Abstract
The recent development of CRISPR-Cas systems as easily accessible and programmable tools for genome editing and regulation is spurring a revolution in biology. Paired with the rapid expansion of personalized and reference genomic sequence information, technologies based on CRISPR-Cas are enabling nearly unlimited genetic manipulation even in previously difficult contexts, including human cells. Although much attention has focused on the potential of CRISPR-Cas to cure Mendelian diseases, the technology also holds promise to transform the development of therapies to treat complex heritable and somatic disorders. Here we discuss how CRISPR-Cas can impact the next generation of drugs through accelerating the identification and validation of high-value targets, uncovering high confidence biomarkers and developing differentiated breakthrough therapies. We focus on the promises, pitfalls and hurdles of this revolutionary gene editing technology, and also discuss key aspects of different CRISPR-Cas screening platforms and offer our perspectives on the best practices in genome engineering.
The central dogma of molecular biology posits a flow of information from gene to messenger RNA to protein1. The genome serves as the blueprint of life, setting the stage for all downstream activity. Although approaches to treat human disease predominantly target the end of the information cascade (for example by inhibiting signalling pathways, supplementing metabolites or interfering with viral polymerases), the discovery and validation of therapeutic targets often takes place at the level of genes and transcripts. The discovery of human mutations directly linked to disease (such as somatic BCR-ABL1 fusions in chronic myeloid leukemia or inherited BRCA1 mutations in breast cancer) or survival benefit (including PCSK9 mutations in minimizing cardiovascular disease) is considered by many to be the gold standard for drug target identification. However, the paucity of scalable genetic engineering tools in mammalian cell culture and model systems has necessitated that many discovery efforts linking genotype with phenotype are either observational, such as genome-wide association studies (GWASs), or take place in genetically malleable invertebrate models such as the fruit fly Drosophila melanogaster and the nematode Caenorhabditis elegans.
The recent development of easily programmable RNA-guided nucleases, derived from microbial adaptive immune systems, has revolutionized the molecular toolbox for mammalian genome engineering2–6. Gene editing technologies in the form of clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR associated (Cas) systems stand poised to transform many stages of drug discovery and development by enabling fast and accurate altering of genomic information in mammalian model systems and human tissues. Additionally, direct somatic editing7 in patients will eventually radically change the druggable space8 by enabling targeting of nearly any entity, including introduction of corrective mutations and modification of regulatory elements or splicing patterns. Following the description of a two-component Cas9-single guide RNA (sgRNA) complex to introduce DNA double-strand breaks (DSBs) in an RNA-guided manner2, many studies have demonstrated ingenious applications and uncovered orthogonal immune systems, together enabling nearly unlimited genome engineering opportunities (Figure 1).
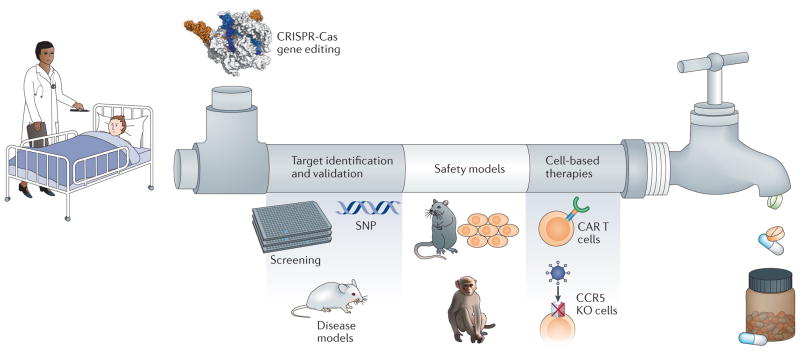
Figure 1. Pipeline of CRISPR-Cas-assisted drug discovery.
Unmet medical needs for numerous diseases and the rapid progress of CRISPR-Cas gene editing can feed into a drug discovery and development pipeline, leading to improved therapies. The CRISPR-Cas system allows for improved target identification and validation, and faster generation of safety models. CRISPR-Cas can also be used to develop cell-based therapies such as CAR T cells for immunotherapy and CCR5 knockout cells for HIV treatment. CRISPR-Cas-assisted drug discovery will yield innovative therapies and treatment paradigms for patients.
The technological domestication of CRISPR-Cas systems and molecular mechanisms of Cas-based genome editing have been thoroughly covered elsewhere9–11. Briefly, a sgRNA directs the Cas9 endonuclease to induce DSBs at homologous sites2. During genome editing, the DSBs are fixed by cellular DNA repair mechanisms, including the predominant error-prone non-homologous end joining (NHEJ)12–14 and less frequent templated homology-directed repair (HDR)15–19 pathways. NHEJ is most often leveraged to disrupt genetic sequences, while HDR can be used to introduce or alter information at a specific locus with properly designed repair templates. Additionally, a catalytically inactive mutant of Cas9 (dCas9) can be fused to various effector domains to activate or repress the transcription of target genes, strategies known as CRISPRa and CRISPRi, respectively20–22. Most studies to date have used Cas9 from Streptococcus pyogenes (SpyCas9), which is the default Cas9 referenced in this review. Cas9s from other species, Cas9-like CRISPR nucleases and engineered Cas9s with novel functions have also been established and can convey particular advantages in various settings (Supplemental Table S1). Although we focus on SpyCas9 and its use in therapeutic discovery and the building of the next generation of transformational drugs, the general outline described here applies to the larger ensemble of CRISPR-Cas tools.
CRISPR-Cas as a tool for drug discovery
Precision cellular models
Advances in DNA sequencing and their large-scale application have provided insight into genetic variation across groups of patients and populations, expanding our understanding of the link between genetic variation and disease predisposition, and between development and treatment response. For example, integrated information from The Cancer Genome Atlas (TCGA)23–28, the Cancer Cell Line Encyclopedia (CCLE)28 and ENCODE29,30 led to improvements in the standard of care for glioblastoma patients, enabling stratification based on MGMT promoter methylation status31. Such advances have stimulated interest in ‘personalized’ or ‘precision’ medicine, which combines classical patient information with personal genetic data to directly inform an individual’s treatment strategies. However, hypotheses generated by large-scale observational ‘omics’ efforts often demand testing with precise genetic models, particularly to evaluate variants of unknown significance, optimize patient stratification, reassign approved drugs to new indications and develop alternative treatment paradigms.
In comparing even a single factor between cells (such as the mutational status of TP53, MYC or KRAS), there are often many confounding features that obscure a direct relationship with a disease phenotype. Currently, researchers often rely on matched patient samples from diseased and normal tissues to tease apart such relationships. However, large collections of matched samples can be difficult to obtain and are not available in many cases. Although overexpression of an appropriate (often mutant) complementary DNA (cDNA) can partially address this issue, such constructs are often expressed at non-native levels and in the presence of the wild type protein. The generation of mutant or knockout clones via classical homologous recombination led to a limited set of isogenic cell lines, in which a derived line differs from the parent by a minimal, defined mutation32–35. These resources have proven incredibly useful, but initial techniques for their generation were very labour intensive and time consuming, hindering their widespread adoption for drug development.
