Abstract
Introduction
Mutation of the HBV precore gene prevents the production of HBeAg, which is an important target for immune responses. Distribution of this mutation varies along with frequency of HBV genotypes in accordance with geographic and ethnic variations. The general objective of this study was to evaluate the prevalence and characteristics of precore mutation in Iran and its correlation with genotypes of hepatitis B.
Methods
In this cross-sectional study, viral DNA of 182 Iranian hepatitis B surface antigen positive patients who were admitted to Bandar Abbas Blood Transfusion Organization in 2012 and 2013 was retrieved from their serum samples. HBeAg, anti-HBe, and anti-HBc IgM diagnostic tests were performed using ELISA kits. Precore and Pre-S regions were amplified using specific primers and PCR thereafter to determine the genotypes; precore mutation, PCR, and restriction fragment length polymorphism (RFLP) methods also were applied. SPSS version 12 was used for data analysis by Mann–Whitney U test, Fisher’s exact probability test, and t-test.
Results
A total of 62 patients (34.1%) had precore mutation (A1896G), and genotype D was the predominant genotype in these patients, which was followed by an unknown genotype that was suspected for genotype B. Interestingly, the relationships between precore mutation and HBeAg (p=0.037) and genotype D (p=0.005) were significant; however, no correlation was observed between this mutation and acute or chronic hepatitis and sex of patients.
Conclusion
This study found high prevalence of precore mutations in southern Iran, which was significantly associated with HBeAg and genotype D.
Keywords: Precore mutants, Genotypes, Hepatitis B, RFLP
1. Introduction
Hepatitis B virus (HBV) is a blood-borne hepatotrophic virus that is the most widely recognized reason for genuine liver disease on the planet. More than 350 million individuals are infected with HBV around the world (1, 2). A few studies have additionally recommended that the hereditary attributes of HBV, including HBV genotype and particular hereditary mutations, are connected with the advancement of genuine liver disease (3–6). The majority of respective mutations mainly occur in precore and core regions in the gen C of HBV. The precore protein encoded by the precore region is further processed in the endoplasmic reticulum to produce secreted HBeAg. The most naturally occurring mutation in the HBV precore gene is substitution of a guanine (G) to adenine (A) at position 1896. The G1896A precore mutation leads to a conversion of codon 28 from TGG (tryptophan) to a premature stop codon TAG. Accordingly, the translation of the precore protein is prevented, and HBeAg production is completely abolished (7–11). The HBe antigen is targeted by immune responses (both cellular and humoral), which leads to disappearing of e antigen. Precore mutations help viruses to avoid detection by the host immune system (12). Single substations and frameshifts in the HBV precore region were infrequent and unclear, as the clinical relationship was demonstrated (13, 14). The prevalence of the precore mutation varies from 20% to 95% and more commonly observed in HBeAg-negative patients (15). HBV genetic is variable both as the emergence of mutations in each infected subject and the evolution of genotypes (16). A genetic classification has defined eight genomic groups, genotypes A to H (17–22). Each HBV genomic group indicate a distinct geographic and ethnic distributions, which have been related to special subsequent complications ranging from mild mental disorders to health-threatening conditions (23–28). Genotype A has a worldwide distribution and is the most common genotype in non-Mediterranean Europe, India, North America, and sub-Saharan Africa. Genotypes B and C are found primarily in Australia and East and Southeast Asia (29). More than half of the HBV carriers are estimated to live in these areas (30). Genotype D is more common in the Middle East and Mediterranean area, and genotype E is more commonly detected in West Africa (25, 29, 31). Genotype F has been reported from Native Americans (25). Genotype G is reported in samples from USA, France, and Mexico (17). Also genotype H is reported from Central and South America (18). There is an association between mutations in the precore region of the HBV and HBV genotype, so the prevalence of infection with G1896A precore mutant is related to HBV genotypes in different geographic areas. Its prevalence is high in the Far East and Mediterranean regions and low in northern Europe and United States (32). However, controversy exists regarding the correlation between hepatitis B genotypes and precore mutation. Although some studies have shown that precore mutation is related with HBV genotypes, the exact kind of relations varied among these studies and remains unclear. G1896A mutation is most commonly associated with genotypes D, E, and B of HBV and is rare in other genotypes of HBV. (16, 32–42). The importance of screening for precore mutation is mainly comprehensible in blood transfusion centers because the transmission of HbeAg negative viruses (precore variants) can lead to severe complications, especially in immunosuppressed or immunocompromised patients who need blood products (43). In addition, there is a lack of studies about the spread of HBV genotypes and precore mutations in Iran, especially in Bandar Abbas City in southern Iran, which is one of the main communication ports of Iran with the Persian Gulf countries. This connection can distinguish various characteristics of this region from other parts of the country (44–46). Thus, the present study was conducted with the aim of determining prevalence and characteristics of precore mutation and its correlation with genotypes of hepatitis B in the blood transfusion center of Bandar Abbas, Iran.
