Abstract
Significant progress has been made in recent years for computer-aided diagnosis of abnormal pulmonary textures from computed tomography (CT) images. Similar initiatives in chest radiographs (CXR), the common modality for pulmonary diagnosis, are much less developed. CXR are fast, cost effective and low-radiation solution to diagnosis over CT. However, the subtlety of textures in CXR makes them hard to discern even by trained eye. We explore the performance of deep learning abnormal tissue characterization from CXR. Prior studies have used CT imaging to characterize air trapping in subjects with pulmonary disease; however, the use of CT in children is not recommended mainly due to concerns pertaining to radiation dosage. In this work, we present a stacked autoencoder (SAE) deep learning architecture for automated tissue characterization of air-trapping from CXR. To our best knowledge this is the first study applying deep learning framework for the specific problem on 51 CXRs (≈ 76.5%, F-score) and a strong correlation with the expert visual scoring (R=0.93, p =< 0.01) demonstrate the potential of the proposed method to characterization of air trapping.
1. INTRODUCTION
Chest radiographs (CXRs) are the most commonly used imaging technique for pulmonary diagnosis; primarily because of speed of acquisition, low cost, portability, and low-radiation dosage compared to other medical imaging modalities such as computed tomography (CT) and magnetic resonance imaging (MRI). However, the progress of adopting computer-aided diagnosis (CAD) methods in the analysis of CXRs has not been as significant as in other modalities. So far the majority of research in the development of CAD techniques for CXRs is mainly focused on lung nodule detection [1]. Nodules are a rare find in lungs, particularly in pediatrics. On the other hand, CXR is the mainstay imaging modality in general, and particularly certain population groups particularly in pediatrics. Air trapping is often used as a surrogate signature of airway obstruction in asthma [2] The use of CAD techniques for air trapping tissue characterization from CXR can translate into clinical settings as an inexpensive, readily available, and safer tool since CXRs are performed routinely in children. Fig. 1 shows typical manifestations of air trapping in CXR which is an increasingly recognized biomarker correlating with clinical severity in asthma [2], and chronic obstructive pulmonary disease [3].
Fig. 1.
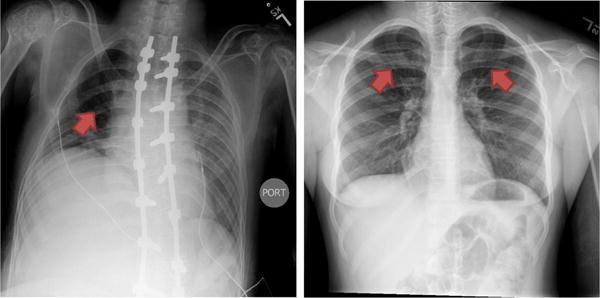
Examples of air trapping manifestation (pointed with red arrows) in CXR image. Air trappings appear as dark lung areas that correspond to abnormal retentions of air that is not fully exhaled.
In this work, we propose a fully automated method for the tissue characterization of air trapping from CXR using the stacked autoencoder (SAE) hierarchical deep learning architecture [4, 5]. Our proposed framework is aimed at leveraging the untapped potential of the deep learning framework as a sensitive and reliable extractor of imaging biomarkers for lung diseases. To the best of our knowledge, this the first major study on characterizing air trapping textured pulmonary abnormalities in pediatrics using CXR and the first study exploring the potential of deep learning architecture in pulmonary tissue characterization from CXR. The proposed automatic characterization method consists of the following major steps: (1) the lung field is segmented (excluding the the retrocardiac space) and the intensities of the CXR are standardized. (2) a probability image map of pixels belonging to air trapping within the lung field is obtained using SAE, (3) differential diagnosis is performed by mapping the SAE probability to the severity score based on the area and density of air trapping. The rest of the paper describes the steps in detail.
2. METHODS
Recent literature has presented deep learning methods for texture classification in the medical imaging after their successful performance for large scale color image classification [6]. For instance, in [7], a modified restricted Boltzmann machine was used for feature classification of lung tissue in CT images. The work in [8] used a convolutional neural network with one convolutional layer and three dense layers to classify lung image patches from CT images. Recently, the authors of [9] proposed a design of convolutional neural network for the classification of interstitial lung disease patterns in CT scans. Specific to CXR, most recently [10] expanded the potential of deep architectures for pathology detection (pleural effusion, enlarged heart, and normal versus abnormal) using CXR.