The advent of CRISPR-Cas genome editing2 has drastically altered this landscape (Figure 2). The generation of isogenic knockout human (and other) cell lines for comparative genomics is now so straightforward that in just 4 years the practice has become commonplace36 and is being carried out by researchers around the globe. Gene knockout via CRISPR-Cas has proven efficacious in virtually all cell types, including induced pluripotent stem cells (iPSCs), cancer-specific organoids and primary immune cells37–40. Knockout-based target discovery efforts are thus no longer limited to specialized cell lines, such as the haploid lines previously used for gene trap experiments41,42, and can instead be performed in the cell type most appropriate for the disease of interest. For example, if a panel of tumor-derived lines are thought to be sensitized to a drug candidate via a genetic lesion, knocking the gene out can directly test the hypothesis of synthetic lethality43–45. Such isogenic knockouts allow researchers to rapidly establish causative roles for oncogenes, tumour suppressors and other factors in a defined context, removing secondary differences.
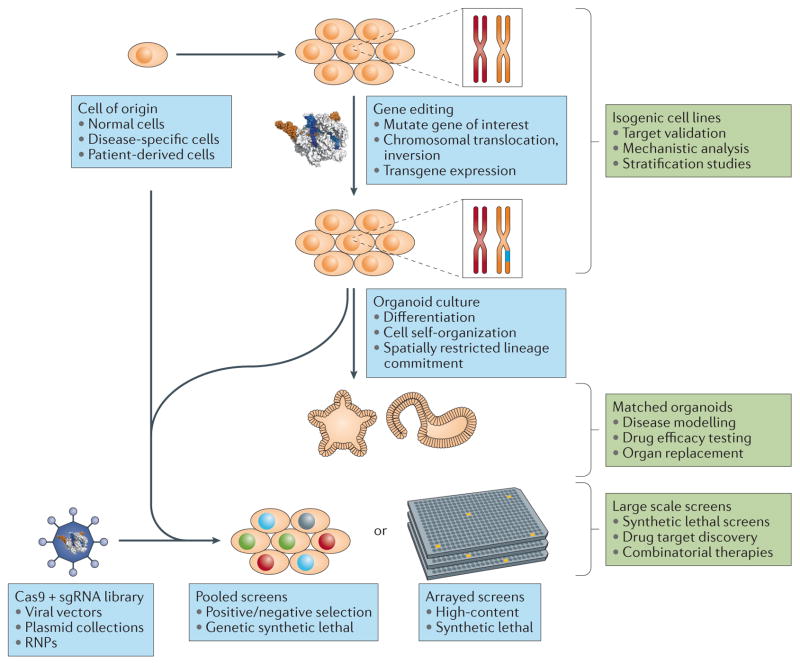
Figure 2. CRISPR-Cas in the generation of cellular models and large-scale screens.
CRISPR-Cas gene editing can be used to generate isogenic cell lines for drug target validation, mechanistic analysis and patient stratification studies. Isogenic cell lines can also be used to generate organoids, which are particularly useful for modelling differentiation and self-organization processes. Large-scale sgRNA libraries can be used for high-throughput pooled or high-content arrayed screens, either on unmodified or CRISPR-Cas-edited cell lines.
Similarly, ‘knocking in’ mutant alleles by HDR allows researchers to test the effects of disease-associated mutations in an isogenic background. For example, HDR can serve to generate mutant allelic series to compare the effects of each variant found across patients, as is the case for oncogenes such as KRAS, PIK3CA and IDH1, or tumor suppressors including TP53, RB1 and VHL46,47. More generally, isogenic series can be used to analyze the effect of mutants on disease development or to query the specificity of mutant-targeting therapeutic candidates. From a technical perspective, HDR requires delivery of the Cas9-sgRNA complex - in the form of a viral vector, plasmid, mRNA/sgRNA or ribonucleoprotein (RNP) complex - along with a DNA repair template. The HDR template can also take on different forms and its exact design substantially changes repair efficiency48,49. In mammalian cells, short single-stranded DNA (ssDNA) oligonucleotides whose design takes advantage of the molecular nature of the Cas9-target architecture have been shown to be advantageous for the introduction of small mutations50. Varying the distance between the DSB and the site of the mutation on the repair template further allows control over the efficiency of mutant introduction and zygosity51.
Despite this promise, although CRISPR-Cas knockouts are effective in nearly any cell, rates of HDR can vary across cell types. As one example, it has been difficult to achieve even moderate levels of HDR in non-mitotic human cells, including neurons. These barriers are particularly frustrating because sequence insertion or replacement in these contexts could be used to model or treat many genetic diseases. New approaches using non-homologous or microhomology-mediated integration of cassettes48,52,53 offer routes to bypass HDR pathways that are inactive in non-mitotic human cells and in organisms in which HDR has proven difficult. Another exciting new development is the engineering of Cas enzymes with additional functionalities to enable precise, template-less introduction of specific mutations by directly altering target bases. A first step towards this goal was the fusion of various cytidine deaminases to Cas9, resulting in hybrid enzymes capable of RNA-guided ‘base editing’54,55, and one can anticipate a future explosion of new Cas derivatives utilizing similar strategies.
Functional screening with CRISPR-Cas
Large-scale functional screening with CRISPR-Cas is simultaneously expanding and evolving, as researchers uncover the advantages and disadvantages of different screening systems. Until recently, systematic loss-of-function studies have focused on genome-wide RNA interference (RNAi) screens56–58 or insertional mutagenesis screens in haploid human cell lines41,42,59,60. CRISPR-Cas screens have rapidly been adopted in a variety of contexts thanks to the simplicity of designing potent sgRNAs and the ability to apply the system to nearly any cell type or tissue (Figure 2). Large-scale screens typically rely upon pooled lentiviral libraries of sgRNAs, often achieving robust hit identification by including 3–10 sgRNAs per gene20,61–65. CRISPR-Cas based screens proceed much like short-hairpin RNA (shRNA) screens. A pool of cells expressing Cas9 and the sgRNA library is subjected to the desired phenotypic selection, and high-throughput DNA sequencing of the sgRNA cassette is used to identify sgRNAs that were enriched or depleted during the treatment.
Genome-scale CRISPR-Cas knockout, inhibition and activation screens have identified essential genes in various cancer cell lines61,62,66–68, uncovered genes involved in the response to small molecule inhibitors58,63 and cellular toxins20,64, and dissected the relative importance of viral host factors69. They have also been used in a xenograft mouse model of tumour growth and metastasis to assay gene phenotypes in cancer evolution70. Although CRISPR-Cas screens for cell growth or survival have been quite successful (except when targeting genetically amplified regions62,66,67) screens for more complex phenotypes are still being optimized. Recent comparisons to microRNA-based shRNA screens have found comparable performance58,68 and the complementary strengths of both approaches should be carefully weighted when choosing a screening platform (Table 1).
Table 1.