2. Materials and methods
2.1. Patient series
In this cross-sectional survey, serum samples were obtained from 182 HBsAg positive patients, which were composed of 144 (79.12%) males and 38 (20.88%) females who were admitted to Bandar Abbas Blood Transfusion Organization in 2012 and 2013. The samples were stored at −20°C. No patient had a positive test for human immunodeficiency virus and hepatitis C and D virus. Patients were excluded if they had received antiviral therapy or had HBV recurrence after liver transplantation.
2.2. Serologic markers
HBe Ag and Ab and anti-HBc IgM were measured using ELISA and DIAPRO kits (Italy). HBe Ag and Ab tests are indicative of viral replication. Further, the anti-HBc IgM is used for detection of chronicity of the disease.
2.3. HBV-DNA extraction and HBV genotyping
A Roche High Pure Viral Nucleic Acid kit (Roche Molecular Biochemicals, Germany) was used for DNA extraction. Restriction fragment length polymorphism (RFLP) was used for determination of HBV genotypes as described by Lindh et al. (47). The pres-S region was amplified and digested by AvaII and MboI restriction enzymes. In short, primers GenoP1 (sense, nt 2823–2845, 5′-TCACCATATTCTTGGGAACAAGA-3′) and GenoP2 (Antisense, nt 80–61, 5′ TTCCTGAACTGGAGCCACCA-3′) were used in PCR to amplify the fragment encompassing nucleotide 80 to 2823, including 479 nucleotides, which contain PRE-S region. 5 μL of the resuspended DNA were added to an amplification mixture containing 5 μL of 10× Taq polymerase buffer, 2μL of 50 mmol/L Mgcl2, 1 μL of 10 mmol/L deoxyribonucleotide triphosphates, 1 μL (5U) of Taq polymerase (Promega, Beijing, China), and 10 pmoL of each primers GenoP1 and GenoP2. The PCR profile was started with 5 min denaturation at 94°C. After that, amplification, annealing, and extension was done. Strand synthesis was completed at 72°C for 7 min. Restriction enzymes including Ava II and MboI (New England Biolabs, Inc., USA) were used for incubation with PCR products. The amplified products were run on 3% agarose gel and visualized under UV. For determination of HBV genotypes, the results were compared with Lindh patterns (47).
2.4. Detection of Precore Mutation
We tested the samples for precore stop codon (G1896A). A similar procedure for HBV genotyping was used in this step. Two HBV primers (Pre1 and Pre2) were used in the first PCR amplification. An aliquot of reaction product was then further amplified in a second reaction with primers, Pre2 and Pre3, which are contained within the region spanned by the first two primers. The primer sequences were pre1, 5′-CACCTCTGCCTAATCATCTC-3′ (sense, nt 1826–1845); pre2, 5′-CTGACTACTAATTCCCTGGATGCTGGGTCT-3′ (antisense, nt 2160–2131); pre3, 5′-CAAGCCTCCAAGCTGTGCCTTGGGTGGCCTT-3′ (antisense, nt 1865–1895). The amplification mixture was contained 2.5 μL of 10× Taq polymerase buffer, 0.5 μL of 25 mmol/L deoxyribonucleotide triphosphates, 2.5μL of 50 mmol/L Mgcl2, 0.4 μL of Taq polymerase (Promega, Beijing, China) and 10 pmoL each of primers Pre1 and Pre2 (total volume of 25 μL). All the procedures of the first-round PCR were similar to those for HBV genotyping. For second-round PCR, 1 μL of the first-round PCR products was used. The condition was the same but with the primers Pre2 and Pre3, with the same procedure. According to Lindh et al., the PCR products were incubated with restriction enzyme Bsu36I that recognize 5′-CCTNAGG-3 sequence and cut C gene into two fragments of 261 and 34 bp in length. Thus, in the absence of PC mutation the production was a fragment of 295 bp in length. The products were visualized by 3% agarose electrophoresis and ethidium bromide staining.
2.5. Statistical Analysis
Data analysis was done using SPSS version 12.0 (SPSS Inc., Chicago, Illinois, USA), and a p-value less than 0.05 was considered to be significant. Mann–Whitney U test and Fisher’s exact probability test were used for comparing the proportions of each factor between groups, and group means were compared using the t-test.
