1 contribute to cell migration by depolymerizing actin filaments and by providing new actin monomers for assembly at barbed ends. Interestingly, although ADF and cofilin-1 knockdown cells were not capable in directional motility on coverslips, they were capable in slowly extending and retracting their lamellipodia. The B16F1 cofilin-1 knockdown cells activated with AlF4- often established continuous lamellipodia, which protruded to multiple directions. This resulted in round, flat cell morphology or a cell exhibiting two separate leading edges, which were protruding to opposite directions. These data indicate that cofilin-1 is required to set the direction of cell migration. Similarly, an increase in the amount of inactive (phosphorylated) ADF/cofilins by overexpression of LIM kinase in fibroblasts has been shown to induce loss of cell polarity and directed motility (Dawe et al., 2003) and local activation of caged cofilin determined the direction of cell migration in MTLn3 cells (Ghosh et al., 2004). It is possible that directional cell migration requires rapid actin filament depolymerization and consequent recycling of actin monomers to the barbed ends. In contrast, transient, nonpolar protrusions of lamellipodia may take place also in the absence of rapid ADF/cofilin-induced actin filament depolymerization. Because ADF/cofilin knockdown cells also contained abnormal actin stress fibers and enlarged focal adhesions, it is possible that the cell motility defects are also partly a result of the strong adhesion of ADF/cofilin-depleted cells to the substrate.
This work demonstrates that the two mammalian nonmuscle ADF/cofilin isoforms play overlapping role in actin dynamics and in various cell processes. Cofilin-1 is expressed in most nonmuscle cells, and we speculate that it is required to maintain actin filament dynamics that is important to most central cellular processes. ADF is expressed especially strongly in neurons and certain epithelial cell types, and we propose that the generation and maintenance of highly polar morphology in these cells may require very rapid actin filament turnover rates. This could be promoted by coexpression of ADF and cofilin-1 in these cells. However, it is possible that the rapid, ADF-induced actin filament depolymerization is also essential for certain cell processes that are specific for neurons and epithelial cells. Therefore, in the future it will be important to compare the localizations and cellular roles of ADF and cofilin-1 in a variety of highly polarized cell types. Furthermore, it will be important to elucidate whether the activities of ADF and cofilin-1 are regulated through same or different signaling pathways.
Supplementary Material
Acknowledgments
This study was supported by grants from the Academy of Finland, Biocentrum Helsinki, Sigrid Jusélius Foundation, Emil Aaltonen Foundation, and the European Molecular Biology Organization Young Investigator Program (to P. L.). Dr. Keith Kozminski is acknowledged for critical reading of the manuscript, Dr. Kimmo Tanhuanpää for advice with the videomicroscope, and Cell Imaging Core of Turku Center for Biotechnology for the help with confocal microscope. Miia Koskinen is acknowledged for excellent technical assistance.
Article published online ahead of print in MBC in Press on November 17, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0555).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abe, H., Obinata, T., Minamide, L. S., and Bamburg, J. R. (1996). Xenopus laevis actin-depolymerizing factor/cofilin: a phosphorylation-regulated protein essential for development. J. Cell Biol. 132, 871-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano, T., Kaji, N., Ohashi, K., and Mizuno, K. (2002). Mitosis-specific activation of LIM motif-containing protein kinase and roles of cofilin phosphorylation and dephosphorylation in mitosis. J. Biol. Chem. 277, 22093-22102. [DOI] [PubMed] [Google Scholar]
- Amano, T., Tanabe, K., Eto, T., Narumiya, S., and Mizuno, K. (2001). LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 354, 149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, S., Barbayannis, F. A., Hanser, H., Schneider, C., Stanyon, C. A., Bernard, O., and Caroni, P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805-809. [DOI] [PubMed] [Google Scholar]
- Ayscough, K. R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D. G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem, C., Wehrle-Haller, B., and Imhof, B. A. (1998). Actin dynamics in living mammalian cells. J. Cell Sci. 111, 1649-1658. [DOI] [PubMed] [Google Scholar]
- Bamburg, J. R., McGough, A., and Ono, S. (1999). Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9, 364-370. [DOI] [PubMed] [Google Scholar]
- Bernstein, B. W., and Bamburg, J. R. (1982). Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil. 2, 1-8. [DOI] [PubMed] [Google Scholar]
- Bertling, E., Hotulainen, P., Mattila, P. K., Matilainen, T., Salminen, M., and Lappalainen, P. (2004). Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell 15, 2324-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, M. F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G. X., Hong, Y., Chua, N. H., and Pantaloni, D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, M. F., Ressad, F., and Pantaloni, D. (1999). Control of actin dynamics in cell motility. Role of ADF/cofilin. J. Biol. Chem. 274, 33827-33830. [DOI] [PubMed] [Google Scholar]
