Abstract
Disruption of PGS1, which encodes the enzyme that catalyzes the committed step of cardiolipin (CL) synthesis, results in loss of the mitochondrial anionic phospholipids phosphatidylglycerol (PG) and CL. The pgs1Δ mutant exhibits severe growth defects at 37°C. To understand the essential functions of mitochondrial anionic lipids at elevated temperatures, we isolated suppressors of pgs1Δ that grew at 37°C. One of the suppressors has a loss of function mutation in KRE5, which is involved in cell wall biogenesis. The cell wall of pgs1Δ contained markedly reduced β-1,3-glucan, which was restored in the suppressor. Stabilization of the cell wall with osmotic support alleviated the cell wall defects of pgs1Δ and suppressed the temperature sensitivity of all CL-deficient mutants. Evidence is presented suggesting that the previously reported inability of pgs1Δ to grow in the presence of ethidium bromide was due to defective cell wall integrity, not from “petite lethality.” These findings demonstrated that mitochondrial anionic lipids are required for cellular functions that are essential in cell wall biogenesis, the maintenance of cell integrity, and survival at elevated temperature.
INTRODUCTION
Cardiolipin (CL), a unique anionic phospholipid with dimeric structure, is ubiquitous in eukaryotes and primarily found in the mitochondrial inner membrane (Schlame et al., 2000). CL plays a key role in mitochondrial bioenergetics (Jiang et al., 2000; Koshkin and Greenberg, 2000, 2002; Schlame et al., 2000; Pfeiffer et al., 2003) and is also involved in mitochondrial biogenesis (Kawasaki et al., 1999; Jiang et al., 2000). Defective remodeling of CL is associated with Barth syndrome, a severe genetic disorder characterized by cardiomyopathy, neutropenia, skeletal myopathy, and respiratory chain defects (Vreken et al., 2000). The phenotype of Barth syndrome is dependent upon multiple factors that are not well understood (Barth et al., 1983, 1996). Elucidation of the functions of CL will help to clarify the abnormalities associated with this disorder.
The biosynthesis of CL is conserved in eukaryotic organisms. It occurs via three enzymatic reactions (Schlame et al., 2000), including formation of phosphatidylglycerolphosphate (PGP) from CDP-DAG and glycerol-3-P, dephosphorylation of PGP to phosphatidylglycerol (PG), and condensation of CDP-DAG and PG to form CL. Disruption of PGS1, the structural gene encoding PGP synthase, results in the complete loss of both PG and CL (Janitor et al., 1996; Chang et al., 1998a). The crd1Δ mutant, which lacks CL synthase, has no detectable CL but accumulates PG (Jiang et al., 1997; Chang et al., 1998b; Tuller et al., 1998; Jiang et al., 2000; Pfeiffer et al., 2003; Zhong et al., 2004). The human taffazin gene (TAZ1), which is associated with Barth syndrome, encodes a transacylase that may be involved in the remodeling of CL (Xu et al., 2003). Deletion of the yeast homolog of this gene, TAZ1, leads to decreased CL, aberrant CL acyl species, and accumulation of monolysocardiolipin (Gu et al., 2004). Mutants deficient in CL biosynthesis exhibit growth defects at elevated temperatures. The taz1Δ mutant is temperature sensitive for growth on ethanol but grows well on other carbon sources at elevated temperature (Gu et al., 2004). The crd1Δ mutant loses viability on both fermentable and nonfermentable carbon sources at elevated temperature, and it does not form colonies from single cells seeded on YPD plates (Jiang et al., 1999, 2000; Zhong et al., 2004). The pgs1Δ mutant exhibits the most severe growth defects and cannot grow at all at 37°C, even on glucose (Chang et al., 1998a; Dzugasova et al., 1998). The temperature-sensitive growth defects observed in CL-deficient mutants suggest that CL plays an essential role in maintaining cell viability at elevated temperature. The greater degree of temperature sensitivity of the pgs1Δ mutant compared with the crd1Δ mutant indicates that PG can substitute for some essential functions of CL. Mitochondria from crd1Δ (Koshkin and Greenberg, 2000, 2002) and taz1Δ (Ma et al., 2004) exhibit defective energetic coupling at elevated temperatures. Although thermal sensitivity of the bioenergetic functions may explain temperature sensitivity of these mutants in nonfermentable medium, the reason for loss of viability on glucose is not known.
In addition to the temperature-sensitive growth defects, CL-deficient mutants exhibit decreased mitochondrial genome stability. Mutant cells of crd1Δ grown in the presence of fermentable or nonfermentable carbon sources segregate large numbers of petites (respiratory incompetent cells) after prolonged culture at elevated temperature (Jiang et al., 2000; Zhong et al., 2004). The pgs1Δ mutant was initially determined to be “petite lethal” because the mutant cells did not survive ethidium bromide mutagenesis, which induces petite formation (Janitor and Subik, 1993; Dzugasova et al., 1998). However, 4′,6-diamidino-2-phenylindole (DAPI) staining of pgs1Δ revealed only the presence of nuclear DNA (Chang et al., 1998b). The absence of mitochondrial DNA (mtDNA) staining was attributed to a lack of elongated mitochondrial structure, but loss of mtDNA was not ruled out.
In a large-scale screen to identify genes involved in cell wall biogenesis, Lussier et al. (1997) reported that disruption of the PGS1 promoter results in several cell wall defects, including decreased glucosamine levels and hypersensitivity to cell wall-perturbing agents such as zymolyase, calcofluor white (CFW), papulacandin, and caffeine. The yeast cell wall is an essential organelle that determines the shape and preserves the osmotic integrity of the cell by counter-acting the internal turgor pressure. Mutants with weakened cell walls tend to form swollen cells with more spherical appearance and larger cell size than the wild-type (Popolo et al., 1993; de Nobel et al., 2000). The yeast cell wall has a two-layered structure. The highly glycosylated mannoproteins form the outer layer. The internal fibrillar layer contains glucans and chitin (Klis et al., 2002). β-1,3-Glucan forms a hollow helical structure that resembles a flexible wire spring. Together with chitin, it confers mechanical strength to the cell wall. β-1,6-Glucan in its mature form is a highly branched water-soluble polymer that interconnects all other cell wall components in a lattice (Klis et al., 2002). Chitin is enriched in the chitin ring in and around bud scars, and a minor portion is also uniformly dispersed in the lateral wall (Molano et al., 1980; Shaw et al., 1991; Cabib et al., 2001). Chitin deposition, however, is increased upon weakening of the cell wall (Popolo et al., 2001; Klis et al., 2002).
The yeast cell wall is a highly dynamic structure. It undergoes a number of modifications during different stages of the cell cycle and in response to environmental changes (Cabib et al., 2001; Smits et al., 2001). Elaborate control mechanisms are strictly coordinated in the regulation of cell wall biogenesis. Stress due to heat shock or ethanol induces the production of trehalose (Attfield, 1987; Hottiger et al., 1987; Neves and Francois, 1992; Hottiger et al., 1994; Singer and Lindquist, 1998) and glycerol (Alonso-Monge et al., 2001), which increase turgor pressure. Defects in the assembly of the cell wall compromise the response to stress and severely threaten cell survival. Supplementation with sorbitol, an osmotic stabilizer, supports the growth of cell wall mutants, presumably by balancing the turgor pressure on the plasma membrane and stabilizing cell wall structure (Popolo et al., 2001; Klis et al., 2002).
To understand the essential functions of CL at elevated temperature, we took the genetic approach of isolating spontaneous suppressor mutants of pgs1Δ that grow at elevated temperatures, one of which was identified to have a loss of function allele of KRE5, which is involved in cell wall biogenesis. In this report, we demonstrated that the absence of mitochondrial anionic phospholipids PG and CL results in defective cell wall assembly. Disruption of KRE5 induces β-1,3-glucan synthesis, strengthening the cell wall structure in pgs1Δ and enabling it to survive at elevated temperature. These data suggest that mitochondrial anionic phospholipids are required for processes that are essential in cell wall biogenesis and the maintenance of cell integrity.
MATERIALS AND METHODS
Materials
All chemicals used were reagent grade or better. The polymerase chain reaction (PCR) was performed using the native pfu enzyme kit from Invitrogen (Carlsbad, CA). The Zymoprep yeast plasmid mini prep kit was from ZymoResearch (Orange, CA). The Wizard Plus Miniprep DNA purification system was from Promega (Madison, WI). All other buffers and enzymes were purchased from Sigma-Aldrich (St. Louis, MO). Glucose, yeast extract, and peptone were purchased from Difco (Detroit, MI).
Yeast Strains and Growth Media
The Saccharomyces cerevisiae strains used in this work are listed in Table 1. Synthetic complete medium (SCD) contained amino acids adenine (20.25 mg/l), arginine (20 mg/l), histidine (20 mg/l), leucine (60 mg/l), lysine (200 mg/l), methionine (20 mg/l), threonine (300 mg/l), tryptophan (20 mg/l), and uracil (20 mg/l), vitamins, salts (essentially components of Difco Vitamin Free Yeast Base without amino acids), inositol (75 μM), and glucose (2%). Synthetic drop out medium (Ura-) contained all ingredients, except uracil for selection. Sporulation medium contained potassium acetate (1%), glucose (0.05%), and the essential amino acids. Complex media contained yeast extract (1%), peptone (2%), and glucose (2%) (YPD) or glycerol (3%) and ethanol (1%) (YPGE). Complex YPDS medium was YPD supplemented with 1 M sorbitol. Solid medium contained agar (2%) in addition to the above-mentioned ingredients.
