Abstract
A variety of studies have implicated the lipid PtdIns(4,5)P2 in endocytic internalization, but how this lipid mediates its effects is not known. The AP180 N-terminal homology (ANTH) domain is a PtdIns(4,5)P2-binding module found in several proteins that participate in receptor-mediated endocytosis. One such protein is yeast Sla2p, a highly conserved actin-binding protein essential for actin organization and endocytic internalization. To better understand how PtdIns(4,5)P2 binding regulates actin-dependent endocytosis, we investigated the functions of Sla2p's ANTH domain. A liposome-binding assay revealed that Sla2p binds to PtdIns(4,5)P2 specifically through its ANTH domain and identified specific lysine residues required for this interaction. Mutants of Sla2p deficient in PtdIns(4,5)P2 binding showed significant defects in cell growth, actin organization, and endocytic internalization. These defects could be rescued by increasing PtdIns(4,5)P2 levels in vivo. Strikingly, mutant Sla2p defective in PtdIns(4,5)P2 binding localized with the endocytic machinery at the cell cortex, establishing that the ANTH-PtdIns(4,5)P2 interaction is not necessary for this association. In contrast, multicolor real-time fluorescence microscopy and particle-tracking analysis demonstrated that PtdIns(4,5)P2 binding is required during endocytic internalization. These results demonstrate that the interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 plays a key role in regulation of the dynamics of actin-dependent endocytic internalization.
INTRODUCTION
Endocytosis is an important process in all eukaryotes. It mediates nutrient uptake, regulates responses to extracellular stimuli, and controls the chemical composition and surface area of the plasma membrane. Previous studies in yeast and mammalian cells have led to the identification of many components required for endocytosis (Geli and Riezman, 1998; Munn, 2001; Engqvist-Goldstein and Drubin, 2003). In addition to proteins, lipids, especially PtdIns(4,5)P2, are known to be important for the a number of steps in receptor-mediated endocytosis (D'Hondt et al., 2000). PtdIns(4,5)P2 is the major polyphosphoinositide found in eukaryotic cell membranes. Early studies identified a role for PtdIns(4,5)P2 as a precursor to the signaling molecules inositol(1,4,5)-triphosphate and diacylglycerol (Berridge and Irvine, 1984). Subsequent studies showed that PtdIns(4,5)P2 itself is important for actin assembly, exocytosis, endocytosis, and intracellular membrane trafficking (Cremona and De Camilli, 2001; Martin, 2001; Takenawa and Itoh, 2001; Wenk and De Camilli, 2004). Because PtdIns(4,5)P2 binding domains are found in many endocytic proteins, the functions of PtdIns(4,5)P2 in endocytosis are thought to be related to its ability to directly bind to specific proteins (Cullen et al., 2001; Itoh and Takenawa, 2002; Lemmon, 2003).
The ENTH (epsin N-terminal homology) domain is a ∼150 amino acid motif that is highly conserved in a variety of proteins present in yeast, oats, rats, mice, frogs, and humans (Kay et al., 1999; Rosenthal et al., 1999). Recently, the ENTH domain and a highly related module, the AP180 N-terminal homology (ANTH) domain, were identified as PtdIns(4,5)P2-binding domains in mammalian cells (Ford et al., 2001; Itoh et al., 2001). The interactions between PtdIns(4,5)P2 and both the ENTH domain of epsin and the ANTH domain of AP180 are essential for endocytic internalization mediated by clathrin-coated pits (Ford et al., 2001; Itoh et al., 2001). The ENTH domain of the yeast epsin homologue Ent1p has also been shown to bind to phosphoinositides and is essential for normal endocytic function and actin cytoskeleton structure (Wendland et al., 1999; Aguilar et al., 2003). Although the ENTH domain and ANTH domain share very similar primary structures (Figure 1A), an important distinction between them is the formation of a critical α-helix within the ENTH domain, α-helix0, upon binding to PtdIns(4,5)P2 (Ford et al., 2002; Stahelin et al., 2003; Legendre-Guillemin et al., 2004). The association of this helix with the lipid bilayer was proposed to displace lipid head groups and impart membrane curvature through lipid remodeling. No such helical structure has been found in the ANTH domain, whose function remains less clear.
Figure 1.

ANTH/ENTH sequence alignment and Sla2p domain structure. (A) Amino acid sequence alignment of ANTH (Sla2p, Hip1R, AP180) and ENTH (Epsin) domains. Conserved motifs are boxed. Four conserved lysines (K14, K24, K26, K62) are marked. K42 in Sla2p is a nonconserved lysine. (B) Schematic representation of Sla2p primary structure.
An ANTH domain is present at the N-terminus of yeast Sla2p and its mammalian homologues Hip1 (Huntingtin Interacting Protein 1) and Hip1R (Huntingtin Interacting Protein 1 Related). SLA2, also known as END4, is required for endocytosis and proper actin organization (Wesp et al., 1997; Yang et al., 1999; Baggett et al., 2003). Like many other yeast endocytic proteins, Sla2p transiently localizes to cortical actin patches during the endocytic internalization pathway. Actin polymerization is required for endocytic internalization and occurs late in the pathway (Kaksonen et al., 2003). Sla2p appears necessary to harness actin polymerization forces for endocytic internalization because actin is continuously nucleated from nonmotile endocytic complexes in sla2Δ cells. An identical phenotype was also observed when the level of Hip1R was reduced in human cells (Engqvist-Goldstein et al., 2004), indicating that Sla2p and Hip1R share a conserved biological role. Previous studies indicated that only the N-terminal region of Sla2p is indispensable for growth, actin organization, and endocytosis and that its proline-rich, coiled-coil, and C terminal talin-like domains are not essential for these processes (Wesp et al., 1997). Because the ANTH domain is at the Sla2p N-terminus (Figure 1B), we speculated that it might be vital to Sla2p's function. The functions of this domain during endocytosis had not been explored in Sla2p, Hip1, or Hip1R. However, recent studies showed that expression of an ANTH domain-deletion mutant of Hip1R induces apoptosis, which implies that the ANTH domain of Hip1R may have a role in cellular survival (Hyun et al., 2004). Whether the ANTH domain in Hip1, Hip1R, or Sla2p binds to phosphoinositides and the biological role of such binding remain to be addressed.
MATERIALS AND METHODS
Lipids
Chloroform solutions of phospholipids and derivatives, except for phosphatidylinositol 4-phosphate (PI4P), phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), and CDP-diacylglycerol (CDP-DAG), were purchased from Avanti Polar Lipids (Birmingham, AL). PI4P, PtdIns(4,5)P2, CDP-DAG, and ergosterol were purchased from Sigma (St. Louis, MO). SYPRO-Red protein stain was purchased from Molecular Probes (Eugene, OR).
Media and Strains
Yeast strains were grown in standard rich media (YPD) or synthetic media (SD) supplemented with the appropriate amino acids. The yeast strains are listed in Table 1. GFP or CFP tags were integrated at the C-terminus of each gene as described previously (Wach et al., 1997). Wild-type and sla2 mutant cells expressing GFP- and/or CFP-tagged proteins had growth properties indistinguishable from wild-type and sla2 mutant cells, respectively.
