Abstract
Measuring panels of protein biomarkers offer a new personalized approach to early cancer detection, disease monitoring and patients’ response to therapy. Multiplex electrochemical methods are uniquely positioned to provide faster, more sensitive, point of care (POC) devices to detect protein biomarkers for clinical diagnosis. Nanomaterials-based electrochemical methods offer sensitivity needed for early cancer detection. This review discusses recent advances in multiplex electrochemical immunosensors for cancer diagnostics and disease monitoring. Different electrochemical strategies including enzyme-based immunoarrays, nanoparticle-based immunoarrays and electrochemiluminescence methods are discussed. Many of these methods have been integrated into microfluidic systems, but measurement of more than 2–4 protein markers in a small single serum sample is still a challenge. For POC applications, a simple, low cost method is required. Major challenges in multiplexed microfluidic immunoassays are reagent additions and washing steps that require creative engineering solutions. 3-D printed microfluidics and paper-based microfluidic devices are also explored.
Keywords: Cancer biomarker protein, Multiplex, Electrochemical Immunosensor, Electrochemiluminescence, Microfluidic
1 Introduction
Biomarkers for cancer are broadly defined as measurable or observable factors that can be evaluated as indicators of normal biological processes, disease processes, or responses to a therapeutic intervention [1–3]. Biomarkers can include physical symptoms, secreted proteins, mutated DNAs and RNAs, cell death or proliferation, and serum concentrations of small molecules such as glucose or cholesterol. This review focuses on protein biomarkers that when present at elevated or depressed concentrations in serum, tissue, or saliva, can be indicative of disease states. The development of low cost, reliable methods for simultaneously measuring panels of protein bio-markers is critically important for early detection of cancer, disease monitoring and personalized cancer therapy [4–6]. Detection of panels of protein markers can minimize false positives and negatives in cancer diagnoses that can arise from measuring a single biomarker [3,5,6,7–12]. For example, PSA, an FDA approved cancer biomarker for prostate cancer, when detected as a single biomarker has positive predictive value ~70%. On the other hand, detecting 5 or more biomarkers for a given cancer by liquid chromatography mass spectrometry (LC-MS) has provided >99% reliable diagnostics [8, 10,13,14].
Another challenge is that many protein biomarkers are correlated to more than one form of cancer or disease conditions. For example, PSA in blood is elevated in some benign prostate diseases as well as prostate cancer [15]. Another biomarker protein, IL-6 is associated with several different cancers, including head and neck squamous cell carcinoma (HNSCC) as well as inflammation. IL-6 mean serum levels in healthy individuals are typically <6 pg mL−1, whereas in patients with HNSCC, the levels ≥20 pg mL−1 [16]. Serum IL-6 levels is also elevated in, gastrointestinal, lung, renal, multiple myeloma, prostate and colorectal cancers [16]. CEA, a biomarker protein most often correlated with colorectal cancer is also found at elevated levels in patients with breast cancer, ovarian cancer, and lung cancer. Health adults have mean CEA serum levels of 3–5 ng mL−1, although CEA serum levels of 10 ng mL−1 have been found in some benign diseases [17]. Thus, carefully selected panels of protein biomarkers with known individual over-expressions for specific cancers are needed for prediction success.
The broad range of clinically relevant concentrations for different biomarkers presents a technical complication in the development of multiplexed electrochemical protein detection. For example, PSA normal levels are typically 0.5 to 2 ng mL−1 and patients with early stage prostate cancer have levels of 4 to 10 ng mL−1 whereas as mentioned above, normal IL-6 mean serum levels are < 6 pg mL−1 compared to > 20 pg mL−1 in oral cancer patients. Thus, a frequent challenge involves measuring some proteins biomarkers in a panel at low levels of pg mL−1 and below, and others at higher levels, e.g. ng mL−1 and above.
Ideal immunosensor arrays for reliable point-of-care (POC) application should be simple, cost effective, fast and capable of measuring the selected panel of proteins [6] (4–10) at ultra-low and elevated levels in the same sample without interference from the thousands of other non-analyte proteins in serum. Enzyme-linked immunosorbent assay (ELISA) [18], has been the gold standard for clinical protein biomarker detection with limits of detection (LOD) 1–3 pg mL−1 [6,15,16,19] for many proteins, but is limited by sample size, relatively expensive test kits and equipment, significant assay time, difficulties in multiplexing, and so is not directly applicable to point of care (POC) diagnostics.
Alternatively, microarrays consisting of analytical spots on a chip with each spot having a selective capture agent for a specific biomarker are being developed. These arrays are simple, highly selective, and allow multiplexed measurement of proteins at reasonable cost [20–25]. The microarrays utilize ELISA strategies where the spots in the array may contain primary antibodies or aptamers to capture the desired analyte proteins, and after washing with detergents or casein to block non-specific binding, a labeled tracer antibody is added to bind to the analyte proteins (Fig. 1). The label provides an optical or electrical signal during measurement. This approach is suitable to early cancer detection and monitoring since it has the potential for relatively rapid high sensitivity determinations of limited panels of biomarkers in serum with good accuracy and precision [6,21].
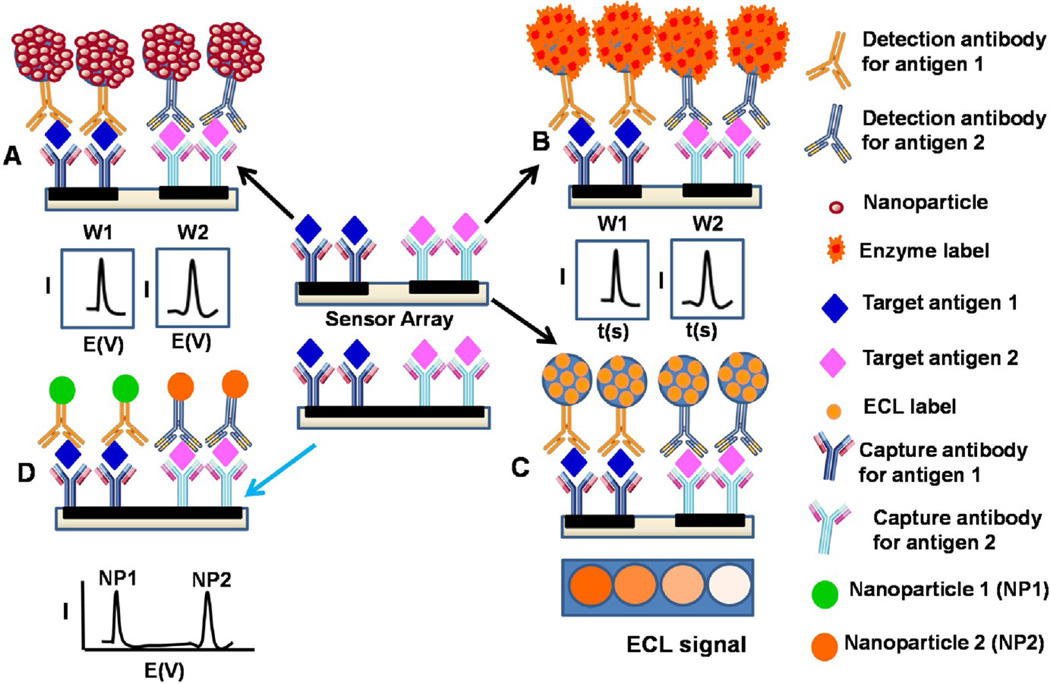
Fig. 1.
Strategies for multiplex electrochemical detection of proteins using a multi-electrode array with separate multiple microwells modified with different capture antibodies for different target biomarker proteins in a sandwich type assay; (A) coupled to multi-nanoparticle labels for enhanced sensitivity, (B) using multi-enzyme labels for enhanced sensitivity (C) Electrochemiluminescence tag and (D) an alternative approach using a single electrode well e.g. GCE or Magnetic beads with different nanocrystal labels or redox probes for distinguishable electrochemical signals. Multi-electrode arrays can also be integrated to microfluidic systems to reduce sample volume, automate the assay process and increase throughput.
This review focuses on different strategies for multiplex electrochemical detection of protein biomarkers, especially those applicable to POC for cancer detection and disease monitoring (see Table 1 for protein biomarkers and correlated cancer types). We begin with a brief overview of different multiplex electrochemical methods using enzyme labels or redox probes followed by a section on multiplex immunoassays using nanoparticle labels. Subsequent sections discuss multiplex immunosensors based on electrochemiluminescence, and microfluidic based immunosensors including paper based analytical methods for multiplex electrochemical detection of biomarker proteins.
Table 1.
Cancer biomarker proteins reported in this review and the most correlated cancer types.
| Cancer biomarker protein | Cancer | Ref. |
|---|---|---|
| Prostate specific antigen (PSA) | Prostate | [45,46] |
| Prostate specific membrane anti- gen (PSMA) |
Prostate | [47] |
| Platelet factor 4 (PF-4) | Prostate | [48] |
| Interleukin-6 (IL-6) | Oral cancers | [16] |
| Interleukin-8 (IL-8) | Oral cancers | [49] |
| Interleukin 1 beta (IL-1β) | Oral cancer mucositis | [50] |
| Carcinoembryonic antigen (CEA) | Colon | [46] |
| α-Fetoprotein (AFP) | Testicular | [45] |
| Vascular endothelial growth factor (VEGF) |
Oral cancer | [51] |
| Cancer antigen 125 (CA 125) | Ovarian | [45,46] |
| Cancer antigen 19-9 (CA 19-9) | Pancreatic | [45,46] |
| Cancer antigen 15-3 (CA 15-3) | Breast | [45,46] |
| β-human Chriogonadotropin (hCG-β) |
Testicular | [46] |
| Tumor necrosis factor (TNF-α) | Inflammation, several cancers including prostate, cervical |
[52] |
| C-reactive protein (CRP) | inflammation, lung cancer |
[53] |
2 Multiplex Electrochemical Immunosensor Arrays
Electrochemical approaches to POC devices offer the inherent ability to achieve high sensitivity with relatively simple measurement systems and electronics. Heineman’ group pioneered enzyme-linked electrochemical immunoassays for proteins [26]. They separated binding of antigens to capture antibodies in space and time from detection [27,28]. Enzyme label alkaline phosphatase produces electroactive products that are transported by a chromatographic or fluidic system to the electrode detector. Inter- digitated electrodes gave the highest sensitivities (pg mL−1 range). This group was among the first to integrate microfluidics into electrochemical protein immunoassays [29].
Two major strategies are being pursued for multiplex electrochemical protein detection. A barcode method where the secondary antibodies are attached to distinct nanoparticles with different characteristics, e.g. different quantum dots, or dissolvable metals that can be detected by stripping analysis. Alternatively, multi-electrode arrays are used in which each electrode is immobilized with a different antibody. Most strategies for multiplex electrochemical immunoassays have adapted the features of enzyme-linked immunosorbent assay (ELISA) (Fig. 1). This approach relies on a sandwich immunoassay whereby primary capture antibodies (Ab1) are attached to sensor elements. Sample is then introduced to the sensor, and the analyte proteins are selectively captured on the antibodies. This is followed by a washing to remove non-specifically bound constituents. Then a labeled secondary antibody (Ab2) is added, which binds selectively to the protein on the sensor (Fig. 1). Nanostructured sensor surfaces with increased electrode surface area can provide 10-fold or more antibody coverage compared to flat surfaces leading to improved sensitivity and detection [22–25,30]. Recent advances in nanomaterials-assisted assays by our research teams [31] and others [30,32–35] have improved multiplexed protein sensitivity up to 1000-fold compared to commercial assays.