Table 1.
Plasmids and yeast strains used in this study
| Plasmid or strain | Characteristics or genotype | Source or reference |
|---|---|---|
| pYES2/CT | 2 μm, URA3 | Invitrogen |
| pRS415-PGS1 | derivative of pYES2/CT, expresses PGS1 from Gal1 promoter | He and Greenberg (2004) |
| Ycp50 | Centromere, URA3 | Rose et al. (1987) |
| Ycp50-KRE5 | derivative of Ycp50, expresses KRE5 from its own promoter | This study |
| GAD74D3A | MAT α, ade8, ura 3, trp1, his3, leu2 | Dzugasova et al. (1998) |
| GAD74D3C | MAT α, ade8, ura 3, trp1, his3, leu2, pgs1::HIS3 | Dzugasova et al. (1998) |
| FGY3 | MAT α, ura 3-52, lys2-801, ade2-101, trp1Δ1, his3Δ200, leu2Δ1 | Jiang et al. (1997) |
| FGY3 (ρ0) | rho0 mutant derived from FGY3 | This study |
| QZY24B (ρ0) | MAT a, ura 3-52, lys2-801, trp1Δ1, his3Δ200, leu2Δ1, pgs1Δ::TRP1 | This study |
| QZY11A (ρ0) | MAT a, ura 3-52, lys2-801, trp1Δ1, his3Δ200, leu2Δ1, pgs1Δ::TRP1, kre5W1166X | This study |
| FGY2 | MAT α, ura 3-52, lys2-801, ade2-101, trp1Δ1, his3Δ200, leu2Δ1, crd1Δ::URA3 | Jiang et al. (1997) |
| 100 | MAT a, ade1, oxi2 (—) | C. Dieckmann |
| 101 | MAT a, ade1, oxi1 (—) | C. Dieckmann |
| 102 | MAT a, ade1, oxi3 (—) | C. Dieckmann |
| 103 | MAT a, ade1, cob (—) | C. Dieckmann |
| 104 | MAT α, met6, oxi2 (—) | C. Dieckmann |
| 105 | MAT α, met6, oxi1 (—) | C. Dieckmann |
| 106 | MAT α, met6, oxi3 (—) | C. Dieckmann |
| 107 | MAT α, met6, cob (—) | C. Dieckmann |
| T158c/S14a | Diploid prototroph S. cerevisiae | ATCC (46427) |
DAPI Stain
Yeast cells were grown to early stationary phase, fixed in 70% ethanol at room temperature for 30 min, and stained with 1 μg/ml DAPI for 5 min. Cells were viewed with an Olympus BX41 epifluorescence microscope, WU filter, and a 100× oil immersion objective. Images were captured with a Q-color3 camera and represent at least 200 observed cells.
Isolation of Extragenic Suppressors of pgs1Δ
Disruption of the PGS1 gene was performed as described previously (Zhong and Greenberg, 2003). Haploid pgs1Δ mutants of opposite mating types were obtained. YPD medium was inoculated from single colonies of pgs1Δ cells and grown for 24 h. About 108 cells from each independent culture were plated on a fresh YPD plate and incubated at 39°C. Single colonies from each plate were reexamined for growth at 39°C. Cells that grew at 39°C were analyzed further. To test the dominant and recessive character of the suppressor mutation, suppressor mutants were crossed to the parent strain, and growth of the diploid cells at 39°C was examined. Genetic complementation analysis was carried out with recessive mutants.
Plasmid Complementation
A yeast genomic DNA library in plasmid YCp50 (Rose et al., 1987) was used to clone the suppressor genes by complementation. Suppressor mutant cells were transformed with library DNA, plated on Ura- plates, and incubated at 25°C until colonies formed. Transformants were replicated onto YPD plates, and growth at 39°C was examined. Plasmid DNA was extracted from transformants that lost the capability to grow at 39°C, amplified in Escherichia coli DH5α, and retransformed into the suppressor mutant to confirm complementation of the suppressor phenotype. The DNA inserts of the positive clones were sequenced using primer YCp50 forward (5′-TTGGAGCCATATCGACTACG-3′) and YCp50 reverse (5′-ATGCGTCCGGCGTAGAGGATC-3′).
Identification of the Suppressor Mutation
Genomic DNA of pgs1Δ (QZY24B) and the suppressor QZY11A was extracted. DNA of the KRE5 region was amplified and sequenced using the following primers F1 (5′-TGTATTGGTTCATACCGGCA-3′); F2 (5′-ATATAGGGTTCTGAATTG-3′); R2 (5′-ATTGGAAGTTAGCGCCACAA-3′); F3 (5′-ACGATATGGCATACCCGAAT-3′); F4 (5′-TTATGGAAGCAATGAATG-3′); R4 (5′-AGAACCCTGGAATTGTGTGGA-3′); F5 (5′-TCCGTACAATTTGCTTACTGC-3′); F6 (5′-CGCCCGTTTAGAAGATAG-3′); R6 (5′-CACCAACAAAGGAAGTATGCA-3′); F7 (5′-GCGTAAGGGACTTATTGCATT-3′); F8 (5′-AAAGGTAAAAAGTCACAC-3′); R8 (5′-GAATCGACAAGTGCTAGGCAT-3′); F9 (5′-TGCCGACACTGGAATTAAACA-3′); F10 (5′-CGGATAAAAAAATTGCTC-3′); R10 (5′-ACCAGCATCTAACTCCCGAAA-3′); F11 (5′-TCAAACGTGCACCTCTAGGA-3′); and R12 (5′-CAGCCCATACCTACTTTCCAT-3′).
K1 Killer Toxin Assay
Sensitivity to K1 killer toxin was evaluated by a seeded plate assay using a modified YPD medium supplemented with 50 mM sodium citrate buffer (pH 3.7-3.8) and 0.003% methylene blue as described previously (Boone et al., 1990).
Sensitivity to Nikkomycin Z
Log phase cells were harvested and resuspended in SCD liquid medium at 1 × 105 cells/ml. Nikkomycin Z was added to a final concentration of 0.1 or 1 mM and incubated at 30°C for 48 h. A550 was measured, and sensitivity was determined by comparing A550 in treated versus untreated cells.
Transmission Electron Microscopy
Cells were grown in YPD media at 30°C to an A550 of 0.5-1 and fixed in 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.2) by addition of an equal amount of double-strength fixative to cells in suspension. Cells were centrifuged into a pellet, left to fix for 24 h, and then washed successively in 100 mM sodium cacodylate and in water, and resuspended in 10% gelatin at 37°C. The cells were pelleted while the gelatin was still warm, and the pellets were left to cool on ice. The gelatin-embedded pellets were cut out of their tubes and sliced into thin strips, which were fixed in 2.5% buffered glutaraldehyde for 1 h. Postfixation, dehydration, and resin infiltration were carried out in a microwave processor using a protocol modified from Giberson and Demaree (1999). A Pelco BioWave Microwave processor (Ted Pella, Redding, CA) was used for all irradiation steps. A flat chamber through which cold water was circulated was placed on the floor of the processor, and specimens were placed on top. The gelatin-embedded yeast cells were placed into glass vials containing ice-cold aqueous 2% osmium tetroxide and irradiated at full power for 40 s at a maximum temperature of 30°C, left at room temperature for 5 min, cooled on ice, and irradiated for an additional 40 s at full power. The osmium tetroxide was removed and replaced with cold water. Acetone dehydration was performed using the following steps: 1 × 50%, 1 × 70%, 1 × 90%, 2 × 100% acetone. Each step was performed in the microwave processor with 100% power for 40 s at a temperature maximum of 37°C. Infiltration with uncatalyzed epoxy resin consisted of full-power irradiation for 15 min at 45°C and full power in 1:1 acetone: resin followed by a similar irradiation in 100% resin. The specimens were then removed from the microwave processor and placed on a rotating table where they were infiltrated for 3 d in epoxy resin, changing the resin each day. Finally, the specimens were embedded in epoxy resin containing a catalyst and left to polymerize overnight at 60°C. Sections were prepared using an Ultracut S ultra-microtome (Leica Microsystems, Deerfield, IL) equipped with a diamond knife (Diatome US, Hatfield, PA). Sections, on metal grids were contrasted with uranyl acetate and lead citrate, and imaged in a CM120 BioTwin transmission electron microscope (FEI, Hillsboro, OR) operating at 80 kV.
