Abstract
The assembly of ribosomes involves the coordinated processing and modification of rRNAs with the temporal association of ribosomal proteins. This process is regulated by assembly factors such as helicases, modifying enzymes, and GTPases. In contrast to the assembly of cytoplasmic ribosomes, there is a paucity of information concerning the role of assembly proteins in the biogenesis of mitochondrial ribosomes. In this study, we demonstrate that the Saccharomyces cerevisiae GTPase Mtg2p (Yhr168wp) is essential for mitochondrial ribosome function. Cells lacking MTG2 lose their mitochondrial DNA, giving rise to petite cells. In addition, cells expressing a temperature-sensitive mgt2-1 allele are defective in mitochondrial protein synthesis and contain lowered levels of mitochondrial ribosomal subunits. Significantly, elevated levels of Mtg2p partially suppress the thermosensitive loss of mitochondrial DNA in a 21S rRNA methyltransferase mutant, mrm2. We propose that Mtg2p is involved in mitochondrial ribosome biogenesis. Consistent with this role, we show that Mtg2p is peripherally localized to the mitochondrial inner membrane and associates with the 54S large ribosomal subunit in a salt-dependent manner.
INTRODUCTION
The assembly of ribosomes requires the coordinate processing and modification of rRNAs with the temporal association of ribosomal proteins. Biogenesis of ribosomes also requires a number of assembly proteins such as helicases, modifying enzymes, and GTPases. In addition to the well characterized Ras-like GTP-binding proteins, all organisms possess a number of conserved GTPases predicted to play roles in translation (Leipe et al., 2002). Direct evidence for a role in ribosome function has been obtained for several of these GTPases, including the bacterial Era and Obg proteins (Tan et al., 2002; Lin et al., 2004; Wout et al., 2004).
The Obg subfamily of GTPases has been identified in all organisms sequenced to date. Prokaryotes have one Obg protein and archaea have two paralogs, whereas eukaryotes such as the yeast Saccharomyces cerevisiae possess four paralogs in three distinct protein classes (Leipe et al., 2002). Obg proteins possess a highly conserved GTP-binding domain, suggesting that they share a common mode of regulation. Mitochondrial and bacterial Obg proteins are likely to be orthologous because they are conserved throughout their protein lengths, in contrast to the other eukaryotic Obg proteins that possess vastly different protein sequences outside of the GTP-binding domain.
In S. cerevisiae, each distinct Obg protein type seems to play a specialized role in ribosome function. The nucleolar Nog1p protein copurifies with pre60S intermediates and is involved in cytosolic ribosome assembly, particularly in biogenesis of the pre60S particle (Fromont-Racine et al., 2003; Kallstrom et al., 2003; Saveanu et al., 2003). In contrast, Rbg1p and Rbg2p, two cytosolic Obg proteins that are 52% similar to one another, associate with translating ribosomes (Wout and Maddock, unpublished data). Interestingly, Rbg1p also may be involved in stress response because it interacts on the translating ribosome with the stress response protein Gcn1p (Marton et al., 1993) through a bridging protein, Gir2p (Wout and Maddock, unpublished data).
Here, we describe the analysis of the fourth S. cerevisiae mitochondrial Obg protein, encoded by YHR168W, and hereafter called Mtg2p for MiTochondrial GTPase 2. We show that deletion of MTG2 leads to loss of mitochondrial DNA, consistent with a role in mitochondrial ribosome function. Mtg2p is found to be peripherally associated with the mitochondrial inner membrane facing the matrix compartment. Moreover, Mtg2p fractionates with the mitochondrial large ribosomal subunit in a salt-dependent manner. Yeast cells harboring a temperature-sensitive allele of MTG2 are defective in translation and have reduced levels of mitochondrial subunits at the nonpermissive temperature. Finally, overexpression of MTG2 partially suppresses a 21S rRNA methyltransferase mutant, Δmrm2. Together, these results suggest that Mtg2p plays a critical role in large mitochondrial ribosome function.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast strains used in this study are listed in Table 1. Complete media used were YEP (1% yeast extract, 2% peptone) containing 2% glucose (YPD), 6% ethanol (YPE), or 3% glycerol (YPG) as carbon source. Synthetic minimal media (0.67% yeast nitrogen base without amino acids containing 2% glucose [SD], 2% galactose [SGal], 0.1% 5-Fluoro-orotic acid, 2% glucose [5FOAD], 0.006% canavanine sulfate, 2% glucose [SCAN]) were supplemented with appropriate amino acids when required, as described previously (Guthrie and Fink, 1991). Semisynthetic lactate medium (0.3% yeast extract, 0.05% glucose, 0.05% CaCl2, 0.05% NaCl, 0.06% MgCl2, 0.1% KH2PO4, 0.1% NH4Cl, 2% dl-lactic acid, and 0.8% NaOH, pH 5.5) was prepared as indicated previously (Glick and Pon, 1995).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| CRY1 | MATa, ura3–52, trp1Δ2, leu2–3112, his3–11, ade2–1, can1–100, ρ+ | Brickner and Fuller, 1997 |
| CRY2 | MATα, ura3–52, trp1Δ2, leu2–3112, his3–11, ade2–1; can1–100, ρ+ | Brickner and Fuller, 1997 |
| SEY6210/SEY6211 | MATa/α, leu2–3112/leu2–3112, ura3–52/ura3–52, his3Δ200/his3Δ200, trp1-Δ901/trp1-Δ901, LYS2/lys2–801, ADE2/ade2–101, suc2-Δ9/suc2-Δ9, ρ+ | Brickner and Fuller, 1997 |
| BY4742 | MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, ρ+ | ATCC |
| JM1837 | MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0, mtg2::kanMX4, ρ0 | EUROSCARF |
| JM1823 | MATa, ura3–52, his3–1, leu2–3112, trp1–289, mrm2::kanMX4, ρ0 | EUROSCARF |
| JM1931 | MATa, ura3–52, trp1Δ2, leu2–3112, his3–11, ade2–1, can1–100, ρ0 | This study |
| JM1914 | MATa/α, ura3Δ0/ura3–52, leu2Δ0/leu2–3112, his3Δ1/his3–11, trp1Δ2/TRP1, LYS2/lys2Δ0, ADE2/ade2–1, CAN1/can1–100, MTG2/mtg2::KanMX4, ρ+ | This study |
| JM1918 | MATα/a, ura3Δ0/ura3–52, leu2Δ0/leu2–3112, his3Δ1/his3–11, trp1Δ2/TRP1, LYS2/lys2Δ0, ADE2/ade2–1, CAN1/can1–100, MTG2/mtg2::KanMX4, pKD5 [CEN, MTG2, URA3], ρ+ | |
| JM3680 | MATa/α, his3/his3Δ1, leu2/leu2Δ0, lys2Δ0/lys2Δ0, ura3/ura3Δ0, ADE2/ade2–1, CAN1/can1–100, mtg2::kanMX4/mtg2::kanMX4, ρ0 | This study |
| JM2195 | MATα, lys2Δ0, ura3, leu2, his3, can1–100, mtg2::kanMX4, pKD5 [CEN, MTG2, URA3], ρ+ | This study |
| JM2575 | MATα, lys2Δ0, ura3, leu2, his3, can1–100, mtg2::kanMX4, pKD7 [CEN, MTG2, HIS3], ρ+ | This study |
| JM2574 | MATα, lys2Δ0, ura3, leu2, his3, can1–100, mtg2::kanMX4, pRS313, ρ+ | This study |
| JM3679 | MATα, lys2Δ0, ura3, leu2, his3, can1–100, mtg2::kanMX4, pKD69 [CEN, mtg2–1, HIS3], ρ+ | This study |
| JM3666 | MATa/α, ura3–52/ura3Δ0, his3–1/his3Δ1, leu2–3112/leu2Δ0, trp1–289/TRP1, LYS2/lys2Δ0, MRM2/mrm2::kanMX4, ρ+ | This study |
| JM3668 | MATα, ura3, his3, leu2, lys2Δ0, trp1–289, mrm2::kanMX4, ρ+ | This study |
| JM3670 | MATα, ura3, his3, leu2, lys2Δ0, trp1–289, mrm2::kanMX4, pKD68 [CEN, MRM2, HIS3], ρ+ | This study |
| JM3671 | MATα, ura3, his3, leu2, lys2Δ0, trp1–289, mrm2::kanMX4, pRS313, ρ+ | This study |
| JM3672 | MATα, ura3, his3, leu2, lys2Δ0, trp1–289, mrm2::kanMX4, pKD7 [CEN, MTG2, HIS3], ρ+ | This study |
Plasmid and Strain Construction
Yeast genomic DNA isolation and yeast transformation were carried out as described previously (Guthrie and Fink, 1991). The MTG2 gene was amplified from the yeast genomic DNA (CRY1) by using polymerase chain reaction (PCR) with oligonucleotides 5′ GGCACGTGCCAAATTTG 3′ and 5′ GCATAGGCCCTGCAAAT 3′. The 2470-bp PCR product was cloned into pCR 2.1-TOPO (Invitrogen, Carlsbad, CA) and the product was confirmed by sequencing in both directions. MTG2 was subcloned into pRS316 (CEN, URA3) as a HindIII-XhoI fragment and into pRS313 (CEN, HIS3) as an EcoRI fragment to yield pKD5 and pKD7, respectively. The MRM2 gene was PCR amplified from the yeast genomic DNA from strain CRY1 with oligonucleotides 5′ GGACTAGTGCACCCGTGCATATTGAAAG 3′ and 5′ CCATCGATTTAGGAGATGATGCAGTCAA 3′. The 1963-bp PCR product was cloned into pCR 2.1-TOPO (Invitrogen). MRM2 was subcloned into pRS313 (CEN, HIS3) as a SpeI-EcoRV fragment to yield pKD68.