Comparison of screening platforms
| Characteristic | CRISPRn | CRISPRi | RNAi (miRNA-based shRNAs) | CRISPRa |
|---|---|---|---|---|
| Effect | Knockout (KO) | Knockdown (KD) | Knockdown (KD) | Activation |
| Mechanism | Indel, mutation | Transcriptional interference | Transcript degradation, translational interference | Transcriptional activation |
| Guide target choice | Anywhere in the genome (PAM) | Transcription start site (TSS; PAM) | Exons | Transcription start site (TSS; PAM) |
| Target selectivity | Can distinguish any target | Depends on TSS, cannot distinguish products derived from the same transcript | Can distinguish splice variants | Depends on TSS, cannot distinguish products derived from the same transcript |
| Highly amplified regions (genes) | Off-target effects: DSBs evoke DNA damage repair, resulting in cell cycle arrest independent of target | Can be targeted if all use the same TSS | Can be targeted | Can be targeted if all use the same TSS |
| Distinguish alternative TSSs | Possible | Yes | Possible | Yes |
| Distinguish transcript splice variants | Possible | No | Possible | No |
| Performance of individual sgRNAs or shRNAs | Most work | Many work | Requires good prediction tools or testing | Many work |
In ‘CRISPR nuclease’ (CRISPRn) screens, stably expressed Cas9 and sgRNA complexes continue to operate on a target site until it is ablated, and can therefore generate homozygous knockout phenotypes at high frequency in most cell types. Conversely, high copy number genomic amplifications can be a barrier to CRISPRn screens, mainly because the large numbers of DNA breaks generated in high copy number regions can lead to reduced cell growth triggered by the DNA damage response and cell cycle arrest, which are activated independently of the targeted gene or genomic region (thus representing a systematic, sequence-independent off-target effect)62,66,67. As CRISPRn generally depends on sequence frame-shifting to generate knockouts, phenotype penetrance can be affected if in-frame deletions are preferentially created. This can be overcome by targeting functional domains43, though doing so requires preexisting knowledge of target proteins. Furthermore, sgRNAs targeting the 5’ end of the coding region may be ineffective if alternative downstream start codons are present66.
CRISPRi screens do not rely on frame-shifting and can offer certain advantages over CRISPRn screens from a drug discovery perspective because knocking down gene expression (using CRISPRi or RNAi) mimics the effects of a small molecule inhibitor more closely than complete gene ablation does71. CRISPRi screens can also identify the contributions of transcripts arising from different transcription start sites (TSSs), whereas RNAi screens can uniquely distinguish different splice variants57,72.
CRISPRa screens, which assess gene targets whose overexpression leads to a given phenotype20,21, are an emerging and particularly exciting area of recent development. They have an array of benefits and trade-offs compared to the complementary DNA (cDNA) screens that have previously been used in this area. Construction and use of cDNA screening resources is labour intensive due to the complex nature of cDNAs. By contrast, the resources necessary to perform CRISPRa screens are similar to those required by CRISPRn or CRISPRi screens20. Moreover, cDNA expression screens can only interrogate the transcripts present in the library, which may lack certain genes or alternatively spliced isoforms. Conversely, by stimulating expression from the endogenous locus, CRISPRa screening can activate expression of alternatively spliced transcript variants as easily as it activates expression of the primary transcript and sgRNAs can be designed to target each TSS within each gene. However, CRISPRa screens are subject to their own set of false-negatives. For example, CRISPRa will have no effect if the target gene contains loss-of-function mutations or is missing entirely in the cell line of interest.
A substantial technical barrier for CRISPRa screening is the activation of highly repressed genes. To overcome this challenge, a range of CRISPRa systems have been developed that recruit multiple and/or diverse transcriptional activation domains to increase the potency of gene activation20,21,73–78. Ultimately, an ideal CRISPRa screening platform would use the fewest exogenous parts necessary to potently activate any gene target; additional developments and systematic comparisons are needed towards this end79.
We expect that CRISPR-Cas based screens will continue to improve, especially as they are used for an increasingly broad array of phenotypes. Most of the pioneering CRISPR screens simply looked for growth advantages and disadvantages, identifying genes essential for proliferation, or resistance or sensitivity to certain toxins. Going forward, there will be more CRISPR screens to examine the sensitivity of cancer cells to candidate therapeutics, resistance to pathogen infection, or the regulation and cellular localization of a gene of interest42,58,69. CRISPR screens in human pathogens can also be used to identify candidate drug targets80. The relatively low cost of sgRNA library design opens the door for creative screening approaches, such as efforts to identify non-coding sequences controlling expression of BCL11A, TP53 and ESR1 using target-tiling CRISPRn screens81,82, and we expect future screens for non-coding regulatory elements to look at even larger regions of DNA sequence. Finally, more systematic analyses are needed to compare CRISPRn, CRISPRi and various types of RNAi screens (including microRNA-based shRNAs). Such comparison will define the relative strengths and weaknesses of each platform and allow researchers to choose the best type of screen to address their question (Table 1).
Rapid generation of animal models
Beyond cell culture applications, genome editing has dramatically altered our ability to generate animal models of disease (Figure 3). It will soon be common for early go/no-go decisions in a drug development campaign to be based on results from mutant animals of the most relevant model species for a disease. Indeed, shortly after their initial development, CRISPR-Cas tools have been used to generate mice with multiple genetic lesions in a single editing step83, as well as for one-step knock-in of reporter and conditional alleles into mouse zygotes84.
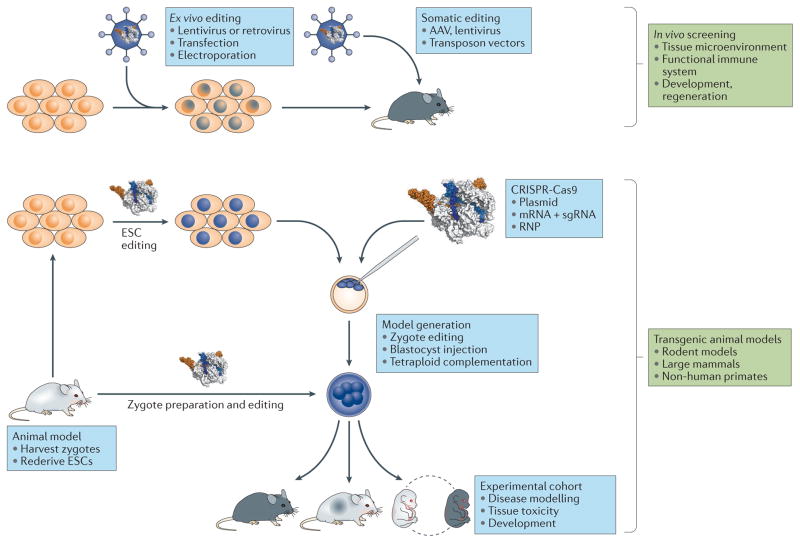
Figure 3. Applications of CRISPR-Cas for in vivo screens and the generation of animal models.
a | Ex vivo editing can be used to generate a library of modified cells for transplantation into recipient animals. Alternatively, editing reagents can be delivered to host animal tissues directly for somatic in situ editing. b | CRISPR-Cas has also revolutionized the generation of transgenic animal models through facile editing of ESCs for traditional gene targeting and by enabling direct zygote editing in most species. Zygote editing can be done ex vivo by electroporating or microinjecting zygotes with CRISPR-Cas constructs in the form of plasmids, RNA preparations or RNPs
Generally, efficient CRISPR-Cas editing, including NHEJ and short HDR, can be achieved by microinjection or simple electroporation of zygotes instead of proceeding through traditional embryonic stem cell (ESC) manipulation85–87. This is a critical development in two ways. First, since multiple genes can be targeted in a single step, double- and triple-mutant mice can be rapidly generated without the need for crossing single-mutant strains, though it must be noted that such alleles follow Mendelian segregation upon breeding. Second, editing in zygotes eliminates the requirement to derive, culture and edit ESCs, which has been a major barrier to widespread genetic tractability in several model organisms relevant to the process of therapeutic discovery, such as rats. Zygote editing also accelerates the generation of additional mutations in pre-existing animal models of disease by eliminating the need for ESC derivation or lengthy back-crossing. Nevertheless, introducing large transgenes or complex multi-component systems via zygote editing remains inefficient; gene targeting in ESCs is likely to remain, for now, the method of choice for generating those animals88,89.