Alkali-Insoluble β-Glucan Quantification
Yeast cells were grown in 50-100 ml of YPD or synthetic Ura- medium to early stationary phase. Cells were harvested and washed once with distilled water. Half the cells were used to determine the dry weight, and the other half were prepared for alkali extraction following the protocol described previously (Boone et al., 1990). The insoluble pellet that remained after zymolase digestion was removed with centrifugation and dialyzed against distilled water using Slide-a-Lyze 7000-Da molecular weight cut-off cassettes (Pierce Chemical, Rockford, IL). Total alkaline insoluble β-1,3 and β-1,6-glucan was determined by analysis of the carbohydrate content of the supernatant before dialysis by using the phenol-sulfuric acid method (Dubois et al., 1956). Analysis of the carbohydrate content of the retained fraction after dialysis determined the proportion of β-1,6 glucan.
Alkali-Soluble β-1,3-Glucan Quantification
Alkali-soluble β-1,3-glucan immunodetection was performed as described previously (Lussier et al., 1998). Briefly, cells were grown to early stationary phase at 30°C in YPD, harvested, and washed once with 5 ml of water. Cell pellets were resuspended in 100 μl of water with 100 μl of glass beads and subjected to five cycles of vortexing for 30 s, interspersed with 30-s incubations on ice. Total cellular protein was determined with the Bradford assay before alkali extraction (1.5 N NaOH, 1 h, 75°C). A set of 1:2 serial dilutions of the alkali-soluble fractions was spotted on nitrocellulose membrane. The immunoblotting was performed in Tris-buffered saline/Tween 20 containing 5% nonfat dried milk powder by using a 1000-fold dilution of anti-β-1,3-glucan primary antibody (Biosupplies Australia, Victoria, Australia), and a 5000-fold dilution of alkaline phosphatase conjugated goat anti-mouse secondary antibody (Promega). The membranes were developed with an AP detection kit (Promega). Dot blots were scanned with a ScanMaker 6800 scanner, and signals were quantitated with Adobe Photoshop software, by using the histogram function.
Chitin Quantification
Yeast cells were grown in 50- to 100-ml cultures to early stationary phase. Chitin levels were determined as described previously (Reissig et al., 1955). Briefly, ∼600-800 mg of cells was harvested. Half the cells were used to determine the cell dry weight, and the other half were transferred to 13 × 100 borosilicate tubes, resuspended in 4 ml of 6% KOH, and incubated at 80°C for 90 min to remove the mannan layer of the cell wall. After alkali treatment, 0.4 ml of glacial acetic acid was added. Cells were centrifuged at 4000 × g for 4 min and washed twice with cold water. Chitinase from Serratia marcescens (0.4 U) was resuspended in 2 ml of 50 mM sodium phosphate buffer (pH 6.3) and added to samples. Digestion was carried out at 30°C overnight, and 400 μl of supernatant was incubated for 1 h at 37°C with cytohelicase (Sigma-Aldrich). A 100-μl portion of each sample, blank or standard, was mixed to 100 μl of 0.27 M potassium-tetraborate pH 9.0, boiled for 3 min, and then cooled on ice. Color was developed by addition of 3 ml of freshly diluted DMAB reagent (Ehrlich's reagent, consisting of 10 g of p-dimethylaminobenzaldehyde in 12.5 ml of concentrated HCl and 87.5 ml of glacial acetic acid, diluted 1:10 with glacial acetic acid). Absorbance at A490 and A585 was measured using a Bio Spec-1601 Shimadzu spectrophotometer after 20-min incubation at room temperature.
Chitin Distribution
Yeast cells grown to early stationary phase were harvested by centrifugation at 2000 × g. Chitin stain was performed using Oregon Green 488 from Molecular Probes following the procedures from the manufacturer and observed using Olympus BX41 NIB filter. Images captured represent at least 200 observed cells.
RESULTS
Disruption of PGS1 Leads to Loss of Mitochondrial DNA
The pgs1Δ mutant in the FGY3 strain background was generated as described previously (Zhong and Greenberg, 2003). Consistent with a previous report (Chang et al., 1998a), the haploid pgs1Δ mutant was viable but did not grow on a nonfermentable carbon source. To determine whether pgs1Δ cells contained a mitochondrial genome, cells were stained with DAPI (Figure 1A). In contrast to isogenic wild-type cells, only nuclear DNA was visible in pgs1Δ cells, whereas mtDNA was not evident. To exclude the possibility that pgs1Δ cells contained incomplete mtDNA, 28 independent haploid pgs1Δ mutant cells derived from sporulation of heterozygous diploids were tested by complementation with ρ- tester strains for growth on nonfermentable medium (YPGE). Representative crosses are shown in Figure 1B. None of the 28 pgs1Δ strains was complemented by any of the ρ- tester strains. As a control, diploid cells that carried complementary ρ- mtDNA mutations grew on YPGE. This demonstrates that pgs1Δ cells grown on glucose medium exhibited loss of mtDNA, even at the optimal growth temperature.
Figure 1.
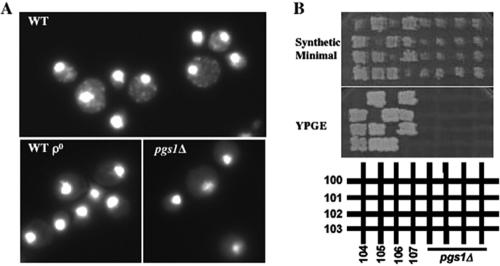
Disruption of PGS1 results in loss of mtDNA. (A) Isogenic wild-type (FGY3), ρ0, and pgs1Δ (QZY24B) cells were grown in YPD to early stationary phase. DNA was visualized by staining with DAPI as described in Materials and Methods. (B) Haploid pgs1Δ and ρ- tester strains (100-107) were crossed on a YPD plate. Diploid cells were selected on synthetic minimal medium. Mitochondrial function was determined by assessing growth on nonfermentable medium (YPGE).
Disruption of KRE5 Suppresses Temperature Sensitivity of pgs1Δ
To gain insight into the role of anionic phospholipids at elevated temperature, we used the genetic approach of isolating spontaneous suppressors of pgs1Δ temperature sensitivity. Eighteen recessive suppressor mutants that grew at nonpermissive temperatures were isolated, and these identified three complementation groups. One of the suppressors, QZY11A, was characterized further. In addition to complementation of growth at 37°C (Figure 2A), the suppressor complemented the enlarged cell size phenotype of the pgs1Δ mutant (Figure 2B). To clone the gene identified by the suppressor mutation, the suppressor was transformed with a genomic library and transformants were screened for inability to grow at nonpermissive temperatures. A plasmid bearing 5.5-kb DNA containing the KRE5 locus complemented the suppressor phenotype (Figure 2A). No other open reading frame was present on the plasmid. Subsequent sequencing analysis revealed a single G-to-A mutation resulting in a nonsense codon at the KRE5 locus of the suppressor mutant, leading to a deduced 201-amino acid deletion from the C terminus of the protein (Figure 3A). We designated this mutation kre5W1166X.
Figure 2.
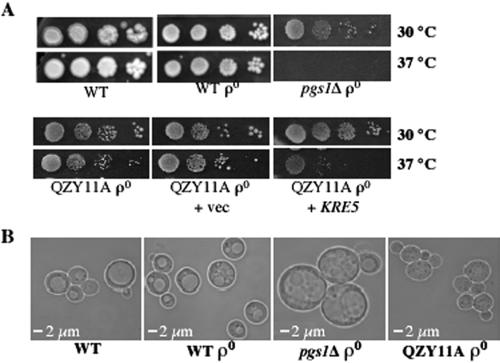
KRE5 complements the suppressor phenotype. (A) Cells from wild-type (FGY3), ρ0, pgs1Δ (QZY24B), the suppressor (QZY11A), and the suppressor transformed with empty vector YCp50 (+vec), or with YCp50 containing KRE5 (+KRE5) were serially diluted, spotted on YPD plates, and incubated at the indicated temperatures. (B) Wild-type (FGY3), ρ0, pgs1Δ (QZY24B), and suppressor (QZY11A) cells were grown to mid-log phase in YPD and examined microscopically.
Figure 3.
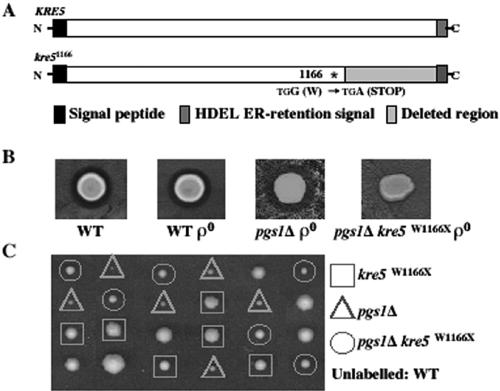
The suppressor mutant carries a loss of function allele of KRE5. (A) DNA of the KRE5 locus from pgs1Δ (QZY24B) and the suppressor (QZY11A) was sequenced as described in Materials and Methods. The single nonsense mutation in the coding sequence of KRE5 causes deletion of 201 amino acids from the C terminus of the protein. (B) K1 killer toxin producing cells (T158c/S14a) were spotted on plates preseeded with wild-type (FGY3), ρ0, pgs1Δ (QZY24B), and suppressor (QZY11A) cells and incubated at 30°C for 2 d. Sensitivity to K1 killer toxin is indicated by the presence of a killing zone surrounding cells. (C) The suppressor mutant (QZY11A) was crossed to the wild-type strain (FGY3). Diploid cells were sporulated, and meiotic tetrad analysis was performed. Genotypes of the haploid spores from six tetrads are shown.