JM2195, a ρ+ strain with a chromosomal mtg2 deletion and a plasmid-borne MTG2 was created as follows: JM1837 (Δmtg2), a ρ0 strain, was mated to strain CRY1 to produce a MTG2/mtg2 ρ+ strain (JM1914). JM1914 was transformed with pKD5 to generate JM1918. After sporulation of JM1918, haploid spores were selected on SCAN. Haploid Δmtg2 ρ+ spores containing pKD5 were screened by patching independent spore products simultaneously on YPD plates supplemented with antibiotic G418 and SD plates lacking uracil to generate JM2195. ρ0 derivatives of CRY1 (JM1931) were obtained by growing cultures in YPD containing 50 μg/ml ethidium bromide.
JM3668, a ρ+ strain with a chromosomal mrm2 deletion, was created as follows: JM1823 (Δmrm2), a ρ0 strain, was mated to BY4742 to produce MRM2/mrm2 ρ+ strain (JM3666). JM3666 was sporulated, and haploid Δmrm2 spores were selected on YPD plates supplemented with antibiotic G418 to generate JM3668.
Generation of mtg2 Temperature-sensitive (ts) Alleles
ts mtg2 alleles were created using error-prone PCR as described previously (Stark, 1998). The entire coding region of MTG2 was amplified using primers 5′ AGATAGAGTTATACATACTT 3′ and 5′ TATATTTACTATTTACAATC 3′. Three independent PCR reactions were carried out in presence of 2 mM MgCl2 containing a 5:1 ratio of (dTTP+dCTP):(dATP+ dGTP). The concentration of MnCl2 was varied from 0.3 to 0.5 mM. The resulting PCR products were cloned into BsrGI and SnaBI digested pKD7 by gap repair. The PCR product and pKD7 (digested with BsrGI and SnaBI) were cotransformed into JM2195, selected on SD plates lacking histidine and uracil, and patched onto 5FOAD to counterselect the URA3 helper plasmid bearing MTG2. Approximately 1800 independent cells were scored for the ability to grow at 23°C but not at 37°C on YPG. Plasmid DNA from potential ts alleles was amplified in Escherichia coli and sequenced. Plasmid DNA was also shuttled into JM2195 to create mgt2-1, and this was confirmed for its ability to grow at 23°C but not at 37°C.
Immunoblot Analysis
Proteins were separated on 10% SDS-PAGE and processed for immunoblot as described previously (Lin et al., 2004). Antibodies to Mtg2p were generated to full-length protein expressed in E. coli from pET28a (Novagen, Madison, WI) (our unpublished data) and used at a dilution of 1:2000 after treatment with acetone powders generated from Δmtg2, as described previously (Harlow and Lane, 1988). The following antibody concentrations were used: Cox2p, 1:50 (Pinkham et al., 1994), F1β, 1:5000 (Emtage and Jensen, 1993), KDH, 1:2000 (Koehler et al., 1998), Pgk1p, 1:300 (Molecular Probes, Eugene, OR), Tim23p, 1:10,000 (Emtage and Jensen, 1993), Mrp7p, 1:200 (Fearon and Mason, 1988), Mrp13, 1:200 (Partaledis and Mason, 1988), and Mrp49, 1:200 (Fearon and Mason, 1992).
Subcellular Localization of Mtg2p
Mitochondria were isolated from 10 l of diploid SEY6210/6211 grown in semisynthetic lactate medium at 30°C to an OD600 of 3. Mitochondria were isolated as described previously (Glick and Pon, 1995), except that cells were incubated with Lyticase (5 mg/gm dry cell weight) in 1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4, at 30°C for 2 h to produce spheroplasts.
Mitochondria were subject to extraction with sodium chloride or sodium carbonate as described previously (Sogo and Yaffe, 1994). Inner and outer membrane vesicles were obtained as described previously (Daum et al., 1982). Mitoplasts were generated by diluting 100 μg of mitochondria (1 mg/ml) in buffer C (0.6 M sorbitol, 20 mM K+ HEPES, pH 7.4) with 20 mM K+ HEPES, pH 7.4, to a final concentration of 0.1 mg/ml followed by incubation on ice for 30 min. Protease digestion of intact mitochondria (in buffer C) or mitoplasts (in 20 mM K+ HEPES, pH 7.4) were carried out on ice for 30 min at the indicated concentration of proteinase K. The reactions were inactivated by addition of trichloroacetic acid (TCA) to 15%.
Separation of Mitochondrial Ribosome
Mitochondrial ribosomes were separated by sucrose density gradient as described previously (Fearon and Mason, 1992) with slight modification. Mitochondria (15 mg) were washed three times in buffer D (10 mM Tris-Cl, pH 7.4, 10 mM magnesium acetate, 100 mM NH4Cl, 7 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride) and resuspended in the same buffer. Mitochondria were incubated on ice for 10 min to allow swelling of the mitochondrial membranes. Mitochondria were recovered by centrifugation at 12,000 × g for 10 min at 4°C, resuspended in buffer D + 2% NP-40, and incubated on ice 10 min to allow further swelling. Lysis was achieved by 30–50 strokes of a tight fitting pestle in a Dounce homogenizer. Mitochondrial detergent lysates were clarified by centrifugation at 40,000 × g for 25 min at 4°C, and UV absorbance at 260 nm (A260) was measured. Equivalent amounts of A260 were layered on a 10 ml of 10–30% linear sucrose gradient containing 10 mM Tris-Cl, pH 7.4, 10 mM magnesium acetate, 7 mM β-mercaptoethanol and 100, 200, or 500 mM NH4Cl, as indicated. The gradients were centrifuged at either 135,000 × g for 4 h or 250,000 × g for 5 h at 4°Cin a Beckman SW41Ti rotor. The gradients were fractionated into 400-μl aliquots, and the UV absorbance at 254 nm was monitored using an ISCO continuous-flow cuvette. Protein samples were precipitated by addition of TCA to 15%, separated by SDS-PAGE, and subjected to immunoblot analysis.