Electrochemical detection can be achieved by measuring electroactive enzyme reaction products or by catalytic reduction of a substrate using an enzyme [23,36–38] like horseradish peroxidase (HRP). Other amplified electrochemical detection strategies have included nanoparticle labels on secondary antibodies that can be dissolved to yield detectable electroactive ions, nanoparticle labels that are used to catalyze subsequent metal deposition, and substrates for enzyme labels that deposit polymer products on the sensor (see Table 2 and 3) [22,23,26–28,39,40]. Detection methods include amperometry, voltammetry, stripping analysis and impedance [41].
Table 2.
Comparative summary of approaches for multiplex electrochemical detection of cancer biomarker proteins
| Approach | Label | Biomarkers | Detection Limit | Linear Range | Ref. |
|---|---|---|---|---|---|
| Amperometry | HRP-Ab2 | IL-8 | 7.4 pg/mL | 0.01–12.5 ng/mL | [54] |
| HRP-Anti fluorescein | IL-8-mRNA | 3.9 fM | 0.005–50 pM | ||
| MWNT-HRP-Ab2 | IL-8 | 8 pg/mL | 8–1000 pg/mL | [55] | |
| PSA | 5 pg/mL | 5–000 pg/mL | |||
| MP-HRP-Ab2 | PSA | 0.23 pg/mL | 0.225–5 pg/mL | [87] | |
| IL-6 | 0.30 pg/mL | 0.3–20 pg/mL | |||
| HRP-Ab2 | CEA | 1.1 µg/L | 1.1–188 µg/L | [57] | |
| AFP | 1.7 µg/L | 1.7–250 µg/L | |||
| β-hCG | 1.2 IU/L | 1.2–256 IU/L | |||
| CA 125 | 1.7 kIU/L | 1.7–334 kIU/L | |||
| ALP-Ab2 | AFP | 1.4 ng/mL | 75–300 ng/mL | [76] | |
| Ferritin | 7.0 ng/mL | 50–200 ng/mL | |||
| CEA | 1.2 ng/mL | 12.5–100 ng/mL | |||
| hCG-β | 1.8 ng/mL | 12.5–100 ng/mL | |||
| CA 15-3 | 0.7 U/mL | 75–300 ng/mL | |||
| CA 125 | 1.2 U/mL | 12.5–100 ng/mL | |||
| CA 19-9 | 1.0 U/mL | 25–100 ng/mL | |||
| HRP-Ab2 | CEA | 0.2 ng/mL | 0.2–200 ng/mL | [85] | |
| CA 15-3 | 5.2 U/mL | 5.2–100 U/mL | |||
| Ab2-MB-HRP | IL-6 | 5 fg/mL | 5–1000 fg/mL | [88] | |
| IL-8 | 10 fg/mL | 10–1000 fg/mL | |||
| VEGF | 5 fg/mL | 5–1000 fg/mL | |||
| VEGF-C | 50 fg/mL | 50–1500 fg/mL | |||
| Ab2-MB-HRP | IL-6 | 5 fg/mL | 5–2500 fg/mL | [89] | |
| IL-8 | 7 fg/mL | 7–3750 fg/mL | |||
| Ab2-MB-HRP | IL-6 | 18 fg/mL | 0.018–4.5 pg/mL | [50] | |
| TNF-α | 10 fg/mL | 0.01–12 pg/mL | |||
| CRP | 15 fg/mL | 0.015–11 pg/mL | |||
| IL-1β | 40 fg/mL | 0.04–22 pg/mL | |||
| Square Wave Voltammetry | AuNR-HRP-Ab2 | p53392 | 5 pM | 0.01–20 nM | [59] |
| p5315 | 20 pM | 0.05–20 nM | |||
| p5346 | 30 pM | 0.1–50 nM | |||
| p53 | 10 pM | 0.05–20 nM | |||
| NSC-Ab2 | CA 125 | 0.08 mU/mL | 0.0001–100 U/mL | [98] | |
| CA 199 | 0.10 mU/mL | 0.0001–100 U/mL | |||
| Cyclic voltammetry | AuNR-HRP-Ab2 | PSA | 0.1 ng/mL | 0.1–10 ng/mL | [60] |
| IL-6 | 5 pg/mL | 5–1000 pg/mL | |||
| PSMA | 0.8 ng/mL | 0.8–400 ng/mL | |||
| NiFe2O4-SiO2-Ab2 | AFP | < 0.5 µg/L | 2–180 µg/L | [84] | |
| CEA | < 0.5 µg/L | 1.5–200 µg/L | |||
| CA 125 | < 0.5 µg/L | 3.2–60 kU/L | |||
| CA 15-3 | < 0.5 µg/L | 2.5–240 kU/L | |||
| NGC-McAb2 | CEA | 0.06 pg/mL | 0.0001–100 ng/mL | [97] | |
| AFP | 0.08 pg/mL | 0.0001–100 ng/mL |
NSC – Nanoporous silver chitosan; ngNGS – Nanoporous gold chitosan; MP – Superparamagnetic particle
Table 3.
Comparative summary of approaches for multiplex electrochemical detection of cancer biomarker proteins
| Approach | Label | Biomarkers | Detection Limit | Linear Range | Ref. |
|---|---|---|---|---|---|
| Electrogenerated Chemiluminescence | QD-SiO2-Ab2 | AFP | 5 pg/mL | 0.01–50 ng/mL | [79] |
| CEA | 6 pg/mL | 0.01–50 ng/mL | |||
| PSA | 3 pg/mL | 0.01–50 ng/mL | |||
| RuBPY-Si-PLL-Ab2 | CA 199 | 0.01 U/L | 0.03–80 U/L | [80] | |
| CEA | 0.02 pg/mL | 0.05–100 pg/mL | |||
| Th-SiO2-Ab2 | AFP | NA | 0.07–14 ng/mL | [81] | |
| PSA | NA | 0.0005–100 ng/mL | |||
| QDs-Ab2 | AFP | 0.4 fg/mL | 1–100 fg/mL | [82] | |
| CEA | 0.4 fg/mL | 1–100 fg/mL | |||
| (P-acid)-NPS | CEA | 0.8 pg/mL | 0.003–20 ng/mL | [96] | |
| PSA | 1 pg/mL | 0.001–10 ng/mL | |||
| RuBPY-SiO2-Ab2 | IL-6 | 0.25 pg/mL | 0.00025-2 ng/mL | [83] | |
| PSA | 1 pg/mL | 0.001–10 ng/mL | |||
| RuBPY-SiO2-Ab2 | IL-6 | 100 fg/mL | 0.0001-5 ng/mL | [92] | |
| PF-4 | 10 fg/mL | 0.0001-5 ng/mL | |||
| PSA | 50 fg/mL | 0.0001-1 ng/mL | |||
| PSMA | 100 fg/mL | 0.0001-10 ng/mL | |||
| RuBPY-SiO2-Ab2 | PF-4 | 420 fg/mL | 0.0005-10 ng/mL | [95] | |
| PSA | 300 fg/mL | 0.0005-10 ng/mL | |||
| PSMA | 535 fg/mL | 0.0005-10 ng/mL | |||
| Differential Pulse Voltammetry | AuNPs-HRP-Ab2 | CA 153 | 0.2 kU/L | 0.4–140 kU/L | [43] |
| CA 125 | 0.5 kU/L | 0.5–330 kU/L | |||
| CA 199 | 0.3 kU/L | 0.8–190 kU/L | |||
| CEA | 0.1 µg/L | 0.1–44 µg/L | |||
| Fc-Au-HRP-Ab2 | AFP | 1 pg/mL | 0.01–200 ng/mL | [44] | |
| Thi-Au-HRP-Ab2 | CEA | 1 pg/mL | 0.01–80 ng/mL | ||
| HRP-Ab2 | CA 19-9 | 0.2 U/mL | 0.2–144 U/mL | [42] | |
| CA 125 | 0.4 U/mL | 0.4–150 U/mL | |||
| CNT-AuNP-GOx-Ab2 | AFP | 2.2 pg/mL | 0.0025–2.5 ng/mL | [56] | |
| CEA | 1.4 pg/mL | 0.0025–2.0 ng/mL | |||
| AuNP-HRP-Ab2 | CA 153 | 0.06 U/mL | 0.084–16 U/mL | [58] | |
| CA 125 | 0.03 U/mL | 0.11–13 U/mL | |||
| CA 199 | 0.10 U/mL | 0.16–15 U/mL | |||
| CEA | 0.04 ng/mL | 0.16–9.2 ng/mL | |||
| SiO2-GOx-Ab2 | CEA | 3.2 pg/mL | 0.005-2 ng/mL | [62] | |
| AFP | 4.0 pg/mL | 0.005-1 ng/mL | |||
| rGO-Au-Thi-Ab2 | CEA | 0.650 pg/mL | 0.01–300 ng/mL | [64] | |
| rGO-Au-PB-Ab2 | AFP | 0.885 pg/mL | 0.01–300 ng/mL | ||
| PtPNPs-Cd-Ab2 | CEA | 0.002 ng/mL | 0.05–200 ng/mL | [71] | |
| PtPNPs-Cu-Ab2 | AFP | 0.05 ng/mL | 0.05–200 ng/mL | ||
| PBNP-Au-Ab2 | CEA | 0.01 ng/mL | 0.01–100 ng/mL | [72] | |
| Cd-Au-Ab2 | AFP | 0.006 ng/mL | 0.01–100 ng/mL | ||
| Stripping Voltammetry | AuNP-Ab2 | CEA | 3.5 pg/mL | 5–5000 pg/mL | [67] |
| AFP | 3.9 pg/mL | 5–5000 pg/mL | |||
| CNT-Ag-Ab2 | CEA | 0.093 pg/mL | 0.1–5000 pg/mL | ||
| AFP | 0.061 pg/mL | 0.1–5000 pg/mL | [70] | ||
| SA-Au-CNH-Ab2 | AFP | 0.024 pg/mL | 0.1–1000 pg/mL | ||
| CEA | 0.032 pg/mL | 0.1–1000 pg/mL | [73] |
2.1 Immunosensor Arrays Based on Enzyme-labels and Redox Probes
Wu et al. demonstrated a low cost, two-analyte disposable immunosensor array for simultaneous electrochemical determination of cancer antigens, CA 19-9 and CA 125 with a limit of detection (LOD) of 0.2 and 0.4 U/ml, respectively [42].
This array was fabricated using cellulose acetate membrane to co-immobilize thionine as a mediator and the two kinds of antigens on two screen printed carbon electrodes (SPCE) followed by a competitive immunoreaction on HRP labelled antibodies captured on the membrane which subsequently generated the electronic signal via catalytic reduction of H2O2 substrate. Serum sample results were in acceptable agreement with a reference method. This group also used colloidal gold nanoparticles with bound horseradish peroxidase (HRP)-labeled with various antibodies that were immobilized on 4 elements of a SPCE with biopolymer/sol-gel to trap their corresponding target antigens from sample solution.