KRE5 encodes an N-glycoprotein of ∼200 kDa that localizes to the endoplasmic reticulum (Levinson et al., 2002). Biochemical activity of Kre5p has not been determined. However, analysis of truncated versions of Kre5p indicated that all major regions of the protein are required for function (Levinson et al., 2002). Disruption of KRE5 results in decreased β-1,6-glucan and resistance to K1 killer toxin, the binding of which requires β-1,6-glucan (Meaden et al., 1990). The kre5W1166X mutant was assayed for K1 killer toxin sensitivity. The absence of a killer zone in the suppressor mutant (Figure 3B) suggests that kre5W1166X is a loss of function allele. To confirm that the suppressor phenotype resulted from the kre5W1166X allele, diploid cells heterozygous for pgs1Δ and kre5W1166X were sporulated, and suppression of temperature sensitivity of the pgs1Δ mutant was analyzed in 18 tetrads. Suppression of temperature sensitivity cosegregated with K1 killer toxin resistance (Figure 3C). Loss of kre5p often leads to extremely slow growth (Meaden et al., 1990) or inviability in some strain backgrounds (Shahinian et al., 1998). Interestingly, the single mutant kre5W1166X in the FGY3 strain background exhibited only slightly compromised growth phenotypes. The above-mentioned genetic analysis demonstrated that the loss of function allele kre5W1166X is an extragenic suppressor of pgs1Δ, which enables it to grow at elevated temperature.
Disruption of PGS1 Leads to a Defective Cell Wall
Identification of a mutant in cell wall synthesis as a suppressor of pgs1Δ temperature sensitivity suggests that pgs1Δ temperature sensitivity results from perturbation of cell wall biosynthesis and/or structure. We therefore examined the cell wall properties of pgs1Δ and the suppressor mutant.
Mutants with defective cell wall structure exhibit sensitivity to cell wall-perturbing agents. Consistent with a previous report (Lussier et al., 1997), pgs1Δ was hypersensitive to caffeine and CFW. In the presence of 1 M sorbitol, an osmotic stabilizer, sensitivity to CFW and caffeine in pgs1Δ was significantly reduced (Figure 4A). Furthermore, the suppressor pgs1Δ kre5W1166X was no longer sensitive to CFW, although sensitivity to caffeine in pgs1Δ was not suppressed. Thus, the suppressor mutation conferred a beneficial effect on the cell wall of pgs1Δ cells (Figure 4A).
Figure 4.
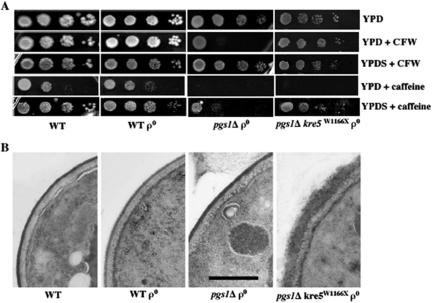
Cell wall properties of pgs1Δ and the suppressor pgs1Δ kre5W1166X. (A) Cells from wild-type (FGY3), ρ0, pgs1Δ (QZY24B), and the suppressor (QZY11A) mutant were serially diluted and spotted on YPD or YPDS plates supplemented with 5 μg/ml CFW or 5 mM caffeine. Cells were incubated at 30°C. (B) Transmission electron microscopy of the cell wall in wild-type (FGY3), ρ0, pgs1Δ (QZY24B), and the suppressor (QZY11A) was performed as described in Materials and Methods. The horizontal bar represents 500 nm.
Deletion of KRE5 leads to a number of changes in the cell wall, such as diffuse cell wall surface, a complete loss of the electron-dense mannoprotein layer (Simons et al., 1998), and enlarged cell wall structure (Levinson et al., 2002). Electron microscopic analysis was performed to compare the ultrastructure of the cell wall of pgs1Δ and the pgs1Δ kre5W1166X suppressor strain. As seen in Figure 4B, the wild-type cell wall has a finely delineated dark-staining mannoprotein layer. In contrast, the cell wall of the suppressor was twice as thick and exhibited a rough appearance, similar to that reported for the kre5 mutant (Simons et al., 1998; Levinson et al., 2002). However, aberrations in cell wall morphology were not observed in pgs1Δ cells.
To gain further insight into how kre5W1166X suppressed pgs1Δ temperature sensitivity, we characterized the cell wall composition of pgs1Δ and the suppressor strain. Levels of the major cell wall components were measured, including β-1,3 and β-1,6-glucan and chitin (Table 2). Because pgs1Δ cells lose mtDNA and are all ρ0 cells (Figure 1), we compared cell wall composition in pgs1Δ and isogenic wild-type ρ0 cells. Surprisingly, the loss of mtDNA from wild-type cells resulted in decreased alkaline-soluble and -insoluble β-1,3-glucan (50 and 12% decrease, respectively) and a decrease (37%) in β-1,6-glucan. Disruption of PGS1 led to an even more pronounced decrease in β-1,3-glucan. Alkaline-soluble and -insoluble β-1,3-glucan was reduced to 76 and 68% of the isogenic ρ0 wild-type levels. In contrast, β-1,3 glucan was dramatically increased in the suppressor mutant to levels greater than those observed in wild-type cells. Alkaline-insoluble β-1,6-glucan was reduced approximately twofold in the pgs1Δ mutant compared with the isogenic ρ0 wild-type and to an even greater extent in the suppressor mutant.
Table 2.
Cell wall composition in pgs1Δ and suppressor mutants
| Alkaline-insoluble glucan
|
Alkaline-soluble
|
|||||
|---|---|---|---|---|---|---|
| Strain | Medium | β-1,6 | β-1,6 + β-1,3 | β-1,3 | β-1,3-glucan | Chitin |
| WT | YPD | 37.8 ± 0.9 | 141.4 ± 4.4 | 103.6 | 100% | 4.65 ± 0.39 |
| WT ρ0 | YPD | 23.7 ± 1.4 | 114.7 ± 8.1 | 91.0 | 50 ± 3% | 4.54 ± 0.26 |
| pgs1Δ ρ0 | YPD | 12.1 ± 1.9 | 74.7 ± 1.4 | 62.6 | 38 ± 2% | 12.36 ± 1.67 |
| pgs1Δ kre5W1166X ρ0 | YPD | 7.7 ± 2.5 | 155.8 ± 1.4 | 148.1 | 174 ± 15% | 12.35 ± 0.39 |
| pgs1Δ ρ0 | YPDS | 21.0 ± 1.6 | 98.8 ± 18.8 | 77.8 | 90 ± 25% | 8.09 ± 1.52 |
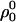 |
Ura−[ρ] | 25.8 ± 5.3 | 131.6 ± 33.5 | 105.8 | 100% | 7.76 ± 0.32 |
| pgs1Δ ρ + vec | Ura−[ρ] | 21.4 ± 2.1 | 69.6 ± 22.7 | 48.2 | 41 ± 6% | 12.04 ± 0.80 |
| pgs1Δ kre5W1166X ρ0 + vec | Ura−[ρ] | 7.4 ± 1.8 | 118.4 ± 17.3 | 111.0 | 151 ± 21% | 12.09 ± 3.14 |
| pgs1Δ kre5W1166X ρ0 + KRE5 | Ura−[ρ] | 27.1 ± 0.9 | 99.3 ± 9.6 | 72.2 | 52 ± 5% | 10.52 ± 0.37 |
Glucan and chitin levels were measured as described in Materials and Methods in wild-type (FGY3), ρ0, pgs1Δ (QZY24B), and suppressor mutant pgs1Δ kre5W1166X (QZY11A) cells grown in YPD or YPDS; pgs1Δ (QZY24B) cells transformed with empty vector pYES2/CT (+vec) or pYES2/CT-PGS1 (+PGS1); and pgs1Δ kre5W1166X suppressor mutant (QZY11A) cells transformed with empty vector YCp50 (+vec) or the genomic clone of KRE5 (+KRE5) grown in synthetic ura− medium. Alkaline insoluble glucan and chitin are expressed as micrograms per milligram of cell dry weight. Alkaline soluble β-1,3-glucan in cells grown in complex medium (top) was expressed as a percentage of that of wild-type (FGY3) cells. Alkaline soluble β-1,3-glucan in cells grown in synthetic ura− medium (bottom) was expressed relative to pgs1Δ (QZY24B) cells transformed with pYES2/CT-PGS1. Data represent three independent experiments.