Analysis of Mitochondrial Translation Products
Cell cultures (15 ml) were grown in SGal with the appropriate prototrophic requirement at 23°C to an OD600 of 1. Cells were shifted to 37°C or maintained at 23°C, as indicated. Mitochondrial translation products were labeled with [35S]methionine as described previously (Fox et al., 1991) except that the cells were labeled with 1 mCi of Trans 35S-label for either 1 h at 23°C or 30 min at 37°C. Mitochondria were isolated as described (Fox et al., 1991). Protein concentration was estimated by the Bradford method (Bradford, 1976). Equal amounts of mitochondrial proteins were separated by SDS-PAGE on a 15% polyacrylamide gel. The gel was dried and exposed to x-ray film to visualize translation products.
Suppression of mrm2 Deletion Strain
Δmrm2 ρ+ cells were transformed with pKD7 (JM3672) or, as controls, either the empty vector (JM3671) or plasmid-borne MRM2 (JM3670). Cells were grown at 37°C in SD-His for 24 h and then plated on either YPD or YPG. The fraction of total cells that was able to grow on YPG represents the percentage of nonpetite cells.
RESULTS
Mtg2p is a member of the Obg family of GTPases that shares sequence similarity with the bacterial proteins along its entire length, although with some noted exceptions. First, Mtg2p possesses 88 N-terminal amino acids not found in the bacterial proteins that are predicted to contain a mitochondrial targeting sequence (PSORT; http://psort.nibb.ac.jp/). Consistent with the cleavage of a mitochondrial import sequence, Mtg2p migrates as an ∼50-kDa band on SDS-PAGE, approximately 5 kDa smaller than the predicted molecular mass of 54.5 kDa (our unpublished data). Second, MTG2 includes two short insertions found in Obg proteins of only a subset of eukaryotes, including fungi and plants. Based on the structure of the B. subtilis Obg protein, these additional domains can be positioned between conserved structural elements within an extended domain called the OBG fold (Buglino et al., 2002). Deletion of a sequence containing both regions results in a nonfunctional protein, suggesting that these insertions are essential for Mtg2p function. Surprisingly, however, relatively large protein insertions such as 3HA (Longtine et al., 1998) and TAP (Rigaut et al., 1999) at the end of this region (at amino acid 294) did not perturb Mtg2p function (our unpublished data). Finally, MTG2 terminates after the conserved GTP-binding domain, whereas many bacterial Obg proteins have additional conserved C-terminal sequences. It also should be noted that the MTG2 sequence previously deposited in GenBank contains a frameshift at amino acid 497; the corrected sequence has been entered into GenBank (AY643812).
MTG2 Is Required for Mitochondrial Translation
To determine the role of Mtg2p in vivo, a phenotypic screen (Hampsey, 1997) was performed on a Δmtg2 strain. Cells lacking MTG2 grew slowly on YPD and were not able to use nonfermentable carbon sources such as glycerol (YPG) and ethanol (YPE) (Figure 1A). To determine whether this phenotype was due to loss of mitochondrial DNA, a Δmtg2 haploid was mated with either wild-type or an ethidium bromide-induced ρ0 tester strain. The Δmtg2/MTG2 heterozygotes grew on both glucose (YPD) and glycerol (YPG) plates, indicating that the Δmtg2 is recessive (Figure 1B). The Δmtg2/MTG2 ρ0 heterozygotes, however, did not grow on YPG (Figure 1B) suggesting that deletion of MTG2 leads to loss of mitochondrial DNA. To verify that this was the case, we examined the distribution of DNA in Δmtg2. 4,6-Diamidino-2-phenylindole (DAPI)-stained wild-type cells display a bright nuclear DNA fluorescence and punctate foci representing mitochondrial DNA. In Δmtg2 cells, however, only the bright nuclear stain was visible (Figure 1C).
Figure 1.
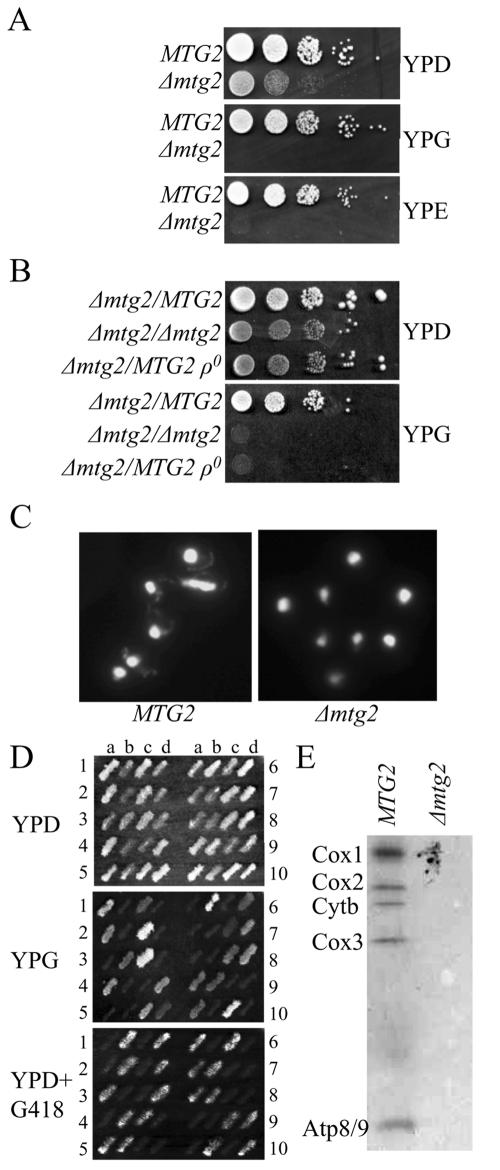
The Δmtg2 mutant lacks mitochondrial DNA. (A) Cells lacking Mtg2p do not grow on nonfermentable carbon sources. Shown are serial spot dilutions of wild-type (MTG2) and Δ mtg2 cells on YPD, YPG, and YPE plates. (B) The Δmtg2 mutant is ρ0. Serial dilutions of the indicated diploids were assayed for growth on YPD and YPG. Strains are as described in the text. (C) Representative fluorescent images of DAPI-stained DNA in wild-type and Δmtg2 haploid cells reveal a lack of mtDNA in Δmtg2 mutants. In each cell, the large focus is the cell nucleus and the smaller punctuate foci represent mitochondrial DNA. (D) Replica plate analysis of dissected tetrads from heterozygous diploid Δmtg2/MTG2ρ+. Haploid spore products (a–d) from dissected tetrads (1–10) were grown on YPD, replica plated onto YPG and YPD with G418, as indicated. (E) [35S]Methionine labeling of newly synthesized mitochondrial gene products in MTG2 or Δmtg2ρ+. Mitochondria were isolated, and equivalent amounts of protein were separated by SDS-PAGE, the gel dried, and the radioactive proteins visualized on x-ray film. The position of Cox1p, Cox2p, Cytbp, Cox3p, and Atp8/9p is indicated.