Founder animals from both zygote editing or conventional blastocyst injection of modified ESCs can exhibit mosaicism (Box 1). Mosaicism in ESC injection studies can be reduced by the tetraploid complementation method90,91 in which modified ESCs are introduced into developmentally compromised blastocysts, though this is a technically complex procedure requiring amenable ESCs. Conversely, in zygote electroporation studies, mosaicism is due to the fact that sometimes the single-cell zygote divides before the editing occurs. Hence, replacing Cas9 mRNA and sgRNA with Cas9-sgRNA ribonucleoprotein (RNP) complexes that can immediately act on their targets increases the fraction of non-mosaic founders85,86. Overall, CRISPR-Cas promises to revolutionize mouse genetics by reducing the time necessary to generate targeted models from years to months or weeks. A large spectrum of models can now be generated in a timescale relevant to early go/no-go decisions in a modern drug discovery campaign. Drug discovery implications of gene editing in additional species are discussed at the end of this section.
Box 1. Mosaicism.
Mosaicism is the presence of cells of multiple different genotypes within a single animal or cell population.
In cell culture
In most cases, a population of edited cells will contain a variety of mutations, even if 100% of alleles within the cell population are edited. This is because DNA double-strand break (DSB) repair by the predominant non-homologous end joining (NHEJ) pathway can lead to different indels in different cells. Depending on the experiment, mosaicism in cultured cells may or may not be problematic. If the editing efficiency is sufficiently high and all mutations cause the same phenotype (for example, loss of function due to mutations in the active site of an enzyme) the mosaicism is functionally irrelevant. In other cases, some indels might result in an in-frame deletion that has no phenotype, leading to a variegated population. Mosaicism can be eliminated by deriving single-cell clones.
In animal models
When edited embryonic stem (ES) cells are injected into a blastocyst for model generation, the resulting animal can be a mosaic of the donor ES cells and the cells of the recipient blastocyst. Tetraploid embryo complementation, a method that renders recipient blastocyst developmentally compromised, can reduce this risk. Mosaicism can also be a result of zygote editing, if editing takes place after the one-cell stage. Hence, editing methods that act on their DNA targets directly upon transduction (such as Cas9-sgRNA RNPs) may reduce mosaicism in founder animals. Mosaicism at a given locus can be eliminated by back-crossing founder animals for a single generation, but can nonetheless be problematic if multiple genes are targeted simultaneously. For example, founder animals with mutations in three targeted genes will not necessarily carry all three mutations in every individual cell. If this is the case, multiple generations of breeding are needed to generate non-mosaic animals with mutations in all three genes.
Pairing CRISPR-Cas with viral or transposon-based vectors has allowed researchers to directly introduce somatic mutations in certain tissues, such as the lung and liver, in adult animals. This approach has been used to create numerous cancer and other disease models92–94, and to correct disease mutations and phenotypes95–98. One illustrative example of the power of CRISPR-Cas tools is the in vivo engineering of oncogenic chromosomal rearrangements that mimic fusion proteins found in patients (e.g. EML4-ALK, KIF5B-RET, CD74-ROS1), leading to in-situ tumor initiation from edited somatic cells99,100. The ability to introduce disease-associated alleles in live animals is particularly transformative when compared to xenograft models that require immunosuppressed recipients and mostly rely on implantation at non-native sites. The in situ introduction of mutations with CRISPR-Cas allows researchers to accurately recapitulate disease initiation, development and maintenance in an autochthonous and immunocompetent setting, including the native microenvironment and tissue structure. This ability will be transformative for many diseases, particularly cancer, where interaction with immune cells can have a drastic effect on disease outcome94,101.
The application of CRISPR-Cas to a large and rapidly growing number of organisms holds great promise, as traditional gene targeting has remained difficult in pre-clinical models other than mice. CRISPR-Cas editing has been performed in rats102, dogs103 and cynomolgus monkeys104, the species most commonly used during pre-clinical drug discovery and development. As with mouse zygote targeting, many of the edited animals exhibit mosaicism. The generation of disease models in primates, such as a model of Duchenne muscular dystrophy in rhesus monkeys105, further emphasizes the benefits of gene editing to not only accelerate therapeutic development but also to test efficacy and safety of therapeutic compounds. CRISPR-Cas may even drive the development of porcine xenotransplant platforms through the inactivation of endogenous retroviruses106. CRISPR-Cas editing will also be a boon to infectious disease research. Many human pathogens are best modelled in hosts other than mouse, such as influenza (ferrets)107, leptospirosis (hamsters)107 and tuberculosis (Guinea pigs)108, and we expect zygote editing to be proven feasible in these organisms in the near future. Optimizing conditions for ESC work was one of the biggest challenges in the genetic manipulation of new mammalian model organisms. CRISPR-Cas zygote editing should soon eliminate this hurdle.
Specificity of CRISPR systems
Although CRISPR-based tools are easily programmed to target basically any genomic location, they can also lead to low rates of off-target editing or sequence-independent cell cycle arrest if highly amplified loci are targeted62,66,67. At first glance, one might assume that a less than perfect gene editing reagent is a deal-breaker. Yet for non-therapeutic use, such stringency might not always be needed and can be compensated for with proper controls. Hence, the most important aspect is a thorough understanding of off-target events, their biological consequences and how these effects can be mitigated.
Sequence-dependent off-target propensities are best understood for the Streptococcus pyogenes Cas9 (SpyCas9) enzyme, for which combinations of systematic and unbiased experiments have begun to shed light on potential liabilities109–111. Several excellent other reviews have extensively discussed CRISPR-Cas off-target effects112–114. Nevertheless, our understanding of the molecular mechanisms through which SpyCas9 can sometimes inappropriately bind and cut off-targets is still in its infancy, and indeed such tolerance may be built into naturally evolved CRISPR-Cas systems as part of the immunological arms race between the phage and its bacterial host. Phenomenological data revealed that the 8–10 nucleotides neighbouring the protospacer adjacent motif (PAM) are most stringently recognized, whereas one or two mismatches can be tolerated in the remaining nucleotides65,115,116.