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| DDY426 | MATα/MAT α his 3-Δ200/his3-Δ200 leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 ade2-1/ADE2 lys2-801/lys2-801 |
| DDY130 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 |
| DDY1810 | MATa, leu2, ura3-52, trp1, prb1-1122, pep4-3, pre1-451 |
| DDY3001 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801sla2Δ::4K-A::URA3 |
| DDY3002 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801sla2Δ::ΔANTH::URA3 |
| DDY2740 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::cgLEU2 |
| DDY3003 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 bar1Δ:cgHIS3 |
| DDY3004 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::ΔANTH::URA3 bar1Δ::cgHIS3 |
| DDY3005 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801sla2Δ::4K-A::URA bar1Δ::cgHIS3 |
| DDY3006 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::cgLEU2 bar1Δ::cgHIS3 |
| DDY3007 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sjl1Δ::cgHIS3 |
| DDY3008 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sjl1Δ::cgHIS3 bar1Δ::cgLEU2 |
| DDY3009 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::4K-A::URA3 sjl1Δ::cgHIS3 |
| DDY3010 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::4K-A::URA3sjl1Δ::cgHIS3 bar1Δ::cgLEU2 |
| DDY2335 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 SLA2-GFP::KanMX6 |
| DDY3011 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 4K-A-GFP::HIS3 |
| DDY3012 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ΔANTH-GFP::HIS3 |
| DDY3013 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 SLA2-GFP::KanMX6 SLA1-CFP::KanMX6 |
| DDY3014 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 SLA1 -GFP::HIS3 ΔANTH -CFP::KanMX6 |
| DDY3015 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 SLA 1-GFP::HIS3 4K-A-CFP::KanMX6 |
| DDY3016 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 LAS17-GFP::KanMX6 SLA2-CFP::KanMX6 |
| DDY3017 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 LAS17 -GFP::HIS3 ΔANTH-CFP::KanMX6 |
| DDY3018 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ade2-1 LAS-GFP::HIS3 4K-A-CFP::KanMX6 |
| DDY3019 | MATαhis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::4K-A::URA ABP1-CFP::KanMX6 SLA1-GFP::HIS3 |
| DDY2737 | MATahis3-Δ200 leu2-3, 112 ura3-52 ABP1-CFP::KanMX6 SLA1-GFP::HIS3 |
| DDY2742 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 ABP1-GFP::HIS3 |
| DDY2743 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::cgLEU2 ABP1-GFP::HIS3 |
| DDY3020 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::4K-A::URA3 ABP1-GFP::HIS3 |
| DDY3021 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::ΔANTH::URA3 ABP1-GFP::HIS3 |
| DDY3022 | MATahis3-Δ200 leu2-3, 112 ura3-52 SLA1-GFP::HIS3 |
| DDY3023 | MATahis3-Δ200 leu2-3, 112 ura3-52 lys2-801 sla2Δ::4K-A::URA3 SLA1-GFP::HIS |
All the strains are derived from DDY426, expect DDY2737, which is derived from DDY1102. cgLEU2, cgURA3 and cgHIS indicate Candida glabrata LEU2 gene, Candida glabrata HIS3 gene, and Candida glabrata URA3, respectively.
Purification of GST-Fusion Proteins from Yeast
GST-fusion proteins were expressed from pEG(KT) (Mitchell et al., 1993) under the galactose-inducible promoter and purified from DDY1810 as described previously (Shang et al., 2003). The yeast cells were induced with galactose at 30°C for 8 h, harvested, frozen in liquid nitrogen, and stored at -80°C. Cell pellets were lysed in liquid nitrogen in a Waring blender and thawed in HEK-T buffer (50 mM HEPES, pH 7.5, 1 mM EDTA, 100 mM KCl, 1% Triton X-100), with 1 mM phenylmethylsulfonyl fluoride and protease inhibitors. The lysate was centrifuged in an SA-600 rotor at 10,000 rpm for 20 min. The supernatant was filtered through cheesecloth and passed over a Q Sepharose Fast Flow column (Amersham Biosciences, Piscataway, NJ). Bound GST-fusion proteins were eluted with 40 ml of HEK500-T buffer (50 mM HEPES, pH 7.5, 1 mM EDTA, 500 mM KCl, 0.5% Triton X-100), and bound to glutathione-agarose (Sigma-Aldrich). Finally, GST-fusion proteins were eluted in elution buffer (20 mM glutathione, 100 mM Tris-HCl, pH 8.0, 120 mM NaCl, 0.5% Triton X-100) and dialyzed into HEKG5 buffer (50 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl, 5% glycerol). The protein concentration was determined using the BCA protein assay reagent kit (Pierce Chemical, Rockford, IL) with bovine serum albumin (BSA; Sigma-Aldrich) as a standard.
Preparation of Liposomes and Binding of Sla2 Proteins
Liposomes were prepared as described by Matusoka et al. (1998). Lipids were hydrated with 20 mM HEPES-KOH (pH 7.5), 0.05 M KCl at room temperature with occasional vortexing. The resulting suspension of multilamellar liposomes was extruded through a polycarbonate filter with a 400-nm pore size and was used for binding experiments.
For Sla2p binding experiments, 12.5 μl of a liposome suspension was mixed with 62.5 μl of a solution containing 0.5 μM Sla2p. After incubation at room temperature for 20 min, 50 μl of 2.5 M sucrose was added and mixed. The mixture, 110 μl, was transferred to a 7 × 20-mm thick-wall polycarbonate tube (Beckman, Fullerton, CA) and overlaid with 100 μl of 0.75 μM sucrose and 20 μl of 20 mM HEPES-KOH (pH 7.5), 50 mM KCl. The resulting step gradient was centrifuged at 100,000 rpm in a Beckman TLA-100 rotor for 10 min at room temperature. Samples of 17.5 μl were collected from the top of the tube. Proteins in the fraction were separated by SDS-PAGE, stained by SYPRO Red, and visualized using a STORM 860 image analyzer (Amersham Biosciences, Piscataway, NJ).
α-factor Uptake Assay
35S-labeled α-factor was prepared as described in Howard et al. (2002). The α-factor uptake assay was performed at 25°C based on a continuous incubation protocol (Sekiya-Kawasaki et al., 2003). Cells were grown in YPD, harvested by centrifugation, and resuspended in internalization media (YPD media with 0.5% casamino acids and 1% BSA). At the indicated time points, aliquots were withdrawn and diluted in ice-cold buffer at pH 6.0 (total α-factor) or pH 1.1 (internalized α-factor). The samples were then filtered, and radioactivity was measured in a scintillation counter. The results were expressed as the ratio of pH 1.1 cpm/pH 6.0 cpm for each time point to represent the percentage of internalization.
Fluorescence Microscopy
Fluorescence microscopy was performed using a Nikon TE300 microscope (Garden City, NY) equipped with a 100×/NA 1.4 objective and Orca-100 - cooled CCD camera (Hamamatsu, Bridgewater, NJ).
For live cell imaging, cells were grown to early log phase at 25°C. The cells in synthetic media were adhered to the surface of a concanavalin A-coated (0.1 μg/ml) coverslip, which was then inverted onto a glass slide, and sealed with vacuum grease (Dow Corning, Midland, MI). All imaging was performed at room temperature.
For single-channel live cell imaging, the excitation light intensity was reduced with neutral density filters and images were acquired continuously at 1-4 frames/s depending on the signal intensity. Two-channel movies were made using a CFP-YFP filter set (JP4, Chroma, Brattleboro, VT) and motorized excitation and emission filter wheels (Sutter Instruments, Novato, CA). The CCD camera and the filter wheels were controlled by Metamorph software (Universal Imaging, West Chester, PA). Fixation and rhodamine phalloidin staining were performed as described (Kaksonen et al., 2003).
Image Analysis
Patch lifetime analyses were done by visually identifying the time points of appearance and disappearance of individual patches from the movies. ImageJ software (http://rsb.info.nih.gov/ij/) was used for general manipulation of images and movies.
Particle-tracking analysis was performed as described by Kaksonen et al. (2003).
RESULTS
Sla2p Binds to PtdIns(4,5)P2 Specifically via its ANTH Domain
To determine if Sla2p can bind to PtdIns(4,5)P2, we tested the ability of GST-Sla2p to bind to liposomes of various compositions. Previous work established a liposome composition (defined as “major-minor mix”, see Materials and Methods) optimized for the recruitment of COPII components (Matsuoka et al., 1998). Liposomes were mixed with Sla2p, and protein-liposome complexes were isolated by flotation through sucrose density gradients. Sla2p was detected in major-minor mix liposomes containing 2% PtdIns(4,5)P2, but not in liposomes containing only PC/PE, or in major-minor mix liposomes lacking PtdIns(4,5)P2 (Figure 2A). As shown in Figure 2B, this binding occurs in a PtdIns(4,5)P2 concentration-dependent manner. Moreover, the interaction was detected when liposomes contained PtdIns(4,5)P2, but not when liposomes contained PtdIns(3,5)P2, demonstrating the stereo specificity of Sla2p's PtdIns(4,5)P2 binding capacity (Figure 2C). Specificity of binding was also studied with PtdIns3P, PtdIns4P, and PtdIns5P. Sla2p showed modest binding to PtdIns3P, but not to PtdIns4P or PtdIns5P (Figure 2D). Because yeast lack a type I PtdIns 3-kinase, which in mammalian cells converts PtdIns(4,5)P2 to PtdIns(3,4,5)P3, yeast cells do not contain detectable amounts of PtdIns(3,4,5)P3 or PI(3,4)P2, and we therefore did not test for binding to these lipids (Hawkins et al., 1993; Dove et al., 1997). Taken together, our results demonstrate that Sla2p binds specifically to PtdIns(4,5)P2.