Loss of mitochondrial DNA in the Δmtg2 mutant indicates that the wild-type gene product either plays a direct role in maintenance of mitochondrial DNA (mtDNA) or an essential role in a distinct mitochondrial function that when disrupted leads to eventual loss of mtDNA. To determine whether the loss of mitochondrial DNA was due to a direct or an indirect consequence of deletion of MTG2, spore products from a Δmtg2/MTG2 ρ+ heterozygous diploid were examined for growth on YPG. All products harboring Δmtg2 failed to grow (Figure 1D). We examined whether Δmtg2 spore products possessed mtDNA by mating them with a MTG2ρ0 tester strain and examined the ability of the subsequent diploid to grow on YPG. Interestingly, 70% of these diploids were able to grow on YPG (our unpublished data), thus indicating a subpopulation of the cells in the newly formed Δmtg2 spore products possessed mtDNA. These newly formed Δmtg2ρ+ spore products lost mtDNA and converted to ρ0 cells after subsequent growth on glucose medium (our unpublished data). To examine whether these Δmtg2ρ+ cells had a translational defect mitochondrial translation products were labeled and compared with wild-type cells. A severe defect in mitochondrial translation was detected in Δmtg2ρ+ cells as determined by the inability to incorporate radiolabeled methionine into newly synthesized mitochondrial protein products (Figure 1E). Thus, we conclude that Mtg2p is required for mitochondrial translation, and it is likely that the ultimate loss of mtDNA in Δmtg2 cells is a consequence of the defect in translation.
Mtg2p Is a Peripherally Associated Mitochondrial Inner Membrane Protein Facing the Matrix
The phenotype of Δmtg2 mutants and the predicted mitochondrial targeting sequence provide strong evidence that Mtg2p is a bona fide mitochondrial protein. To determine the cellular location of Mtg2p, we performed a series of biochemical fractionations. Mtg2p was enriched in the mitochondrial fraction, as was Cox2p, an integral membrane subunit of the cytochrome c oxidase encoded by the mitochondrial genome (Pinkham et al., 1994), but not in fractions that were enriched with Pgk1p, a cytosolic marker (Figure 2A). To determine the submitochondrial localization of Mtg2p, mitochondria were fractionated into membrane pellet and soluble matrix. Mtg2p cofractionated with Cox2p (Pinkham et al., 1994) but not with a soluble mitochondrial matrix protein KDH (Koehler et al., 1998) (Figure 2B), indicating that Mtg2p is associated with the mitochondrial membrane. To examine the nature of association of Mtg2p with the mitochondrial membrane, mitochondria were treated with 0.1 M Na2CO3, 1 M NaCl, or buffer. Sodium carbonate dislodges peripheral membrane proteins but leaves integral membrane proteins attached (Fujiki et al., 1982). Like the membrane protein Cox2p and the peripheral membrane protein F1β (Emtage and Jensen, 1993), Mtg2p remained in the membrane pellet when treated with either the buffer control or sodium chloride (Figure 2C), indicating a tight association with the membrane. In contrast, when mitochondria were treated with sodium carbonate, Cox2p was present in the pellet fraction, whereas Mtg2p and F1β were found in the soluble fraction (Figure 2C). Therefore, Mtg2p is a peripheral membrane protein, consistent with absence of predicted transmembrane domain(s) within Mtg2p.
Figure 2.
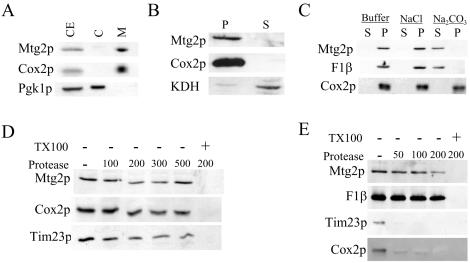
Mtg2p is enriched on the mitochondrial inner membrane facing the matrix. Localization of Mtg2p in mitochondria. (A) Yeast cell extracts (CE) were fractionated into cytosol (C) and mitochondria (M). Proteins were separated by SDS-PAGE and subjected to immunoblot analysis. (B) Mitochondria were disrupted by sonication and further fractionated into membrane (P) and soluble matrix fractions (S) by centrifugation at 100,000 × g. Equivalent amounts of protein were separated by SDS-PAGE and subjected to immunoblot analysis. (C) Purified mitochondria were either treated with 0.1 M Na2CO3, 1 M NaCl or buffer as indicated. Soluble (S) and membrane (P) fractions were subjected to SDS-PAGE and immunoblot analysis. (D) Intact mitochondria were treated with 0–500 μM/ml proteinase K in the presence or absence of 1% Triton X-100 (TX100), as indicated. Samples were TCA precipitated, separated by SDS-PAGE, and subjected to immunoblot analysis. (E) Mitoplasts were generated by subjecting purified mitochondria to osmotic shock to disrupt the outer membrane. Mitoplasts were then treated with 0–200 μM/ml proteinase K at the indicated concentration in the presence or absence of 1% Triton X-100 as indicated. Samples were TCA precipitated, separated by SDS-PAGE, and subjected to immunoblot analysis. Samples were analyzed using antibodies against Mtg2p, Cox2p, Pgk1p, F1β, KDH, and Tim23p. Cox2p and Tim23p are integral inner membrane proteins having antibody epitopes facing the intermembrane space of the mitochondria. F1β is a peripherally associated inner membrane protein facing the mitochondrial matrix, whereas KDH is a soluble matrix protein. Pgk1p is a cytosolic marker protein.
To determine whether Mtg2p was associated with the outer or inner mitochondrial membrane, a series of protease sensitivity assays were performed. To examine whether Mtg2p was associated with the outer membrane, intact mitochondria were treated with increasing concentrations of proteinase K in the presence or absence of TritonX-100. Inner membrane proteins (such as Cox2p and Tim23p) were inaccessible to exogenously added protease in the absence of detergent, indicating that the outer membrane was intact in our preparation (Figure 2D). Mtg2p, also remained protease protected in intact mitochondria (Figure 2D), indicating that Mtg2p does not have domains facing the cytosol. Furthermore, when inner membrane vesicles were separated from outer membrane vesicles, Mtg2p was present predominantly with the inner membrane vesicles, as was F1β (our unpublished data).
We next determined whether Mtg2p faces the intermembrane space or the matrix compartment. Mitochondria were subjected to hypo-osmotic shock, which disrupts the outer membrane of the mitochondria, thereby making the intermembrane space accessible to protease. The resulting mitoplasts were treated with increasing concentrations of proteinase K in presence or absence of Triton X-100. In absence of any detergent, F1β, a peripheral inner membrane protein facing the matrix compartment remains protease protected, confirming that the inner membrane remained intact (Figure 2E). As expected Cox2p and Tim23p, inner membrane proteins with epitopes facing the intermembrane space (Emtage and Jensen, 1993; Pinkham et al., 1994), were progressively degraded by increasing concentrations proteinase K (Figure 2E). Under these conditions, however, Mtg2p remained protease protected, mimicking the properties of a matrix protein (Figure 2E). Together, these studies demonstrate that Mtg2p is a mitochondrial matrix protein tightly associated with the inner membrane.
Mtg2p Fractionates with Mitochondrial Ribosomal Proteins in a Salt-dependent Manner
In mitochondria, translation takes place on ribosomes associated with the inner membrane (Spithill et al., 1978; Marzuki and Hibbs, 1986). Mutations in genes involved in mitochondrial ribosome function lead to either complete loss or large deletions of mitochondrial DNA (Myers et al., 1985). The peripheral association of Mtg2p with the inner membrane (Figure 2), the loss of mitochondrial DNA in the Δmtg2 strain (Figure 1), and the predicted role of GTPases in translation (Leipe et al., 2002) strongly suggest a role for Mtg2p in ribosome function. To examine whether Mtg2p associates with the mitochondrial ribosome, ribosomal subunits were separated on sucrose gradients. The identities of individual subunits were confirmed by subjecting fractions to immunoblot analysis by using antibodies to the small subunit (37S) protein Mrp13p (Partaledis and Mason, 1988) and the large subunit (54S) protein Mrp7p (Fearon and Mason, 1988). In buffer containing 100 mM NH4Cl, Mtg2p cofractionated with the large subunit marker Mrp7p (Figure 3A). Increasing NH4Cl concentration to 200 mM, however, resulted in reduced Mtg2p fractionation with the large ribosomal subunit (our unpublished data) and that association was further diminished in 500 mM NH4Cl (Figure 3B). We conclude that Mtg2p is associated with the large ribosomal subunit but is not a core ribosomal protein.