In contrast to β-glucans, chitin levels were not affected by loss of mtDNA (Table 2). In both pgs1Δ and the suppressor mutant, chitin levels were ∼3 times higher than in wild-type cells (Table 2). Examination of the distribution of chitin by using specific fluorescent probes revealed the presence of chitin predominantly in the bud scars of wild-type cells (Figure 5A). In pgs1Δ and suppressor mutants, however, chitin staining was uniformly distributed to the lateral cell wall, and the bud scar was hardly distinguishable from the rest of the cell wall. Hyperaccumulation of chitin in cell wall mutants is mediated by chitin synthase III (CSIII) (Osmond et al., 1999; Valdivieso et al., 2000). When grown in the presence of 0.1 mM Nikkomycin Z, a known inhibitor of CSIII (Gaughran et al., 1994), viability of the pgs1Δ mutant was reduced by 40% (our unpublished data). In contrast, the wild-type and suppressor mutant tolerated 1 mM Nikkomycin Z with no obvious loss of viability. Hypersensitivity to Nikkomycin Z suggested that increased chitin is essential for the survival of pgs1Δ. We wished to determine whether a further increase in chitin synthesis led to increased viability of pgs1Δ cells at 37°C. Glucosamine was recently shown to induce chitin synthesis via CSIII in wild-type cells as well as in cell wall mutants (Bulik et al., 2003). As shown in Figure 5B, in the presence of 10 mM glucosamine, growth of pgs1Δ was slightly improved at 30°C. Chitin levels in pgs1Δ were doubled at 37°C compared with 30°C in the presence of 10 mM glucosamine (our unpublished data). However, at 37°C, pgs1Δ cells exhibited only limited growth and lost viability after 20 h (Figure 5B). Thus, 10 mM glucosamine did not restore growth to levels observed in the presence of the suppressor mutation. Supplementation with 5-20 mM glucosamine led to a shortened lag in growth and increased saturation optical density of the pgs1Δ mutant at 30°C, but was not sufficient to support growth at elevated temperature (our unpublished data).
Figure 5.
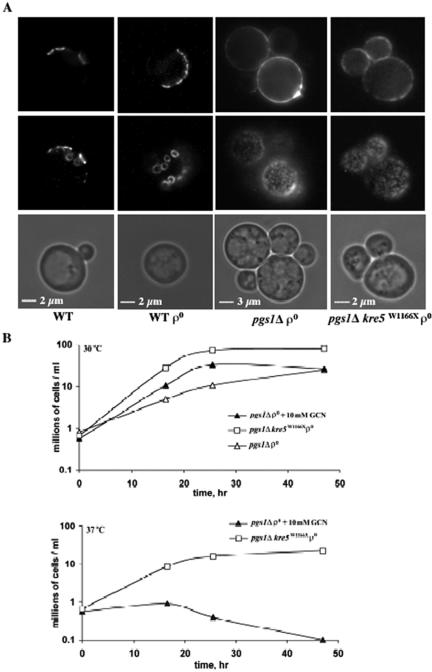
Increased chitin deposition in pgs1Δ and the suppressor mutant. (A) Cells from wild-type (FGY3), ρ0, pgs1Δ (QZY24B), and the suppressor (QZY11A) were grown in YPD to early stationary phase. Chitin was visualized by staining with Oregon Green 488 as described in Materials and Methods. Chitin distribution was visualized by focusing on two planes. (B) Cells from pgs1Δ (QZY24B) and the suppressor mutant (QZY11A) were grown in YPD in the presence or absence of 10 mM glucosamine at the indicated temperatures. Viable cells were determined by serial dilution and plating.
When transformed with a plasmid-borne copy of the PGS1 gene under the control of the PGAL1 promoter, β-1,3-glucan levels in the pgs1Δ mutant significantly increased and chitin decreased (Table 2). This was observed even during growth in glucose, in which low levels of expression of PGS1 from this plasmid are sufficient to restore growth of pgs1Δ at elevated temperature and synthesis of PG and CL (He and Greenberg, 2004). Expression of the plasmid-borne copy of KRE5 in the suppressor mutant restored β-1,6-glucan and decreased β-1,3-glucan levels (Table 2). Together, these experiments indicate that disruption of PGS1 leads to multiple cell wall defects resulting in defective growth at elevated temperature. Increasing cell wall synthesis, particularly β-1,3-glucan and chitin, restored growth at 37°C.
Osmotic Stabilization of the Cell Wall Restores Growth of pgs1Δ at Elevated Temperature
Suppression of pgs1Δ temperature sensitivity by increased β-1,3-glucan synthesis suggested that osmotic stabilization of the cell wall might alleviate cell wall stress and support growth of pgs1Δ at elevated temperature. To address this possibility, we examined the effect of sorbitol on cell wall composition and growth of pgs1Δ at 37°C. Mutant cells of pgs1Δ grown in the presence of 1 M sorbitol contained 1.8 and 2.4-fold increased alkaline soluble β-1,3-glucan and alkaline insoluble β-1,6-glucan levels, respectively, compared with levels observed in pgs1Δ cells grown in YPD lacking sorbitol, whereas chitin levels were significantly reduced (Table 2). Consistent with this finding, the majority of pgs1Δ cells displayed the wild-type pattern of chitin staining of bud scars and exhibited normal cell size in the presence of sorbitol (data not shown). Supplementation with sorbitol restored growth of pgs1Δ in two strain backgrounds, FGY3 and GA74D (Figure 6A). Sorbitol also supported colony formation of crd1Δ on YPD (Figure 6B), as well as growth of taz1Δ on ethanol (data not shown) at 37°C.
Figure 6.
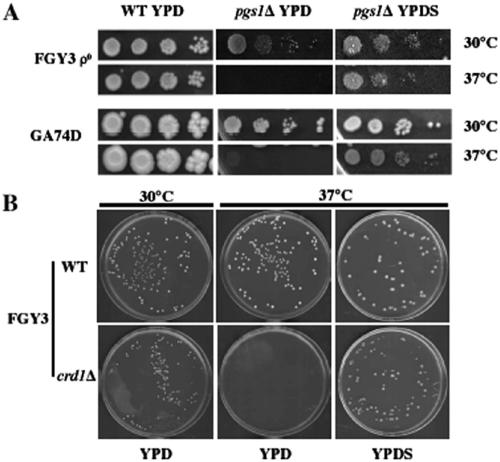
Supplementation with sorbitol supports growth of CL-deficient mutants at 37°C. (A) Isogenic wild-type and pgs1Δ mutant cells from two genetic backgrounds (FGY3 ρ0 and GA74D ρ+) were serially diluted and spotted on YPD or YPDS plates and incubated at the indicated temperature. (B) Wild-type (FGY3) and crd1Δ (FGY2) cells were grown overnight in YPD. Single cells were seeded on YPD or YPDS plates and incubated at the indicated temperature.
As this report has shown, pgs1Δ cells in the FGY3 background grown on YPD are all ρ0 cells. However, the pgs1Δ mutant in the GA74D strain background, GA74D3C, was previously thought to be “petite lethal”, because the mutant cells did not survive ethidium bromide mutagenesis, which induces loss of mtDNA (Janitor and Subik, 1993; Dzugasova et al., 1998). A likely explanation for the inability of pgs1Δ cells to grow in the presence of ethidium bromide is that pgs1Δ ρ0 cells in the GA74D strain background fail to survive due to loss of cell wall integrity. To resolve this discrepancy, we examined the effects of ethidium bromide on pgs1Δ cells in the presence or absence of osmotic support. Consistent with the previous report (Janitor and Subik, 1993; Dzugasova et al., 1998), pgs1Δ (GA74D3C) cells failed to grow on plates containing 25 μg/ml ethidium bromide. However, when supplemented with 1 M sorbitol, the cells grew in the presence of ethidium bromide (Figure 7). Failure to complement ρ- tester strains for growth on YPGE (our unpublished data) confirmed the loss of mtDNA. The pgs1Δ ρ0 mutant in this genetic background was viable on YPD but exhibited slower growth than the isogenic ρ+ strain. Consistent with these observations, a greater decrease in alkaline-soluble and -insoluble β-1,3-glucan was observed in pgs1Δ ρ0 cells than in pgs1Δ ρ+ cells (our unpublished data). These data suggest that pgs1Δ cells can survive ethidium bromide treatment in the presence of increased osmotic support.
Figure 7.
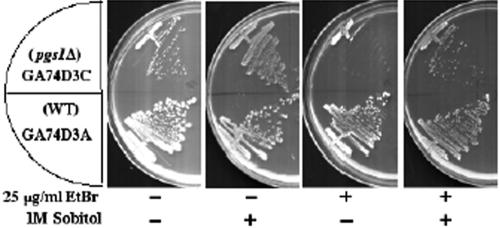
Pgs1Δ survives ethidium bromide mutagenesis in the presence of osmotic stabilizer. Wild-type GA74D3A and isogenic pgs1Δ mutant GA74D3c cells were streaked on synthetic minimal plates with ethidium bromide or sorbitol as indicated and incubated at 30°C.
In summary, disruption of PGS1 leads to a defective cell wall and inability to grow at elevated temperature. Stabilization of the cell wall, either by increased synthesis of β-1,3-glucan or increased osmotic support, restores growth at 37°C. These findings show that mitochondrial anionic phospholipids are essential in cellular functions required for cell wall biogenesis and maintenance of cell integrity.