3. Results
3.1. Demographics and viral characteristics
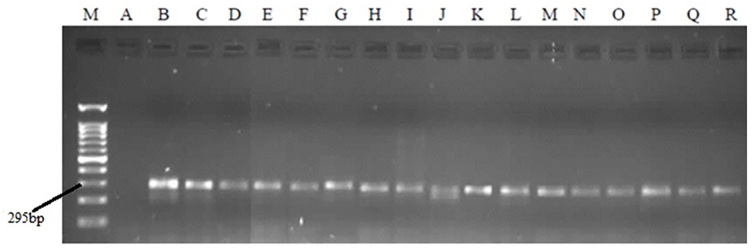
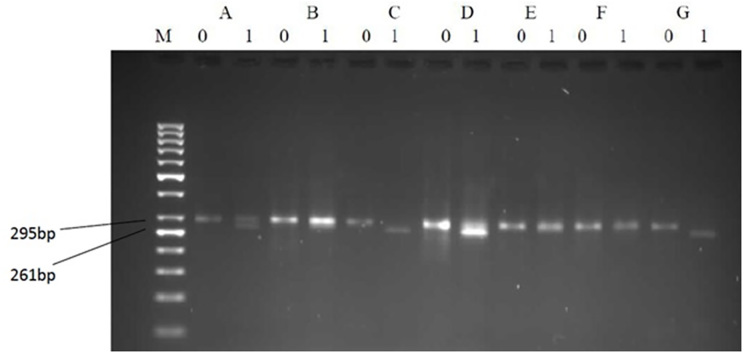
A total of 182 serums were enrolled in our study. Thirty-eight (20.88%) of patients were female, and 144 (79.12%) were male. Precore (G1896A) mutation was found in 34.1% (62 samples) of the study population, which produced two fragments of 261 and 34 bp in length, which were observable by electrophoresis (Figures 1, 2). Three RFLP patterns were identified in relation to HBV genotypes in the present study, two patterns were belonged to genotype D, and one pattern was not matched with any of Lindh patterns (47). Serological tests showed that 169 samples (92.9%) were HBeAg negative and 13 samples (7.1%) were HBeAg positive. Anti-HBe test also indicated replication of the virus was positive in 159 sample (87.4%) and negative in 23 samples (12.6%). This difference between HBeAg and HBeAb status could be due to a small proportion of patients, which are positive for both of the HBeAg and HBeAb tests, simultaneously. Finally, Anti-HBc IgM were detected in four (2.2%) samples that indicated acute hepatitis in these cases (Table 1).
Figure 1.
PCR products in relation to precore mutation on agarose gel electrophoresis. M: Marker 100bp. A: negative control. B-R: replicated regions in serum samples.
Figure 2.
Representative patterns on agarose gel electrophoresis illustrating the restriction enzyme-based detection of precore mutants and variants. Each analysis is represented by two lanes: (1) the left without and (2) the right with a restriction enzyme Bsu36I added. B and E show TGG (wild-type); C, D, and G show TAG (mutant); and A shows a mixture of TGG/TAG at codon 28; M: Marker 50bp.
Table 1.
Epidemiologic and viral Characteristics and Associations
| Variables | n (%) | Precore status | p-value | ||
|---|---|---|---|---|---|
| Mutant | Wild type | ||||
| Sex | Male | 144 (79.12) | 71 (49) | 73 (51) | > 0.05 |
| Female | 38 (20.88) | 20 (51) | 18 (49) | ||
| HBeAg | Positive | 13 (7.1) | 1 (7.7) | 12 (92.3) | 0.037 |
| Negative | 169 (92.9) | 61 (36.1) | 108 (63.9) | ||
| HBeAb | Positive | 159 (87.4) | 73 (46) | 86 (54) | > 0.05 |
| Negative | 23 (12.6) | 11 (47.8) | 12 (52.2) | ||
| Anti-HBc IgM | Positive | 4 (2.2) | 0 (0) | 4 (100) | > 0.05 |
| Negative | 178 (97.8) | 62 (34.83) | 116 (65.17) | ||
3.2. Relationship between preC mutant and HBV genotypes
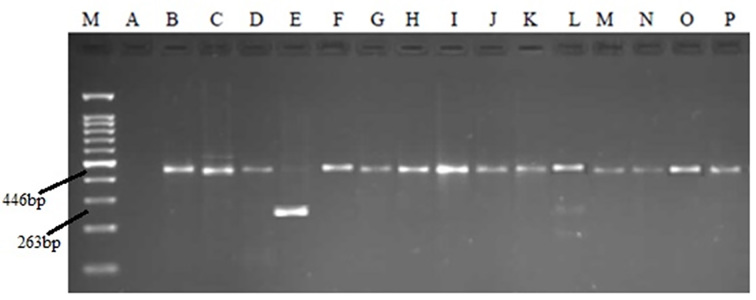
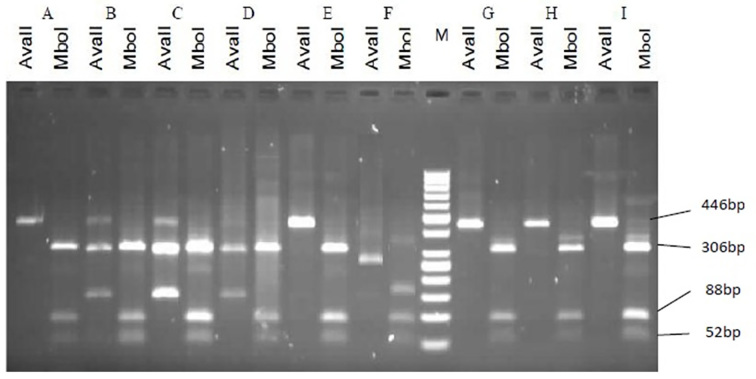
As described, PCR products were incubated with restriction enzymes, the product restriction patterns were compared with Lindh patterns, and genotypes were determined. Three different patterns were found: two patterns were consistent with Lindh patterns and Belonged to genotype D. However, one pattern was not matched with Lindh patterns. All sequences of genotype D in NCBI had a deletion in their Pre-S region, which produced a fragment of 446 bp in length instead of a fragment of 479 bp in length. The first pattern in the current study had no excision site for AvaII. However, it was cut to three fragments by the MboI enzyme (306 nt, 88 nt, 52 nt). This pattern was similar to the D2 pattern, which was obtained by Lindh. The second pattern had a deletion of 183 nucleotides; thus, a 263 nt fragment was amplified, which had no site for AvaII and two sites for MboI (123 nt, 88 nt, 52 nt). This pattern was similar to the Ddel pattern, which was obtained by Lindh. The third pattern, which was not matched with Lindh patterns, was a 446bp amplified fragment that was cut into three fragments by MboI (306 nt, 88 nt, 52 nt). It was excised into two fragments of 300 bp and 146 bp in length by AvaII, which were not matched with any of patterns achieved by Lindh (Figure 3, 4). However, it seems that this pattern belonged to the B genotype based on a comparison with Lindh patterns. Data analysis indicated a strong correlation between HBV genotype D and precore variants (p=0.005).