Figure 3.
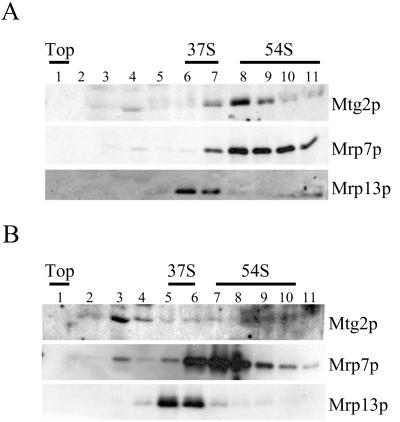
Mtg2p cofractionates with the large ribosomal subunit. Mitochondrial lysates from wild-type cells were separated on 10–30% sucrose gradients containing either 100 mM NH4Cl (A) or 500 mM NH4Cl (B) at 135,000 × g for 4 h. Gradient fractions were TCA precipitated, separated by SDS-PAGE, and subjected to immunoblot analysis. Antibodies used were against Mtg2p, Mrp7p, and Mrp13p. The top of the gradient and the migration of the small 37S and large 54S subunit peaks, based on immunoblots, are indicated.
Cells Harboring mtg2ts Allele Are Defective in Mitochondrial Translation
To define the role of Mtg2p in mitochondrial ribosome function, temperature-sensitive alleles of mtg2 were generated and expressed episomally in a Δmtg2 background. A number of temperature-sensitive alleles that grew well on YPG at 23°C but not 37°C were isolated. One such allele, mtg2-1, was further characterized.
At the permissive temperature, cells expressing either MTG2 or mtg2-1 grew at equivalent rates on either YPD or YPG plates (Figure 4A). At the nonpermissive temperature, however, mtg2-1 cells grew slowly on YPG (Figure 4A). Under these conditions, the levels of mutant protein were slightly reduced (Figure 6). Interestingly, although cells expressing mtg2-1 were impaired for growth at the nonpermissive temperature, they remained ρ+, even after 48–72 h at 37°C (our unpublished data), unlike Δmtg2 that were ρ0 (Figure 1). One likely possibility is that the mtg2-1 expresses a partially functional protein as evidenced by the reduction, but not complete elimination, of expression of cytochrome c oxidase subunit I and III (Cox1p and Cox3p) and cytochrome b (Cytbp) (see below).
Figure 4.
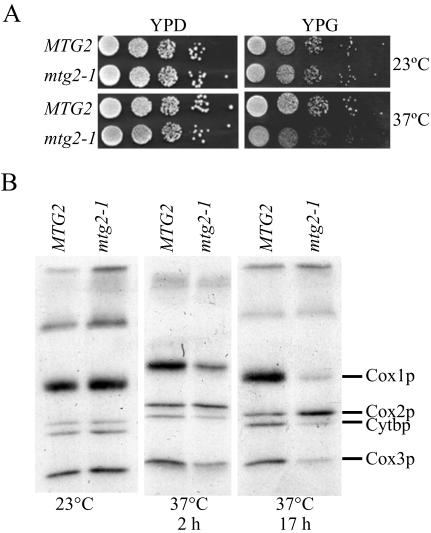
The temperature-sensitive mtg2-1 mutant displays aberrant mitochondrial translation. (A) The growth of mtg2-1 cells on YPG is impaired at 37°C. Shown are serial dilutions of cells expressing MTG2 or mtg2-1 on YPD and YPG plates at 23°C and 37°C, as indicated. (B) Newly synthesized mitochondrial protein products were labeled in vivo in cells expressing MTG2 or mtg2-1, with [35S]methionine, at either 23°C or after shift to 37°C for either 2 or 17 h. Mitochondria were isolated, and equivalent amounts of protein were separated by SDS-PAGE, the gel dried, and the radioactive proteins visualized on x-ray film. The position of Cox1p, Cox2p, Cytbp, and Cox3p is indicated.
Figure 6.
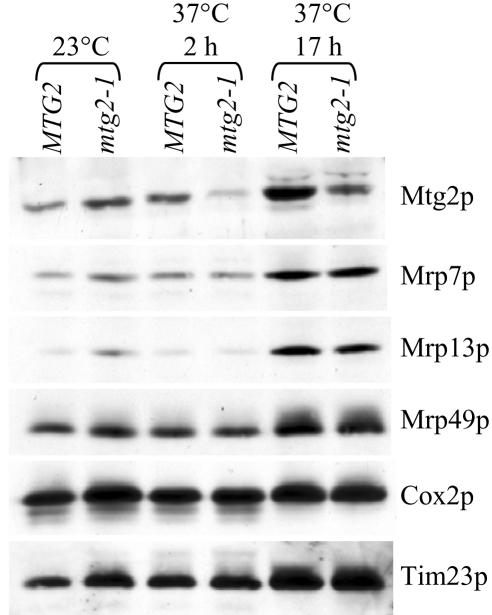
The stability of early assembling large ribosomal proteins is not affected in mtg2–1. Mitochondrial protein extracts obtained from cells expressing MTG2 or mtg2–1 allele grown at either 23°C or 37°C (for 2 or 17 h), as indicated, were separated by SDS-PAGE and subjected to immunoblot analysis. The relative levels of Mtg2p, the early assembling proteins Mrp49p and Mrp13p, and the late assembling protein Mrp7p were detected using protein specific antibodies. Loading controls include Cox2p and Tim23p, two proteins that should remain unaffected in this assay.
We next assayed whether Mtg2p was important for mitochondrial translation. Cells expressing MTG2 or mtg2-1 allele had equivalent amounts of mitochondrial translation products when grown at 23°C (Figure 4B). In contrast, after a shift to 37°C the synthesis of Cox1p, Cox3p, and Cytbp was reduced in mtg2-1 cells, and synthesis of Cox2p was elevated, as determined by incorporation of radiolabeled methionine (Figure 4B). The reduction of incorporation of [35S]methionine into Cox1p, Cox3p, and Cytbp in mtg2-1 at 37°C was likely due to defects in protein synthesis rather than protein stability, because radiolabeled protein levels were lower after an extended shift to the nonpermissive temperature before addition of [35S]methionine (Figure 4B). To confirm that reduced levels of translation product were a consequence of a defect in protein synthesis rather than protein stability, cells were pulse labeled at 23°C and chased for 6 h at 37°C. In this case, the level, and therefore the stability, of labeled proteins were comparable in both MTG2 and mtg2-1 cells (our unpublished data). Thus, although preexisting translation products are equally stable at 37°C in mtg2-1 and MTG2 cells, cells harboring the mtg2-1 allele are defective in mitochondrial translation at the nonpermissive temperature.