Off-targets are determined by the nuclease and the sgRNA sequence. Thus, several algorithms have been developed to predict sgRNA efficiency and off-target sites65,117–121, so far focusing on SpyCas9. Although comparison to unbiased genome-wide assessment of off-target sites has revealed the limited predictive power of many algorithms for distantly related off-targets109, likely off-target sites and clearly risky sgRNAs can still be identified. From the perspective of research use for target identification and validation, any candidate identified through a CRISPR-Cas knockout experiment should be validated with orthogonal strategies to rule out off-target effects. These might include the use of multiple sgRNAs, isolation of multiple clonal lines, validation by alternative transcript knockdown methods (CRISPRi, RNAi), and cDNA or CRISPRa complementation studies. This mirrors required follow-up experiments for comparable RNAi approaches. In a research setting, the ability to perform such validation experiments makes the extensive identification of rare off-targets relatively superfluous. Off-target analyses and de-risking strategies are far more critical for therapeutic CRISPR-Cas gene editing than for pre-clinical investigations.
Much effort has been put into the development of strategies to systematically minimize CRISPR-Cas off-target effects. One elegant tactic requires two Cas9 nickases (Cas9-D10A or Cas9-H840A2, that cleave or ‘nick’ only a single DNA strand) to bind at neighbouring sites, thereby increasing the effective stringency due to the low probability of adjacent off-target sites within a genome122,123. Similarly, a dimerizing FokI nuclease domain (used by other DNA editing enzymes such as zinc finger nucleases (ZFNs) and TALENs) has been fused to a catalytically inactive Cas9 (dCas9) in order to require paired binding to induce DSBs124,125. Although they reduce off-target events, paired-nickase strategies also reduce the targetable space by requiring two sgRNAs to bind within a relatively narrow stretch of DNA. In pooled screening scenarios, paired-nickases also require a combinatorial library or tandem sgRNA vectors.
A second strategy to reduce off-target events relies upon sgRNA or protein engineering to enforce higher specificity. Truncated guide RNAs can remove a few of the relatively permissive bases from the 5’ end of the guide RNA, resulting in both decreased on- and off-target activity109,126; the specific mechanism through which this occurs is not yet fully understood. SpyCas9 has also been mutagenized to more specifically recognize only a single PAM127, or to abrogate nonspecific binding and thereby reduce the cleavage of non-target sequences128,129; though again the mechanism of action is still under investigation.
A third strategy to reduce off-target events adds strict control over the amount of active Cas9 in cells. So far, such approaches have used tightly regulated induction of Cas9 activity130 and even reversible small molecule- or photo-induced Cas9 activity131,132. These methods reduce off-target effects and also enable temporal control of genome editing. In appropriate scenarios, use of carefully titrated amounts of Cas9-sgRNA RNP complexes, which are rapidly degraded, can have similar benefits. Ultimately, many of the strategies outlined here might even be modularly combined for further gains in specificity, though this has yet to be experimentally tested. Overall, SpyCas9 already shows a naturally high fidelity, and a variety of approaches have been able to improve its specificity. It is easy to foresee how additional engineering approaches, combined with a more detailed mechanistic understanding of the conformational changes occurring during target binding and cleavage, will advance editing precision.
Using CRISPR-Cas to make therapeutics
In addition to generating powerful research tools, genome editing with CRISPR-Cas also holds great promise to make therapeutic agents or as a therapeutic itself. In principle, any DNA editing technology could be used for the therapeutic strategies described in this section. Although ZFNs have advanced the furthest in clinical trials to date, there is currently insufficient evidence to declare whether the clinical utility of CRISPR-Cas, ZFNs or TALENs will be superior. The fast and inexpensive reprogramming of Cas9 gives it a clear advantage in contexts in which rapid experimental iteration is beneficial or in which many different loci need to be targeted. Here we briefly discuss the current state of therapeutic gene editing (mostly in the context of ZFNs and TALENs), and how CRISPR-Cas can contribute to the field. We focus on therapeutic applications other than in vivo gene editing, as this topic has been covered by several recent reviews.
Creating CAR-T cellular therapies with gene editing
The application of gene editing for somatic diseases has begun to overlap with the exploding field of cancer immunotherapy, with immediate interest centering on the production of next-generation chimeric antigen receptor T cells (CAR-Ts). These modified T cells, which express tumour-targeting receptors, have shown promise in the treatment of various leukemias and lymphomas, and may eventually be used to treat solid cancers133. Chimeric antigen receptors (CARs) comprise an extracellular binding domain (currently a single chain variable fragment), which recognizes an antigen that is strongly expressed on - and specific to - tumour cells, and an intracellular chimeric signalling domain that activates the T cell upon receptor engagement, promoting T-cell-mediated tumor cell killing. The first battery of CAR-T therapies targeted CD19, an antigen expressed by B cells and related cancer cells; several have entered clinical trials (clinicaltrials.gov identifiers: Juno Therapeutics: NCT02535364, NCT02631044; Kite: NCT02601313, NCT02348216; Novartis: NCT02030834, NCT02445248).
Currently, most CAR-T cells are generated using each patient’s own T cells, an expensive and time-consuming process that involves isolating, modifying and expanding T cells for every new patient. This process is limited by current manufacturing capabilities. Hence, the economics of CAR-Ts are less favourable than antibody-based checkpoint cancer immunotherapies such as ipilimumab, pembrolizumab and nivolumab. CAR-T therapy could become much faster and less expensive if universal donor CAR-Ts could be generated, as ’off-the-shelf’ cells would substantially increase the number of patients that could be treated by a single CAR-T cell product. However, graft-versus-host disease (GvHD) and host rejection, caused by recognition of recipients’ cells by the CAR-T cells and recognition of the CAR-T cells by the host, respectively, remain major barriers to an off-the-shelf approach. In this context, ZFNs and TALENs have been used to knock out endogenous TCR genes in T cells, which could prevent unwanted graft-versus-host reactivity134,135 (clinicaltrials.gov identifiers: Institut de Recherches Internationales Servier: NCT02808442). Genome editing strategies could also be used to prevent or delay the rejection of CAR-T cells by the recipient’s immune system by eliminating or decreasing expression of histocompatibility antigens on the donor T cells136.
In addition to enabling off-the-shelf CAR-T cells, genome editing could be used to boost CAR-T-cell efficacy by knocking out T cell inhibitory receptors or signaling molecules, such as CTLA4 or PD1137,138. Indeed, the NIH Recombinant DNA Advisory Committee (RAC) recently approved a University of Pennsylvania clinical trial in which Cas9 will be used to knock out PD1 and the endogenous T cell receptor in melanoma-targeting CAR-T cells (see Further information). China has also approved a clinical trial using Cas9 to knock out PD1 in the T cells of lung cancer patients, although no chimeric antigen receptor will be introduced in that trial139. Similar trials with PD1 knockout T cells for prostate and bladder cancer, as well as renal cell carcinoma are also being initiated (see Further information). In the future, gene editing might even be used to introduce the chimeric antigen receptor itself via HDR. Site-specific knock-in would eliminate the need for randomly integrating viral delivery vectors and allow for control over where the CAR integrates140,141. Future CAR-T cell therapies could benefit from combined modification of endogenous T cell receptor genes, histocompatibility genes and components of signalling pathways. Still, it will be important to establish that the removal of inhibitory signals does not enable uncontrolled proliferation of the CAR-T cells.