Electrochemical signals decreased with antigen binding enabling simultaneous detection of cancer antigens, CA153, CA 125, carbohydrate antigen 199, with LODs of 0.2–0.5 kU/L and carcinoembryonic antigen, CEA, LOD of 0.1 µg/L, in clinical serum samples [43].
Tang et al. demonstrated a flow through multiplex immunoassay for CEA and AFP using biofunctionalized magnetic graphene nanosheets (MGO) immunosensor platform and (HRP)-thionine and HRP-ferrocene nano-gold hollow microspheres (GHS-Ab1 and GHS-Ab2) tags to provide distinguishable signal [44]. The assay was based on the catalytic reduction of H2O2 at the various peak potentials in the presence of the corresponding mediators. The flow-through detection cell was coupled to an external magnet control and provided a wide linear range of 0.01–200 ng mL−1 for AFP and 0.01–80 ng mL−1 for CEA and a LOD of 1 pg mL−1 for both analytes in serum samples. Clinical serum sample results gave good correlation with an ECL reference method.
Wong et al. demonstrated an electrochemical immunosensor for oral cancer detection based on multiplex detection of two salivary biomarkers: interleukin (IL)-8 mRNA and IL-8 protein [45]. The sensor was fabricated on a 16 element chip where each unit of the array features sensor, counter, and reference electrodes. Biotin and fluorescein dual labeled hairpin probe for IL-8 mRNA and biotinylated capture antibody for IL-8 were immobilized on different sensors coated with streptavidin modified dendrimer nanoparticles on a conducting polymer under layer. Target analytes were detected in a sandwich type sensor featuring HRP conjugated anti-IL8 and anti-fluorescein-HRP. The LOD of salivary IL-8 mRNA were 3.9 fM and 7.4 pg mL−1 for IL-8 protein in saliva.
Wan et al. demonstrated a similar strategy consisting of a 16 channel SPCE with each element containing a carbon working electrode, a carbon counter and a silver reference electrode, and used it for multiplex electrochemical detection of PSA and IL-8 cancer protein biomarkers (Fig. 2) [46]. The capture antibodies were immobilized on different electrochemically activated working electrode elements by EDC/NHS bioconjugation chemistry. A sandwich type immunoassay was coupled to a multi-labeled nanoprobe consisting of MWNT loaded with HRP and goat anti-rabbit IgG (secondary antibody, Ab2). This method demonstrated a detection limit of 5 pg mL−1 for PSA and 8 pg mL−1 for Interleukin 8 (IL-8).
Fig. 2.
Illustration of SPCE array with multiple elements for multiplex electrochemical detection of proteins. The working electrode elements are modified with different capture antibodies for a sandwich type assay coupled to universal MWNT bioconjugates with multi-enzyme labels for enhanced sensitivity. Adapted with permission from ref. [55]. Copyright 2011 Elsevier.
Lai et al. demonstrated a multiplex immunoarray based on glucose oxidase loaded Au/SWNT bioconjugate coupled to nanostructured disposable SPCE array [47]. The array consisted of 2 working carbon electrodes sharing a carbon counter and Ag/AgCl reference electrode. The sandwich immunoarray was fabricated by coating layer-by-layer colloidal Prussian blue (PB), gold nanoparticles, and capture antibodies on screen-printed carbon electrodes. The PB acts as a mediator in the catalytic reduction of H2O2. The immunoassay gave LODs of 1.4 for CEA and 2.2 pg mL−1 for alpha feto-protein, AFP. Results in serum samples were in good agreement with a reference method.
Wu et al. used an 8 element SPCE array coated with a redox mediator in a biopolymer film in a flow cell electrochemical system to simultaneously detect carcinoembryonic antigen, α-fetoprotein, β-human choriogonadotropin, and carcinoma antigen 125 as model cancer biomarkers [48]. The protein analytes mixed with redox mediator and biopolymer were immobilized on the electrode and used to capture corresponding HRP labeled antibodies in competitive immunoreactions with target biomarker proteins in a sample. This multiplex immunosensor approach had a throughput of 60 samples/h and provided LODs of 1.1 µg/L, 1.7 µg/L, 1.2 IU/L, and 1.7 kIU/L in serum samples for carcinoembryonic antigen, α-fetoprotein, beta-human choriogonadotropin, and carcinoma antigen 125, respectively.
Wu et al. also reported a fast, simple, sensitive, low-cost method for multianalyte immunosensing by combining electric field-driven incubation with a SPCE array [49]. Different HRP-labeled antibodies modified with gold nanoparticles in biopolymer/sol-gel were immobilized on 4 different sensors of the array. A mixture of target ana-lytes was added on each spot on the array and an electric field applied to shorten incubation to 2 min. Responses from HRP catalyzed reduction of H2O2 decreased with analyte concentration. This enabled simultaneous detection of carbohydrate antigens 153, 125, and 199 and CEA in less than 5 min, with LODs 0.06, 0.03, and 0.10 U mL−1 and 0.04 ng mL−1, respectively. Du et al. used similar electrical driven incubation to shorten assay time, and demonstrated a multiplexed electrochemical immunoassay for detection of phosphorylated p53 at Ser392 (phospho-p53392), Ser15 (phospho-p5315), Ser46 (phospho-p5346), and total p53 simultaneously [50]. A disposable 4-sensor array was coated with different capture antibodies on each spot. The sandwich immunoassay was facilitated by coupling captured target analytes with gold nanorods (AuNRs) carrying multiple HRP and detection antibodies (Ab2). Reduction of HRP oxidized thionine in the presence of hydrogen peroxide provided the response. This less than 5 min immunoassay simultaneously detected phospho-p53392, phospho-p5315, phospho-p5346, and total p53 with LODs of 5–30 pM. Accuracy was determined by good correlation with ELISA in serum samples.
Liu et al. reported multiplexed immunoarray on a flexible polydimethylsiloxane (PDMS) slice deposited with 8 × 8 nano-Au film electrodes for simultaneous detection of prostate specific antigen (PSA), prostate specific membrane antigen (PSMA), and interleukin-6 (IL-6) [51]. Primary antibodies linked with magnetic beads (Ab1-MBs) were magnetically collected on the nano-Au film electrodes. Upon binding the corresponding antigens, gold nanorods conjugated with HRP, detection antibodies were bound, and the H2O2 reduction signal generated. LODS were PSA (0.1 ng mL−1), IL-6 (5 pg mL−1), and PSMA (0.8 ng mL−1) in serum samples.
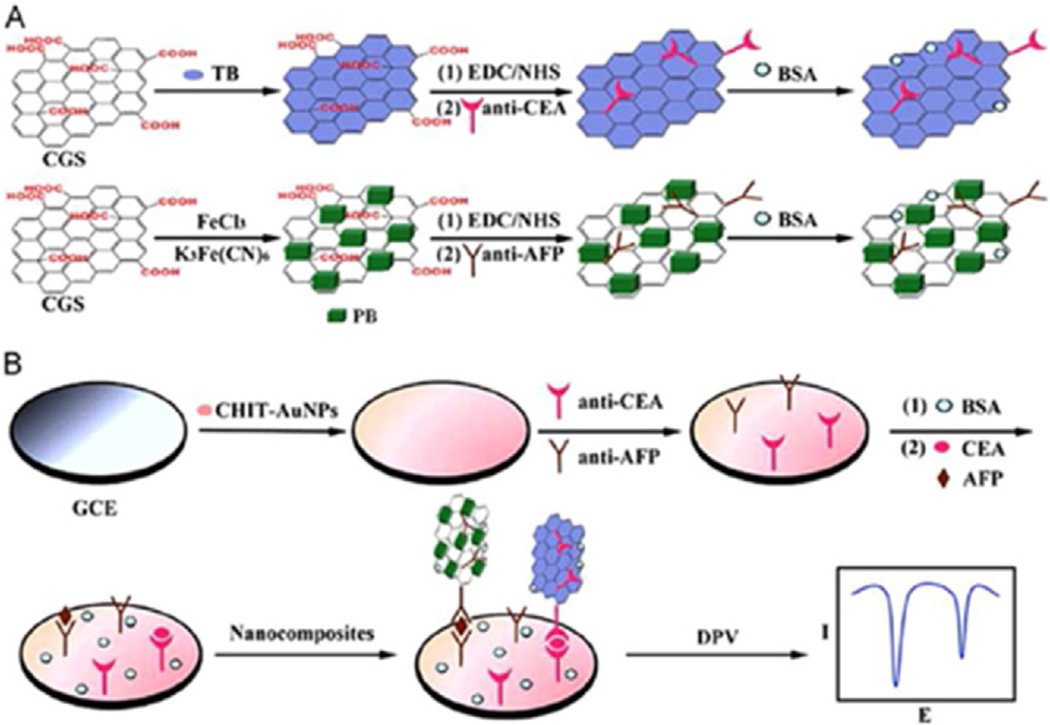
Chen et al. demonstrated a sandwich electrochemical immunosensor system for simultaneous determination of the carcinoembryonic antigen (CEA) and alpha fetoprotein (AFP) utilizing bio-functional carboxyl graphene nanosheets as immunosensor probes. These probes were created by immobilizing toluidine blue and anti-CEA, for detection of CEA antigen, and Prussian blue and anti-AFP, for detection of AFP antigen, respectively (Fig. 3).
Fig. 3.
Multiplex electrochemical detection using a single well glassy carbon electrode (GCE) modified with a mixture of capture antibodies for different target biomarker proteins. (A) preparation of biofunctional graphene tag containing 2 different redox probes, toluidine blue and Prussian blue for distinguishable signals, and (B) multiplexed electrochemical immunoassay protocol. Reprinted from ref. [61]. Copyright 2013 Elsevier.
The mixed capture antibodies for both CEA and AFP were immobilized on Chitosan Au nanoparticles coated on glassy carbon electrode (GCE). Linear range was 0.5–60 ng mL−1 for both CEA and AFP with LOD of 0.1 ng mL−1 for CEA and 0.05 ng mL−1 for AFP (S/N = 3). Results for human serum sample gave good correlation with standard ELISA [52].
Lai et al. fabricated a multiplex immunosensor for electrochemical detection of CEA and AFP using silica nanoparticles loaded with glucose oxidase labels [53]. Capture antibodies were attached to 2 elements on a carbon screen printed electrode array modified with gold nanoparticles assembled on carbon nanotubes-chitosan. The signal was generated via mediated catalytic oxidation of glucose.
The immunoassay in the microwells gave LODs of 3.2 pg mL−1 for CEA and 4.0 pg mL−1 for AFP with good agreement for serum sample results with a reference method.
Xu et al. demonstrated a multiplexed sandwich immunosensor for the simultaneous detection of three bio-markers using square wave voltammetry. The biosensor platform was built on a GCE modified with ionic liquid reduced graphene oxide, IL-GO and polystyrenesulfonate layer upon which a mixture of three capture antibodies specific to the target biomarkers, AFP, CEA and PSA were immobilized. After addition of the target biomarkers, electrochemical detection was archived using carbon gold nanocomposite (CGN) bioconjugates containing the respective secondary antibodies and different redox probes, thionine, 2,3-diaminophenazine (DAP) and Cd2+ which gave discrete signals for the three target biomarkers. This method gave LODs of 2.7 pg mL−1 for CEA, 4.8 pg mL−1 for PSA and 3.1 pg mL−1 for AFP. Serum samples also gave good correlation with ELISA [54].