Figure 2.
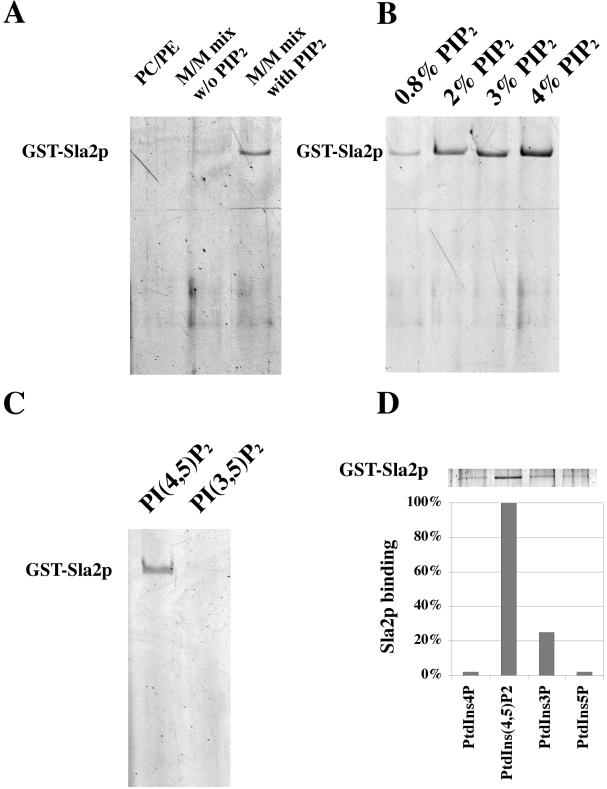
Sla2p binding to liposomes in a PtdIns(4,5)P2 concentration-dependent and specific manner in vitro. (A) Sla2p binding to liposomes of various lipid compositions was tested. GST-Sla2p, 0.5 μM, was incubated with liposomes in a 70-μl reaction. Liposome-associated proteins were resolved by SDS-PAGE and were stained with SYPRO Red. PC/PE: 77 mol% DOPC, 22 mol% DOPE. Major-minor mix without PtdIns(4,5)P2: 51 mol% DOPC, 21 mol% DOPE, 9.8 mol% PI, 8 mol% DOPS, 5 mol% DOPA, 4.2 mol% PI4P, 2 mol% CDP-DAG. Major-minor mix with PtdIns(4,5)P2: 51 mol% DOPC, 21 mol% DOPE, 7.8 mol% PI, 8 mol% DOPS, 5 mol% DOPA, 4.2 mol% PI4P, 2 mol% CDP-DAG, 2 mol% PtdIns(4,5)P2. (B) Sla2p binding to liposomes composed of various PtdIns(4,5)P2 concentrations. Major-minor mix with (0.8-4 mol% PtdIns(4,5)P2): 51 mol% DOPC, 21 mol% DOPE, 9-5.8 mol% PI, 8 mol% DOPS, 5 mol% DOPA, 2.2 mol% PI4P, 2 mol% CDP-DAG, 0.8-4 mol% PtdIns(4,5)P. The Sla2p concentration in each assay was 0.5 μM. (C) Preferential binding of Sla2p to PtdIns(4,5)P2 versus PtdIns(3,5)P2. Liposomes made from Major-minor mix with 2 mol% PtdIns(4,5)P2 or 2 mol% PtdIns(3,5)P2 were tested for the binding of GST-Sla2p. The Sla2p concentration of each assay was 0.5 μM. (D) Binding of Sla2p to PtdIns3P, PtdIns4P, and PtdIns5P. Liposomes made from Major-minor mix with 2 mol% PtdIns(4,5)P2 or 2 mol% PtdIns3P, or PtdIns4P, or PtdIns5P. The Sla2p concentration in each assay was 0.5 μM.
To determine whether the ANTH domain is responsible for the Sla2p-PtdIns(4,5)P2 interaction, we purified a recombinant Sla2p in which the ANTH domain (amino acids 1-125) was completely deleted (GST-ANTHΔ). As shown in Figure 3A, GST did not bind to PtdIns(4,5)P2-containing, major-minor mix liposomes. Compared with binding of the intact fusion protein, binding of GST-ANTHΔ was significantly reduced (Figure 3A). A previous study showed that the ANTH domain of AP180 bound to PtdIns(4,5)P2 via a lysine-rich motif, K(X)9KX(K/R)(H/Y) (Ford et al., 2001). K76 in the ENTH domain of epsin was also reported to be an essential residue for PtdIns(4,5)P2 binding (Itoh et al., 2001). Sequence alignment revealed that these lysine-rich motifs are well conserved in Sla2p's ANTH domain (Figure 1A). We therefore tested the requirement of these residues for PtdIns(4,5)P2 binding. We used site-directed mutagenesis to change lysines into alanines, generating K14A, K24A K26A, K62A, K14A K24A K26A, and K14A K24A K26A K62A mutants. K42 in the ANTH domain of Sla2p is not conserved, and we predicted that the K42A mutant would not affect Sla2p's interaction with PtdIns(4,5)P2 or other functions. Strikingly, all these mutants, except K42A, failed to bind to PtdIns(4,5)P2 (Figure 3B). Together, these data indicate that Sla2p binds to PtdIns(4,5)P2 specifically through the ANTH domain and that several conserved lysine residues in the ANTH domain are required for this interaction.
Figure 3.
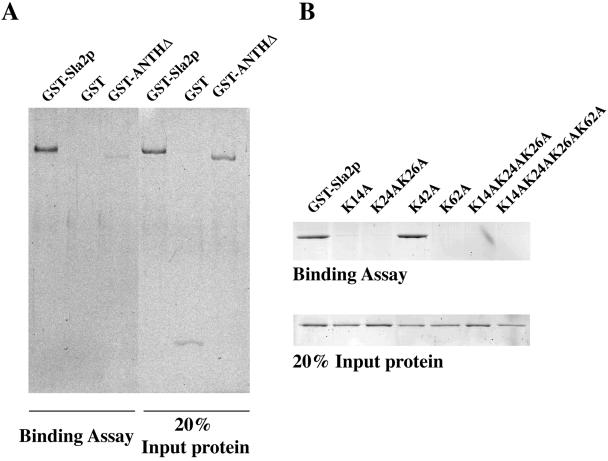
Sla2p binds to PtdIns(4,5)P2 specifically through the ANTH domain. (A) Liposomes made from major-minor lipid mix with 2 mol% PtdIns(4,5)P2 were tested for binding to GST-Sla2p (0.5 μM), GST only (0.5 μM), and GST-ANTHΔ (0.5 μM) in which the ANTH domain (amino acids 1-125) was deleted. (B) Binding of Sla2p mutants to liposomes made from major-minor lipid mix with 2 mol% PtdIns(4,5)P2. The protein concentration in each assay is 0.5 μM. K14, K24, K26, and K62 are conserved residues in the ANTH domain (Figure 1A). K42 is not conserved and was mutated as a control.
The Interaction of Sla2p with PtdIns(4,5)P2 Is Important for Cell Growth and Endocytic Internalization
To investigate the function of the Sla2p-PtdIns(4,5)P2 interaction in vivo, we generated two different sla2 mutant strains. One is sla2 ANTHΔ, in which the ANTH domain of Sla2p was completely deleted, and the other is sla2 4K-A, in which the four conserved lysine residues K14, 24, 26, and 62 were changed to alanines. Each mutant was integrated into the SLA2 chromosomal locus. Whole-cell protein extracts from the different mutants grown at 25°C or shifted for 2 h to 37°C were analyzed by SDS-PAGE and immunoblotted with polyclonal antisera against Sla2p (Figure 4A). Each mutant expressed a protein of the predicted size. The sla2 4K-A mutant was expressed at wild-type levels, whereas the sla2 ANTHΔ mutant appeared to be expressed at slightly lower levels. Although sla2Δ mutant cells were unable to grow at 34°C or above, sla2 ANTHΔ was able to grow at 34°C but not at 37°C (Figure 4B). Compared with wild-type, the sla2 4K-A mutant grew much slower at 37°C (Figure 4B). A similar temperature-sensitive growth defect was observed for sla2 3K-A(K14AK24AK26A) cells, but not for sla2 K14A cells, sla2 K24AK26A cells, or sla2 K62A cells (unpublished data). These results show that the ANTH domain of Sla2p is important for cell growth.