Cells Harboring mtg2ts Allele Are Not Defective in Early Assembly of Ribosomal Subunits
Bacterial Obg proteins seem to play a role in the late assembly of the large ribosomal subunit (Tan et al., 2002; Lin et al., 2004; Wout et al., 2004). Given the sequence similarity of Mtg2p and the bacterial Obg proteins, one possibility is that the translational defect observed in mtg2-1 cells is an indirect consequence of improper ribosome assembly. To determine whether this was the case, we first examined whether there were gross alterations of mature mitochondrial ribosomal subunit ratios in mtg2-1 cells at the nonpermissive temperature. Cells harboring either MTG2 or mtg2-1 at 23°C have similar ratios of 54S to 37S subunits, as well as similar total levels of ribosomal subunits (Figure 5A). Growth of either MTG2 or mtg2-1 at 37°C for 2 or 8 h leads to a slight accumulation of 54S relative to 37S, although there are no significant differences between these strains (Figure 5, B and C). When mtg2-1 cells were shifted to 37°C for an extended period (17 h), however, the total level of ribosomal subunits was reduced but the relative ratio of 54S to 37S was unchanged (Figure 5D), consistent with the translational defects observed under similar conditions (Figure 4B). We conclude that Mtg2p does not play a critical role in the early assembly of either mature 37S or 54S ribosomal complexes.
Figure 5.
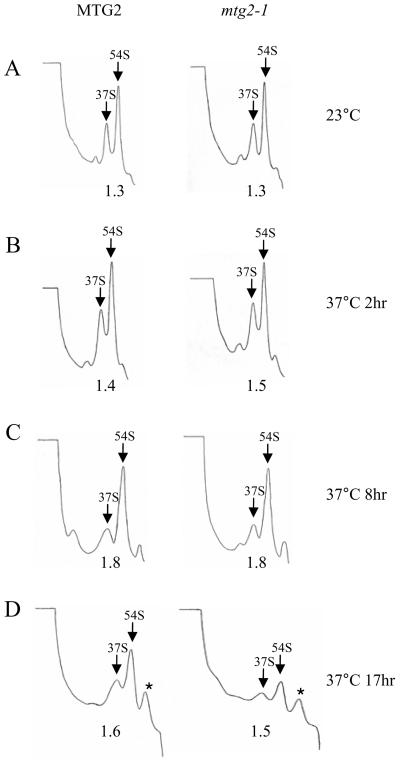
The levels of both 37S and 54S ribosomal particles are reduced in mgt2-1 mutants at the nonpermissive temperature. Mitochondrial ribosome profiles from cells expressing either MTG2 or mtg2–1 grown (A) continuously at 23°C or shifted to 37°C for (B) 2h, (C) 8h or (D)17h. Mitochondrial ribosomal subunits were fractionated on 10–30% linear sucrose gradients containing 200 mM NH4Cl at 250,000 × g for 5 h and monitored by UV absorbance at 254 nm. The positions of the 37S and 54S peaks are labeled. The relative ratios of the 54S/37S peak areas are listed below each profile. Asterisks indicate contaminating peaks that do not correlate with mitochondrial ribosomal proteins (data not shown).
To confirm that Mtg2p does not function in early ribosome assembly, we examined the steady-state levels of both early and late-assembling mitochondrial ribosomal proteins. Stable accumulation of some, but not all mitochondrial ribosomal proteins requires the presence of full-length rRNA (Fearon and Mason, 1988; Partaledis and Mason, 1988; Dang et al., 1990; Fearon and Mason, 1992; Pan and Mason, 1995, 1997; Biswas and Getz, 1999). It has been proposed that the difference in stability of these ribosomal proteins is due to differences in their temporal association during mitochondrial ribosome assembly, with early assembly proteins being unstable in mutants defective for early steps of ribosomal assembly (Fearon and Mason, 1988, 1992; Biswas and Getz, 1999). For example, MRP7, a protein thought to associate at a late step in large ribosome assembly, accumulates stably in mitochondria from ρ0 cells lacking both 21S and 15S rRNA (Fearon and Mason, 1988), whereas two early assembling proteins, MRP13 and MRP49, do not accumulate in either early ribosome assembly mutants or in strains lacking rRNA (Partaledis and Mason, 1988; Fearon and Mason, 1992; Biswas and Getz, 1999). We show here that the levels of Mrp7p, Mrp13p, and Mrp49p remained constant in mitochondria from cells harboring either a MTG2 or mtg2-1 allele after shift to 37°C (Figure 6), confirming that Mtg2p is not involved in a critical step in early ribosome synthesis.
Multiple Copies of MTG2 Partially Suppress a Methyltransferase Mutant, Δmrm2
In E. coli, overexpression of the Obg protein CgtAE suppresses both the growth and ribosome assembly defects of ΔrrmJ mutant, providing evidence that CgtAE is involved in the late assembly of the large ribosomal subunit (Tan et al., 2002). RrmJ is an rRNA methyltransferase responsible for methylation of U2552 within the peptidyl transferase center of the 23S rRNA (Bugl et al., 2000; Caldas et al., 2000). Nucleotide modification of the large rRNA molecule within the peptidyl transferase center is a universally conserved feature in prokaryotic, eukaryotic, as well as mitochondrial translation system. The 21S mitochondrial rRNA contains three modified nucleotides: one pseudouridine (Ψ2819) and two 2′-O-methylated nucleotides (Gm2270 and Um2791) (Sirum-Connolly and Mason, 1993; Ansmant et al., 2000; Pintard et al., 2002). In S. cerevisiae, the RrmJ ortholog Mrm2p is responsible for methylation of U2791 in 21S rRNA (analogous to position U2552 in E. coli) both in vivo and in vitro (Pintard et al., 2002). S. cerevisiae cells lacking MRM2 rapidly lose mitochondrial DNA, becoming ρ0 at 37°C (Pintard et al., 2002), a phenotype consistent with a role in late 54S assembly.
To provide evidence that Mtg2p functions late in 54S assembly, we tested whether multiple copies of MTG2 would suppress the Δmrm2 mutant. To do this, we monitored the rapid conversion from ρ+ to ρ0 in Δmrm2 cells incubated at 37°C in cells harboring either complementing MRM2 or MTG2, on plasmids. The addition of MTG2 on a CEN plasmid resulted in an approximately twofold increase in Mtg2p levels (Figure 7A). As shown previously (Pintard et al., 2002), Δmrm2 cells harboring a control plasmid rapidly lost mitochondrial DNA at 37°C and became ρ0 at a high frequency (monitored by growth on YPG), whereas Δmrm2 mutants complemented with MRM2 remained ρ+ (Figure 7, B and C). Interestingly, the introduction of MTG2 partially suppressed the loss of mitochondrial DNA in Δmrm2 mutants (Figure 7, B and C). The expression of MTG2 from a 2μ plasmid did not result in further suppression (our unpublished data). Thus, the introduction of extra copies of MTG2 partially suppresses the thermosensitive loss of mitochondrial DNA of Δmrm2 and is consistent with a role of Mtg2p in late ribosome assembly.
Figure 7.
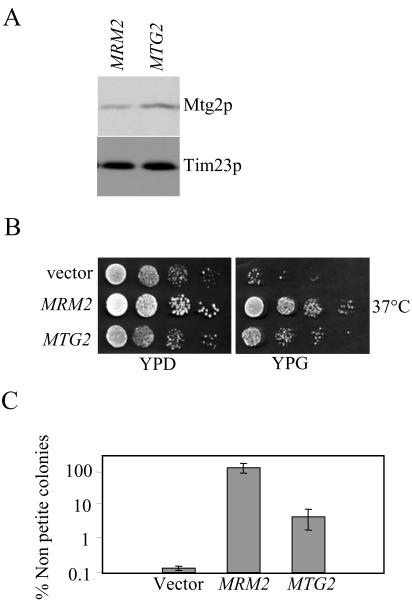
Partial suppression of Δmrm2 by multiple copies of MTG2. (A) Mitochondrial protein extracts from Δmrm2 cells harboring either MRM2 or MTG2 on a CEN plasmid were grown at 30°C, separated by SDS-PAGE, and subjected to immunoblot analysis by using antibodies against Mtg2p and Tim23p (as a loading control). (B) MTG2 partially suppresses the formation of ρ0 cells in Δmrm2 mutants at 37°C. Serial dilution of Δmrm2 cells harboring an empty vector, MRM2, or MTG2 were assayed for growth on YPD and YPG plates at 37°C as indicated. (C) Quantitative analysis reveals the partial suppression of Δmrm2 by MTG2. Multiple cultures of Δmrm2 cells harboring an empty vector, MRM2 or MTG2 were grown in glucose media at 37°C for 24 h and plated on YPD and YPG plates. The number of nonpetite cells, as assayed by growth on both YPG and YPD, was counted. Shown are the average ± SD of the results corresponding to six independent starting cultures.