Our proposed framework for tissue characterization from CXR using the deep learning framework is shown in Fig. 2. The method consists of lung field delineation and intensity standardization as pre-processing steps followed by tissue classification using the SAE framework. The lung field is then partitioned into four quadrants per lung and mapped to an air trapping severity score based on expert training. The details of individual modules of the proposed method are discussed in greater details in the following subsections.
Fig. 2.
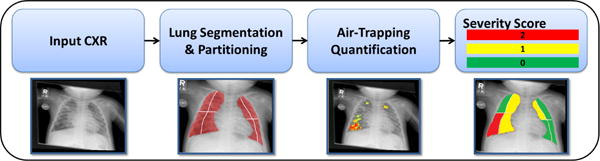
The pipeline for automated tissue characterization from CXR (localization and severity quantification).
2.1. Data
Fifty-one pediatric posterior-anterior CXRs from patients with viral chest infections were collected at Children’s National Health System using multiple scanners. The radiographs had dimensions ranging from 660×987 to 660×4240 pixels having a spatial resolution ranges between 0.1mm×0.1mm to 0.14mm×0.14mm and a digital resolution of 12-bits. The ground truth of air trapping was performed through visual inspection by two expert pulmonologists. Each lung was divided into 4 regions (distal/proximal, upper/lower) and the severity of air trapping was recorded on a scale of 0–2 (0 = no air trapping, 1 = mild air trapping, and 2 = dense air trapping) for each region. The ground truth for training the deep learning classifier was prepared manually by two experienced raters using the ITK-Snap interactive software (http://www.itksnap.org/).
2.2. Lung Field Segmentation using Weighted Partitioned ASM
The first step in our proposed framework is the delineation of lung field from CXR. For air trapping tissue characterization task, we excluded the the retrocardiac space (region of the lung field behind the heart) from the lung field, since air trapping and other pathological patterns are not clearly discernible in those regions using CXR. For the lung field segmentation, we used our previous method described in [11]. Briefly, the method uses a weighted-shape partitioning approach by dividing the lung field into a set of partial shapes and learning a statistical shape model for each partition separately. The approach allows local flexibility in the model to adapt to shape variability. Then, the lung field landmarks are divided into various shape and appearance consistent partitions using fuzzy c-means clustering. Next, a local appearance model consisting of three appearance features is constructed for every partition: (i) normalized intensity derivatives, (ii) three class fuzzy c-means, and (iii) elongated rib structure probability using the vesselness filter. The presence of pathologies and various acquisition parameters makes certain shape landmarks less reliable than others; therefore, each landmark is assigned a relative weight based on their reliability calculated using the local appearance features. At the next step, statistical shape model fitting is performed individually for each partition and the optimal position for each landmark was determined by minimizing the Mahalanobis distance. Lastly, the final lung field segmentation is computed by adding the different partitions and averaging the shape parameters of the overlapping landmarks.
2.3. Image Intensity Standardization
Due to the non-standard nature of CXR image intensities, images collected using different scanners and protocol settings exhibit different intensity ranges that pose a challenge for automated classification techniques. Using intensity standardization (mapping the acquired data to a predefined intensity profile) as a pre-processing step can alleviate this challenge. In [12], we presented a generic method for the intensity standardization of 2D/3D medical images using the multiscale curvelet transform [13]. During the training phase, the reference data are first decomposed into scale and orientation localized subbands using the multiscale curvelet transform, followed by calculating a reference energy value for each subband. During the testing stage, the localized energy of each subband is iteratively scaled to the reference localized energy value from the training stage. In our proposed approach for air trapping tissue characterization, we use the method to standardize the intensity of test CXRs within the extracted lung field region only.
2.4. Stacked Autoencoder Hierarchical Deep Learning
A deep learning paradigm begins by learning low-level features and hierarchically building up comprehensive high-level features in a layer-by-layer fashion. SAE is a deep neural network consisting of multiple layered sparse autoencoders (AE) in which the output of every layer is wired to the input of the successive layer. An AE consists of two components: the encoder and the decoder. The encoder aims to fit a non-linear mapping that can project higher-dimensional observed data (f (xn)∈ ℝn) into its lower dimension feature representation. The decoder aims at recovering the observed data from the feature representation with minimal recovery error. For a detailed description of AE, SAE, and related deep learning concepts, reader is encouraged to review [4,5].