Figure 4.
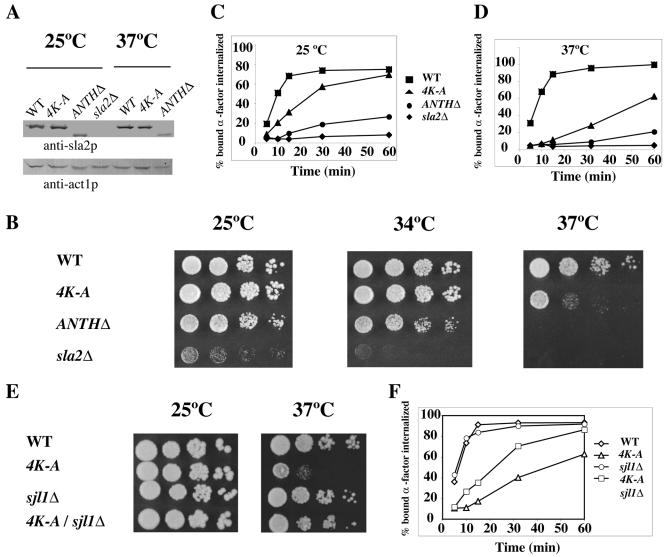
The Sla2p-PtdIns(4,5)P2 interaction is important for cell growth and the endocytic internalization. (A) Extracts of cells grown at 25°C or after 2 h at 37°C were prepared and analyzed by Western blotting with an anti-Sla2p antibody or an anti-actin antibody. (B) The ANTH domain is essential for cell growth at elevated temperatures. Dilution series of wild-type, sla2 4K-A, sla2 ANTHΔ, and sla2Δ cells were plated on YPD medium and incubated for 2 d at 25, 34, or 37°C, respectively. (C and D) Receptor-mediated internalization of [35S]methionine-labeled α-factor in sla2 mutants after a 5-min preincubation at either 25 or 37°C. Each assay was performed three times, and similar results were obtained each time. (E) Deletion of SJL1 suppressed the temperature-sensitive growth defect of the sla2 4K-A mutant. Dilution series of wild-type, sla2 4K-A, sjl1Δ, and sla2 4K-A sjl1Δ cells were plated on YPD medium and incubated for 2 d at either 25 or 37°C. (F) Deletion of SJL1 suppressed the endocytic internalization defect of the sla2 4K-A mutant. Endocytosis of [35S]methionine-labeled α-factor was measured after a 5-min preincubation at 37°C. This assay was performed three times, and similar results were obtained each time.
Because Sla2p is required for endocytic internalization, we next examined internalization rates in each mutant using an [35S]methionine-labeled α-factor uptake assay. The sla2 4K-A mutant, as well as the sla2 3K-A(K14AK24AK26A) mutant, showed a significant endocytic defect at 25°C, and this defect was even more pronounced at 37°C (Figure 4, C and D, and unpublished data). Like the sla2Δ mutant, sla2 ANTHΔ mutants showed pronounced defects at both temperatures (Figure 4, C and D). Similar results were also obtained using a Lucifer Yellow uptake assay (unpublished data).
As a test of whether the growth and endocytic defects of the sla2 4K-A mutant are caused by decreased PtdIns(4,5)P2 binding in vivo, we used a genetic approach. Sjl1p (synaptojanin-like protein 1), also referred to as Inp51p (inositol polyphosphate 5-phosphatase 1), was reported to be the main phosphatase acting on PtdIns(4,5)P2 (Stolz et al., 1998b). Moreover, the level of PtdIns(4,5)P2 in sjl1Δ is reported to be two times higher than in wild-type cells (Stolz et al., 1998a). We generated a double mutant sla2 4K-A sjl1Δ by crossing sla2 4K-A with sjl1Δ. The higher PtdIns(4,5)P2 levels in this strain may enhance PtdnIns(4,5)P2-Sla2 4K-A protein binding and rescue the growth and endocytosis defects seen in the sla2 4K-A strain. Consistent with the hypothesis that the phenotype results from reduced PtdIns(4,5)P2 binding, the temperature-sensitive growth defect of sla2 4K-A strain at 37°C was significantly suppressed in the double mutant (Figure 4E). In contrast, sjl1Δ could not suppress the temperature-sensitive growth defect of the sla2 ANTHΔ or sla2Δ mutants (unpublished data). In addition, the [35S]methionine-labeled α-factor uptake assay revealed that sjl1Δ also suppresses the endocytic internalization defect of the sla2 4K-A strain at 37°C (Figure 4F). These results strongly suggest that the interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is important for both cell growth and endocytosis.
Sla2 4K-A and Sla2 ANTHΔ Are Recruited to Endocytic Patches
Data presented above show that Sla2p binds to PtdnIns(4,5)P2 through its ANTH domain in vitro and that this binding is required for endocytic internalization in vivo. These results focused our attention on addressing how this interaction affects endocytic internalization.
In wild-type cells, Sla2p localizes to punctate cortical structures at the cell cortex that we believe represent individual endocytic sites (Figure 5A). One possibility is that Sla2p is recruited to the plasma membrane via the interaction between Sla2p's ANTH domain and PtdIns(4,5)P2 and that when PtdIns(4,5)P2 is dephosphorylated to PtdIns(4)P, this interaction is terminated. We speculated that the endocytic and growth defects of the sla2 4K-A strain might be the result of its inability to efficiently localize Sla2p to endocytic patches. To test this hypothesis, we tagged the Sla2 ANTHΔ protein and Sla2 4K-A protein with GFP and analyzed the localization of these fusion proteins. Surprisingly, both Sla2 ANTHΔ-GFP and Sla2 4K-A GFP localized to cortical patches to the same extent as Sla2-GFP (Figure 5A). Similar results were obtained when untagged Sla2 proteins were localized using immunofluorescence (unpublished data).
Figure 5.
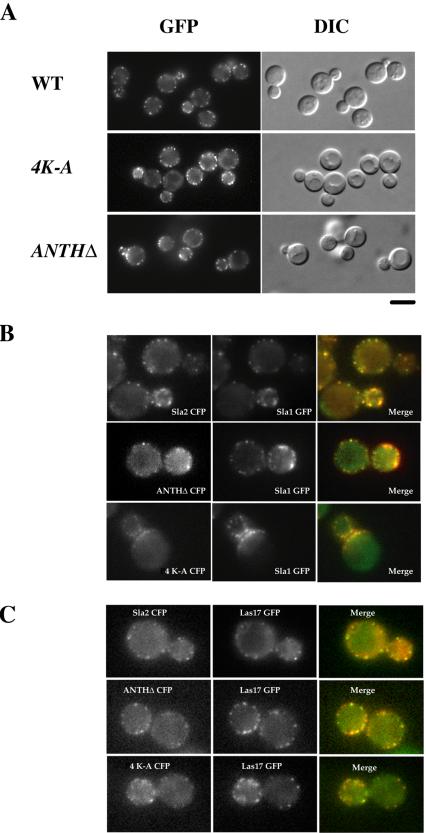
Sla2 4K-A and Sla2 ANTHΔ localize to endocytic patches. (A) The intracellular localization of wild-type and mutant Sla2p. Cells expressing Sla2-GFP, or Sla2 4K-A-GFP, or Sla2 ANTHΔ-GFP were observed by fluorescence microscopy. (B) Colocalization of the sla2 mutant proteins and Sla1p. sla2 mutant cells expressing Sla1 GFP and either Sla2-CFP, or ANTHΔ-CFP, or 4K-A- CFP, respectively, were fixed and observed by fluorescence microscopy using a YFP/CFP filter set. (C) Colocalization of the sla2 mutant proteins and Las17p. sla2 mutant cells expressing Las17 GFP and either Sla2 CFP, or ANTHΔ-CFP, or 4K-A-CFP, respectively, were fixed and observed by fluorescence microscopy using a YFP/CFP filter set.