Jia et al. developed a label free multiplex electrochemical immunosensor for detection of CEA and AFP utilizing indium tin oxide modified with graphene nanocomposites. Anti CEA and Anti AFP capture antibodies were immobilized on separate units on the ITO coated with rGO/thionine/AuNP and rGO/Prussian blue/AuNP respectively to generate distinguishable signals. This immunosensor approach was based on the fact that due to the formation of antibody–antigen immunocomplex, the decreased response currents of thionine and Prussian Blue were directly proportional to the concentrations of corresponding antigens. This system showed a linear working range of 0.01–300 ng mL−1 for both antigens with LODs for CEA and AFP of 0.650 pg mL−1 and 0.885 pg mL−1 respectively. Accuracy was assessed by measurements of CEA and AFP in clinical patient samples that gave good correlation with ELISA [55].
2.2 Nanoparticle-Labeled Immunosensor Arrays
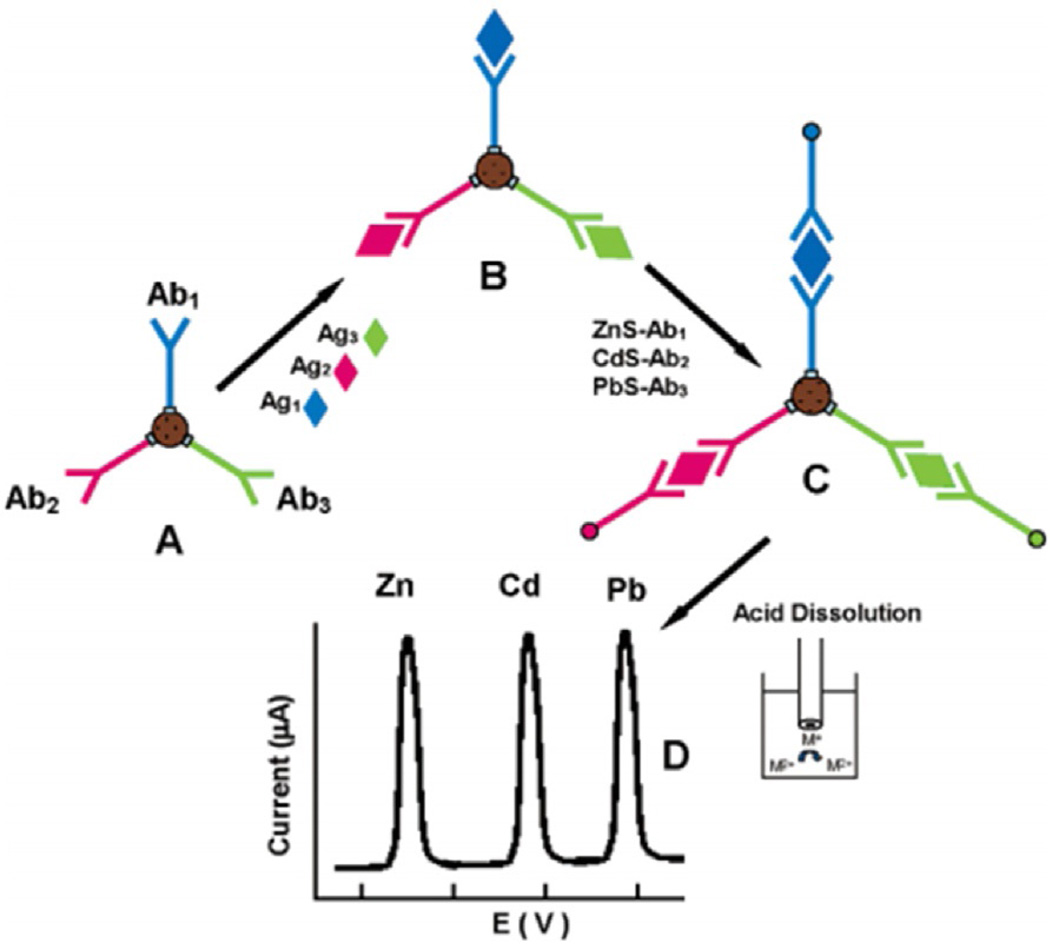
Unique properties of nanoparticles including large surface-to-volume ratio enable higher binding capacities in biomedical applications. Multiplex immunoassays based on nanoparticle labels rely on either the barcode approach where detection antibodies (Ab2) are modified with different metal nanoparticles or nanocrystals that can be later dissolved and the metal ions detected by anodic stripping voltammetry. Alternatively, multi-electrode arrays with different antibodies can be coupled to secondary antibodies with metal nanoparticles that are detected by linear sweep voltammetry without dissolution. Nanoparticles can greatly enhance sensitivity. In a pioneering approach, Wang et al. demonstrated an electrochemical immunoassay for simultaneous measurement of 4 proteins. Individual secondary antibodies labeled with zinc sulfide, cadmium sulfide, copper sulfide or lead sulfide quantum dots (Qdots) were used to detect four different proteins by sandwich immunoassay (3 proteins shown in Fig. 4) [56]. A magnetic bead immunocomplex consisted of the four Qdots, each attached to a specific antibody for a different protein. After antigen binding, Qdots were dissolved in acid and the electroactive metals ions detected by stripping voltammetry. This approach gives distinguishable stripping peak potentials for each target protein similar to single analyte protein [57]. The 4 protein panel consisting of C-reactive protein, IgG, bovine serum albumin and β2-microglobulin were simultaneously measured with ultra-low LODs in the femtomole range.
Fig. 4.
Multiplex electrochemical detection based on Qdots for distinguishable signals. (A) Different capture antibodies attached to a magnetic bead, followed by (B) binding of the corresponding target antigens on the magnetic beads; (C) Binding of the Qdot-labeled secondary antibodies to form a sandwich type assay (D) acid dissolution of Qdots and subsequent electrochemical stripping detection. Adapted from ref. 65. Copyright 2004 American chemical society.
Lai et al. demonstrated a sensitive immunosensor for multiplex electrochemical detection of CEA and AFP protein biomarkers based on stripping of silver nanoparticles (AgNP). Capture antibodies were attached on 2 separate sensors on a chitosan modified SPCE array. Then, in a sandwich type assay, secondary antibody-functionalized AuNP were used to induce silver deposition for enhanced sensitivity measured by anodic stripping analysis. This method showed linear ranges of three orders of magnitude with LODs of 3.5 pg mL−1 for CEA and 3.9 pg mL−1, for AFP [58]. This group also demonstrated a multiplexed immunoassay with electrochemical stripping analysis of silver nanoparticles catalytically deposited by gold nanoparticles and enzymatic reaction [59]. Using the same platform to attach the capture antibodies the immunoreaction was coupled to ALP-Ab2/Au NPs that catalyzed hydrolysis of 3-indoxyl phosphate producing an indoxyl intermediate to reduce Ag+. Silver deposition was catalyzed by both ALP and AuNPs to amplified the signal. Human and mouse IgG as model analytes were simultaneously measured by stripping voltammetry with LODs of 4.8 and 6.1 pg mL−1, respectively.
Wang et al. demonstrated a multiplex sandwich immnosensor based on poly-l-lysine (PLL) nanotag coupled with dual antibodies captured on magnetic beads for electrochemical detection of AFP and CEA cancer biomarkers. The PLL was activated with Au nanoparticles upon which metallic apoferritin, Cd-APO and Pb-APO, along with detection antibodies, Anti-AFP and Anti-CEA, were immobilized. Electrochemical stripping of the metallic components produced distinguishable peaks at different potentials enabling simultaneous detection of AFP and CEA biomarkers within solution. This approach gave a dynamic range of 0.01–50 ng mL−1 for both AFP and CEA with LODs of 4 pg mL−1 and good correlation of serum sample results with standard ELISA method [60].
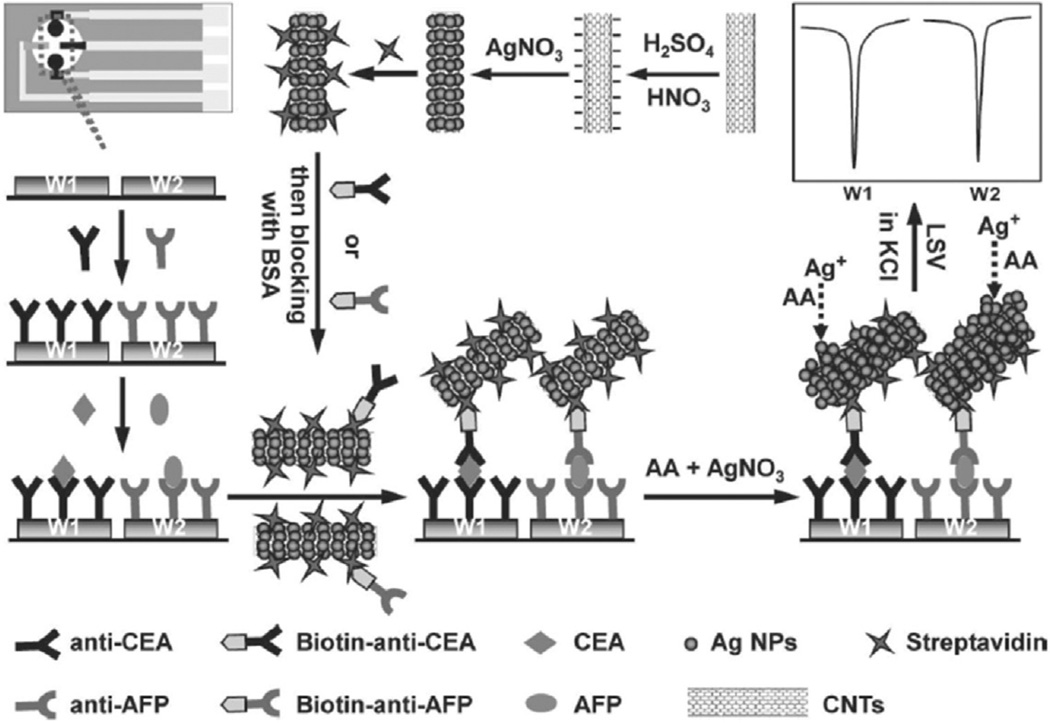
Lai et al. reported a streptavidin-functionalized silver-nanoparticle-enriched carbon nanotube (CNT/Ag NP) tag for ultrasensitive multiplexed measurements of tumor markers using a SPCE immunosensor array [61]. Capture antibodies were covalently attached to different chitosan-modified SPCE and coupled to CNT/AgNP tag in a sandwich assay to measure AFP and CEA biomarker proteins (Fig. 5). High sensitivity electrochemical stripping signal of Ag NP was achieved via numerous Ag NPs that are captured onto every single immunoreaction and was further enhanced using silver enhancing solution. This multi-analyte electrochemical detection approach gave a wide linear range over four orders of magnitude with LODs down to 0.093 pg mL−1 for CEA and 0.061 pg mL−1 for AFP. Results from serum samples gave good correlation with ELISA method.
Fig. 5.