DISCUSSION
Ribosome biogenesis, in general, is a complex process requiring numerous factors that, although they interact with precursor ribosome intermediates and aid in their maturation into functional subunits, often are not a part of the mature ribosome. For example, in S. cerevisiae the assembly of cytoplasmic ribosomes requires the coordinated action of ∼170 assembly proteins (Fromont-Racine et al., 2003). Many auxiliary proteins are required for temporal processing and modification of rRNA, coordinated loading of ribosomal proteins onto rRNA to form functional subunits and ribosomal export from the nucleus. These assembly factors include nucleases, ATP-dependent RNA helicases, and modifying enzymes, as well as a number of recently identified GTPases (Kressler et al., 1999; Venema and Tollervey, 1999; Fatica and Tollervey, 2002; Fromont-Racine et al., 2003). In E. coli, although ribosomal subunits can be reconstituted in vitro by using purified components, this process is slow and requires nonphysiological conditions (Herold and Nierhaus, 1987; Culver and Noller, 2000). In vivo, however, additional nonribosomal proteins (rRNA-modifying enzymes, chaperones, and GTPases) aid in efficient assembly of bacterial ribosomal subunits (Bugl et al., 2000; Tan et al., 2002; Inoue et al., 2003).
The assembly of mitochondrial ribosomes requires the coordinated synthesis of mitochondrially encoded rRNAs and nuclear encoded proteins as well as the import of these cytosolic proteins into the mitochondria and association into functional subunits. Interestingly, only few mitochondrial ribosome assembly factors have been identified (Sirum-Connolly and Mason, 1993, 1995; Sirum-Connolly et al., 1995; Pintard et al., 2002; Barrientos et al., 2003). These include the 21S rRNA-modifying enzymes that are responsible for two 2′-O-methylated nucleotides (Gm2270 and Um2791) and one pseudouridine (Ψ2819) that form a part of the conserved peptidyl transferase center (Sirum-Connolly and Mason, 1993; Ansmant et al., 2000; Pintard et al., 2002). More significantly, two novel GTPases, Mss1p and Mtg1p, have been identified that functionally interact with 15S and 21S rRNA, respectively (Decoster et al., 1993; Barrientos et al., 2003). In addition, Mtg1p, is involved in biogenesis of the large ribosomal subunit (Barrientos et al., 2003). Mtg1p is a member of the YawG/YlqF subfamily of GTPases (Leipe et al., 2002) that includes the 60S biogenesis GTPases Nug1p and Nog2p.
In this study, we show that another putative mitochondrial GTPase, Mtg2p, also seems to play a role in the assembly of the large ribosomal subunit. We demonstrated that, as expected for a ribosome assembly factor, Mtg2p is localized to the mitochondria and peripherally associated with the inner membrane facing the mitochondrial matrix (Figure 2). Furthermore, deletion of mtg2 led to a decrease in mitochondrial translation and subsequent loss of mitochondrial DNA (Figure 1), a phenotype commonly found in cells defective in translation (Myers et al., 1985; Fearon and Mason, 1992).
Although the precise function of Mtg2p is unknown, it is likely that Mtg2p is either involved directly in the biogenesis of the mitochondrial large ribosomal subunit or plays an as yet undefined role in translation. We favor the former hypothesis for several reasons. First, Mtg2p is required for mitochondrial translation, as shown indirectly by the loss of mtDNA in a Δmtg2 mutant (Figure 1) and directly in the analysis of mitochondrial translation products in the temperature-sensitive mtg2-1 mutant (Figure 4). Second, Mtg2p associates specifically with the 54S subunit but is not a core ribosomal protein, as demonstrated by its dissociation in 500 mM NH4Cl. Third, Mtg2p is not necessary for early ribosome assembly, because yeast cells harboring the mtg2-1 allele did not display a drop in the stability of large ribosomal proteins that assemble early (Figure 6). Fourth, mtg2-1 mutants do not exhibit an altered ribosomal subunit ratio, but it did show reduced levels of both 34S and 54S ribosomal subunits at the nonpermissive temperature (Figure 5). It has been proposed that the relative levels of mitochondrial ribosomal subunits are coordinately controlled (Biswas and Getz, 1999). Thus, a reduction in the assembly of the 54S subunit may coordinately lower 37S subunit to maintain the optimal subunit ratio. Finally, extra copies of Mtg2p partially suppress the thermosensitive formation of petite cells in a Δmrm2 mutant (Figure 7). Mrm2p is responsible for 2′-O-ribose methylation of U2791 on the 21S rRNA which is, in turn, required for optimal mitochondrial ribosome function (Pintard et al., 2002). In vitro, Mmr2p methylates 21S rRNA that has already been assembled into the large 54S ribosomal subunit but not free rRNA, indicating that it is required for a late step in ribosome assembly (Pintard et al., 2002). Thus, suppression of growth defects of Δmrm2 by overexpression of Mtg2p is consistent with a role of Mtg2p in a late step of ribosome biogenesis.
A role for Mtg2p in mitochondrial ribosome function is also consistent with the proposed function of Obg proteins in general, as well as the association of bacterial Obg proteins with the 50S subunit (Lin et al., 2004; Wout et al., 2004) and the implied role of these proteins in a late step of 50S subunit biogenesis (Tan et al., 2002; Lin et al., 2004; Wout et al., 2004), specifically. Interestingly, in addition to a role in ribosome maturation, the bacterial Obg proteins also interact with proteins involved in stress response (Scott and Haldenwang, 1999; Scott et al., 2000; Wout et al., 2004), a function that has not been reported for and is unlikely in mitochondria. One possibility is that the cytosolic Obg proteins Rbg1p and Rbg2p are involved in stress response (Wout and Maddock, unpublished data) whereas the nucleolar (Fromont-Racine et al., 2003; Kallstrom et al., 2003; Saveanu et al., 2003) and mitochondrial Obg proteins (Nog1p and Mtg2p, respectively) may be exclusively involved in ribosome assembly. We are currently pursuing studies to determine the specific molecular role of Mtg2p in ribosomal synthesis.
Acknowledgments
We are extremely grateful to Drs. Michael P. Yaffe for antibodies against F1β, Robert E. Jensen for antibodies against Tim23p, Carla Koehler for antibodies against KDH, and Thomas L. Mason for antibodies against Cox2p. We also thank Dr. Suzanne Lybarger and Raji Janakiraman for technical assistance and Susan Sullivan for both technical assistance and critical reading of this manuscript. This work was supported by National Science Foundation MCB-0316357 and National Institutes of Health GM-55133 to J.R.M. and from a University of Michigan Rackham Merit fellowship to J.L.F.
Article published online ahead of print in MBC in Press on December 9, 2004 (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0622).