We next used several endocytic proteins as markers to investigate whether the sla2 mutants localize to endocytic patches. We coexpressed GFP- and CFP-tagged pairs of proteins and imaged the fixed cells. Sla1p was reported to serve as an endocytic targeting adaptor to link NPFX(1,2) d-containing cargo to the endocytic machinery (Howard et al., 2002). In wild-type cells, Sla2p colocalizes with Sla1p (Figure 5B). As shown in Figure 5B, both Sla2 ANTHΔ-GFP and Sla2 4K-A GFP showed nearly complete colocalization with Sla1-CFP (Figure 5B). Similar results were also obtained with other endocytic proteins, such as Las17p and Pan1p, which are activators of the Arp2/3 complex (Figure 5C and unpublished data).
Taken together, these data demonstrate that sla2 4K-A and sla2 ANTHΔ mutants are still able to localize to endocytic patches. Thus, the ANTH-PtdIns(4,5)P2 interaction is not necessary for Sla2p recruitment to endocytic complexes, but provides yet-to-be-identified function during endocytic internalization.
The PtdIns(4,5)P2-ANTH Domain Interaction Is Required for the Normal Turnover of Actin Cortical Patches
Sla2p and its mammalian homologue Hip1R are believed to function at the interface between actin and the endocytic machinery (Engqvist-Goldstein et al., 1999, 2001, 2004; Kaksonen et al., 2003). Indeed, instead of forming normal transient punctate cortical actin patches, we found that actin comet tails associated with the cell cortex persistently in sla2Δ cells (Kaksonen et al., 2003; Figure 6A, Supplementary Movie 1). This observation suggested that Sla2p plays an important role in the productive turnover of actin cortical patches. To address whether the ANTH domain is important for Sla2's role in regulating the coupling of actin polymerization to endocytic internalization, we examined F-actin structures in sla2 4K-A and sla2 ANTHΔ mutants. The cells were fixed, stained with rhodamine-phalloidin, and observed by confocal laser microscopy. Both mutants displayed similar defects with varying severity (Figure 6B). Compared with wild-type cells, actin patches in the mutants are depolarized, more abundant, and are elongated in shape (Figure 6B). Interestingly, the severity of actin organizational defects in these mutants correlated with the severity of the endocyotic defects, sla2Δ being the most severe, and sla2 4K-A the least severe (Figures 4C and 6B).
Figure 6.
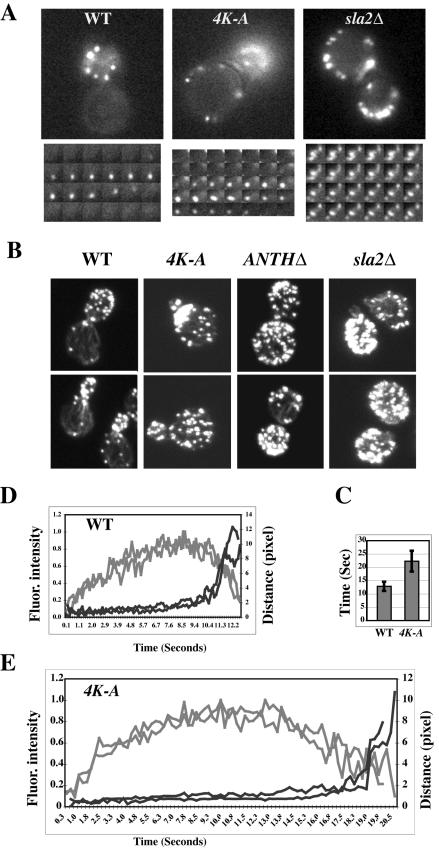
PtdIns(4,5)P2 binding is required for the normal turnover of actin cortical patches. (A) Time-lapse videomicroscopy of wild-type cells or sla2 mutants expressing Abp1-GFP. Bottom panels show selected frames of single patches at 1-s intervals. (B) Actin organization is defective in sla2 mutants. Wild-type, sla2 4K-A, sla2 ANTHΔ, and sla2Δ strains were grown on YPD to log phase at 25°C. Cells were harvested, fixed, stained with rhodamine-phalloidin, and observed by confocal laser microscopy. (C) Lifetime for Abp1 patches in wild-type or sla2 4K-A cells ± SD; n = 20. (D and E) Correlation of the formation of Abp1p patches with their movement in wild-type or sla2 4K-A mutant cells. Fluorescence intensity and distance were measured from the site of patch formation for Abp1-GFP patches over time. Each curve represents data from one patch. Fluorescence intensity over time was corrected for photobleaching. Data for two patches are shown in D and E. Gray lines represent fluorescence intensity, and black lines represent distance.
We also performed live cell imaging to analyze the dynamics of actin patches in the sla2 mutant strains. As a marker for actin, we expressed Abp1-GFP in each strain. Abp1p is an actin-binding protein that colocalizes intimately with cortical actin patches (Drubin et al., 1988). We confirmed that Abp1-GFP faithfully reports actin organization in the sla2 mutants by rhodamine-phalloidin staining (unpublished data). In the sla2 4K-A mutant, actin still assembles and disassembles at the membrane, but the actin forms somewhat elongated rather than punctate structures (Figure 6A, Supplementary Movie 1). Abp1-GFP patch lifetime in the sla2 4K-A mutant is ∼22 s, almost twice that of Abp1p patches in wild-type cells (Figure 6C). In sla2 ANTHΔ cells, most of the Abp1-GFP was present in comet tail structures, similar to those seen in sla2Δ cells, and the tails had lifetimes longer than 8 min (Supplementary Movie 2). Because we could, in contrast to the situation in sla2Δ cells, still observe formation of new actin patches in sla2 ANTHΔ cells, we speculate that actin patches can still assemble and disassemble, but at a much slower rate than normal.
Previously we showed that the fluorescence intensity of the Abp1p patches develops in a very regular manner in wild-type cells (Figure 6D; Kaksonen et al., 2003). Fast movement of Abp1p patches away from the site of formation starts suddenly and only after the Abp1-GFP intensity has already fallen considerably (Figure 6D). We speculate that this movement reflects release of the endocytic vesicle. Strikingly, in the sla2 4K-A mutant, Abp1p patches still show this behavior, except that much more time passes before the start of the fast movement (free vesicle) phase (Figure 6E). This result indicates that the PtdIns(4,5)P2-ANTH domain interaction is required for the productive coupling of actin assembly to endocytic vesicle release.
Spatiotemporal Colocalization of Sla1p and Abp1p in sla2 4K-A Cells
We recently defined a pathway of formation, internalization, and disassembly of the budding yeast endocytic complex by using live cell imaging (Kaksonen et al., 2003). In this 30-40-s process, proteins known to be involved in endocytic internalization localize to an endocytic site at different times in a highly regular manner. The initial step in the pathway is the assembly of a nonmotile complex at the plasma membrane containing Sla1p, Sla2p, Las17p, and Pan1p. After ∼20s, the Arp2/3 complex, actin and Abp1p join the patches; upon which time Sla1p, Sla2p, and Pan1p begin a slow directed inward movement. The initial patch complex is then disassembled, and the late patch complexes containing actin, Abp1p, and the Arp2/3 complex undergo a marked increase in motility. Data presented thus far in this study showed that the Sla2 4K-A protein and Sla2 ANTHΔ protein can still localize with the other early proteins, Sla1p, Pan1p, and Las17p, but that the later arriving Abp1p, actin, and Arp2/3 proteins have a longer patch life span in these sla2 mutants than in wild-type cells (Figures 5 and 6). Because Sla2p functions at the interface between actin and the endocytic machinery, we decided to further study the spatiotemporal relationship between actin-associated proteins and the early patch proteins in the sla2 mutants.