Schematic representation of sandwich type nanoparticle-based immunoarray fabricated on 2 separate microwells with different capture antibodies for different target biomarker detection. Schematic shows preparation of multi-nanoparticles trace tag, and detection strategy by linear-sweep stripping voltammetric analysis of AgNPs on the immunosensor surface without acid dissolution. Adapted from reference 70. Copyright 2011 Wiley.
Ma et al. used porous platinum nanoparticles hybrid (PtPNPs) with attached metal ion labels for simultaneous detection of 2 cancer biomarkers, CEA and AFP in a sandwich type immimosensor with the capture antibodies attached to Ionic liquid reduced graphene oxide (IL-rGO) modified glassy carbon electrode [62]. In this approach, 2 separate batches of PtPNP were labeled with Cd2+ and Cu2+ and bioconjuated to Anti-CEA and Anti-AFP respectively to provide distinguishable signals upon binding on the target proteins bound on to the capture antibodies. The metal ions corresponding to the analytes were detected by DPV without acid dissolution. This immunoassay gave linear range, 0.05 ng mL−1 to 200 ng mL−1 for both CEA and AFP and with LOD of 0.002 ng ml−1 for CEA and 0.050 ng mL−1 for AFP, (S/N = 3). Serum sample results gave good correlation with ELISA. This group also used Au-ionic liquid functionalized reduced graphene oxide nanocomposite (IL-rGO-Au) coated on GCE to develop a sandwich immunoarray for simultaneous detection of CEA and AFM tumor markers [63]. Mixed primary antibodies were attached on the electrode and target proteins captured. In sandwich immunoassay, mixed secondary antibody, Ab2 conjugates containing chitosan (CS) coated Prussian blue/Au nanoparticles (PB-CS-Au) and cadmium hexacyanoferrate/Au nanoparticles (Cd-CS-Au) for the 2 target proteins were used to provide distinguishable signals. This method gave a LOD of 0.01 ng mL−1 for CEA and 0.006 ng mL−1 for AFP. Furthermore, serum sample results were in good agreement to ELISA.
Zhao et al. demonstrated an ultrasensitive multiplexed immunoassay using streptavidin/nanogold/carbon nanohorn (SA/Au/CNH) as a novel signal tag to induce silver enhancement for signal amplification [64]. The sandwich immunosensor array was prepared on disposable SPCE where the capture antibodies were immobilized on 2 separate electrode elements. Upon binding of the specific cancer biomarker protein, antigens on the capture antibodies, biotinylated antibody conjugates were added to allow for biotin-streptavidin affinity reaction, involving SA/Au/CNH tag specially designed to induce silver deposition to amplify the electrochemical stripping signals. This immunosensor gave a wide linear range with LODs of AFP (0.024 pg mL−1) and CEA (0.032 pg mL−1) with good correlation of serum samples results with a reference method.
Wilson et al. developed an immunosensor array where pairs of iridium oxide electrodes were used to covalently attach capture antibodies for two proteins detection [65]. A 2.5 mm gap between the working electrodes eliminated cross talk enabling simultaneous electrochemical immunoassay using alkaline phosphatase-labeled Ab2 and detection of the electroactive enzyme product hydroquinone.
Detection limits were ~1 ng mL−1 for cancer biomarkers carcinoembryonic antigen (CEA), α-fetoprotein (AFP), goat IgG, and mouse IgG. Additionally, this concept was extended to an 8-electrode immunosensor array for simultaneous electrochemical detection [66] of goat IgG, mouse IgG, human IgG, and chicken IgY with LODs of ~3 ng mL−1. The method was validated vs. ELISA method using synthetic sera containing the 4 proteins. A chip-based sensor consisting of an array of 8 iridium oxide sensing electrode was fabricated in to a 12-well plate and used for simultaneous electrochemical detection of a panel of seven tumor markers, [67] AFP, ferritin, CEA, hCG-β, CA 15-3, CA 125, and CA 19-9 with LODs<2 ng mL−1 and excellent precision and accuracy (1.9–8.1% interassay CV). A different antigen was immobilized on each electrode and used for electrochemical detection of specific protein cancer biomarker in an enzyme-based competitive immunoassay.
3 Electrochemiluminescence (ECL) Methods
ECL is a method in which light is emitted from a high energy dye during a special electrochemical reaction featuring a co-reactant [68,69]. ECL methods offer advantages including low cost, ease of use, high sensitivity and selectivity, and the lack of need for individual electrically addressed sensors. The detection of biomarkers can be done using an immunochemical sandwich assay featuring a nanoparticle ECL tag. ECL tags vary but one commonly used ECL label for multiplex ECL-based cancer biomarker protein detection is (2,2’-bipyridyl) ruthenium (II) (RuBPY) doped silica nanoparticles modified with secondary antibodies [25]. RuBPY emits a photon of light when a voltage is applied due to a complex redox pathway with a suitable co-reactant such as tripropylamine (TPrA). This amplification strategy provides a large ECL signal for a small concentration of antigen, which allows very low LODs. Qdots can also serve as ECL tags. These nanoparticles emit light when excited and the wavelength of the emitted light is dependent on the particle’s composition and size, typically between 2–6 nm. The variation of emitted wavelength can be used to distinguish one QD from another and thus can be used to multiplex an immunoarray by using different sized QDs for different biomarkers. ECL signal capture is generally done using a charged coupled device (CCD) camera or a photomultiplier tube (PMT). Commercially available ECL systems for the detection of biomarkers are available [25] but there are issues for clinical use such as sensitivity and high cost.
One strategy for a multiplexed ECL-based immunoarray was developed by Zhang et al. for the simultaneous detection of CEA, PSA and AFP. They used carbon Qdots on silica nanoparticles labeled with secondary antibodies as a detection bioconjugate [70]. A flow injected sandwich assay was constructed on separate indium tin oxide glass arrays as the detection platform. The ECL signal was detected using a photomultiplier giving LODs for CEA (6 pg mL−1), PSA (3 pg mL−1) and α-AFP (5 pg mL−1) in diluted serum samples.
Jiang’s group reported an ECL detection method that utilized screen-printed electrodes to perform a multiplexed assay of CEA and CA199 [71]. The sensor was modified with AuNPs and the antigen bound to it by the electro-polymerization of the electrode with the biomarker using the dopamine monomer. RuBPY silica nanoparticles modified with poly-L-lysine (PLL) were used to attach gold nanoparticles. PLL served as a bridging agent for AuNPs as well as a co-reactant for the RuBPY. ECL was detected by a PMT. LODs were 0.02 pg mL−1 for CEA and 0.01U/L for CA199. Patient’s serum sample results were in good agreement with ELISA method.
Wu et al. demonstrated a multiplexed biosensor using a closed bipolar electrode (BPE) [72]. ITO electrodes were used with Au film modification on the cathodes. Thionine-doped silica nanoparticles was used as the ECL tag for signal amplification and as a recognition probes for mediating the ECL of Ru(bpy)32+/tripropylamine (TPA) on the anodes of BPE array. The system used an antibody sandwich assay to detect AFP and PSA, and also as a DNA based aptamer set up to detect ATP and thrombin. This system provided a linear detection region of 70 pg mL−1 to 14 ng mL−1 for AFP and 0.5 pg mL−1 to 100 ng mL−1 for PSA. Results of PSA in human serum gave good correlation with a reference method, chemiluminescent analyzer (Abbott i2000 system).
Guo et al. reported utilizing multicolor Qdots for multiplexed detection of cancer biomarkers, [73] with emission λmax values 525 nm and 625 nm. These Qdots were decorated with AFP and CEA antibodies, respectively. This assay distinguished the signal of one antigen from another based on wavelength of the ECL. The array used a GCE electrode modified with graphene to increase electron transfer to the QDs. This assay was able to detect both biomarkers at ultralow LODs of 0.4 fg mL−1 in serum. It uses a single well in which the concentration of two analytes can be determined simultaneously by distinguishing the wavelengths emitted.
Rusling’s group has worked extensively with RuBPY/ TPrA ECL reactions with using a CCD camera for ECL signal detection. Earlier work described the fabrication of an ECL device on a pyrolytic graphite chip, featuring carboxylic acid functionalized single walled carbon nanotube forests constructed in the bottom of microwells (~ 10 µL) on the PG chip. Capture antibodies were attached to the SWNTs and a RuBPY-silica nanoparticle decorated with secondary antibodies was used for detection. This system detected PSA and IL-6 biomarkers simultaneously in 16 micro wells. Therefore, controls and multiple sample replicates could be run simultaneously. LODS of 1 pg mL−1 and 0.25 pg mL−1 in diluted calf serum were obtained for PSA and IL-6 respectively using this system [74].
4 Microfluidic Immunoarrays and Micro Paper Based Analytical Devices
For POC applications, overall assay time, sample volume, and automation are important factors that need to be addressed. Integration of immunoassay systems with micro-fluidics can decrease the overall assay times, sample volumes, and for introducing pump-free reagent delivery. Several groups have made significant progress towards this research area.
Tang et al. fabricated an electrochemical magnetically controlled microfluidic device consisting of magnet core/ shell NiFe2O4/SiO2 nanoparticles for detection of 4 tumor markers, CEA, AFP, CA125 and CA15–3 [75]. The microfluidic system is fabricated on glass substrate consisting of 5 gold disk working electrodes modified with NiFe2O4/SiO2 nanoparticle for enhanced sensitivity and an Ag/AgCl reference electrode. Capture antibodies are immobilized on the NiFe2O4/SiO2 nanoparticle to measure a specific tumor marker using noncompetitive immunoassay. The detection principal is based on potential shift before and after antibody-antigen interaction. Under optimal conditions, the multiplex immunoassay enabled the simultaneous detection of 4 tumor markers with LODs of <0.5 µg mL−1.
Kellner et al. developed a fully automated microfluidic system for simultaneous electrochemical detection of breast cancer biomarkers, CEA and CA15-3. This computer-controlled system integrates reagent storage with electrochemical detection making it simple, accurate and more efficient than manually operated microfluidic systems. They compared the analytical performance of a manual microfluidic system to the fully housed and automated system by simultaneous measurement of carcinoembryonic antigen (CEA) and cancer antigen-15-3 (CA-15-3), in human serum. The fully automated system provided enhanced accuracy, and reproducibility as indicated by a reduction of LOD for CEA and CA15-3 detection when compared to the manual driven system. Results for CEA spiked serum samples also gave excellent correlation with ELISA [76].
Hwang et al. demonstrated a proof of concept of an immunosensor array based on a microfluidic biochip fabricated on glass substrate for simultaneous electrochemical detection of MMP2 and MMP7 biomarkers for ovarian and colorectal cancers. The biochip is integrated with a Au working electrode and a Pt counter/reference electrode within photoresist channels etched on the glass substrate. The system relies on unlabeled peptides specific to the 2 MMPs which are immobilized onto the sulfur activated Au working electrodes and impedance of the system is then measured. Upon injection of MMP samples in the system, the corresponding peptides are cleaved by enzyme hydrolysis and then the impedance is measured again. The difference between the impedances before and after injection of the MMPs can be directly correlated to the concentration of the MMPs present in the sample. The overall assay time was reduced to approximately one hour and the system yielded a detection range of 0.1–400 ng mL−1 for MMP2 and 0.001–100 ng mL−1 for MMP7 [77].