Compared to other gene editing reagents, such as ZFNs and TALENs, CRISPR-Cas allows for extremely rapid testing of any newly proposed candidate genetic modifications. Several industry partnerships have been announced between developers of CAR-T cell therapies and companies specializing in gene editing, including Novartis’ collaboration with Intellia Therapeutics and Caribou Biosciences, and Juno Therapeutics’ collaboration with Editas Medicine. CAR-T producer Cellectis acquired a license to use TALENs from the University of Minnesota.
Therapeutic ex-vivo gene editing
Drug delivery to the appropriate cells or tissue in situ is challenging in many fields, and is certainly a major limitation for therapeutic applications of Cas9. Ex vivo manipulation of target cells circumvents this issue. The haematopoietic system is an excellent target for ex vivo gene editing because cells are readily obtained from peripheral blood samples and can be re-injected after manipulation and expansion. Therapeutic ex vivo gene editing of hematopoietic stem cells has previously been explored using ZFNs and TALENs, and some of these therapies are showing promise in clinical trials. The most advanced strategy uses ZFNs to target the CCR5 gene in cells from HIV patients142. CCR5 is a co-receptor for HIV entry, and individuals with loss-of-function mutations in CCR5 are highly resistant to HIV infection but are otherwise healthy. Importantly, transplantation of bone marrow from a CCR5-deficient donor to a leukaemia patient infected with HIV, known as the ’Berlin patient’, reduced the patient’s viral load to undetectable143. While CCR5-deficient, HLA-matched donors are too rare for cell transplantation to be a broadly applicable treatment, they served as a proof-of-principle for the targeted disruption of CCR5 to cure HIV.
Researchers have used ZFNs to disrupt the CCR5 gene in T cells isolated from HIV patients, followed by expansion and re-injection of the edited T cells, to create a pool of HIV-resistant autologous T cells within the patient142,144. Phase 1/2 clinical trials of this approach are currently underway. Although mutations in the CCR5 gene in T cells are permanent, the T cells themselves may not be. Researchers have recently focused on disrupting CCR5 in haematopoietic stem cells (HSCs) in order to produce long-term self-renewing HIV-resistant cells145. CRISPR-Cas9 could also be applied to the same workflow of extracting, editing and re-implanting, and several groups have edited CCR5 with Cas96,146,147.
The haemoglobinopathies sickle cell disease (SCD) and beta-thalassaemia have been targeted for ex vivo gene correction instead of disruption. All patients with sickle cell disease carry the same causal mutation in the HBB gene, which causes a glutamate (Glu) to valine (Val) substitution. ZFNs have been used to correct the sickle allele in HSCs via HDR using an integrase-defective lentiviral vector or a single-stranded oligonucleotide donor148. CRISPR-Cas has rapidly caught up to ZFNs, demonstrating correction of the sickle allele using either an AAV6 or oligonucleotide donor149,150. In contrast, beta-thalassaemia is caused by a variety of null or hypomorphic mutations in HBB151, requiring a plethora of case-specific targeting complexes and repair donors. CRISPR-Cas could be superior to other nucleases in such situations, as designing new sgRNAs is much faster and cheaper than engineering new TALENs or ZFNs. The regulatory landscape surrounding such personalized approaches is in flux. Currently, even though multiple editing reagents might revert mutations to the same sequence, they would be classified as a separate investigational new drugs (INDs).
Regardless, individually correcting all disease-causing HBB mutations could be unnecessary, as beta-thalaessemia and SCD may be correctable by reactivation of fetal globin expression. The transcription factor BCL11A represses fetal globin in adults, and Sangamo and Biogen initially sought to systemically disrupt it to increase fetal globin expression in patients with beta-thalassemia. However, several groups have now used TALENs, ZFNs and CRISPR-Cas to identify an erythroid-specific enhancer controlling BCL11A expression81,152,153. Notably, tiling sgRNA libraries took advantage of CRISPR-Cas’ easy reprogramming to probe over 500 sites in the enhancer region, identifying a minimal target sequence for disruption81. As disruption of the enhancer leads to an erythroid-specific decrease in BCL11A and an increase in fetal globin production, Biogen and Sangamo have combined their BCL11A programmes to focus on mutating the enhancer (see Further information)81. These efforts currently use ZFNs, but various companies are exploring CRISPR-Cas for clinical disruption of the BCL11A enhancer.
Defining a path to the clinic
The path by which new gene editing therapies advance to the clinic will undoubtedly be shaped by Sangamo’s use of ZFNs to disrupt CCR5. Establishing the specificity of the nuclease was an important early hurdle and the methods used to predict and measure off-target DNA breaks are similar to the tools for assessing Cas9 specificity discussed above. However, safety testing of gene editing therapies must extend well beyond establishing the specificity of a nuclease. IND-enabling safety studies of ZFN-treated T cells and HSCs, which are setting the stage for future CRISPR-Cas therapies, aim to demonstrate that edited T cells and HSCs will not lead to leukemia. These studies have included karyotype analysis, soft-agar transformation assays and tumorigenicity studies of whole-patient doses of cells in immunodeficient mice (see Further information). Yet, the capacity of in vitro and animal models to predict adverse events in humans will always raise concerns. Ultimately, patients, clinicians and regulatory agencies must discuss the level of risk that is acceptable under each circumstance and develop appropriate safety measures.
Even if effective DNA editing reagents are developed and the treated cells are shown to be non-tumorigenic, substantial hurdles can remain for advancing a therapy into the clinic. Producing ex vivo edited cells at clinical scale (a dose of 1010 ZFN-treated T cells was used in the first trial of CCR5 editing to treat HIV-positive patients142) under conditions compliant with good manufacturing practice is a major challenge. It is also important that the phenotype of gene-edited cells is only changed by editing, and not epigenetically altered through ex vivo culture. Many assays, such as the capacity of HSCs to engraft and differentiate into a spectrum of leukocyte subsets145, can assess the healthy function of edited T cells and HSCs, though whether such assays can fully recapitulate behaviour in humans is unclear. It will thus be crucial to build deep phenotypic characterization protocols for all gene-editing therapies.
Last but not least, newly expressed or corrected proteins may be recognized as foreign by the recipient’s immune system. For example, haemophilia patients can develop neutralizing antibodies against replacement blood clotting factors154. Furthermore, edited cells could, in principle, be recognized and eliminated by cytotoxic T lymphocytes155. However, rejection of edited cells due to recognition of the transgene has not yet been an issue in clinical trials of anti-sickling globin gene therapy (see ASH meeting abstracts in Further information), suggesting that this may not be a problem for all edited genes or cell types.
Conclusions
CRISPR-Cas tools have been developed for a variety of cells and organisms in which genetic manipulation was previously relatively intractable, from human ESCs to the malaria parasite. Particularly in mammalian model systems and human cells, these technologies can accelerate functional genomics to uncover cellular mechanisms and identify or validate new drug targets. Applying CRISPR-Cas editing to animals could lead to better models of human disease, more predictive safety testing, and improved stratification and treatment regimens for patients. Rapid gene editing and regulation also promises to enable innovative therapies for non-genetic diseases through the generation of customized autologous cellular treatments, including cancer-seeking T cells and reprogrammed induced pluripotent stem cells (iPSCs). Although CRISPR-Cas systems will undoubtedly further improve, and new complementary or orthogonal methods will be developed to deliver reagents and edit somatic tissues directly in humans, we believe gene editing will have one of its most immediate impacts in drug development. CRISPR-Cas-aided discovery, validation and safety testing allows acceleration and improvement of known protocols and pipelines, without the need to solve delivery or redefine administrative procedures. CRISPR-Cas will be key to the next generation of transformational therapies and treatment paradigms.