DISCUSSION
The pgs1Δ mutant, which lacks mitochondrial anionic phospholipids PG and CL, exhibits severe temperature sensitivity for growth even on fermentable carbon sources (Chang et al., 1998a; Dzugasova et al., 1998), suggesting that anionic phospholipids are required for essential cellular functions. This report shows that mutants lacking these lipids are defective in cell wall biogenesis. Reorganization of the cell wall in the pgs1Δ kre5W1166X suppressor or osmotic stabilization with sorbitol restores growth at elevated temperature. The pgs1Δ mutant exhibited a marked decrease in β-1,3-glucan (Table 2), the lack of which greatly impairs the mechanical strength of the cell wall and severely threatens viability of cells at elevated temperature (Klis et al., 2002). As a result, pgs1Δ mutant cells become enlarged and rounded (Figure 2), resembling another cell wall mutant, gas1Δ, which is also defective in β-glucan synthesis (Popolo et al., 1993; Ram et al., 1998). Likely as a result of dramatically increased β-1,3-glucan in the pgs1Δ kre5W1166X suppressor strain (Table 2), the cell wall structure was thicker (Figure 4B), and cell size and growth at elevated temperature were restored (Figure 2). Consistent with these findings, osmotic stabilization with sorbitol suppressed temperature sensitivity of all CL deficient mutants at elevated temperature (Figure 6), suggesting that deficiency in mitochondrial anionic phospholipids results in cell wall defects that lead to a temperature-sensitive growth phenotype.
Cell wall biogenesis is a highly regulated process. β-1,3-Glucan is synthesized by β-1,3-glucan synthase (GS) localized on the plasma membrane (Qadota et al., 1996). GS is composed of a catalytic subunit encoded by the two homologous genes FKS1 and FKS2 (Inoue et al., 1995; Mazur et al., 1995), and a regulatory subunit, the small GTPase, Rho1p (Drgonova et al., 1996; Qadota et al., 1996). The Rho-type GTPase is generally regulated by switching between a GDP-bound inactive state and a GTP-bound active state (Wei et al., 1997; Ihara et al., 1998). Various factors are involved in the regulation of β-1,3-glucan synthesis by Rho1p in yeast cells. The putative cell surface sensor protein Wsc1p plays a critical role in stimulating nucleotide exchange of Rho1p through the GDP/GTP exchange factor, Rom2p (Philip and Levin, 2001). Exchange of GDP for GTP stimulates Rho1p, leading to activation of GS activity. Lrg1p, a GTPase-activating protein, promotes formation of GDP-bound Rho1p, thus negatively regulating β-1,3-glucan synthesis (Watanabe et al., 2001). Thus, overexpression of ROM2 or WSC1 (Sekiya-Kawasaki et al., 2002), or loss of function of LRG1 (Watanabe et al., 2001) restores the impaired β-1,3-glucan synthesis observed in GS mutants. In addition, posttranslational modification of Rho1p by the geranylgeranyl group is required for binding of Rho1p to GS and activation of GS activity (Inoue et al., 1999). Other factors affect β-1,3-glucan synthesis by regulation of the catalytic subunit of GS. Movement of Fks1p driven by actin is required for the construction of a uniform and solid cell wall (Utsugi et al., 2002). Transcription of FKS2 is up-regulated in response to cell wall stress induced by heat, cell wall mutations, and cell wall-perturbing agents (Zhao et al., 1998; de Nobel et al., 2000; Lagorce et al., 2003; Garcia et al., 2004). Deletion of KRE5 leads to a 114-fold up-regulation of FKS2 in response to an impaired cell wall (Kapteyn et al., 1999). Restored β-1,3-glucan levels in pgs1Δ mutant cells in the presence of the kre5W1166X suppressor mutation (Table 2) could be mediated by up-regulation of FKS2 expression. In fact, increased β-1,3-glucan is a general characteristic shared by several kre mutants, along with defective β-1,6-glucan synthesis (Roemer et al., 1994; Dijkgraaf et al., 1996; Shahinian et al., 1998; Shahinian and Bussey, 2000).
Decreased β-glucan levels in ρ0 cells and in mutants lacking mitochondrial anionic lipids suggest the existence of a regulatory link between mitochondrial biogenesis and cell wall synthesis. It has been suggested that cytoplasmic petite mutants isolated after ethidium bromide mutagenesis have altered cell wall assembly (Wauters et al., 2001). In this study, we have shown for the first time that loss of mtDNA alone led to a significant decrease in β-glucan. These defects were exacerbated in the pgs1Δ mutant (Table 2). Our findings that greater cell wall defects were observed in pgs1Δ than in the wild-type ρ0 cells suggests that, along with oxidative phosphorylation, other mitochondrial functions requiring PG and/or CL may be required for cell wall biogenesis. A link between mitochondrial functions and cell wall biogenesis has been implicated in several previous studies as well. In addition to pgs1, Lussier et al. (1997) reported that mutations in four other genes with mitochondrial associated functions, IMP2′, IFM1, SMP2, and COX11 have cell wall defects. Three of these genes (IFM1, SMP2, and COX11) are required for mtDNA stability (Vambutas et al., 1991; Irie et al., 1993; Tzagoloff et al., 1993). A genome-wide screen for deletion mutants that exhibit increased resistance to K1 killer toxin, which indicates alterations in the cell surface, identified 17 deletion mutants affecting genes for respiration and ATP metabolism (Page et al., 2003). All of the mutants are respiratory deficient, and four are involved in mitochondrial genome maintenance.
The identification of cell wall defects in mutants with mitochondrial dysfunction suggests that mitochondria may play a general role in the regulation of cell wall biogenesis. Several enzymes involved in β-1,3-glucan synthesis were found to have dual localization in the plasma membrane and mitochondria. Both the catalytic and the regulatory subunit of GS are localized on the plasma membrane at the site of cell wall synthesis (Qadota et al., 1996). Interestingly, both Fks1p and Rho1p are also present in mitochondria (Sickmann et al., 2003). Gas1p, a putative β-1,3-glucan-remodeling enzyme (Popolo and Vai, 1999; Mouyna et al., 2000), the loss of which also results in reduced β-1,3-glucan in the cell wall (Popolo et al., 1993; Ram et al., 1998), is attached to the plasma membrane via a glycosyl-phosphatidylinositol anchor (Conzelmann et al., 1988; Nuoffer et al., 1991). Gas1p also is found in mitochondria (Grandier-Vazeille et al., 2001; Sickmann et al., 2003). It is not known whether dual plasma membrane/mitochondria localization of these enzymes has any physiological relevance. It is tempting, however, to assume that mitochondria are required for the maturation or modification of those enzymes, in which case mitochondrial dysfunction would result in decreased enzyme activity and cell wall defects. Alternatively, mitochondrial dysfunction may trigger signals that prevent proper mobilization of those enzymes to the site of cell wall biosynthesis.
Our finding that pgs1Δ in the FGY3 strain background exhibited loss of mtDNA even at optimal growth temperature suggests that PG and CL are required for maintaining mtDNA. Mutants lacking only CL exhibit a strain-dependent decrease in mtDNA stability at elevated temperature (Jiang et al., 2000; Zhong et al., 2004). We have noticed that crd1Δ mutants from different strain backgrounds differ greatly with respect to the temperature at which growth is defective and mtDNA becomes unstable (Zhong et al., 2004). In addition, crd1Δ was less thermotolerant on synthetic medium than on rich medium (Zhong et al., 2004). Although pgs1Δ in the GA74D strain background does not lose mtDNA on YPD at 30°C, it cannot grow on synthetic medium with glycerol and ethanol as carbon source (Dzugasova et al., 1998), suggesting that it may lose mtDNA under this condition. Furthermore, the results presented here show that pgs1Δ is not “petite lethal,” and the previously reported inability of pgs1Δ to survive ethidium bromide (Janitor and Subik, 1993; Dzugasova et al., 1998) was due to defective cell wall integrity. Pgs1Δ ρ0 cells in the GA74D strain background were obtained after ethidium bromide treatment in the presence of osmotic support and were viable on YPD (Figure 7B). Those petite cells exhibited slower growth than the isogenic ρ+ strain, which presumably resulted from the exacerbated cell wall defects caused by loss of mtDNA. The further compromised growth observed in pgs1Δ ρ0 cells suggests a “synthetic sick” interaction between pgs1Δ and the ρ0 mutation. This interaction predicts that the number of pgs1Δ cells surviving the loss of mtDNA would be low. This seems paradoxical in light of our finding that lack of PG and CL in the pgs1Δ mutant strain resulted in 100% petite formation on YPD. Interestingly, mutations in ATP15 and ATP16, two structural genes encoding ε and δ subunit of F1-ATPase, lead to similar phenotypes. On one hand, atp15 and atp16 exhibited an extremely high frequency of petite formation. However, the petite mutants have severe growth defects. Like PGS1, ATP15 and ATP16 also were thought to be essential in a petite background (Giraud and Velours, 1997; Lai-Zhang et al., 1999; Contamine and Picard, 2000).
In summary, we have isolated and identified an extragenic suppressor of pgs1Δ, the loss of function allele of KRE5, kre5W1166X. Characterization of pgs1Δ and the suppressor strain strongly suggests that temperature sensitivity of CL-deficient mutants and the previously reported “petite lethal” phenotype of pgs1Δ mutant cells were primarily due to defective cell wall integrity. This work is the first demonstration of defective cell wall biosynthesis in mutants lacking mitochondrial anionic phospholipids PG and CL. Our findings thus provide new insights into the essential functions of these lipids and point to a regulatory role of mitochondria in cell wall biogenesis.