The encoder step
Given an L–dimensional observed data , the encoder maps to an M –dimensional activation vector , through a deterministic mapping , where W∈ ℝM×L is the mapping matrix is the bias vector, and σ is the logistic sigmoid function .
The decoder step
The activation vector is decoded back to the observed data vector using the reverse deterministic mapping, i.e., where en∈ ℝM is reconstruction error and is the bias vector. Furthermore, sparsity constraints on the M hidden nodes can lead to a set of compact representative features. By switching, a hidden node m as “active” if hn(m)→1 or “inactive” if hn (m) → 0, a compact set of input can be achieved for each training input . Finally, by integrating the sparsity constraint, the objective function for AE can be formulated as an optimization problem:
| (1) |
where is the average activation of hidden layers and β denotes the strength of sparsity constraint.
2.5. Image Patch Sampling and Augmentation
Next we extract patches from the intensity standardized lung field for training the SAE. The training patches are divided into two categories: (1) a positive category, if the center pixel belongs to the air trapping region according to the ground-truth label and (2) a negative category from areas of lung field not belonging to air trapping. Since negative samples outnumber the positive samples, the class imbalance is accommodated using the random sampling. Moreover, data augmentation is employed in the feature classification literature to artificially enlarge the dataset. In our framework we performed augmentation by extracting patches and their horizontal reflection from the training [6].
2.6. Severity Scoring
One of the main reason most CAD methods do not translate to clinical settings is their inability to fit into existing clinical practices and nomenclatures. For instance, air trapping pathologies in CXR are assessed in clinics using subjective visual scores on the scale of 0–2 for each lung quadrant [14]. In order to provide a translation of air trapping tissue characterization performed by our framework to expert visual score, we translate the severity characterization to visual score (0–2) per lung quadrant using the training data and based on the area of detected air trapping as shown in Fig. 3.
Fig. 3.
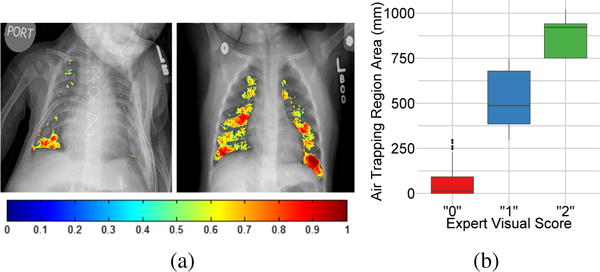
(a) Lung field air trapping tissue characterization from CXR using our method. The quantified air trapping is shown as a color map with colors indicating the density of air trapping. (b) Box plots demonstrating the correlation between the air trapping area obtained using the proposed deep learning framework and the expert pulmonologist visual score.
3. RESULTS
Our method was implemented using MATLAB-based Toolbox for Deep Learning (https://github.com/rasmusbergpalm/DeepLearnToolbox). Routines not involving deep learning were also implemented using MATLAB. All experiments were performed on 64-bit Windows 7 operating system using a machine with Intel E5-1607 CPU @3.00 GHz and 16.0 GB of RAM.
To test our framework, the dataset was split randomly at patient level to 31 training and 20 validation CXRs. An average Dice similarity coefficient (DSC) of 0.91±0.17 was obtained for lung field segmentation. For air-trapping characteization, the patch size was set to 11×11 for both training and validation and 3 layers of SAE were employed. The sparsity penalty was set to 0.1 and the number of epochs used were 100. The image patch size as well as the parameters of the SAE classifier were adjusted based on the empirically obtained best performance on the training data set. Fig. 3(a) shows the probability maps of air trapping obtained using the proposed method and overlaid over the CXR image. Fig. 3(b) shows the box plots of expert visual scores against the amount of air trapping detected using the proposed method over the entire CXR validation data. A correlation coefficient R = 0.93 with the expert visual assessment of air trapping from CXR was obtained. Furthermore, the proposed configuration yielded an average F-score of 0.7648 based on two classes (air trapping and everything else within the lung field).
| (2) |
where and
4. CONCLUSION
We demonstrated the use of stacked autoencoder hierarchical deep learning architecture for the automated pulmonary tissue characterization from chest radiographs (CXR). The study is the first in proposing the use of low-radiation CXR images with image processing to detect and classify air trapping as an alternative to commonly used CT scans. Our study identified that regional air trapping can be detected and quantified in infants using CXRs with image analysis. Moreover, the proposed framework, evaluated against the expert visual score, showed promising results for the air trapping characterization and holds great potential to be extended to additional textural pulmonary patterns. Our future work include performing an extensive investigation on the correlation of quantification obtained using the proposed framework with CT data and extending the framework to other pulmonary pathologies.