Supplementary Material
Key points.
CRISPR-Cas tools are easily programmable RNA-guided nucleases, derived from microbial adaptive immune systems, that enable rapid genome engineering in vitro and in vivo.
Paired with the rapid expansion of genomic information, CRISPR-Cas enables facile genetic manipulation even in previously difficult contexts such as human cells. Gene knockout via error-prone repair works well in nearly all cell types, whereas knockin via homology-directed repair is more variable.
CRISPR-Cas holds the promise to transform the discovery and development of therapies to treat complex heritable and somatic disorders. Early applications in the field of cancer immunotherapy are entering clinical trials.
CRISPR-Cas gene editing expedites the generation of accurate cellular and animal models of human disease to facilitate drug discovery and validation. CRISPR-Cas is applicable to all major species used during a typical pre-clinical drug development campaign.
The low barrier to deploying CRISPR-Cas technology has enabled its rapid spread throughout the scientific community and is revolutionizing biomedical research. CRISPR-Cas systems are excellent tools for large-scale functional screens using gene knockout (CRISPRn), inhibition (CRISPRi) and activation (CRISPRa).
Further evolution of CRISPR-Cas9 may enable cures for Mendelian diseases in somatic tissues by directly correcting the underlying disease-causing mutations. Pioneering work with ZFN and TALEN based therapies will inform the path to therapeutic gene editing with CRISPR-Cas.
Acknowledgments
We thank members of the Doudna and Corn labs, as well as Fyodor Urnov, for insightful comments and discussions. C.F. is supported by a K99/R00 Pathway to Independence Award (K99GM118909) from the National Institutes of Health (NIH), National Institute of General Medical Sciences (NIGMS). The IGI is supported by the Li Ka Shing Foundation.
Glossary
- CRISPRa
fusing transcriptional activators to catalytically inactivated Cas9 to increase the expression of an RNA
- CRISPRi
fusing transcriptional repressors to catalytically inactivated Cas9 to decrease the expression of an RNA
- CRISPRn
targeting a DNA sequence with catalytically active Cas9 to generate a double stranded break or a nick
- Protospacer adjacent motif (PAM)
Short genomic sequence adjacent to the sequence targeted by the guide RNA that is required for recognition by Cas effectors. This sequence varies based on the effector’s identity (e.g. Cas9 vs. Cpf1) and species (e.g. Streptococcus pyogenes vs. Francisella novicida)
Biographies
Christof Fellmann is a Postdoctoral Fellow in the Doudna Lab at UC Berkeley and an NIH Pathway to Independence Awardee of the National Institute of General Medical Sciences (NIGMS). His research focuses on the molecular mechanism and therapeutic application of RNA-guided immune systems, including RNA interference and CRISPR-Cas tools. He is a co-founder and former Chief Scientific Officer of Mirimus, a start-up company developing genetically engineered mouse models of human disease for early stage drug discovery. Christof enjoys the outdoors and is a passionate triathlete.
Benjamin Gowen is an IGI-AstraZeneca Postdoctoral Fellow in Jacob Corn’s lab at the Innovative Genomics Initiative, University of California, Berkeley, Berkeley, USA. His research at the Innovative Genomics Initiative uses high-throughput screens to understand gene regulation in T cells. Benjamin received his PhD in Molecular and Cell Biology from UC Berkeley in 2015, where he worked with Professor David Raulet to study the regulation of cell surface proteins that activate natural killer cells. Benjamin gets all of his good ideas while walking his dog.
Pei-Chun Lin was a postdoctoral fellow in the Department of Molecular and Cell Biology, University of California, Berkeley, USA, and is currently supported by the University of California, San Francisco Dermatology T32 grant. Her research uses next generation genome editing and regulation technologies to develop a comprehensive system biology approach to immunotherapy for squamous cell carcinomas (SCCs), using a mouse skin tumour model system with heterogeneous mutational load that mimics clinical progression of the equivalent human tumors. Pei-Chun received her PhD from Cornell University, and worked as an IGI-AstraZeneca Postdoctoral Fellow from 2014 to early 2016, where she worked with Dr. Jacob Corn and used CRISPR-Cas9 mediated high-throughput screens in lung cancer cells. Pei-Chun likes traveling and spending time with her family. She is now at the Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, USA.
Jacob Corn is the Scientific Director of the Innovative Genomics Institute. His lab studies the development and application of next-generation genome editing and regulation technologies to tackle inherited and somatic diseases. Over the last fifteen years, Jacob has bridged academia and industry, using biophysics, computational biology, biochemistry, and cell biology in therapeutic areas that include oncology and neurodegeneration. Jacob is committed to the improvement of human health through better fundamental understanding of disease mechanisms. Jacob likes being outside and is an avid backpacker.
Jennifer Doudna is an HHMI Investigator and the Li Ka Shing Chancellor’s Chair and professor of Molecular and Cell Biology and Chemistry at University of California, Berkeley, USA. Her lab studies the biology and molecular mechanisms of CRISPR adaptive bacterial immune systems, development and applications of CRISPR systems for genome editing and RNA-mediated regulation of cellular protein synthesis. She is an advocate for responsible use of gene editing and she fosters research and educational partnerships through her role as Executive Director of the Innovative Genomics Initiative at UC Berkeley and UCSF.
Footnotes
Further information
Clinical trials: www.clinicaltrials.gov
NIH RAC on clinical trials of CRISPR-edited CAR-T cells: http://osp.od.nih.gov/office-biotechnology-activities/event/2016-06-21-123000-2016-06-22-164500/rac-meeting
Clinical trials with CRIPSR: https://clinicaltrials.gov/ct2/results?term=CRISPR&Search=Search
Biogen and Sangamo’s programme for BCL11A: http://investor.sangamo.com/releasedetail.cfm?ReleaseID=912987
NIH RAC on safety of CRISPR-edited CAR-T cells: http://osp.od.nih.gov/office-biotechnology-activities/event/2013-09-11-120000-2013-09-12-213000/rac-meeting, http://osp.od.nih.gov/sites/default/files/1240_Zaia.pdf
ASH 57th Annual Meeting, 2015. Abstracts 202: https://ash.confex.com/ash/2015/webprogramscheduler/Paper78442.html and 3233: https://ash.confex.com/ash/2015/webprogramscheduler/Paper78470.html
Competing Interests Statement
J.A.D. is employed by HHMI and works at the University at California, Berkeley. UC Berkeley and HHMI have patents pending for CRISPR technologies on which J.A.D. and J.E.C. are inventors. J.A.D. is the executive director and J.E.C is the scientific director of the Innovative Genomics Initiative at UC Berkeley and UCSF. J.A.D. is a co-founder of Editas Medicine, Intellia Therapeutics and Caribou Biosciences, and a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics and Driver. J.E.C. is a consultant to or has funded research collaborations with AstraZeneca, CRISPR Therapeutics, Editas Medicine, Genentech, Intellia, and Pfizer.