- Chan, A. Y., Bailly, M., Zebda, N., Segall, J. E., and Condeelis, J. S. (2000). Role of cofilin in EGF-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148, 531-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Godt, D., Gunsalus, K., Kiss, I., Goldberg, M., and Laski, F. A. (2001). Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat. Cell Biol. 3, 204-209. [DOI] [PubMed] [Google Scholar]
- Coue, M., Brenner, S. L., Spector, I., and Korn, E. D. (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, 316-318. [DOI] [PubMed] [Google Scholar]
- Dawe, H. R., Minamide, L. S., Bamburg, J. R., and Cramer, L. P. (2003). ADF/cofilin controls cell polarity during fibroblast migration. Curr. Biol. 13, 252-257. [DOI] [PubMed] [Google Scholar]
- Des Marais, V., Ichetovkin, I., Condeelis, J., and Hitchcock-DeGregori, S. E. (2002). Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115, 4649-4660. [DOI] [PubMed] [Google Scholar]
- Dong, C. H., Xia, G. X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N. H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Ghosh, M., Song, X., Mouneimne, G., Sidani, M., Lawrence, D. S., and Condeelis, J. S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743-746. [DOI] [PubMed] [Google Scholar]
- Gunsalus, K. C., Bonaccorsi, S., Williams, E., Verni, F., Gatti, M., and Goldberg, M. L. (1995). Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 131, 1243-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne, P., Sechi, A., Benesch, S., and Small, J. V. (2001). Scar/WAVE is localized at the tips of protruding lamellipodia in living cells. FEBS Lett. 492, 215-220. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., Grant, W., and Condeelis, J. (2002). Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12, 79-84. [DOI] [PubMed] [Google Scholar]
- Ikeda, S., Cunningham, L. A., Boggess, D., Hobson, C. D., Sundberg, J. P., Naggert, J. K., Smith, R. S., and Nishina, P. M. (2003). Aberrant actin cytoskeleton leads to accelerated proliferation of corneal epithelial cells in mice deficient for destrin (actin depolymerizing factor). Hum. Mol. Genet. 12, 1029-1037. [DOI] [PubMed] [Google Scholar]
- Kaji, N., Kazumasa, O., Shuin, M., Niwa, R., Uemura, T., and Mizuno, K. (2003). Cell cycle-associated changes in Slingshot phosphatase activity and roles in cytokinesis in animal cells. J. Biol. Chem. 278, 33450-33455. [DOI] [PubMed] [Google Scholar]
- Lappalainen, P., and Drubin, D. G. (1997). Cofilin promotes rapid actin filament turnover in vivo. Nature 388, 78-82. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., Matheson, C., Marra, M. A., Wakarchuk, M. F., and Baillie, D. L. (1994). The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242, 346-357. [DOI] [PubMed] [Google Scholar]
- Mies, B., Rottner, K., and Small, J. V. (1998). Multiple immunofluorescence microscopy of the cytoskeleton. In: Cell Biology: A Laboratory Handbook, 2nd ed., ed. J. E. Celis, New York: Academic Press, 469-476.
- Moon, A. L., Janmey, P. A., Louie, K. A., and Drubin, D. G. (1993). Cofilin is an essential component of the yeast cortical cytoskeleton. J. Cell Biol. 120, 421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka, R., Abe, H., Kusano, K., and Obinata, T. (1995). Concentration of cofilin, a small actin-binding protein, at the cleavage furrow during cytokinesis. Cell Motil. Cytoskeleton 30, 1-7. [DOI] [PubMed] [Google Scholar]
- Niwa, R., Nagata-Ohashi, K., Takeichi, M., Mizuno, K., and Uemura, T. (2002). Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108, 233-246. [DOI] [PubMed] [Google Scholar]
- Ono, S., Baillie, D. L., and Benian, G. M. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., Minami, N., Abe, H., and Obinata, T. (1994). Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 269, 15280-15286. [PubMed] [Google Scholar]
- Ono, S., and Ono, K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni, D., Le Clainche, C., and Carlier, M. F. (2001). Mechanism of actin-based motility. Science 292, 1502-1506. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., Blanchoin, L., and Mullins, R. D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545-576. [DOI] [PubMed] [Google Scholar]
- Roovers, K., Klein, E. A., Castagnino, P., and Assoian, R. K. (2003). Nuclear translocation of LIM kinase mediates Rho-Rho kinase regulation of cyclin D1 expression. Dev. Cell 5, 273-284. [DOI] [PubMed] [Google Scholar]
- Rosenblatt, J., Agnew, B. J., Abe, H., Bamburg, J. R., and Mitchison, T. J. (1997). Xenopus actin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeria monocytogenes tails. J. Cell Biol. 136, 1323-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma, M. P., Fasulo, B., Cenci, G., Cundari, E., and Gatti, M. (2002). Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13, 2448-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen, M., Ojala, P. J., Auvinen, P., Peränen, J., and Lappalainen, P. (2000). Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol. Cell. Biol. 20, 1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen, M. K., Mustonen, T., Mattila, P. K., Ojala, P. J., Thesleff, I., Partanen, J., and Lappalainen, P. (2002). The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 13, 183-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E., and Mizuno, K. (1998). Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393, 809-812. [DOI] [PubMed] [Google Scholar]
- Yeoh, S., Pope, B., Mannherz, H. G., and Weeds, A. (2002). Determining the differences in actin binding by human ADF and cofilin. J. Mol. Biol. 315, 911-925. [DOI] [PubMed] [Google Scholar]
- Zebda, N., Bernard, O., Bailly, M., Welti, S., Lawrence, D. S., and Condeelis, J. S. (2000). Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151, 1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