Acknowledgments
We thank Professor Július Subík for providing the pgs1Δ strain in the GA74D strain background. We also thank Shuliang Chen and Guiling Li for help initiating this study and Dr. Deirdre Vaden for advice. This work was supported by grant HL-62263 from the National Institutes of Health.
Article published online ahead of print in MBC in Press on November 24, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0808).
Abbreviations used: CFW, calcoflour white; CL, cardiolipin; CSIII, chitin synthase III; GS, glucan synthase; mtDNA, mitochondrial DNA; PGP, phophatidylglycerolphosphate; PG, phosphatidylglycerol; Ura-, synthetic drop out medium without uracil; YPD, yeast extract, peptone, and dextrose; YPGE, yeast extract, glycerol and ethanol; YPDS, YPD supplemented with 1 M sorbitol.
References
- Alonso-Monge, R., Real, E., Wojda, I., Bebelman, J. P., Mager, W. H., and Siderius, M. (2001). Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol. Microbiol. 41, 717-730. [DOI] [PubMed] [Google Scholar]
- Attfield, P. V. (1987). Trehalose accumulates in Saccharomyces cerevisiae during exposure to agents that induce heat shock response. FEBS Lett. 225, 259-263. [DOI] [PubMed] [Google Scholar]
- Barth, P. G., Scholte, H. R., Berden, J. A., Van der Klei-Van Moorsel, J. M., Luyt-Houwen, I. E., Van 't Veer-Korthof, E. T., Van der Harten, J. J., and Sobotka-Plojhar, M. A. (1983). An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 62, 327-355. [DOI] [PubMed] [Google Scholar]
- Barth, P. G., Van den Bogert, C., Bolhuis, P. A., Scholte, H. R., van Gennip, A. H., Schutgens, R. B., and Ketel, A. G. (1996). X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): respiratory-chain abnormalities in cultured fibroblasts. J. Inherit. Metab. Dis. 19, 157-160. [DOI] [PubMed] [Google Scholar]
- Boone, C., Sommer, S. S., Hensel, A., and Bussey, H. (1990). Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J. Cell Biol. 110, 1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik, D. A., Olczak, M., Lucero, H. A., Osmond, B. C., Robbins, P. W., and Specht, C. A. (2003). Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2, 886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib, E., Roh, D. H., Schmidt, M., Crotti, L. B., and Varma, A. (2001). The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276, 19679-19682. [DOI] [PubMed] [Google Scholar]
- Chang, S. C., Heacock, P. N., Clancey, C. J., and Dowhan, W. (1998a). The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9829-9836. [DOI] [PubMed] [Google Scholar]
- Chang, S. C., Heacock, P. N., Mileykovskaya, E., Voelker, D. R., and Dowhan, W. (1998b). Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273, 14933-14941. [DOI] [PubMed] [Google Scholar]
- Contamine, V., and Picard, M. (2000). Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64, 281-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann, A., Riezman, H., Desponds, C., and Bron, C. (1988). A major 125-kd membrane glycoprotein of Saccharomyces cerevisiae is attached to the lipid bilayer through an inositol-containing phospholipid. EMBO J. 7, 2233-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel, H., Ruiz, C., Martin, H., Morris, W., Brul, S., Molina, M., and Klis, F. M. (2000). Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146, 2121-2132. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf, G. J., Brown, J. L., and Bussey, H. (1996). The KNH1 gene of Saccharomyces cerevisiae is a functional homolog of KRE9. Yeast 12, 683-692. [DOI] [PubMed] [Google Scholar]
- Drgonova, J., Drgon, T., Tanaka, K., Kollar, R., Chen, G. C., Ford, R. A., Chan, C. S., Takai, Y., and Cabib, E. (1996). Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272, 277-279. [DOI] [PubMed] [Google Scholar]
- Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350-356. [Google Scholar]
- Dzugasova, V., Obernauerova, M., Horvathova, K., Vachova, M., Zakova, M., and Subik, J. (1998). Phosphatidylglycerolphosphate synthase encoded by the PEL1/PGS1 gene in Saccharomyces cerevisiae is localized in mitochondria and its expression is regulated by phospholipid precursors. Curr. Genet. 34, 297-302. [DOI] [PubMed] [Google Scholar]
- Garcia, R., Bermejo, C., Grau, C., Perez, R., Rodriguez-Pena, J. M., Francois, J., Nombela, C., and Arroyo, J. (2004). The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279, 15183-15195. [DOI] [PubMed] [Google Scholar]
- Gaughran, J. P., Lai, M. H., Kirsch, D. R., and Silverman, S. J. (1994). Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J. Bacteriol. 176, 5857-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giberson, R. T., and Demaree, R.S.J. (1999). Microwave processing techniques for electron microscopy: a four-hour protocol. Methods Mol. Biol. 117, 145-158. [DOI] [PubMed] [Google Scholar]
- Giraud, M. F., and Velours, J. (1997). The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho- yeast cells by a lack of assembly of the catalytic sector F1. Eur. J. Biochem. 245, 813-818. [DOI] [PubMed] [Google Scholar]
- Grandier-Vazeille, X., Bathany, K., Chaignepain, S., Camougrand, N., Manon, S., and Schmitter, J. M. (2001). Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry 40, 9758-9769. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Valianpour, F., Chen, S., Vaz, F. M., Hakkaart, G. A., Wanders, R. J., and Greenberg, M. L. (2004). Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol. Microbiol. 51, 149-158. [DOI] [PubMed] [Google Scholar]
- He, Q., and Greenberg, M. L. (2004). Post-translational regulation of phosphatidylglycerolphosphate synthase in response to inositol. Mol. Microbiol. 53, 1243-1249. [DOI] [PubMed] [Google Scholar]
- Hottiger, T., Boller, T., and Wiemken, A. (1987). Rapid changes of heat and desiccation tolerance correlated with changes of trehalose content in Saccharomyces cerevisiae cells subjected to temperature shifts. FEBS Lett. 220, 113-115. [DOI] [PubMed] [Google Scholar]
- Hottiger, T., De Virgilio, C., Hall, M. N., Boller, T., and Wiemken, A. (1994). The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur. J. Biochem. 219, 187-193. [DOI] [PubMed] [Google Scholar]
- Ihara, K., Muraguchi, S., Kato, M., Shimizu, T., Shirakawa, M., Kuroda, S., Kaibuchi, K., and Hakoshima, T. (1998). Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 273, 9656-9666. [DOI] [PubMed] [Google Scholar]
- Inoue, S. B., Qadota, H., Arisawa, M., Watanabe, T., and Ohya, Y. (1999). Prenylation of Rho1p is required for activation of yeast 1,3-beta-glucan synthase. J. Biol. Chem. 274, 38119-38124. [DOI] [PubMed] [Google Scholar]
- Inoue, S. B., Takewaki, N., Takasuka, T., Mio, T., Adachi, M., Fujii, Y., Miyamoto, C., Arisawa, M., Furuichi, Y., and Watanabe, T. (1995). Characterization and gene cloning of 1,3-beta-D-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231, 845-854. [DOI] [PubMed] [Google Scholar]
- Irie, K., Takase, M., Araki, H., and Oshima, Y. (1993). A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283-288. [DOI] [PubMed] [Google Scholar]
- Janitor, M., Obernauerova, M., Kohlwein, S. D., and Subik, J. (1996). The pel1 mutant of Saccharomyces cerevisiae is deficient in cardiolipin and does not survive the disruption of the CHO1 gene encoding phosphatidylserine synthase. FEMS Microbiol. Lett. 140, 43-47. [DOI] [PubMed] [Google Scholar]
- Janitor, M., and Subik, J. (1993). Molecular cloning of the PEL1 gene of Saccharomyces cerevisiae that is essential for the viability of petite mutants. Curr. Genet. 24, 307-312. [DOI] [PubMed] [Google Scholar]
- Jiang, F., Gu, Z., Granger, J. M., and Greenberg, M. L. (1999). Cardiolipin synthase expression is essential for growth at elevated temperature and is regulated by factors affecting mitochondrial development. Mol. Microbiol. 31, 373-379. [DOI] [PubMed] [Google Scholar]
- Jiang, F., Rizavi, H. S., and Greenberg, M. L. (1997). Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26, 481-491. [DOI] [PubMed] [Google Scholar]
- Jiang, F., Ryan, M. T., Schlame, M., Zhao, M., Gu, Z., Klingenberg, M., Pfanner, N., and Greenberg, M. L. (2000). Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387-22394. [DOI] [PubMed] [Google Scholar]
- Kapteyn, J. C., Van Egmond, P., Sievi, E., Van Den Ende, H., Makarow, M., and Klis, F. M. (1999). The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and beta 1,6-glucan-deficient mutants. Mol. Microbiol. 31, 1835-1844. [DOI] [PubMed] [Google Scholar]
- Kawasaki, K., Kuge, O., Chang, S. C., Heacock, P. N., Rho, M., Suzuki, K., Nishijima, M., and Dowhan, W. (1999). Isolation of a Chinese hamster ovary (CHO) cDNA encoding phosphatidylglycerophosphate (PGP) synthase, expression of which corrects the mitochondrial abnormalities of a PGP synthase-defective mutant of CHO-K1 cells. J. Biol. Chem. 274, 1828-1834. [DOI] [PubMed] [Google Scholar]
- Klis, F. M., Mol, P., Hellingwerf, K., and Brul, S. (2002). Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26, 239-256. [DOI] [PubMed] [Google Scholar]
- Koshkin, V., and Greenberg, M. L. (2000). Oxidative phosphorylation in cardiolipin-lacking yeast mitochondria. Biochem. J. 347, 687-691. [PMC free article] [PubMed] [Google Scholar]