Acknowledgments
This project was funded by NIH grants UL1TR000075/KL2TR000076 and a gift from the Government of Abu Dhabi to the Children’s National Health System.
References
- 1.van Ginneken B, Hogeweg L, Prokop M. Computer-aided diagnosis in chest radiography: Beyond nodules. European Journal of Radiology. 2009;72(2):226–230. doi: 10.1016/j.ejrad.2009.05.061. [DOI] [PubMed] [Google Scholar]
- 2.Busacker A, Newell JD, Keefe T, Hoffman EA, Granroth JC, Castro M, Fain S, Wenzel S. A multivariate analysis of risk factors for the air-trapping asthmatic phenotype as measured by quantitative CT analysis. CHEST Journal. 2009;135(1):48–56. doi: 10.1378/chest.08-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castaldi PJ, Dy J, Ross J, Chang Y, Washko GR, Curran-Everett D, Williams A, Lynch DA, Make BJ, Crapo JD, et al. Cluster analysis in the COPD gene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014 doi: 10.1136/thoraxjnl-2013-203601. pp. thoraxjnl–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengio Y, Lamblin P, Popovici D, Larochelle H, et al. Greedy layer-wise training of deep networks. Advances in Neural Information Processing Systems. 2007;19:153. [Google Scholar]
- 5.Larochelle H, Erhan D, Courville A, Bergstra J, Bengio Y. An empirical evaluation of deep architectures on problems with many factors of variation. Proceedings of the 24th International Conference on Machine Learning ACM. 2007:473–480. [Google Scholar]
- 6.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012:1097–1105. [Google Scholar]
- 7.van Tulder G, de Bruijne M. Learning features for tissue classification with the classification restricted Boltzmann machine. Medical Computer Vision: Algorithms for Big Data Springer. 2014:47–58. [Google Scholar]
- 8.Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M. Medical image classification with convolutional neural network. Control Automation Robotics & Vision (ICARCV), 2014 13th International Conference on; IEEE. 2014; pp. 844–848. [Google Scholar]
- 9.Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Transactions on Medical Imaging. 2016;(99):1–1. doi: 10.1109/TMI.2016.2535865. vol. PP. [DOI] [PubMed] [Google Scholar]
- 10.Bar Y, Diamant I, Wolf L, Greenspan H. Deep learning with non-medical training used for chest pathology identification. SPIE Medical Imaging International Society for Optics and Photonics. 2015:94 140V–94 140V. [Google Scholar]
- 11.Okada K, Marzieh G, Mansoor A, Perez GF, Pancham K, Khan A, Nino G, Linguraru MG. Severity quantification of pediatric viral respiratory illnesses in chest X-ray images. Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE. 2015:165–168. doi: 10.1109/EMBC.2015.7318326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansoor A, Cerrolaza J, Idrees R, Biggs E, Alsharid M, Avery R, Linguraru MG. Deep learning guided partitioned shape model for anterior visual pathway segmentation. IEEE Transactions on Medical Imaging. 2016;(99):1–1. doi: 10.1109/TMI.2016.2535222. vol. PP. [DOI] [PubMed] [Google Scholar]
- 13.Candes E, Demanet L, Donoho D, Ying L. Fast discrete curvelet transforms. Multiscale Modeling & Simulation. 2006;5(3):861–899. [Google Scholar]
- 14.Lucidarme O, Grenier PA, Cadi M, Mourey-Gerosa I, Benali K, Cluzel P. Evaluation of air trapping at CT: Comparison of continuous-versus suspended-expiration CT techniques. Radiology. 2000;216(3):768–772. doi: 10.1148/radiology.216.3.r00se21768. [DOI] [PubMed] [Google Scholar]