Figure 3.
PCR products in relation to HBV genotyping on agarose gel electrophoresis. M: Marker 100bp. B: negative control. C-P: replicated regions in serum samples.
Figure 4.
Representative patterns on agarose gel electrophoresis illustrating the restriction enzyme-based detection of HBV genotypes. Each analysis is represented by two lanes: (1) the left with a restriction enzyme AvaII; (2) the right with a restriction enzyme MboI added. M: Marker 50bp. First pattern: Samples A, E, H, I, and G, which are matched with a D2 pattern of Lindh patterns that were cut into three fragments of 52nt, 88nt, and 306nt by MboI and had no excision site for AvaII (446bp). Second pattern: Sample F, which is matched with Ddel pattern of Lindh patterns that were cut into three fragments of 52nt, 88nt, and 123nt by MboI and had no excision site for AvaII, which produced a 263bp fragment. The third pattern belonged to samples B, C, and D, which were not matched with any of the Lindh patterns.
3.3. Relationship between HBeAg/Ab and Anti-HBc IgM status and preC mutation
Mutations in the precore region were found in 61 samples (36.1%) among 169 HBeAg-negative and one sample (7.7%) among 13 HBeAg-positive patients, respectively. Hbe Ag negative patients had higher prevalence of preC mutation compare with HBe Ag negative patients (36.1% vs. 7.7%, p=0.037). Thus, our findings indicated statistically significant association between HBeAg/Ab status and precore mutation. However, there was no precore mutant among anti-HBc IgM-positive patients, and no statistically significant association was observed (p > 0.05).
3.4. Relationship between preC mutation and Gender
The frequency of precore mutation was 49% in male patients and 51% in female patients. Thus, no correlation was found between precore mutation and gender of patients (p > 0.05).
4. Discussion
In this study, we have assessed the relationship between preC mutant and HBV genotypes, HBeAg/Ab status, anti-HBc IgM test, and gender of patients. Mutation of G1896A in PreC region shuts off the HBe Ag synthesis by introducing a stop codon, (9, 48). In the present study, precore variants were found in 34.1% of HBsAg positive samples, which is in agree,emnt with findings of a recent study on Brazilian patients with chronic hepatitis B (49) Lots of previous studies, with adults in particular, reported HBeAg stop codon mutants in 20%–95% of investigated patients (15, 50–54). Despite the fact that HBV genotypes were identified in 1988 (20), there are only several reports on a small numbers of patients focusing on the relationship between HBV genotypes and the HBeAg/anti-HBe status and PC mutations (16, 55–58). We found association between HBV genotypes and precore variants in this study. In fact, findings of this study demonstrated that the prevalence of preC mutant is high among HBV with genotype D. Similar findings were reported by Li JS et al. in a study conducted in France (39). Two other studies also indicated the G1896A precore mutation was more frequent in the group of patients with genotype D than in the patients with genotype A (38, 49). These findings were in agreement with the theory that the relation between G1896A precore mutation and the HBV genotypes is due to different stabilization energy of each genotype as a result of the base pairing of the stem loop structure of the encapsidation sequence of pregenome RNA. This stabilization is the result of best pairing in the presence of these mutations. The G1896A mutation occurs when nucleotide 1858 is composed of the thymine–nitrogen base, which leads to better stabilization in lower stem. This mutation increases genotype D virus replication. This leads to a positive selection of preC mutants the high prevalence of these mutations in genotype D (16, 36, 39, 59). This explains the high prevalence of G1896A in Asia and the Mediterranean region where the predominant HBV genotype is D (35, 36, 60, 61), and its low prevalence in North America and Europe (62). In another study, Orito et al. also observed that PreC mutation (G1896A) is restricted to HBV strains with T1858 and hardly occurs in those with C1858 (37). HBV has low diversity in our study, which is compatible with the study by Magnius et al., and suggests that genotype D has a short evolution in Iran, as represented in their study (19). To our knowledge, all previous studies in Iran could detect only genotype D of HBV. Based on these results, genotype D is the most common HBV genotype in Iran, which is in agreement with entire previous studies. However, one of the patterns in the present study suspected to the B genotype should be confirmed by DNA sequencing in our further studies. Based on the