Abstract
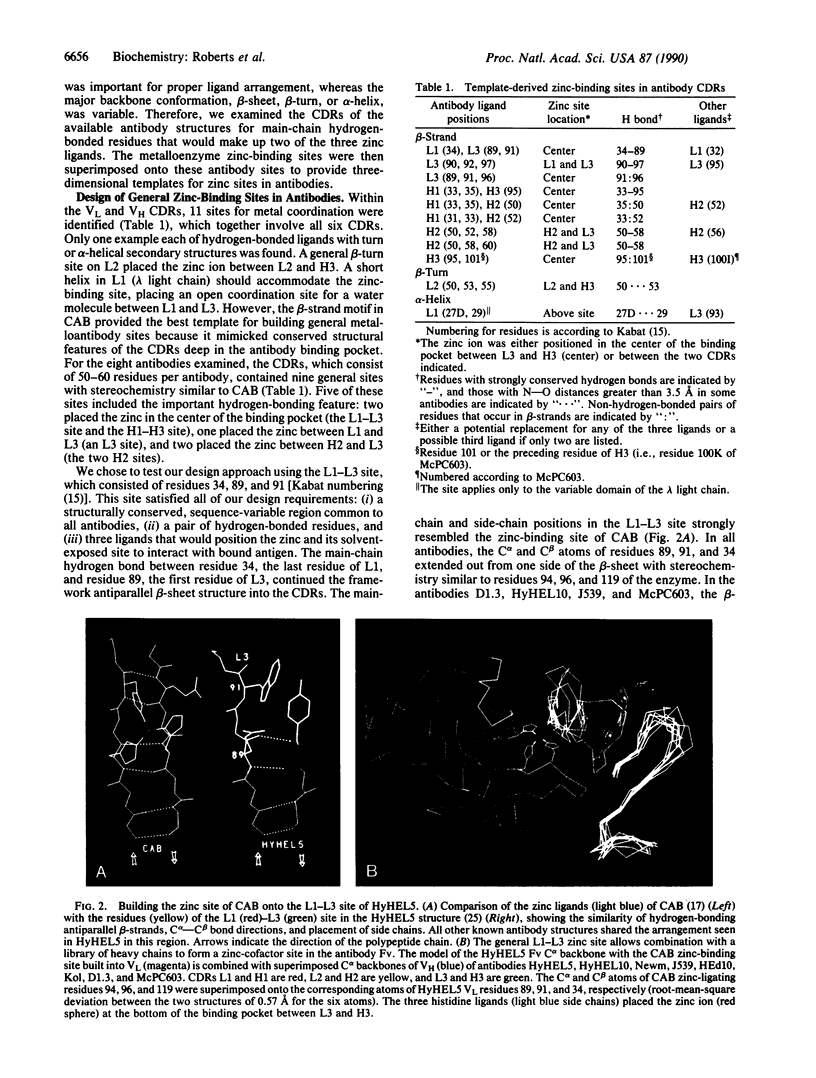
To develop a general approach to designing cofactor-binding sites for catalytic antibodies, we characterized structural patterns in the binding sites of antibodies and zinc enzymes. Superposition of eight sets of antibody light- and heavy-chain variable domains identified structurally conserved sites within the sequence-variable complementarity determining regions. The pattern for catalytic zinc sites included two ligands close in sequence, a sequence-distant ligand, and a main-chain hydrogen bond joining two ligands. In both the light- and heavy-chain variable domains, the stereochemistry of five structurally conserved sites general to all known antibody structures matched that of the zinc ligands of carbonic anhydrase: three residues on two hydrogen-bonded antiparallel beta-strands. For one such general site, an antibody model replacing residue 34 on the first complementarity determining region of the light chain (L1) and residues 89 and 91 on the third complementarity determining region of the light chain (L3) with histidine ligands formed a zinc-binding site with an open coordination position at the bottom of the antibody binding pocket. For the anti-fluorescein antibody 4-4-20, this L1-L3 site placed the zinc ion about 4 A from the bound fluorescein, an indicator for metal binding. This predicted zinc-binding mutant was created in the single-chain variable domain construct, expressed, and found by fluorescence quenching to bind metal ion with an affinity constant of 10(6) M-1. Thus, our template-based multisite design proved successful for remodeling an antibody to contain a cofactor-binding site, without requiring further mutagenesis and screening. Combination of a specific light or heavy chain containing a catalytic metal site with a library of complementary chains raised to potential substrates or transition state analogs should greatly improve the production of catalytic antibodies with desired activities and specificities.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amit A. G., Mariuzza R. A., Phillips S. E., Poljak R. J. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986 Aug 15;233(4765):747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- Argos P., Garavito R. M., Eventoff W., Rossmann M. G., Brändén C. I. Similarities in active center geometries of zinc-containing enzymes, proteases and dehydrogenases. J Mol Biol. 1978 Dec 5;126(2):141–158. doi: 10.1016/0022-2836(78)90356-x. [DOI] [PubMed] [Google Scholar]
- Baldwin E., Schultz P. G. Generation of a catalytic antibody by site-directed mutagenesis. Science. 1989 Sep 8;245(4922):1104–1107. doi: 10.1126/science.2672338. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bird R. E., Hardman K. D., Jacobson J. W., Johnson S., Kaufman B. M., Lee S. M., Lee T., Pope S. H., Riordan G. S., Whitlow M. Single-chain antigen-binding proteins. Science. 1988 Oct 21;242(4877):423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- Chien N. C., Roberts V. A., Giusti A. M., Scharff M. D., Getzoff E. D. Significant structural and functional change of an antigen-binding site by a distant amino acid substitution: proposal of a structural mechanism. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5532–5536. doi: 10.1073/pnas.86.14.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987 Aug 20;196(4):901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- Getzoff E. D., Geysen H. M., Rodda S. J., Alexander H., Tainer J. A., Lerner R. A. Mechanisms of antibody binding to a protein. Science. 1987 Mar 6;235(4793):1191–1196. doi: 10.1126/science.3823879. [DOI] [PubMed] [Google Scholar]