References
- Ansmant, I., Massenet, S., Grosjean, H., Motorin, Y., and Branlant, C. (2000). Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of psi(2819) in 21S mitochondrial ribosomal RNA. Nucleic Acids Res. 28, 1941-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos, A., Korr, D., Barwell, K. J., Sjulsen, C., Gajewski, C. D., Manfredi, G., Ackerman, S., and Tzagoloff, A. (2003). MTG1 codes for a conserved protein required for mitochondrial translation. Mol. Biol. Cell 14, 2292-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, T. K., and Getz, G. S. (1999). The single amino acid changes in the yeast mitochondrial S4 ribosomal protein cause temperature-sensitive defect in the accumulation of mitochondrial 15S rRNA. Biochem. 38, 13042-13054. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- Brickner, J. H., and Fuller, R. S. (1997). SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139, 23-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugl, H., Fauman, E. B., Staker, B. L., Zheng, R., Kushner, S. R., Saper, M. A., Bardwell, J. C., and Jakob, U. (2000). RNA methylation under heat shock control. Mol. Cell 6, 349-360. [DOI] [PubMed] [Google Scholar]
- Buglino, J., Shen, V., Hakimian, P., and Lima, C. D. (2002). Structural and biochemical analysis of the Obg GTP-binding protein. Structure 10, 1581-1592. [DOI] [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., Costa, A., Desgres, J., and Richarme, G. (2000). The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275, 16414-16419. [DOI] [PubMed] [Google Scholar]
- Culver, G. M., and Noller, H. F. (2000). In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 318, 446-460. [DOI] [PubMed] [Google Scholar]
- Dang, H., Franklin, G., Darlak, K., Spatola, A. F., and Ellis, S. R. (1990). Discoordinate expression of the yeast mitochondrial ribosomal protein MRP1. J. Biol. Chem. 265, 7449-7454. [PubMed] [Google Scholar]
- Daum, G., Bohni, P. C., and Schatz, G. (1982). Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028-13033. [PubMed] [Google Scholar]
- Decoster, E., Vassal, A., and Faye, G. (1993). MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J. Mol. Biol. 232, 79-88. [DOI] [PubMed] [Google Scholar]
- Emtage, J. L., and Jensen, R. E. (1993). MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J. Cell Biol. 122, 1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., and Tollervey, D. (2002). Making ribosomes. Curr. Opin. Cell Biol. 14, 313-318. [DOI] [PubMed] [Google Scholar]
- Fearon, K., and Mason, T. L. (1988). Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP7, a protein of the large subunit of the mitochondrial ribosome. Mol. Cell. Biol. 8, 3636-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon, K., and Mason, T. L. (1992). Structure and function of MRP20 and MRP49, the nuclear genes for two proteins of the 54 S subunit of the yeast mitochondrial ribosome. J. Biol. Chem. 267, 5162-5170. [PubMed] [Google Scholar]
- Fox, T. D., Folley, L. S., Mulero, J. J., McMullin, T. W., Thorsness, P. E., Hedin, L. O., and Costanzo, M. C. (1991). Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 194, 149-165. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Senger, B., Saveanu, C., and Fasiolo, F. (2003). Ribosome assembly in eukaryotes. Gene 313, 17-42. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A. L., Fowler, S., and Lazarow, P. B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, B. S., and Pon, L. A. (1995). Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213-223. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. R. (1991). Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1-863. [PubMed] [Google Scholar]
- Hampsey, M. (1997). A review of phenotypes in Saccharomyces cerevisiae. Yeast 13, 1099-1133. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: a laboratory manual, 633.
- Herold, M., and Nierhaus, K. H. (1987). Incorporation of six additional proteins to complete the assembly map of the 50S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262, 8826-8833. [PubMed] [Google Scholar]
- Inoue, K., Alsina, J., Chen, J., and Inouye, M. (2003). Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48, 1005-1016. [DOI] [PubMed] [Google Scholar]
- Kallstrom, G., Hedges, J., and Johnson, A. (2003). The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23, 4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C. M., Merchant, S., Oppliger, W., Schmid, K., Jarosch, E., Dolfini, L., Junne, T., Schatz, G., and Tokatlidis, K. (1998). Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17, 6477-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., Linder, P., and de la Cruz, J. (1999). Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe, D. D., Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41-72. [DOI] [PubMed] [Google Scholar]
- Lin, B., Thayer, D. A., and Maddock, J. R. (2004). The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186, 481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie 3rd, A., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Marton, M. J., Crouch, D., and Hinnebusch, A. G. (1993). GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol. 13, 3541-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzuki, S., and Hibbs, A. R. (1986). Are all mitochondrial translation products synthesized on membrane-bound ribosomes? Biochim. Biophys. Acta 866, 120-124. [DOI] [PubMed] [Google Scholar]
- Myers, A. M., Pape, L. K., and Tzagoloff, A. (1985). Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4, 2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C., and Mason, T. L. (1995). Identification of the yeast nuclear gene for the mitochondrial homologue of bacterial ribosomal protein L16. Nucleic Acids Res. 23, 3673-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C., and Mason, T. L. (1997). Functional analysis of ribosomal protein L2 in yeast mitochondria. J. Biol. Chem. 272, 8165-8171. [DOI] [PubMed] [Google Scholar]
- Partaledis, J. A., and Mason, T. L. (1988). Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP13, a protein of the small subunit of the mitochondrial ribosome. Mol. Cell. Biol. 8, 3647-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham, J. L., Dudley, A. M., and Mason, T. L. (1994). T7 RNA polymerase-dependent expression of COXII in yeast mitochondria. Mol. Cell. Biol. 14, 4643-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard, L., Bujnicki, J. M., Lapeyre, B., and Bonnerot, C. (2002). MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J. 21, 1139-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- Saveanu, C., Namane, A., Gleizes, P.-E., Lebreton, A., Rousselle, J.-C., Noaillac-Depeyre, J., Gas, N., Jacquier, A., and Fromont-Racine, M. (2003). Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23, 4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J. M., and Haldenwang, W. G. (1999). Obg, and essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. J. Bacteriol. 181, 4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J. M., Ju, J., Mitchell, T., and Haldenwang, W. G. (2000). The Bacillus subtilis GTP binding protein Obg and regulators of the σB stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 182, 2771-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirum-Connolly, K., and Mason, T. L. (1993). Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science 262, 1886-1889. [DOI] [PubMed] [Google Scholar]
- Sirum-Connolly, K., and Mason, T. L. (1995). The role of nucleotide modifications in the yeast mitochondrial ribosome. Nucleic Acids Symp. Ser. 33, 73-75. [PubMed] [Google Scholar]
- Sirum-Connolly, K., Peltier, J. M., Crain, P. F., McCloskey, J. A., and Mason, T. L. (1995). Implications of a functional large ribosomal RNA with only three modified nucleotides. Biochimie 77, 30-39. [DOI] [PubMed] [Google Scholar]
- Sogo, L. F., and Yaffe, M. P. (1994). Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126, 1361-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spithill, T. W., Trembath, M. K., Lukins, H. B., and Linnane, A. W. (1978). Mutations of the mitochondrial DNA of Saccharomyces cerevisiae which affect the interaction between mitochondrial ribosomes and the inner mitochondrial membrane. Mol. Gen. Genet. 164, 155-162. [DOI] [PubMed] [Google Scholar]
- Stark, M.J.R. (1998). Studying essential genes: generating and using promoter fusions and conditional alleles. In Methods in Microbiology, ed. A. J. Brown and M. Tuite, San Diego: Academic Press, 83-99.
- Tan, J., Jakob, U., and Bardwell, J. C. (2002). Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184, 2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., and Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261-311. [DOI] [PubMed] [Google Scholar]
- Wout, P., Pu, K., Sullivan, S. M., Reese, V., Zhou, S., Lin, B., and Maddock, J. R. (2004). The Escherichia coli GTPase, CgtAE, cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 186. [DOI] [PMC free article] [PubMed]