literature, genotype B is frequent in Southeast Asia, so the existence of B genotype could be due to immigrants of Southeast Asia who are frequent in Bandar Abbas due to the seaway between southern Iran and Southeast Asia. These results are compatible with available information about geographic distribution of HBV genotypes (63, 64). In a recent study, R.E.F. Rezende et al. also reported that genotype D is the most frequent genotype in Brazilian patients (49). However, Halfon et al. observed the distribution of HBV genotypes in France on 262 patients with chronic HBV infection. Their findings revealed that the most frequent genotypes D, A, E, C, and B were the most common genotype with the prevalence of 27%, 24%, 13%, 12%, and B 7%, respectively (65). The frequency of the G1896A mutation was significantly higher in the HBe Ag negative patients in our study, which was compatible with results of previous studies. These results indicate that the appearance of the preC mutation is directly associated with HBe Ag seroconversion, which is in favor of the fact that PreC mutation turns off the synthesis of HBe Ag (29, 35, 54). High prevalence of preC mutation in a later phase of HBV infection is associated with viral replication and hepatic necro-inflammatory activity (12, 32). Wintermeyer et al. also reported that precore variants were more common in anti-HBe positive children, but the difference did not reach statistical significance (66). However, in another study, Chang MH et al. denied any relationship between anti-HBe status and PC mutation, which suggest at least no strong evidence for the hypothesis that the mutant is selected by host immune pressure (67). Finally, no correlation was found between precore mutation and gender of patients based on our findings. Although, in a recent study, Pivert et al. indicated that the mutated and mixed preC viruses were more common among males, several studies denied this relationship (68, 69).
5. Conclusions
In line with previous scientific evidence, preC variants were more common in patients who were negative for HbeAg in this study. Additional findings of our study were compatible with previous studies, which claimed that Iranian HBV viruses belong to genotype D and indicated significant association of the preC G1896A mutation with this genotype of HBV. In the future studies, it is suggested to determine the distribution of HBV genotypes using more sensitive molecular assays in large multicenter projects.
Acknowledgments
This article is extracted from the author’s postgraduate thesis. The authors would like to thank the Student Research Committee of Hormozgan University of Medical Sciences for their help and support.
Footnotes
iThenticate screening: August 03, 2016, English editing: February 10, 2017, Quality control: March 02, 2017
Conflict of Interest:
There is no conflict of interest to be declared.
Authors’ contributions:
All authors contributed to this project and article equally. All authors read and approved the final manuscript.
References
- 1.Takahashi K, Aoyama K, Ohno N, Iwata K, Akahane Y, Baba K, et al. The precore/core promoter mutant (T1762A1764) of hepatitis B virus: clinical significance and an easy method for detection. J Gen Virol. 1995;76(Pt 12):3159–64. doi: 10.1099/0022-1317-76-12-3159. [DOI] [PubMed] [Google Scholar]
- 2.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337(24):1733–45. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 3.Garcia PD, Ou JH, Rutter WJ, Walter P. Targeting of the hepatitis B virus precore protein to the endoplasmic reticulum membrane: after signal peptide cleavage translocation can be aborted and the product released into the cytoplasm. J Cell Biol. 1988;106(4):1093–104. doi: 10.1083/jcb.106.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen IH, Huang CJ, Ting LP. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69(6):3647–57. doi: 10.1128/jvi.69.6.3647-3657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen HS, Kew MC, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, et al. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J Virol. 1992;66(9):5682–4. doi: 10.1128/jvi.66.9.5682-5684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 7.Akahane Y, Yamanaka T, Suzuki H, Sugai Y, Tsuda F, Yotsumoto S, et al. Chronic active hepatitis with hepatitis B virus DNA and antibody against e antigen in the serum: Disturbed synthesis and secretion of e antigen from hepatocytes due to a point mutation in the precore region. Gastroenterology. 1990;99(4):1113–9. doi: 10.1016/0016-5085(90)90632-B. [DOI] [PubMed] [Google Scholar]