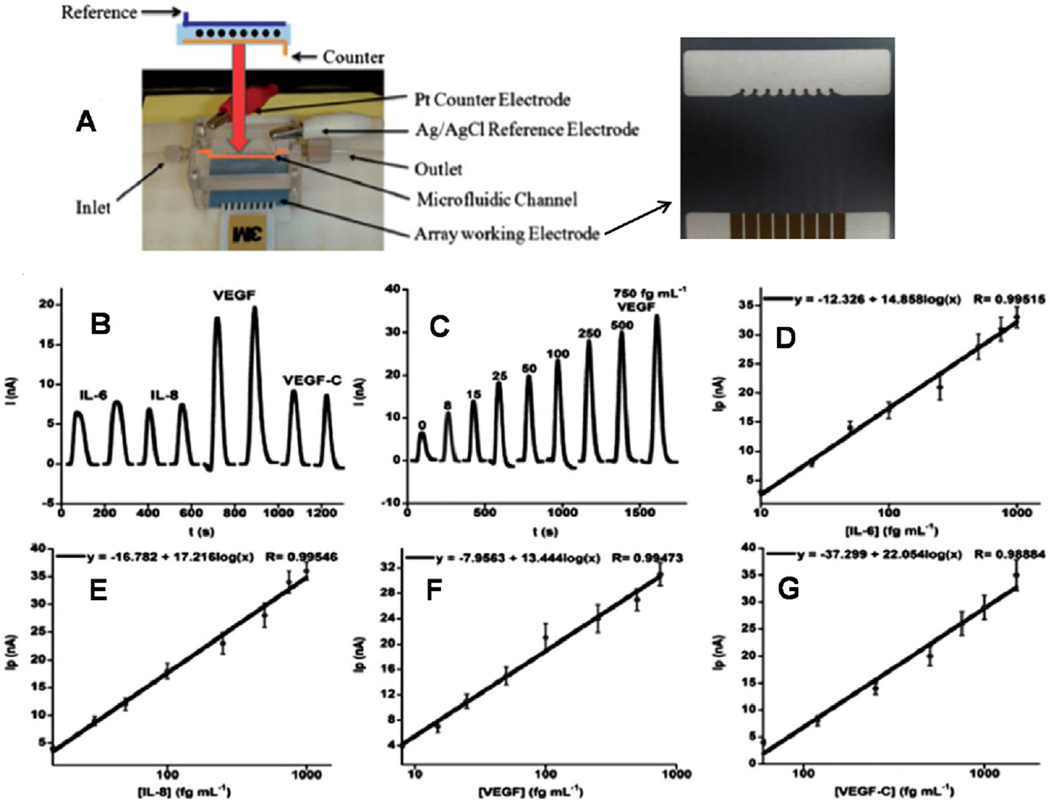
Rusling et al. integrated a partly automated microfluidic system featuring a capture and a detection channel made from PDMS encased in polymethylmethacrylate plastic. The 8-sensor array was positioned in a PDMS microfluidic channel containing a platinum (Pt) wire as the counter electrode and a silver/silver chloride (Ag/AgCl) wire reference electrode (Fig. 6). This semi-automatic detection device featured off-line capture of analytes by heavily-HRP-labeled 1 µm superparamagnetic particle (MP)-antibody bioconjugates and capture antibodies attached to an 8-electrode measuring chip modified with 5 nm AuNPs to enhance sensitivity. The immunosensor with 1.15 h assay time was used for simultaneous detection of cancer biomarker proteins, PSA and interleukin-6 (IL-6) in serum with LODs of 0.23 pg mL−1 and 0.30 pg mL−1 in diluted serum, respectively. Results also gave excellent correlation with standard enzyme-linked immunosorbent assays (ELISA) for patient samples [78]. In another microfluidic immunosensor, a SWNT modified 8 electrode array with capture antibodies was coupled to a massively labeled bioconjugate and used for multiplexed electrochemical detection of IL-6, IL-8, VEGF and VEGF-C in diluted serum samples with ultra-low LODs of 5–50 fg mL−1 (see Fig. 6). The high sensitivity in this 50 min immunosensor assays was achieved by using off-line protein capture by magnetic beads carrying 400,000 enzyme labels and 120,000 antibodies [79]. The high enzyme/antibody loading was realized via streptavi-din-biotin interactions. The accuracy of this approach was assessed by good correlations with ELISA for protein determinations in conditioned cancer cell media. A Normalized Mean concentration derived from the 4 protein levels in patient serum samples (78 oral cancer and 49 controls) reveled high diagnostic utility giving clinical sensitivity of 89 % and specificity of 98 % for cancer prediction. Recently, Rusling et al. included a protein capture chamber into the above microfluidic immunoarray [80]. Target proteins in serum are delivered to and captured on HRP-Ab2-magnetic beads in a stirred PDMS capture chamber controlled by an external magnet. Removal of the magnet and valve switching enabled magnetic bead protein bioconjugates to flow into the 8 antibody-decorated gold nanoparticle sensor array and were detected amperometrically. This 50 min semi-automated microfluidic system allowed simultaneous detection of IL- 6 and IL-8 with ultralow LODs of 5 fg mL−1 and 7 fg mL−1 in serum respectively. Good correlation with standard ELISA was demonstrated by measuring IL-6 and IL-8 in conditioned media from oral cancer cell lines. Sensitivity could be traded for assay time demonstrated by simultaneous electrochemical detection of IL-6 and IL-8 in serum with clinically relevant LODs of 5 pg mL−1 in 8 min [90]. Rusling et al. further demonstrated an online capture microfluidic system incorporating all immunoassay steps in a microfluidic chip for a multiplexed detection of IL-1β, IL6, tumor necrosis factor (TNF) α, and C-reactive protein (CRP) with detection limits as low as 10–40 fg mL−1 in 30 min assays with a good correlation to ELISA [50].
Fig. 6.
Electrochemical proteins detected in serum by multi-electrode array integrated into microfluidic system featuring a capture and a detection channel made from PDMS encased in polymethylmethacrylate plastic (A). The electrochemical sensor array was positioned in a PDMS microfluidic channel containing a platinum (Pt) wire as the counter electrode and a silver/silver chloride (Ag/ AgCl) wire reference electrode. Amperometry signal was generated upon incubation with Ab2-MB-HRP-analytes in the measurement chamber for 20 min, then injecting a mixture of H2O2 and HQ: (B) duplicate responses in simultaneous array measurements on a standard mixture of 10 fg mL−1 IL-6, 15 fg mL−1 IL-8, 25 fg mL−1 VEG, and 60 fg mL−1 VEGFC illustrating reproducibility, (C) responses to human VEGF in mixtures of biomarker proteins (peaks for VEGF were extracted from four-protein determinations and presented together), (D–G) corresponding Array calibration plots of standard mixtures in calf serum for IL-6 (D), IL-8 (E), VEGF (F), VEGF-C (G). Standard deviations correspond to 2 sensors each on three separate arrays (n = 6). Adapted from ref. 88. Copyright 2012 American Chemical Society.
This protein biomarker panel is associated with oral cancer mucositis, a serious therapy side effect [82].
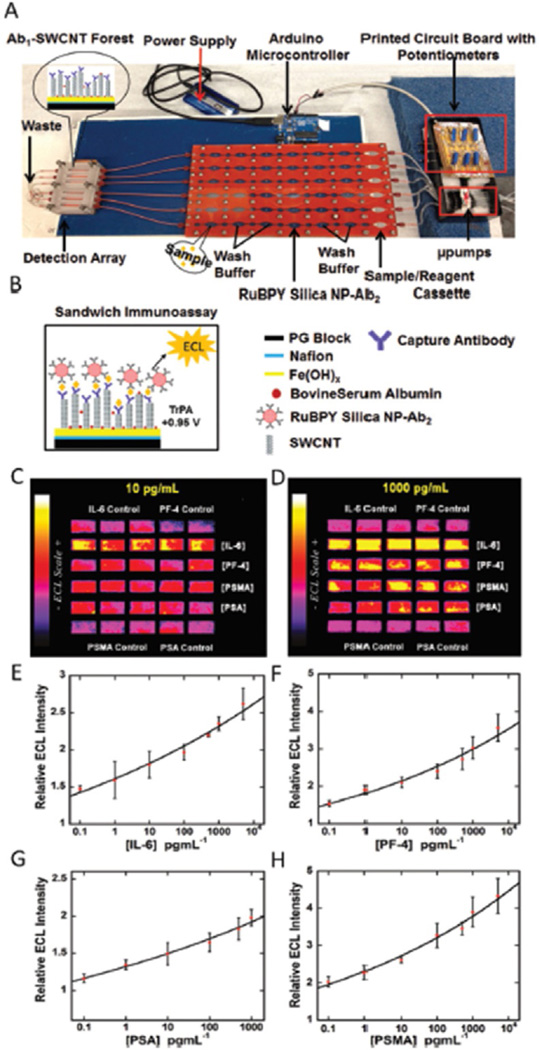
Rusling et al. have also combined earlier ECL based strategies to a microfluidic systems to create an automated detection system and high throughput (Fig. 7). This system used flow injection pumps controlled by a microprocessor [83]. This device used thirty printed microwells on a pyrolytic graphite chip to detect four cancer bio-markers: PSA, PSMA, PF-4 and IL-6. in 36 min. LODs were 10–100 fg mL−1 for the four biomarkers in serum. Integrated systems hold tremendous commercial potential, however there are still challenges associated with building and housing individual components into a single automated machine for POC.
Fig. 7.
Automated ECL Immunoarray: (A) microprocessor-controlled microfluidic immunoarray featuring a 30 microwell SWCNT modified detection array fed with sample/immunore-agents from a reagent cassette (red) using inexpensive micro-pumps. (B) automated immunoassay steps that are controlled by a microprocessor. ECL results (C–D) for a panel of four cancer biomarkers, IL-6, PF4, PSMA, PSA, at different target protein concentrations and E-H showing corresponding immunoassay calibration curves for IL-6, PF4, PSA, and PSMA. Adapted from ref. 92. Copyright 2015 American Chemical Society.
Vaidyanathan et al. demonstrated a microfluidic based multiplex immunosensor utilizing alternating current electrohydrodynamics (ac-EHD) induced surface sheer force (nanoshearing) to minimize NSB events and allow ultrasensitivity detection without the need for external pumps and valves [84]. The group showed that by utilizing this alternating current field they can significantly enhance capture efficiency by increasing the target protein collision rate with the sensor modified with capture antibodies. The tunable nature of the shear forces allow preferential selection of strongly (specifically) bound proteins over more weakly (nonspecifically) bound proteins on the sensor surfaces thus minimizing NSB. A proof of concept for this approach was demonstrated by integrating long array of planar asymmetric Au electrode pairs with 3 capture antibodies specific for Her2, PSA and IgG in a 3 channel microfluidic device. Spiked serum samples with target proteins and large excess of non-analyte protein were injected in each channel, then a mixture of Ab2-HRP specific for each protein was injected followed by naked eye detection or UV/Vis absorption after addition of TMB substrate. This method allowed sensitive detection of multiple protein biomarkers at concentrations as low as 100 fg mL−1 in human serum.