- Koshkin, V., and Greenberg, M. L. (2002). Cardiolipin prevents rate-dependent uncoupling and provides osmotic stability in yeast mitochondria. Biochem. J. 364, 317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorce, A., Hauser, N. C., Labourdette, D., Rodriguez, C., Martin-Yken, H., Arroyo, J., Hoheisel, J. D., and Francois, J. (2003). Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278, 20345-20357. [DOI] [PubMed] [Google Scholar]
- Lai-Zhang, J., Xiao, Y., and Mueller, D. M. (1999). Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J. 18, 58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson, J. N., Shahinian, S., Sdicu, A. M., Tessier, D. C., and Bussey, H. (2002). Functional, comparative and cell biological analysis of Saccharomyces cerevisiae Kre5p. Yeast 19, 1243-1259. [DOI] [PubMed] [Google Scholar]
- Lussier, M., Sdicu, A. M., Shahinian, S., and Bussey, H. (1998). The Candida albicans KRE9 gene is required for cell wall beta-1, 6-glucan synthesis and is essential for growth on glucose. Proc. Natl. Acad. Sci. USA 95, 9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier, M., et al. (1997). Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Vaz, F. M., Gu, Z., Wanders, R. J., and Greenberg, M. L. (2004). The human TAZ gene complements mitochondrial dysfunction in the yeast taz1delta mutant. Implications for Barth syndrome. J. Biol. Chem. 279, 44394-44399. [DOI] [PubMed] [Google Scholar]
- Mazur, P., Morin, N., Baginsky, W., el-Sherbeini, M., Clemas, J. A., Nielsen, J. B., and Foor, F. (1995). Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase. Mol. Cell. Biol. 15, 5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaden, P., Hill, K., Wagner, J., Slipetz, D., Sommer, S. S., and Bussey, H. (1990). The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1-6)-beta-D-glucan synthesis and normal cell growth. Mol. Cell. Biol. 10, 3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molano, J., Bowers, B., and Cabib, E. (1980). Distribution of chitin in the yeast cell wall. An ultrastructural and chemical study. J. Cell Biol. 85, 199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouyna, I., Fontaine, T., Vai, M., Monod, M., Fonzi, W. A., Diaquin, M., Popolo, L., Hartland, R. P., and Latge, J. P. (2000). Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275, 14882-14889. [DOI] [PubMed] [Google Scholar]
- Neves, M. J., and Francois, J. (1992). On the mechanism by which a heat shock induces trehalose accumulation in Saccharomyces cerevisiae. Biochem. J. 288, 859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer, C., Jeno, P., Conzelmann, A., and Riezman, H. (1991). Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11, 27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond, B. C., Specht, C. A., and Robbins, P. W. (1999). Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA 96, 11206-11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, N., et al. (2003). A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163, 875-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, K., Gohil, V., Stuart, R. A., Hunte, C., Brandt, U., Greenberg, M. L., and Schagger, H. (2003). Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873-52880. [DOI] [PubMed] [Google Scholar]
- Philip, B., and Levin, D. E. (2001). Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21, 271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo, L., Gualtieri, T., and Ragni, E. (2001). The yeast cell-wall salvage pathway. Med. Mycol. 39 (suppl) 1, 111-121. [PubMed] [Google Scholar]
- Popolo, L., and Vai, M. (1999). The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta 1426, 385-400. [DOI] [PubMed] [Google Scholar]
- Popolo, L., Vai, M., Gatti, E., Porello, S., Bonfante, P., Balestrini, R., and Alberghina, L. (1993). Physiological analysis of mutants indicates involvement of the Saccharomyces cerevisiae GPI-anchored protein gp115 in morphogenesis and cell separation. J. Bacteriol. 175, 1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., Python, C. P., Inoue, S. B., Arisawa, M., Anraku, Y., Zheng, Y., Watanabe, T., Levin, D. E., and Ohya, Y. (1996). Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 272, 279-281. [DOI] [PubMed] [Google Scholar]
- Ram, A. F., Kapteyn, J. C., Montijn, R. C., Caro, L. H., Douwes, J. E., Baginsky, W., Mazur, P., van den Ende, H., and Klis, F. M. (1998). Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of beta1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180, 1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissig, J. L., Storminger, J. L., and Leloir, L. F. (1955). A modified colorimetric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 217, 959-966. [PubMed] [Google Scholar]
- Roemer, T., Paravicini, G., Payton, M. A., and Bussey, H. (1994). Characterization of the yeast (1->6)-beta-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J. Cell Biol. 127, 567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., Novick, P., Thomas, J. H., Botstein, D., and Fink, G. R. (1987). A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60, 237-243. [DOI] [PubMed] [Google Scholar]
- Schlame, M., Rua, D., and Greenberg, M. L. (2000). The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39, 257-288. [DOI] [PubMed] [Google Scholar]
- Sekiya-Kawasaki, M., Abe, M., Saka, A., Watanabe, D., Kono, K., Minemura-Asakawa, M., Ishihara, S., Watanabe, T., and Ohya, Y. (2002). Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics 162, 663-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinian, S., and Bussey, H. (2000). beta-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 35, 477-489. [DOI] [PubMed] [Google Scholar]
- Shahinian, S., Dijkgraaf, G. J., Sdicu, A. M., Thomas, D. Y., Jakob, C. A., Aebi, M., and Bussey, H. (1998). Involvement of protein N-glycosyl chain glucosylation and processing in the biosynthesis of cell wall beta-1,6-glucan of Saccharomyces cerevisiae. Genetics 149, 843-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J. A., Mol, P. C., Bowers, B., Silverman, S. J., Valdivieso, M. H., Duran, A., and Cabib, E. (1991). The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114, 111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann, A., et al. (2003). The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100, 13207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J. F., Ebersold, M., and Helenius, A. (1998). Cell wall 1,6-beta-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17, 396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. A., and Lindquist, S. (1998). Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol. 16, 460-468. [DOI] [PubMed] [Google Scholar]
- Smits, G. J., van den Ende, H., and Klis, F. M. (2001). Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147, 781-794. [DOI] [PubMed] [Google Scholar]
- Tuller, G., Hrastnik, C., Achleitner, G., Schiefthaler, U., Klein, F., and Daum, G. (1998). YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421, 15-18. [DOI] [PubMed] [Google Scholar]
- Tzagoloff, A., Nobrega, M., Gorman, N., and Sinclair, P. (1993). On the functions of the yeast COX10 and COX11 gene products. Biochem. Mol. Biol. Int. 31, 593-598. [PubMed] [Google Scholar]
- Utsugi, T., Minemura, M., Hirata, A., Abe, M., Watanabe, D., and Ohya, Y. (2002). Movement of yeast 1,3-beta-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7, 1-9. [DOI] [PubMed] [Google Scholar]
- Valdivieso, M. H., Ferrario, L., Vai, M., Duran, A., and Popolo, L. (2000). Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182, 4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vambutas, A., Ackerman, S. H., and Tzagoloff, A. (1991). Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur. J. Biochem. 201, 643-652. [DOI] [PubMed] [Google Scholar]
- Vreken, P., Valianpour, F., Nijtmans, L. G., Grivell, L. A., Plecko, B., Wanders, R. J., and Barth, P. G. (2000). Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279, 378-382. [DOI] [PubMed] [Google Scholar]
- Watanabe, D., Abe, M., and Ohya, Y. (2001). Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 18, 943-951. [DOI] [PubMed] [Google Scholar]
- Wauters, T., Iserentant, D., and Verachtert, H. (2001). Impact of mitochondrial activity on the cell wall composition and on the resistance to tannic acid in Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 47, 21-26. [DOI] [PubMed] [Google Scholar]
- Wei, Y., Zhang, Y., Derewenda, U., Liu, X., Minor, W., Nakamoto, R. K., Somlyo, A. V., Somlyo, A. P., and Derewenda, Z. S. (1997). Crystal structure of RhoA-GDP and its functional implications. Nat. Struct. Biol. 4, 699-703. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Kelley, R. I., Blanck, T. J., and Schlame, M. (2003). Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 278, 51380-51385. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Jung, U. S., Garrett-Engele, P., Roe, T., Cyert, M. S., and Levin, D. E. (1998). Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18, 1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Q., Gohil, V. M., Ma, L., and Greenberg, M. L. (2004). Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 279, 32294-32300. [DOI] [PubMed] [Google Scholar]
- Zhong, Q., and Greenberg, M. L. (2003). Regulation of phosphatidylglycerophosphate synthase by inositol in Saccharomyces cerevisiae is not at the level of PGS1 mRNA abundance. J. Biol. Chem. 278, 33978-33984. [DOI] [PubMed] [Google Scholar]