- Getzoff E. D., Tainer J. A., Lerner R. A., Geysen H. M. The chemistry and mechanism of antibody binding to protein antigens. Adv Immunol. 1988;43:1–98. doi: 10.1016/s0065-2776(08)60363-6. [DOI] [PubMed] [Google Scholar]
- Herron J. N., He X. M., Mason M. L., Voss E. W., Jr, Edmundson A. B. Three-dimensional structure of a fluorescein-Fab complex crystallized in 2-methyl-2,4-pentanediol. Proteins. 1989;5(4):271–280. doi: 10.1002/prot.340050404. [DOI] [PubMed] [Google Scholar]
- Holmes M. A., Matthews B. W. Structure of thermolysin refined at 1.6 A resolution. J Mol Biol. 1982 Oct 5;160(4):623–639. doi: 10.1016/0022-2836(82)90319-9. [DOI] [PubMed] [Google Scholar]
- Huse W. D., Sastry L., Iverson S. A., Kang A. S., Alting-Mees M., Burton D. R., Benkovic S. J., Lerner R. A. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989 Dec 8;246(4935):1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- Iverson B. L., Lerner R. A. Sequence-specific peptide cleavage catalyzed by an antibody. Science. 1989 Mar 3;243(4895):1184–1188. doi: 10.1126/science.2922606. [DOI] [PubMed] [Google Scholar]
- Kannan K. K., Notstrand B., Fridborg K., Lövgren S., Ohlsson A., Petef M. Crystal structure of human erythrocyte carbonic anhydrase B. Three-dimensional structure at a nominal 2.2-A resolution. Proc Natl Acad Sci U S A. 1975 Jan;72(1):51–55. doi: 10.1073/pnas.72.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauptit R. A., Karlsson R., Picot D., Jenkins J. A., Niklaus-Reimer A. S., Jansonius J. N. Crystal structure of neutral protease from Bacillus cereus refined at 3.0 A resolution and comparison with the homologous but more thermostable enzyme thermolysin. J Mol Biol. 1988 Feb 5;199(3):525–537. doi: 10.1016/0022-2836(88)90623-7. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Lewis M., Lipscomb W. N. Refined crystal structure of carboxypeptidase A at 1.54 A resolution. J Mol Biol. 1983 Aug 5;168(2):367–387. doi: 10.1016/s0022-2836(83)80024-2. [DOI] [PubMed] [Google Scholar]
- Riechmann L., Clark M., Waldmann H., Winter G. Reshaping human antibodies for therapy. Nature. 1988 Mar 24;332(6162):323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- Sheriff S., Silverton E. W., Padlan E. A., Cohen G. H., Smith-Gill S. J., Finzel B. C., Davies D. R. Three-dimensional structure of an antibody-antigen complex. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokat K. M., Leumann C. J., Sugasawara R., Schultz P. G. A new strategy for the generation of catalytic antibodies. Nature. 1989 Mar 16;338(6212):269–271. doi: 10.1038/338269a0. [DOI] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Alexander H., Houghten R. A., Olson A. J., Lerner R. A., Hendrickson W. A. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984 Nov 8;312(5990):127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Richardson J. S., Richardson D. C. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983 Nov 17;306(5940):284–287. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Active-site zinc ligands and activated H2O of zinc enzymes. Proc Natl Acad Sci U S A. 1990 Jan;87(1):220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E. S., Güssow D., Griffiths A. D., Jones P. T., Winter G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature. 1989 Oct 12;341(6242):544–546. doi: 10.1038/341544a0. [DOI] [PubMed] [Google Scholar]