Emerging low cost 3D printing methods can in principle print complete devices including fluidic components and may offer solutions to some of the problems with achieving POC devices for protein-based cancer diagnostics [85]. Rusling et al. recently demonstrated a simple, a low cost 3D–printed ECL-based immunoarray for multiplex detection a panel of 3 cancer biomarker proteins [86]. This device uses a supercapacitor as power supply that is rechargeable with a low cost solar panel. This system demonstrated multiplex ECL detection of PSA, PSMA and PF4 with LODs of 300, 535, 420 fg mL−1, in under 35 min. The multiplexed array uses a simple gravity reagent delivery system and screen-printed electrodes to achieve a low cost system using simple technology requiring little expertise to run.
Paper-based system can reduce cost of analysis and eliminate external pumps, but often at the expense of sensitivity. Yu et al. have demonstrated a quick, cheap microfluidic paper-based analytical device (µ-PAD), for simultaneous detection of PSA and CEA cancer biomarker proteins [87]. The µ-PAD is operated using a voltage tunable device powered by a 3 V lithium battery with a PMT for ECL detection. The µ-PAD was constructed with wax to insulate and create reservoirs. Screen printed carbon sensors were used with a Ag/AgCl auxiliary electrode. The sensors were modified via layer-by-layer assembly of chitosan and AuNP-modified graphene. CEA capture antibodies were attached to one working electrode and PSA antibodies to the other.
Manual changing of the cathode clamp from one working electrode to the other allows for nearly simultaneous detection of both proteins. The detection bioconjugate was a composite of [4,4-(2,5-dimethoxy-1,4-phenylene)bi-s(ethyne-2,1-diyl) dibenzoic acid] (P-acid) and nanoporous silver (NPS) with secondary antibodies attached via EDC/NHS crosslinking. P-acid under goes a redox reaction with TPrA in solution to produce ECL at +1.0 V (vs. Ag/AgCl). The LOD was 1.0 pg mL−1 for PSA and 0.8 pg mL−1 for CEA.
Micro-paper analytical devices, µ-PAD offer advantages including low cost, ease of operation, and ability to function without external pumps. A simple and sensitive 3D microfluidic origami multiplex electrochemical immunodevice was developed using nanoporous Ag-modified paper working electrode as a sensor platform and metal ion functionalized nanoporous gold–chitosan as a tracer [88]. The nanoporous silver platform provided a larger effective surface to enhance conductivity and sensitivity. The LOD for CEA and AFP tumor markers were 0.06 and 0.08 pg mL−1, respectively.
Li et al. demonstrated another paper based microfluidic system featuring cuboid silver modified paper working electrode (CS-PWE) sensor platform and different metal ions-coated nanoporous silver-chitosan (NSC) as labels [89]. The CS-PWE was fabricated through a seed-mediated growth approach and used to attach capture antibodies. The tracer metal ions in the sandwich assay gave distinguishable peaks and were detected directly through square wave voltammetry without metal preconcentration. This approach gave a LOD of 0.08 and 0.10 mU mL−4 for CA125 and CA199 in human serum samples, respectively.
Su et al. demonstrated a simple multiplexed paper-based system for rapid sequential detection of two cancer biomarkers, CEA and AFP on living MCF-7 cancer cells using a paper-based electrode array. In this sandwich immunosensor approach, capture antibodies were bound to Au-paper working electrode (Au-PWE). Upon binding of the target biomarkers, measurement of the two antigens at the cell surface was based on the use of CdTe QDs and luminol groups doped AuPd nanostructures as ECL probes. H2O2 was used as co-reactant to generate the ECL signals under applied scanning potential. This strategy demonstrated large detection ranges of 0.005–200 ng mL−1 for both CEA and AFP biomarkers [90].
Sun et al. developed a multiplexed system utilizing printed paper-based multi-electrode array system. The paper based sandwich immunosensor array was modified by reduced graphene oxide that incorporated ZnO nanorods to improve conductivity and binding sites for capture antibodies on the electrode. The detection tags for the three target biomarkers features a secondary antibodies, Ab2 attached to reduced graphene oxide sheets containing BSA and silver nanoparticles labels to provide a catalytic reduction of H2O2 in solution. Further amplification was achieved by binding silver particles to the signal tag after the sandwich assay is constructed. This system gave LODs of 0.0007 mIU mL−1, 0.35 pg mL−1 0.33 pg mL−1 for hCG, PSA, and CEA biomarkers in serum samples respectively [91]. Results of clinical samples also gave good correlation to referee method.
5 Conclusions
We have summarized above recent advances in multiplex electrochemical detection of cancer biomarker proteins aimed at real diagnostic applications. Electrochemical detection is based on enzyme or nanoparticle labels. Electrochemiluminescence immunoarrays using ECL labels are also being pursued due to their advantages including low costs, ease of use and high sensitivity and selectivity. Most strategies feature nanostructured electrode arrays coupled to multi-labels bioconjugate to enhance sensitivity and detection limits. Low cost microfluidic systems seem to be important for future success in these endeavors. In spite of the current advances, multiplexing beyond 4 proteins for biomedical samples is still a challenge. The integration of EC and ECL immunoassays into low cost microfluidic format can create versatile platforms for fabrication of low-cost, reliable, portable devices for clinical diagnostics, particularly for POC Full automation at low cost is a definite need, and is starting to be addressed by some researchers. The development and use of these systems hold great potential for a future device that can enable fast clinical decision making leading to reduced healthcare cost and patient stress. However, the related important issue of validation of reliable panels for cancer diagnostics for each type of cancer also needs to be addressed.
Acknowledgments
Preparation of this article and the author’s research described therein was financially supported by grant no. 2P20GM103430 from National Institute of General Medical Science, NIGMS/NIH (BSM) and by grant Nos. EB016707 and EB014586 from the National Institute of Biomedical Imaging and Bioengineering NIBIB/NIH (JFR). Contents are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NIBIB or NIH.
References
- 1.Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, G Peck C, Schooley RT, Spilker BA. Clin. Pharmacol. Ther. 2001;69:89–95. [Google Scholar]
- 2.Hawkridge AM, Muddiman DC. Annu. Rev. Anal. Chem. 2009;2:265–277. doi: 10.1146/annurev.anchem.1.031207.112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulasingam V, Diamandis EP. Nat. Clin. Pract. Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Nat. Rev. Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 6.Kingsmore SF. Nat. Rev. Drug Discovery. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanash SM, Pitteri SJ, Faca VM. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 8.Ebert MAP, Korc M, Malfertheiner P, Rçcken C. J. Proteome. Res. 2006;5:19–25. doi: 10.1021/pr050271e. [DOI] [PubMed] [Google Scholar]
- 9.Weston AD, Hood L. J. Proteome. Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 10.Tothill IE. Semin. Cell. Dev. Biol. 2009;20:55–62. doi: 10.1016/j.semcdb.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Stevens EV, Liotta LA, Kohn EC. Int. J. Gynecol. Cancer. 2003;13:133–139. doi: 10.1111/j.1525-1438.2003.13358.x. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DS, Nock S. Angew. Chem. Int. Ed. 2003;42:494–500. doi: 10.1002/anie.200390150. [DOI] [PubMed] [Google Scholar]
- 13.Wagner PD, Verma M, Srivastava S. Ann. N Y Acad. Sci. 2004;1022:9–16. doi: 10.1196/annals.1318.003. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Clin. Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 15.Lilja H, Ulmert D, Vickers AJ. Nat. Rev. Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 16.Riedel F, Zaiss I, Herzog D, Gçtte K, Naim R, Hçrman K. Anticancer Res. 2005;25:2761–2766. [PubMed] [Google Scholar]
- 17.Mujagic Z, Mujagic H, Prnjavorac B. Med. Arh. 2004;58:23–26. [PubMed] [Google Scholar]
- 18.Findlay JW, Smith WC, Lee JW, Nordblom GD, Das I, DeSilva BS, Khan MN, Bowsher RR. J. Pharm. Biomed. Anal. 2000;21:1249–1273. doi: 10.1016/s0731-7085(99)00244-7. [DOI] [PubMed] [Google Scholar]
- 19.Williams TI, Toups KL, Saggese DA, Kalli KR, Cliby WA, Muddiman DC. J. Proteome. Res. 2007;6:2936–2962. doi: 10.1021/pr070041v. [DOI] [PubMed] [Google Scholar]
- 20.Wilson DS, Nock S. Angew. Chem. Int. Ed. 2003;42:494–500. doi: 10.1002/anie.200390150. [DOI] [PubMed] [Google Scholar]
- 21.Rasooly A, Jacobson J. Biosens. Bioelectron. 2006;21:1851–1858. doi: 10.1016/j.bios.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Rusling JF, Kumar CV, Gutkind JS, Patel V. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusling JF, Munge BS, Sardesai NP, Malhotra R, Chikkaveeraiah BV. In: Nanobioelectrochemistry. Cres-pilho F, editor. Berlin, Heidelberg, Germany: Springer; 2012. pp. 1–26. [Google Scholar]
- 24.Rusling JF. Anal. Chem. 2013;85:5304–5310. doi: 10.1021/ac401058v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusling JF, Bishop GW, Doan N, Papadimitrakopoulos F. J. Mater. Chem. B. 2014;2:12–30. doi: 10.1039/C3TB21323D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heineman WR, Halsall HB. Anal. Chem. 1985;75:1321A–1331A. doi: 10.1021/ac00289a804. [DOI] [PubMed] [Google Scholar]
- 27.Vijayawardhana CA, Halsall HB, Heineman WR. Milestones of Electrochemical Immunoassay at Cincinnati. In: Chambers JQ, Bratjer-Toth A, editors. Electroanalytical Methods for Biological Materials. Marcel Dekker, NY: 2002. pp. 195–231. [Google Scholar]
- 28.Ronkainen-Matsuno NJ, Thomas JH, Halsall HB, Heineman WR. Trends Anal. Chem. 2002;21:213–225. [Google Scholar]