First, we measured the lifetime of endocytic proteins in patches in the sla2 mutants using fluorescence microscopy. Sla2 4K-A-GFP patches showed a lifetime of ∼60 s, which is almost twice as long as for wild-type Sla2-GFP (Figure 7A, Supplementary Movie 3). The lifetimes of Sla2 ANTHΔ-GFP patches showed more variation. Some patches persisted for an entire 4-min movie. Some patches existed in the first frame and disappeared in the middle of the movie, and other patches appeared during the movie, indicating that Sla2 ANTHΔ-GFP patches still can turnover, but at a very slow rate (Supplementary Movie 4). We also obtained very similar results for the lifetime in these mutants by using 4D deconvolution microscopy (unpublished data). In addition, consistent with the colocalization result (Figure 5B), Sla1-GFP showed a similar increase in lifetime to that observed in the sla2 4K-A mutant (Figure 7A, Supplementary Movie 5). Furthermore, the increased lifetime of Sla1p in the sla2 4K-A mutant (59 ± 8 s) was partially suppressed by sjl1Δ (49 ± 4s; p < 0.01). These data indicate that perturbation of the association between Sla2p and PtdIns(4,5)P2 extends the lifespan of endocytic complexes at the plasma membrane.
Figure 7.
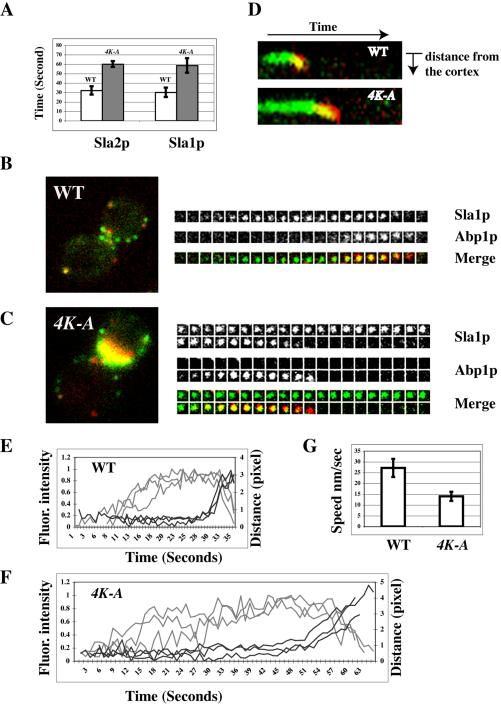
The spatiotemporal relationship between actin patches and Sla1p in sla2 4K-A mutants. (A) Lifetime for different patch proteins in wild-type or sla2 4K-A mutant cells (±SD); n = 20. Wild-type, open bars; sla2 4K-A, shaded bars. (B and C) Localization of GFP- and CFP-tagged patch proteins in wild-type and sla2 4K-A mutant cells. (B and C) Left panels: single frames from Sla1 GFP (green) and Abp1 CFP (red) merged image; right panels: time series showing composition of a single patch at 2-s intervals. Top and middle panels show separate channels. Bottom: merged images. (D) Kymograph representation of Sla1-GFP (green) and Abp1-CFP (red) in a single patch over time in a wild-type and an sla2 4K-A mutant cell. Note curvature at end of the kymograph as the patch moves off the cortex, toward the cell center. The slope of the curvature is directly proportional to the patch speed. (E and F) Correlation of the formation of Sla1p patches with their movement in wild-type or sla2 4K-A mutant cells. Fluorescence intensity and distance were measured from the site of Sla1-GFP patch formation over time. Each curve represents data from one patch. Fluorescence intensity over time was corrected for photobleaching. Data for three patches is shown in E and F. Gray line represents fluorescence intensity and black line represents distance. (G) Speed of Sla1p movement in wild-type or sla2 4K-A mutant cells (±SD). n = 10.
Next, we coexpressed pairs of GFP- and CFP-tagged proteins to reveal the spatiotemporal relationship between the late patch proteins (Abp1p, actin, and the Arp2/3 complex) and the early patch proteins (Sla1p, Sla2p, Las17p, and Pan1p) in the sla2 mutants. Because Sla1-GFP gave the brightest signal, we used it to represent the early class of proteins. In the sla2 4K-A mutant, Sla1p appears at the cell cortex early, is joined by Abp1p later, and then disappears, which is the similar order of events observed in wild-type cells (Figure 7, B and C, Supplementary Movie 6). However, the whole process in sla2 4K-A mutants takes much longer than in wild-type cells (Figure 7, B and C). Kymographs of the two-color image revealed that Sla1p exists in a nonmotile state in the sla2 4K-A mutant roughly twice as long as in wild-type cells (Figure 7D). Moreover, Abp1p and Sla1p coexisted on the cortex longer in the sla2 4K-A mutant than it in wild-type cells (Figure 7D). Thus, in the sla2 4K-A mutant the time between appearance of the endocytic complex and the onset of actin assembly is delayed. Furthermore, once actin begins to assemble, the normally very transient stage at which early and late proteins (e.g., Sla1p and Abp1p) coexist in patches lasts longer before the transition to the fast movement stage thought to represent movement of the free vesicle.
Interestingly, the slope of the curve at the end of the kymograph in the sla2 4K-A mutant is less than in wild-type cells, indicating that in the mutant the slow inward movement thought to represent membrane invagination progresses more slowly than normal (Figure 7D). To further investigate the speed of the slow movement, we performed particle-tracking analysis on Sla1p patches in both wild-type and sla2 4K-A mutant (Figure 7, E and F). The rate of Sla1p movement in sla2 4K-A mutant is ∼15 nm/s, which is significantly slower than the 27 nm/s seen in wild-type cells (Figure 7G).
Taken together, these results show that association of Sla2p and PtdIns(4,5)P2 is required for the actin-dependent slow inward movement of endocytic complex, which likely reflects membrane invagination.
DISCUSSION
Although it is now generally accepted that PtdIns(4,5)P2 plays an important role during endocytosis, the specific mechanisms of its action remain to be addressed. The importance of PtdIns(4,5)P2 in the recruitment of endocytic proteins to the plasma membrane has been confirmed by a variety of functional studies (Lemmon, 2003). In addition, recent studies revealed that epsin directly modifies membrane curvature by binding to PtdIns(4,5)P2 through its ENTH domain. To better understand the function of PtdIns(4,5)P2 on endocytosis, we focused on SLA2, which is a conserved gene essential for endocytic internalization. In this study, we characterized the interaction between Sla2p's ANTH-domain and PtdIns(4,5)P2 by using biochemical, genetic and cell biological approaches. Our data establish that the interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is not essential for recruitment to the membrane, but is crucial for the regulation of the proper timing of events during actin-dependent endocytic internalization.
ANTH Domain: Liposome binding assay
Sla2p is the founding member of a conserved family of proteins that function at the interface between actin and the endocytic machinery (Wesp et al., 1997; Engqvist-Goldstein et al., 1999, 2001, 2004; Yang et al., 1999; Baggett et al., 2003; Kaksonen et al., 2003). Amino acid sequence analysis reveals that these proteins contain ANTH domains and that each of the lysine residues implicated in PtdIns(4,5)P2 binding is conserved (Ford et al., 2001; Itoh et al., 2001). In our study, we examined whether Sla2p could bind to phosphoinositides. We adopted for these studies the protein-lipid binding assay that was used previously to study COPII vesicle formation (Matsuoka et al., 1998). Using the described conditions, we demonstrated that Sla2p binds preferentially to PtdIns(4,5)P2 through its ANTH domain. No binding signal was detected in the single lysine mutants, demonstrating that each of the four conserved lysines is important for PtdIns(4,5)P2 binding. Interestingly, the K14A, K24AK26A, and K62A mutants do not affect growth, endocytosis, or actin organization in vivo, whereas the triple and quadruple mutants (3K-A, 4K-A) are affected. It is possible that the K-A single mutations have additive effects in vivo because each mutation incrementally reduces the lipid-binding affinity.
The ANTH Domain-PtdIns(4,5)P2 Interaction Plays a Crucial Role in Sla2p Function
Previous studies indicated that only Sla2p's N-terminal region is indispensable for its roles in actin organization and endocytosis (Wesp et al., 1997). Although our subsequent analysis (Yang et al., 1999) contradicted some of these conclusions, we have repeated our analysis and the results agree with those of Wesp et al. (1997). Thus, further analysis of Sla2p's N-terminal region is required to understand the mechanism of Sla2p function. Wesp et al. showed that a mutant with a deletion of Sla2p amino acids 114-284, referred to as ΔN1, led to a severe endocytic defect (Wesp et al., 1997). However, there is no obvious motif in this region. The ANTH domain appears just before this region. We speculated that the ANTH domain may actually posses the critical activity revealed by the ΔN1 mutant, which may cause a conformational change in the ANTH domain. We integrated the sla2 ANTHΔ mutant, in which the ANTH domain is completely and precisely deleted, into its genomic locus. This sla2 ANTHΔ mutant showed a very similar phenotype to the ΔN1 mutant, supporting our hypothesis. Moreover, we also integrated into the SLA2 genomic locus the sla2 4K-A mutant, in which only the four conserved lysine residues were changed to alanines. The sla2 4K-A mutant showed significant defects on endocytic internalization at both 25 and 37°C. Together, these data established that the ANTH domain plays a key role in Sla2p's biological functions.