- 8.Okada K, Kamiyama I, Inomata M, Imai M, Miyakawa Y, Mayumi M. e antigen and anti-e in the serum of asymptomatic carrier mothers as indicators of positive and negative transmission of hepatitis B virus to their infants. N Eng J Med. 1976;294(14):746–9. doi: 10.1056/NEJM197604012941402. [DOI] [PubMed] [Google Scholar]
- 9.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, et al. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2(8663):588–91. doi: 10.1016/S0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 10.Brunetto MR, Giarin MM, Oliveri F, Chiaberge E, Baldi M, Alfarano A, et al. Wild-type and e antigen-minus hepatitis B viruses and course of chronic hepatitis. Proc Natl Acad Sci U S A. 1991;88(10):4186–90. doi: 10.1073/pnas.88.10.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunetto M. Identification of HBV variants which cannot produce pre-core derived HBeAg and may be responsible for severe hepatitis. Ital J Gastroenterol. 1989;21:151–4. Available from: http://ci.nii.ac.jp/naid/10006179330/ [Google Scholar]
- 12.Zuckerman AJ, Zuckerman JN. Molecular epidemiology of hepatitis B virus mutants. J med virol. 1999;58(3):193–5. doi: 10.1002/(SICI)1096-9071(199907)58:3<193::AID-JMV1>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.López JL, Mbayed VA, Telenta PF, González JE, Campos RH. ‘Hbe minus’ mutants of hepatitis B virus. Molecular characterization and its relation to viral genotypes. Virus Res. 2002;87(1):41–9. doi: 10.1016/S0168-1702(02)00078-3. [DOI] [PubMed] [Google Scholar]
- 14.Günther S, Meisel H, Reip A, Miska S, Krüger DH, Will H. Frequent and rapid emergence of mutated pre-C sequences in HBV from e-antigen positive carriers who seroconvert to anti-HBe during interferon treatment. Virology. 1992;187(1):271–9. doi: 10.1016/0042-6822(92)90315-G. [DOI] [PubMed] [Google Scholar]
- 15.Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res. 1999;52:25. doi: 10.1016/S0065-3527(08)60298-5. [DOI] [PubMed] [Google Scholar]
- 16.Lindh M, Hannoun C, Dhillon AP, Norkrans G, Horal P. Core promoter mutations and genotypes in relation to viral replication and liver damage in East Asian hepatitis B virus carriers. J Infect Dis. 1999;179(4):775–82. doi: 10.1086/314688. [DOI] [PubMed] [Google Scholar]
- 17.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, et al. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81(Pt 1):67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 18.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83(8):2059–73. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 19.Magnius LO, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38(1–2):24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69(Pt 10):2575–83. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 21.Norder H, Hammas B, Löfdahl S, Couroucé AM, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol. 1992;73(Pt 5):1201–8. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 22.Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198(2):489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 23.Guo W, Chen M, Yen TS, Ou JH. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13(1):443–8. doi: 10.1128/MCB.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo WT, Bell KD, Ou JH. Characterization of the hepatitis B virus EnhI enhancer and X promoter complex. J Virol. 1991;65(12):6686–92. doi: 10.1128/jvi.65.12.6686-6692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norder H, Hammas B, Lee SD, Bile K, Couroucé AM, Mushahwar IK, et al. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;(Pt 7):1341–8. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- 26.Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34(3):590–4. doi: 10.1053/jhep.2001.27221. [DOI] [PubMed] [Google Scholar]
- 27.Ashouri FP, Rasekhi S. A Review on Medical Students Mental Health Problems and Proposed Solutions. International Electronic Journal of Medicine. 2015;4(1):23–31. [Google Scholar]
- 28.Ashouri FP, Rasekhi S. Correlation between Religious Beliefs with Mental Health and Academic Performance in Medical Students. International Electronic Journal of Medicine. 2016;5(1):1–6. [Google Scholar]
- 29.Pivert A, Servant-Delmas A, Lunel-Fabiani F, Guillou-Guillemette L, Laperche S, Ducancelle A. Correlation between the promoter basal core and precore mutations and HBsAg quantification in French blood donors infected with hepatitis B virus. J Med Virol. 2015;87(3):529–35. doi: 10.1002/jmv.24064. [DOI] [PubMed] [Google Scholar]
- 30.Shiina S, Fujino H, Uta Y, Tagawa K, Unuma T, Yoneyama M, et al. Relationship of HBsAg subtypes with HBeAg/anti-HBe status and chronic liver disease. Part I: Analysis of 1744 HBsAg carriers. Am J Gastroenterol. 1991;86(7):866–71. [PubMed] [Google Scholar]