- 29.Bange A, Halsall HB, Heineman WR. Biosens. Bioelectron. 2005;20:2488–2503. doi: 10.1016/j.bios.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Nature Nanotech. 2014;9:969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusling JF, Bishop GW, Doan N, Papadimitrakopoulos F. J. Materials Chem. B. 2014;2:12–30. doi: 10.1039/C3TB21323D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Guo Y, Xianyu Y, Chen W, Zhao Y, Jiang X. Adv. Materials. 2013;25:3802–3819. doi: 10.1002/adma.201301334. [DOI] [PubMed] [Google Scholar]
- 33.Das J, Kelley SO. Anal. Chem. 2011;83:1167–1172. doi: 10.1021/ac102917f. [DOI] [PubMed] [Google Scholar]
- 34.Lam B, Holmes RD, Live L, Sage A, Sargent EH, Kelley SO. Nature Comm. 2013;4 doi: 10.1038/ncomms3001. [DOI] [PubMed] [Google Scholar]
- 35.Meissner EG, Decalf J, Casrouge A, Masur H, Kottilil S, Albert ML, et al. PLoS ONE. 2015;10(7):e0133236. doi: 10.1371/journal.pone.0133236. doi:10.1371/journal.pone.0133236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wa ng J. Analyst. 2005;130:421–426. doi: 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Katz E, Willner I. In: Biomaterial - Nanoparticle Hybrid Systems for Sensing and Electronic Devices. Bio-electronics: From Theory to Applications. Katz E, Willner I, editors. Weinheim, Germany: Wiley-VCH; 2005. pp. 231–264. [Google Scholar]
- 38.Wang J. Biosens. Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 39.Chikkaveeraiah BV, Bhirde AA, Morgan NY, Eden HS, Chen X. ACS Nano. 2012;6:6546–6561. doi: 10.1021/nn3023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanez-Sedeno P, Riu J, Pingarron JM, Rius FX. Trends Anal. Chem. 2010;29:939–953. [Google Scholar]
- 41.Tkac J, Davis JJ. In: Engineering the Bioelectronic Interface: Applications to Analyte Biosensing and Protein Detection. Davis JJ, editor. Cambridge, U.K: RSC Publishing; 2009. pp. 193–224. [Google Scholar]
- 42.Wu J, Zhang Z, Fu Z, Ju H. Biosens. Bioelectron. 2007;23:111–120. doi: 10.1016/j.bios.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Yan F, Zhang X, Yan Y, Tang J, Ju H. Clinical Chemistry. 2008;54:91481–91488. doi: 10.1373/clinchem.2007.102350. [DOI] [PubMed] [Google Scholar]
- 44.Tang J, Tang D, Niessner R, Chen G, Knopp D. Anal. Chem. 2011;83:5407–5414. doi: 10.1021/ac200969w. [DOI] [PubMed] [Google Scholar]
- 45.Fuzery AK, Levin J, Chan MM, W Chan D. Clin. Proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry NL, Hayes DF. Mol. Oncol. 2012;6:140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madu CO, Lu Y. J. Cancer. 2010;1:150–77. doi: 10.7150/jca.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Y Kim J, Song HJ, Lim HJ, G Shin M, Kim JS, Kim HJ, Y Kim B, W Lee S. Mol. Cell. Proteomics. 2008;7:431–441. doi: 10.1074/mcp.M700194-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Gokhale RI, Haddad AS, Cavacini LA, et al. Oral Oncol. 2005;41:70–76. doi: 10.1016/j.oraloncology.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Krause EC, Otieno BA, Bishop GW, Phadke G, Choquette L, Lalla RV, Peterson DE, Rusling JF. Anal. Bioanal. Chem. 2015;407:7239–7243. doi: 10.1007/s00216-015-8873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang ZJ, Li JR, Li ZB. Int. J. Oral Maxillofac. Surg. 2002;31:495–498. doi: 10.1054/ijom.2002.0284. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Lin Y. Acta Pharmacol Sin. 2008;29:1275–1288. doi: 10.1111/j.1745-7254.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allin KH, G Nordestgaard B. Crit Rev Clin Lab Sci. 2011;48:155–70. doi: 10.3109/10408363.2011.599831. [DOI] [PubMed] [Google Scholar]
- 54.Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho C, Wong DT. Clin. Cancer Res. 2009;15:4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan Y, Deng W, Y Su, Zhu X, Peng C, Hu H, Peng H, Song S, Fan C. Biosensors and Bioelectronics. 2011;30:93–99. doi: 10.1016/j.bios.2011.08.033. [DOI] [PubMed] [Google Scholar]
- 56.Lai G, Yan F, Ju H. Anal. Chem. 2009;81:9730–9736. doi: 10.1021/ac901996a. [DOI] [PubMed] [Google Scholar]
- 57.Wu J, Yan F, Tang J, Zhai C, Ju H. Clinical Chemistry. 2007;53:1495–1502. doi: 10.1373/clinchem.2007.086975. [DOI] [PubMed] [Google Scholar]
- 58.Wu J, Yan Y, Yan F, Ju H. Anal. Chem. 2008;80:6072–6077. doi: 10.1021/ac800905k. [DOI] [PubMed] [Google Scholar]
- 59.Du D, Wang J, Lu D, Dohnalkova A, Lin Y. Anal. Chem. 2011;83:6580–6585. doi: 10.1021/ac2009977. [DOI] [PubMed] [Google Scholar]
- 60.Liu J, C Lu, Zhou H, Xu J, Chen H. ACS Appl. Mater. Interfaces. 2014;6:20137–20143. doi: 10.1021/am505726b. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Jia X, Han J, Ma J, Ma Z. Biosens. Bioelectron. 2013;50:356–361. doi: 10.1016/j.bios.2013.06.054. [DOI] [PubMed] [Google Scholar]
- 62.Lai G, Wu J, Leng C, Ju H, Yan F. Biosens. Bioelectron. 2011;26:3782–3787. doi: 10.1016/j.bios.2011.02.032. [DOI] [PubMed] [Google Scholar]
- 63.Xu T, Liu N, Yuan J, Ma Z. Biosens. Bioelectron. 2015;70:161–166. doi: 10.1016/j.bios.2015.03.036. [DOI] [PubMed] [Google Scholar]
- 64.Jia X, Liu Z, Liu N, Ma Z. Biosens. Bioelectron. 2014;53:160–166. doi: 10.1016/j.bios.2013.09.050. [DOI] [PubMed] [Google Scholar]
- 65.Liu G, Wang J, Kim J, Jan MR, Collins GE. Anal. Chem. 2004;76:7126–7130. doi: 10.1021/ac049107l. [DOI] [PubMed] [Google Scholar]
- 66.Wang J. Electroanalysis. 2007;19:769–776. [Google Scholar]
- 67.Lai G, Wang L, Wu J, Ju H, Yan F. Anal. Chim. Acta. 2012;721:1–6. doi: 10.1016/j.aca.2012.01.048. [DOI] [PubMed] [Google Scholar]
- 68.Lai G, Yan F, Wu J, Leng C, Ju H. Anal. Chem. 2011;83:2726–2732. doi: 10.1021/ac103283p. [DOI] [PubMed] [Google Scholar]
- 69.Wang D, Li T, Gan N, Zhang H, Long N, F Hu, Cao Y, Jiang Q. S. Jiang. Electrochim. Acta. 2015;163:238–245. [Google Scholar]
- 70.Lai G, Wu J, Ju H, Yan F. Adv. Funct. Mater. 2011;21:2938–2943. [Google Scholar]
- 71.Wang Z, Liu N, Ma Z. Biosens. Bioelectron. 2014;53:324–329. doi: 10.1016/j.bios.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 72.Liu N, Ma Z. Biosens. Bioelectron. 2014;51:184–190. doi: 10.1016/j.bios.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 73.Zhao C, Wu J, Ju H, Yan F. Analytica Chimica Acta. 2014;847:37–43. doi: 10.1016/j.aca.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 74.Wilson MS. Anal. Chem. 2005;77:1496–1502. doi: 10.1021/ac0485278. [DOI] [PubMed] [Google Scholar]
- 75.Wilson MS, Nie W. Anal. Chem. 2006;78:2507–2513. doi: 10.1021/ac0518452. [DOI] [PubMed] [Google Scholar]
- 76.Wilson MS, Nie W. Anal. Chem. 2006;78:6476–6483. doi: 10.1021/ac060843u. [DOI] [PubMed] [Google Scholar]
- 77.Bard AJ, editor. Electrogenerated Chemiluminescence. New York: Marcel Dekker; 2004. [Google Scholar]
- 78.Forster RJ, Bertoncello P, Keyes TE. Electrogenerated Chemiluminescence. Annu. Rev. Anal. Chem. 2009;2:359–385. doi: 10.1146/annurev-anchem-060908-155305. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Liu W, Ge S, Yan M, Wang S, Yu J, N Li, Song X. Biosens. Bioelectron. 2013;41:684–690. doi: 10.1016/j.bios.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 80.Feng X, Gan N, Zhou J, Li T, Cao Y, Hu F, Yu H, Jiang Q. Electrochim. Acta. 2014;139:127–136. [Google Scholar]
- 81.Wu M, Liu Z, Shi H, Chen H, Xu J. Anal. Chem. 2015;87:530–537. doi: 10.1021/ac502989f. [DOI] [PubMed] [Google Scholar]
- 82.Guo Z, Hao T, Du S, Chen B, Wang Z, Li X, Wang S. Biosens. Bioelectron. 2013;44:101–107. doi: 10.1016/j.bios.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 83.Sardesai N, Barron J, Rusling JF. Anal. Chem. 2011;83:6698–6703. doi: 10.1021/ac201292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang D, Yuan R, Chai Y. Clinical Chem. 2007;53:1323–1329. doi: 10.1373/clinchem.2006.085126. [DOI] [PubMed] [Google Scholar]
- 85.Kellner C, Botero ML, Latta D, Drese K, Fragoso A, O’Sullivan CK. Electrophoresis. 2011;32:926–930. doi: 10.1002/elps.201000667. [DOI] [PubMed] [Google Scholar]
- 86.Hwang SY, Seo IJ, Lee SY, Ahn Y. J. Electroanal. Chem. 2015;756:118–123. [Google Scholar]
- 87.Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Biosens. Bioelectron. 2011;26:4477–4483. doi: 10.1016/j.bios.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, Abraham MT, Dey DK, Gutkind JS, Rusling JF. Anal. Chem. 2012;84:6249–6255. doi: 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Otieno BA, Krause CE, Latus A, Chikkaveeraiah BV, Faria RC, Rusling JF. Biosens. Bioelectron. 2014;53:268–274. doi: 10.1016/j.bios.2013.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krause CE, Otieno BA, Latus A, Faria RC, Patel V, Gutkind JS, Rusling JF. Chemistry Open. 2013;2:141–145. doi: 10.1002/open.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dixit CK, Kadimisetty K, Otieno BA, Tang C, Malla S, Krause CE, Rusling JF. Analyst. 2016;141:536–547. doi: 10.1039/c5an01829c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kadimisetty K, Malla S, Sardesai N, Joshi A, Faria R, Lee N, Rusling JF. Anal. Chem. 2015;87:4472–4478. doi: 10.1021/acs.analchem.5b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Va idyanathan R, Leeuwen L, Rauf LS, Shiddiky M, Trau M. Scientific Reports. 2015;5:9756. doi: 10.1038/srep09756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leigh SJ, Bradley RJ, Purssell CP, Billson DR, Hutchins DA. PLoS ONE. 2012;7:e49365. doi: 10.1371/journal.pone.0049365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kadimisetty K, Mosa I, Malla S, Satterwhite-Warden J, Kuhns T, Faria R, Lee N, Rusling JF. Biosens. Bioelectron. 2016;77:188–193. doi: 10.1016/j.bios.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li W, Li M, Ge S, Yan M, Huang J, Yu J. Anal. Chim. Acta. 2013;767:66–74. doi: 10.1016/j.aca.2012.12.053. [DOI] [PubMed] [Google Scholar]
- 97.Li W, Li L, Li M, Yu J, Ge S, Yan M, Song X. Chem. Commun. 2013;49:9540–9542. doi: 10.1039/c3cc44955f. [DOI] [PubMed] [Google Scholar]
- 98.Li W, Li L, Ge S, Song X, Ge L, Ya n M, Yu J. Biosens. Bioelectron. 2014;56:167–173. doi: 10.1016/j.bios.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 99.Su M, Liu H, Ge S, Ren N, Ding L, Yu J, Song X. RCS Adv. 2016;6:16500–16506. [Google Scholar]
- 100.Sun G, Zang L, Zang Y, Yang H, Ma C, Ge S, Yan M, Yu J, Song X. Biosens. Bioelectron. 2015;71:30–36. doi: 10.1016/j.bios.2015.04.007. [DOI] [PubMed] [Google Scholar]