Another important question we wanted to address is whether the ANTH domain plays these critical roles through an interaction with PtdIns(4,5)P2. Yeast express three synaptojanin-like proteins, Sjl1p, Sjl2p, and Sjl3p, also named as Inp51p (inositol polyphosphate 5-phosphatase 1), Inp52p, Inp53p. None of these proteins is essential by itself for growth or endocytosis (Singer-Kruger et al., 1998). However, sjl1Δ sjl2Δ double mutants exhibit severe defects in receptor-mediated and fluid-phase endocytosis (Singer-Kruger et al., 1998). Most relevant to our studies was the observation that the cellular PtdIns(4,5)P2 level in sjl1Δ cells is increased two- to threefold compared with wild-type cells. On the other hand, sjl2Δ or sjl3Δ cells had no detectable changes in PtdIns(4,5)P2 levels (Stolz et al., 1998a). Previous studies showed that mutants of SJL1 have genetic interactions with mutants of other endocytic genes, such as PAN1 (Arp2/3 complex activator) and SAC6 (actin filament bundling protein; Singer-Kruger et al., 1998; Wendland and Emr, 1998). These results implicate Sjl1p in actin-dependent endocytosis. Strikingly, in our study, sjl1Δ suppressed not only the temperature-sensitive growth defect of the sla2 4K-A mutant, but also suppressed its endocytic defect at 37°C (Figure 4, E and F). In contrast, a Lucifer yellow uptake assay and rhodamine phalloidin staining revealed that deletion of SJL1 did not suppress the endocytotic and actin defects of the sla2 ANTHΔ or sla2Δ mutants at 37°C (unpublished data). These results suggest that the ANTH domain-PtdIns(4,5)P2 interaction is important for this rescue. In wild-type cells the cellular level of PtdIns(4,5)P2 has been shown to be higher at elevated temperatures than at room temperature (Stefan et al., 2002). We therefore speculate that sjl1Δ may lead an even more dramatic accumulation of PtdIns(4,5)P2 at 37°C, partially restoring the sla2 4K-A-PtdIns(4,5)P2 interaction. Further support for this possibility is the observation that overexpression of Mss4p, which is the only 1-phosphatidylinositol-4-phosphate-5-kinase in yeast, also modestly suppressed the temperature-sensitive growth defect of the sla2 4K-A mutant (Sun and Drubin, unpublished observation).
We cannot exclude the possibility that alternative pathways regulated by PtdIns(4,5)P2 play a role in the rescue. Additionally, the different severity of the sla2 4K-A mutant and the sla2 ANTHΔ mutant suggests that some feature in addition to the PtdnIns(4,5)P2 binding may also be important for Sla2p function. For example, several recent studies have suggested the existence of other ENTH/ANTH binding partners (Hyman et al., 2000; Hussain et al., 2003). Nevertheless, the fact that the sla2 4K-A mutant, but not the sla2 ANTHΔ mutant, could be suppressed, supports the conclusion that the ANTH domain-PtdnIns(4,5)P2 interaction plays a key role in Sla2p function during endocytic internalization.
The Function of the ANTH Domain-PtdnIns(4,5)P2 Interaction during Endocytosis
What is the exact role of PtdIns(4,5)P2 during endocytosis? Because PtdIns(4,5)P2 is involved in multiple important cellular processes, it is difficult to deduce specific roles by perturbing PtdIns(4,5)P2 levels. However, genetic studies of the function of a PtdIns(4,5)P2 binding domain can help circumvent this challenge. Indeed, many such studies have made significant contributions to our understanding of PtdIns(4,5)P2-protein interactions, which are made by many endocytic proteins. The N-terminal 80 amino acid region of the AP-2 α subunit, for example, is responsible for its membrane recruitment (Gaidarov and Keen, 1999). Also, membrane recruitment and activity of dynamin requires a PtdIns(4,5)P2 interaction mediated by a PH domain (Barylko et al., 1998). ENTH and ANTH domains are closely related PtdIns(4,5)P2-binding modules of ∼150 amino acids present in many proteins that function in receptor-mediated endocytosis (Kay et al., 1999; Rosenthal et al., 1999). Recent important findings by Ford et al. (2002) showed that ENTH domains but not ANTH domains can induce membrane curvature. Thus, in addition to providing a membrane recruitment function, PtdIns(4,5)P2-binding may also lead “membrane-remodeling” activity. Our results presented here suggest that the ANTH domain in Hip1R/Sla2p family proteins has a regulatory rather than a membrane recruitment function. We found that the ANTH domain is not essential for membrane localization of Sla2p, because the sla2 ANTHΔ mutant protein localizes to endocytic complexes on the plasma membrane. In addition to the ANTH domain, Sla2p also has several coiled-coiled domains, which bind to several endocytic proteins (such as Sla1p, Scd5p, Pan1p, and Clc1p; Henry et al., 2002; Baggett et al., 2003; Gourlay et al., 2003). These other interactions likely function in recruitment of Sla2p to endocytic sites on the plasma membrane.
Recently, using multicolor real-time fluorescence microscopy, we observed that the cells lacking Sla2p form a stable association between actin comet tails, endocytic cargo, and adaptors, indicating that Sla2p is a key regulator of actin-dependent internalization (Kaksonen et al., 2003). In the presence of a member of the Sla2p/Hip1R family, actin normally appears only transiently at an endocytic site at the time of internalization (Merrifield, 2004). Here, we extended the same type of analysis to an sla2 mutant deficient in PtdIns(4,5)P2 binding (sla2 4K-A). We found that the interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is required for the productive turnover of actin cortical patches. First, Sla2 4K-A-GFP patches exhibited twice the normal lifetime. Second, actin also showed a significantly increased lifetime in the sla2 4K-A mutant. Third, in sla2 4K-A mutant cells two-color live-cell imaging showed that actin coexisted with endocytic complexes much longer than in wild-type cells and that the speed of the slow inward movement of the endocytic complex was much reduced. Finally, the increased lifetime of Sla1p in the sla2 4K-A mutant was partially suppressed by sjl1Δ. Although the interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is not required for recruitment to endocytic sites, this interaction might orient the protein properly for interaction with other factors, such as the Arp2/3 complex activator Pan1p, or it might cause a conformational change of Sla2p necessary for such interactions. Recent findings from Wendland's group suggest that the ENTH domain interaction of the yeast epsin homologue Ent1p with membrane lipids cooperates with the binding of the membrane-associated ubiquitin moieties (Aguilar et al., 2003). Further biochemical and genetic analysis of the Sla2p complex should provide more insights into the mechanism by which the ANTH-PtdIns(4,5)P2 interaction regulates actin-dependent endocytic internalization.
Supplementary Material
Acknowledgments
We thank Ching Shang, Claire X. Zhang, Åsa E.Y. Engqvist-Goldstein, Sebastien Carreno, and Mariko Sekiya-Kawasaki for stimulating discussions; Eugene Futai and Sebastien Carreno for technical advice; and Chris Toret, Adam Martin, Voytek Okreglak, and Linda Lee for critical reading of the manuscript. This work was supported by National Institutes of Health grants GM42759 and GM50399 to D.G.D. and by Sigrid Jusélius Foundation and Finnish Cultural Foundation fellowships to M.K.