- 31.Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83(Pt 6):1267–80. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 32.Hadziyannis SJ, Vassilopoulos D. Hepatitis B e antigen–negative chronic hepatitis B. Hepatology. 2001;34(4 Pt 1):617–24. doi: 10.1053/jhep.2001.27834. [DOI] [PubMed] [Google Scholar]
- 33.Grandjacques C, Pradat P, Stuyver L, Chevallier M, Chevallier P, Pichoud C, et al. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J Hepatol. 2000;33(3):430–9. doi: 10.1016/S0168-8278(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 34.Lindh M, Furuta Y, Vahlne A, Norkrans G, Horal P. Emergence of precore TAG mutation during hepatitis B e seroconversion and its dependence on pregenomic base pairing between nucleotides 1858 and 1896. J Infect Dis. 1995;172(5):1343–7. doi: 10.1093/infdis/172.5.1343. [DOI] [PubMed] [Google Scholar]
- 35.Ayed K, Gorgi Y, Ayed-Jendoubi S, Aouadi H, Sfar I, Najjar T, et al. Hepatitis B virus genotypes and precore/core-promoter mutations in Tunisian patients with chronic hepatitis B virus infection. J Infect. 2007;54(3):291–7. doi: 10.1016/j.jinf.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Lok AS, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc Natl Acad Sci U S A. 1994;91(9):4077–81. doi: 10.1073/pnas.91.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, et al. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33(1):218–23. doi: 10.1053/jhep.2001.20532. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Frias F, Buti M, Jardi R, Cotrina M, Viladomiu L, Esteban R, et al. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology. 1995;22(6):1641–7. doi: 10.1002/hep.1840220605. [DOI] [PubMed] [Google Scholar]
- 39.Li JS, Tong SP, Wen YM, Vitvitski L, Zhang Q, Trépo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67(9):5402–10. doi: 10.1128/jvi.67.9.5402-5410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meldal BH, Moula NM, Barnes IH, Boukef K, Allain JP. A novel hepatitis B virus subgenotype, D7, in Tunisian blood donors. J Gen Virol. 2009;90(Pt 7):1622–8. doi: 10.1099/vir.0.009738-0. [DOI] [PubMed] [Google Scholar]
- 41.Bahri O, Cheikh I, Hajji N, Djebbi A, Maamouri N, Sadraoui A, et al. Hepatitis B genotypes, precore and core promoter mutants circulating in Tunisia. J Med Virol. 2006;78(3):353–7. doi: 10.1002/jmv.20554. [DOI] [PubMed] [Google Scholar]
- 42.Borchani-Chabchoub I, Gargouri A, Mokdad-Gargouri R. Genotyping of Tunisian hepatitis B virus isolates based on the sequencing of preS2 and S regions. Microbes Infect. 2000;2(6):607–12. doi: 10.1016/S1286-4579(00)00365-8. [DOI] [PubMed] [Google Scholar]
- 43.Bodhiphala P, Chaturachumroenchai S, Chiewsilp P, Pruksananonda P. Detection of HBV genome by gene amplification method in HBsAg negative blood donors. J Med Assoc Thai. 1999;82(5):491–5. [PubMed] [Google Scholar]
- 44.Alam-mehrjerdi Z, Mokri A, Dolan K. Methamphetamine use and treatment in Iran: a systematic review from the most populated Persian Gulf country. Asian J Psychiatr. 2015;16:17–25. doi: 10.1016/j.ajp.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 45.Hamadiyan H, Pour Ashouri F, Rasekhi S. Growth of Head Circumference, Weight and Height from Birth to 18 Months in the South of Iran. International Electronic Journal of Medicine. 2015;4(1):1–5. [Google Scholar]
- 46.Yadollahi S, Hamadiyan H, Pour Ashouri F, Fallahi S, Parvizpanah A, Rasekhi S. Trends in the Lipid Profile, Mean Age and Fatality of Patients with Myocardial Infarction in the South of Iran from 2008 to 2014. Int J Med Res Health Sci. 2016;5(6):221–8. [Google Scholar]
- 47.Lindh M, Gonzalez JE, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72(2):163–74. doi: 10.1016/S0166-0934(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 48.Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, et al. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol. 1990;64(3):1298–303. doi: 10.1128/jvi.64.3.1298-1303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rezende RE, Fonseca BA, Ramalho LN, Zucoloto S, Pinho JR, Bertolini DA, et al. The precore mutation is associated with severity of liver damage in Brazilian patients with chronic hepatitis B. J Clin Virol. 2005;32(1):53–9. doi: 10.1016/j.jcv.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Vivekanandan P, Abraham P, Sridharan G, Chandy G, Shaji RV, Daniel D, et al. High frequency of the 1896 precore mutation in patients and blood donors with hepatitis B virus infection from the Indian subcontinent. Mol Diagn. 2004;8(1):51–6. doi: 10.1007/BF03260047. [DOI] [PubMed] [Google Scholar]
- 51.Funk ML, Rosenberg DM, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J Viral Hepatitis. 2002;9(1):52–61. doi: 10.1046/j.1365-2893.2002.00304.x. [DOI] [PubMed] [Google Scholar]