References
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–3. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 7.Cox DBT, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–31. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 9.Wright AV, Nuñez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 11.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–73. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–23. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 16.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981;78:6354–8. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 18.Lin FL, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–34. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasin M, de Villiers J, Weber F, Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell. 1985;43:695–703. doi: 10.1016/0092-8674(85)90242-9. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–8. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.TCGA. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.TCGA, Cancer, T., Atlas, G., Network, T. C. & TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2013;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 30.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thon N, Kreth S, Kreth FW. Personalized treatment strategies in glioblastoma: MGMT promoter methylation status. Onco Targets Ther. 2013;6:1363–72. doi: 10.2147/OTT.S50208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 33.Sur S, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 35.Yun J, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumann K, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112:10437–42. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grobarczyk B, Franco B, Hanon K, Malgrange B. Generation of Isogenic Human iPS Cell Line Precisely Corrected by Genome Editing Using the CRISPR/Cas9 System. Stem Cell Rev. 2015;11:774–787. doi: 10.1007/s12015-015-9600-1. [DOI] [PubMed] [Google Scholar]
- 39.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–62. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 40.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 41.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–5. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 42.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–3. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–7. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasap C, Elemento O, Kapoor TM. DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets. Nat Chem Biol. 2014;10:626–8. doi: 10.1038/nchembio.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smurnyy Y, et al. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat Chem Biol. 2014;10:623–5. doi: 10.1038/nchembio.1550. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Ye Y, Sun H, Shi G. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer Chemother Pharmacol. 2013;71:265–72. doi: 10.1007/s00280-012-2005-9. [DOI] [PubMed] [Google Scholar]
- 47.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paix A, et al. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 2014;198:1347–56. doi: 10.1534/genetics.114.170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paix A, Schmidt H, Seydoux G. Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res. 2016;44:e128. doi: 10.1093/nar/gkw502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–44. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 51.Paquet D, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–9. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 52.Nakade S, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakuma T, Nakade S, Sakane Y, Suzuki KIT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–33. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 54.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–4. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishida K, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 56.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol. 2014;16:10–8. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deans RM, et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016;12:361–6. doi: 10.1038/nchembio.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jae LT, et al. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science. 2014;344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–6. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 61.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–26. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 62.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 65.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Munoz DM, et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- 67.Aguirre AJ, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–6. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–63. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–60. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mandegar MA, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18:541–53. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc Natl Acad Sci U S A. 2015;112:E3384–91. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–46. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–7. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zalatan JG, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–8. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chakraborty S, et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem cell reports. 2014;3:940–7. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–71. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chavez A, et al. Comparison of Cas9 activators in multiple species. Nat Methods. 2016;13:563–7. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sidik SM, et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell. 2016;166:1423–1435.e12. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–7. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–8. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen S, Lee B, Lee AYF, Modzelewski AJ, He L. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J Biol Chem. 2016;291:14457–67. doi: 10.1074/jbc.M116.733154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang W, et al. Delivery of Cas9 Protein into Mouse Zygotes through a Series of Electroporation Dramatically Increases the Efficiency of Model Creation. J Genet Genomics. 2016;43:319–27. doi: 10.1016/j.jgg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin W, et al. Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease. Genetics. 2015;200:423–30. doi: 10.1534/genetics.115.176594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Premsrirut PKK, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 90.Nagy A, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–21. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 91.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–4. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanchez-Rivera FJ, Jacks T, Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–11. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–3. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–7. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–7. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DuPage M, Jacks T. Genetically engineered mouse models of cancer reveal new insights about the antitumor immune response. Curr Opin Immunol. 2013;25:192–9. doi: 10.1016/j.coi.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li D, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681–3. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 103.Zou Q, et al. Generation of gene-target dogs using CRISPR/Cas9 system. J Mol Cell Biol. 2015;7:580–3. doi: 10.1093/jmcb/mjv061. [DOI] [PubMed] [Google Scholar]
- 104.Niu Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–43. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 105.Chen Y, et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet. 2015;24:3764–74. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang L, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–4. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 107.Matsuoka Y, Lamirande EW, Subbarao K. The ferret model for influenza. Curr Protoc Microbiol. 2009;Chapter 15(Unit 15G.2) doi: 10.1002/9780471729259.mc15g02s13. [DOI] [PubMed] [Google Scholar]
- 108.Clark S, Hall Y, Williams A. Animal models of tuberculosis: Guinea pigs. Cold Spring Harb Perspect Med. 2015;5:a018572. doi: 10.1101/cshperspect.a018572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–97. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–5. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–83. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 112.Gori JL, et al. Delivery and Specificity of CRISPR-Cas9 Genome Editing Technologies for Human Gene Therapy. Hum Gene Ther. 2015;26:443–51. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 113.O’Geen H, Yu AS, Segal DJ. How specific is CRISPR/Cas9 really? Curr Opin Chem Biol. 2015;29:72–8. doi: 10.1016/j.cbpa.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bolukbasi MF, Gupta A, Wolfe SA. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Methods. 2016;13:41–50. doi: 10.1038/nmeth.3684. [DOI] [PubMed] [Google Scholar]
- 115.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–85. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS One. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–5. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–3. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 120.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–6. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–82. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–5. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–8. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316–8. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotech. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 132.Oakes BL, et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotechnol. 2016;34:646–51. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maus MV, Grupp SA, Porter DL, June CH. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123:2625–35. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Torikai H, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119:5697–705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qasim W, et al. First Clinical Application of Talen Engineered Universal CAR19 T Cells in B-ALL. Blood. 2015;126:2046 LP-2046. [Google Scholar]
- 136.Torikai H, et al. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood. 2013;122:1341–9. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lloyd A, Vickery ON, Laugel B. Beyond the antigen receptor: editing the genome of T-cells for cancer adoptive cellular therapies. Front Immunol. 2013;4:221. doi: 10.3389/fimmu.2013.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15:235–47. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 139.Cyranoski D. Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535:476–477. doi: 10.1038/nature.2016.20302. [DOI] [PubMed] [Google Scholar]
- 140.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12:51–8. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 141.Kalos M, June CH. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity. 2013;39:49–60. doi: 10.1016/j.immuni.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–10. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hütter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–8. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 144.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–16. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Holt N, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–47. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–61. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mandal PK, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–52. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hoban MD, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang J, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.DeWitt MA, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cao A, Galanello R. Beta-thalassemia. Genet Med. 2010;12:61–76. doi: 10.1097/GIM.0b013e3181cd68ed. [DOI] [PubMed] [Google Scholar]
- 152.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–7. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vierstra J, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12:927–30. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roberts SA, et al. Engineering Factor Viii for Hemophilia Gene Therapy. J Genet Syndr gene Ther. 2011:1. doi: 10.4172/2157-7412.S1-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stripecke R, et al. Immune response to green fluorescent protein: implications for gene therapy. Gene Ther. 1999;6:1305–12. doi: 10.1038/sj.gt.3300951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.