Article published online ahead of print in MBC in Press on December 1, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-08-0740).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aguilar, R. C., Watson, H. A., and Wendland, B. (2003). The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J. Biol. Chem. 278, 10737-10743. [DOI] [PubMed] [Google Scholar]
- Baggett, J. J., D'Aquino, K. E., and Wendland, B. (2003). The Sla2p talin domain plays a role in endocytosis in Saccharomyces cerevisiae. Genetics 165, 1661-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko, B., Binns, D., Lin, K. M., Atkinson, M. A., Jameson, D. M., Yin, H. L., and Albanesi, J. P. (1998). Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J. Biol. Chem. 273, 3791-3797. [DOI] [PubMed] [Google Scholar]
- Berridge, M. J., and Irvine, R. F. (1984). Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature 312, 315-321. [DOI] [PubMed] [Google Scholar]
- Cremona, O., and De Camilli, P. (2001). Phosphoinositides in membrane traffic at the synapse. J. Cell Sci. 114, 1041-1052. [DOI] [PubMed] [Google Scholar]
- Cullen, P. J., Cozier, G. E., Banting, G., and Mellor, H. (2001). Modular phosphoinositide-binding domains—their role in signalling and membrane trafficking. Curr. Biol. 11, R882-R893. [DOI] [PubMed] [Google Scholar]
- D'Hondt, K., Heese-Peck, A., and Riezman, H. (2000). Protein and lipid requirements for endocytosis. Annu. Rev. Genet. 34, 255-295. [DOI] [PubMed] [Google Scholar]
- Dove, S. K., Cooke, F. T., Douglas, M. R., Sayers, L. G., Parker, P. J., and Michell, R. H. (1997). Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390, 187-192. [DOI] [PubMed] [Google Scholar]
- Drubin, D. G., Miller, K. G., and Botstein, D. (1988). Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107, 2551-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., and Drubin, D. G. (2003). Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19, 287-332. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., Kessels, M. M., Chopra, V. S., Hayden, M. R., and Drubin, D. G. (1999). An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J. Cell Biol. 147, 1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., Warren, R. A., Kessels, M. M., Keen, J. H., Heuser, J., and Drubin, D. G. (2001). The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J. Cell Biol. 154, 1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., Zhang, C. X., Carreno, S., Barroso, C., Heuser, J. E., and Drubin, D. G. (2004). RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol. Biol. Cell 15, 1666-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, M. G., Mills, I. G., Peter, B. J., Vallis, Y., Praefcke, G. J., Evans, P. R., and McMahon, H. T. (2002). Curvature of clathrin-coated pits driven by epsin. Nature 419, 361-366. [DOI] [PubMed] [Google Scholar]
- Ford, M. G., Pearse, B. M., Higgins, M. K., Vallis, Y., Owen, D. J., Gibson, A., Hopkins, C. R., Evans, P. R., and McMahon, H. T. (2001). Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, 1051-1055. [DOI] [PubMed] [Google Scholar]
- Gaidarov, I., and Keen, J. H. (1999). Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J. Cell Biol. 146, 755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli, M. I., and Riezman, H. (1998). Endocytic internalization in yeast and animal cells: similar and different. J. Cell Sci. 111(Pt 8), 1031-1037. [DOI] [PubMed] [Google Scholar]
- Gourlay, C. W., Dewar, H., Warren, D. T., Costa, R., Satish, N., and Ayscough, K. R. (2003). An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J. Cell Sci. 116, 2551-2564. [DOI] [PubMed] [Google Scholar]
- Hawkins, P. T., Stephens, L. R., and Piggott, J. R. (1993). Analysis of inositol metabolites produced by Saccharomyces cerevisiae in response to glucose stimulation. J. Biol. Chem. 268, 3374-3383. [PubMed] [Google Scholar]
- Henry, K. R., D'Hondt, K., Chang, J., Newpher, T., Huang, K., Hudson, R. T., Riezman, H., and Lemmon, S. K. (2002). Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of sla2p in yeast. Mol. Biol. Cell 13, 2607-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. P., Hutton, J. L., Olson, J. M., and Payne, G. S. (2002). Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 157, 315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, N. K., Yamabhai, M., Bhakar, A. L., Metzler, M., Ferguson, S. S., Hayden, M. R., McPherson, P. S., and Kay, B. K. (2003). A role for epsin N-terminal homology/AP180 N-terminal homology (ENTH/ANTH) domains in tubulin binding. J. Biol. Chem. 278, 28823-28830. [DOI] [PubMed] [Google Scholar]
- Hyman, J., Chen, H., Di Fiore, P. P., De Camilli, P., and Brunger, A. T. (2000). Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF). J. Cell Biol. 149, 537-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, T. S., Rao, D. S., Saint-Dic, D., Michael, L. E., Kumar, P. D., Bradley, S. V., Mizukami, I. F., Oravecz-Wilson, K. I., and Ross, T. S. (2004). HIP1 and HIP1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin N-terminal homology domains. J. Biol. Chem. 279, 14294-14306. [DOI] [PubMed] [Google Scholar]
- Itoh, T., Koshiba, S., Kigawa, T., Kikuchi, A., Yokoyama, S., and Takenawa, T. (2001). Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291, 1047-1051. [DOI] [PubMed] [Google Scholar]
- Itoh, T., and Takenawa, T. (2002). Phosphoinositide-binding domains: functional units for temporal and spatial regulation of intracellular signalling. Cell Signal. 14, 733-743. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D. G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475-487. [DOI] [PubMed] [Google Scholar]
- Kay, B. K., Yamabhai, M., Wendland, B., and Emr, S. D. (1999). Identification of a novel domain shared by putative components of the endocytic and cytoskeletal machinery. Protein Sci. 8, 435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin, V., Wasiak, S., Hussain, N. K., Angers, A., and McPherson, P. S. (2004). ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 117, 9-18. [DOI] [PubMed] [Google Scholar]
- Lemmon, M. A. (2003). Phosphoinositide recognition domains. Traffic 4, 201-213. [DOI] [PubMed] [Google Scholar]
- Martin, T. F. (2001). PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13, 493-499. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., Orci, L., Amherdt, M., Bednarek, S. Y., Hamamoto, S., Schekman, R., and Yeung, T. (1998). COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell 93, 263-275. [DOI] [PubMed] [Google Scholar]
- Merrifield, C. J. (2004). Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 14, 352-358. [DOI] [PubMed] [Google Scholar]
- Mitchell, D. A., Marshall, T. K., and Deschenes, R. J. (1993). Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9, 715-722. [DOI] [PubMed] [Google Scholar]
- Munn, A. L. (2001). Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim. Biophys. Acta 1535, 236-257. [DOI] [PubMed] [Google Scholar]
- Rosenthal, J. A., Chen, H., Slepnev, V. I., Pellegrini, L., Salcini, A. E., Di Fiore, P. P., and De Camilli, P. (1999). The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J. Biol. Chem. 274, 33959-33965. [DOI] [PubMed] [Google Scholar]
- Sekiya-Kawasaki, M. et al. (2003). Dynamic phosphoregulation of the cortical actin cytoskeleton and endocytic machinery revealed by real-time chemical genetic analysis. J. Cell Biol. 162, 765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, C., Hazbun, T. R., Cheeseman, I. M., Aranda, J., Fields, S., Drubin, D. G., and Barnes, G. (2003). Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell 14, 3342-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger, B., Nemoto, Y., Daniell, L., Ferro-Novick, S., and De Camilli, P. (1998). Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J. Cell Sci. 111(Pt 22), 3347-3356. [DOI] [PubMed] [Google Scholar]
- Stahelin, R. V., Long, F., Peter, B. J., Murray, D., De Camilli, P., McMahon, H. T., and Cho, W. (2003). Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 278, 28993-28999. [DOI] [PubMed] [Google Scholar]
- Stefan, C. J., Audhya, A., and Emr, S. D. (2002). The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell 13, 542-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz, L. E., Huynh, C. V., Thorner, J., and York, J. D. (1998a). Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 148, 1715-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz, L. E., Kuo, W. J., Longchamps, J., Sekhon, M. K., and York, J. D. (1998b). INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J. Biol. Chem. 273, 11852-11861. [DOI] [PubMed] [Google Scholar]
- Takenawa, T., and Itoh, T. (2001). Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533, 190-206. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. (1997). Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065-1075. [DOI] [PubMed] [Google Scholar]
- Wendland, B., and Emr, S. D. (1998). Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, B., Steece, K. E., and Emr, S. D. (1999). Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 18, 4383-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk, M. R., and De Camilli, P. (2004). Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proc. Natl. Acad. Sci. USA 101, 8262-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesp, A., Hicke, L., Palecek, J., Lombardi, R., Aust, T., Munn, A. L., and Riezman, H. (1997). End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Cope, M. J., and Drubin, D. G. (1999). Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol. Biol. Cell 10, 2265-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