- 52.Vray M, Debonne JM, Sire JM, Tran N, Chevalier B, Plantier JC, et al. Molecular epidemiology of hepatitis B virus in Dakar, Senegal. J Med Virol. 2006;78(3):329–34. doi: 10.1002/jmv.20544. [DOI] [PubMed] [Google Scholar]
- 53.Biswas A, Chandra PK, Datta S, Panigrahi R, Banerjee A, Chakrabarti S, et al. Frequency and distribution of hepatitis B virus genotypes among eastern Indian voluntary blood donors: Association with precore and basal core promoter mutations. Hepatol Res. 2009;39(1):53–9. doi: 10.1111/j.1872-034X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- 54.Davidson F, Lycett C, Sablon E, Petrik J, Dow BC. Hepatitis B virus genotypes and precore mutations in Scottish blood donors. Vox Sang. 2005;88(2):87–92. doi: 10.1111/j.1423-0410.2005.00597.x. [DOI] [PubMed] [Google Scholar]
- 55.Bläckberg J, Kidd-Ljunggren K. Genotypic differences in the hepatitis B virus core promoter and precore sequences during seroconversion from HBeAg to anti-HBe. J Med Virol. 2000;60(2):107–12. doi: 10.1002/(SICI)1096-9071(200002)60:2<107::AID-JMV1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 56.Lindh M, Andersson AS, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus—large-scale analysis using a new genotyping method. J Infect Dis. 1997;175(6):1285–93. doi: 10.1086/516458. [DOI] [PubMed] [Google Scholar]
- 57.Chan HL, Hussain M, Lok AS. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis B e antigen seroconversion. Hepatology. 1999;29(3):976–84. doi: 10.1002/hep.510290352. [DOI] [PubMed] [Google Scholar]
- 58.Friedt M, Gerner P, Lausch E, Trübel H, Zabel B, Wirth S. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology. 1999;29(4):1252–8. doi: 10.1002/hep.510290418. [DOI] [PubMed] [Google Scholar]
- 59.Laskus T, Rakela J, Persing DH. The stem-loop structure of the cis-encapsidation signal is highly conserved in naturally occurring hepatitis B virus variants. Virology. 1994;200(2):809–12. doi: 10.1006/viro.1994.1247. [DOI] [PubMed] [Google Scholar]
- 60.Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J Viral Hepatitis. 2005;12(5):456–64. doi: 10.1111/j.1365-2893.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 61.Arankalle VA, Murhekar KM, Gandhe SS, Murhekar MV, Ramdasi AY, Padbidri VS, et al. Hepatitis B virus: predominance of genotype D in primitive tribes of the Andaman and Nicobar islands, India (1989–1999) J Gen Virol. 2003;84(Pt 7):1915–20. doi: 10.1099/vir.0.18943-0. [DOI] [PubMed] [Google Scholar]
- 62.Kim H, Jee Y, Mun HS, Song BC, Park JH, Hyun JW, et al. Comparison of full genome sequences between two hepatitis B virus strains with or without preC mutation (A1896) from a single Korean hepatocellular carcinoma patient. J Microbiol Biotechnol. 2007;17(4):701–4. [PubMed] [Google Scholar]
- 63.Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Mahboudi F, Sabahi F, Adeli A, Noorinayer B, et al. Genotype characterization and phylogenetic analysis of hepatitis B virus isolates from Iranian patients. J Med Virol. 2005;75(2):227–34. doi: 10.1002/jmv.20261. [DOI] [PubMed] [Google Scholar]
- 64.Taghavi SA, Tabibi M, Eshraghian A, Keyvani H, Eshraghian H. Prevalence and clinical significance of hepatitis B Basal core promoter and precore gene mutations in southern Iranian patients. Hepat Mon. 2010;10(4):294–7. [PMC free article] [PubMed] [Google Scholar]
- 65.Halfon P, Bourliere M, Pol S, Benhamou Y, Ouzan D, Rotily M, et al. Multicentre study of hepatitis B virus genotypes in France: correlation with liver fibrosis and hepatitis B e antigen status. J Viral Hepat. 2006;13(5):329–35. doi: 10.1111/j.1365-2893.2005.00692.x. [DOI] [PubMed] [Google Scholar]
- 66.Wintermeyer P, Gerner P, Gehring S, Karimi A, Wirth S. Prevalence of hepatitis B virus precore stop codon mutations in chronically infected children. World J Gastroenterol. 2006;12(14):2235–8. doi: 10.3748/wjg.v12.i14.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang MH, Hsu HY, Ni YH, Tsai KS, Lee PI, Chen PJ, et al. Precore stop codon mutant in chronic hepatitis B virus infection in children: its relation to hepatitis B e seroconversion and maternal hepatitis B surface antigen. J Hepatol. 1998;28(6):915–22. doi: 10.1016/S0168-8278(98)80337-1. [DOI] [PubMed] [Google Scholar]
- 68.Gandhe SS, Chadha MS, Arankalle VA. Hepatitis B virus genotypes and serotypes in western India: lack of clinical significance. J Med Virol. 2003;69(3):324–30. doi: 10.1002/jmv.10292. [DOI] [PubMed] [Google Scholar]
- 69.Abbas Z, Muzaffar R, Siddiqui A, Naqvi SA, Rizvi SA. Genetic variability in the precore and core promoter regions of hepatitis B virus strains in Karachi. BMC Gastroenterol. 2006;6:20. doi: 10.1186/1471-230X-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]