Abstract
Background:
Congenital heart disease (CHD), plus cleft lip and palate (CLP) are currently the most common types of structural malformation in infants. Many genes have been investigated for their involvement in CHD with CLP. Targeted next-generation sequencing can analyze large amounts of genetic information rapidly, and thus address this question.
Methods:
The authors designed a targeted, next-generation sequencing gene panel for 455 genes previously implicated in CHD or CLP. A single-subject patient served as a genetic source. Variants that affect protein-coding regions were classified into silico and filtered through databases, such as the Single-Nucleotide Polymorphism Database, Yan Huang, the Exome Sequencing Project, and the 1000 Genomes Project. The authors then predicted the function of gene mutations by PolyPhen-2, SIFT, and Mutation Taster. To confirm the related disease genes, the authors surveyed relevant literature on PubMed. Finally, the variant was verified by Sanger sequencing.
Results:
A total of 1520 mutations were successfully found in a patient using combined tetralogy of Fallot and CLP by the targeted next-generation sequencing. However, there were 6 gene mutations (ZNF528, PVRL2, methylenetetrahydrofolate reductase [MTHFR], EVC2, DAND5, CCDC39) that were not found on Single-Nucleotide Polymorphism Database, Yan Huang, Exome Sequencing Project, and 1000 Genomes Project. Four genes (ZNF528, PVRL2, EVC2, CCDC39) were all predicted to be “tolerated,” “benign,” or “polymorphic” by SIFT, PolyPhen-2, and Mutation Taster. The DAND5 gene was predicted to be “possibly damaging” and “disease causing” respectively by PolyPhen-2 and Mutation Taster, but the SIFT program predicted this mutation to be “tolerated.” Likewise, the MTHFR gene mutation was predicted to be “damaging,” “possibly damaging,” and “disease causing” respectively by SIFT, PolyPhen-2, and Mutation Taster. There is no relevant report about MTHFR gene mutation (c.G586A, p.G196S) on PubMed.
Conclusion:
Using targeted, next-generation sequencing technology, the authors identified for the first time a mutation (c.G586A, p.G196S) in the MTHFR gene as a possible cause of TOF and CLP in a patient.
Keywords: Cleft lip and palate, targeted next-generation sequencing, tetralogy of Fallot, the MTHFR gene
Congenital heart diseases (CHD), cleft lip and cleft palate (CLP) comprise the highest incidence of birth defects in the world. The results of epidemiological investigation show a rising incidence of CHD and CLP year by year.1,2 Congenital heart diseases include ventricular septal defect, atrial septal defect, tetralogy of Fallot (TOF), and so on. Tetralogy of Fallot consist of pulmonary artery stenosis, ventricular septal communication, rightward deviation of the aorta's origin, and hypertrophy of the right ventricle. However, due to the complex and long cycle of treatment procedures for CHD and CLP, these diseases can bring a heavy burden to family and society and seriously affect neonatal health. The high heritability of CHD, estimated to be between 0.6 and 0.7, suggests a strong genetic component and numerous genes, which have been linked to syndromic and nonsyndromic forms of CHD.3 Together, the above factors drive the need to identify the disease genes responsible for CHD with CLP.
Many genes have been implicated in the development of congenital heart disease or cleft lip and palate. With the rapid development of technology, many methods also had been used to find these disease genes, such as Array-SNP, CNVs, targeted, next-generation sequencing, and whole exome sequencing. Targeted, next-generation sequencing rapidly analyzes large amounts of genetic information. In this paper, we wanted to find disease genes through targeted next-generation sequencing.
METHODS
Patient
The study protocol was approved by the Review Board at Second Xiangya Hospital of Central South University (China), and the related study subject gave informed consent. All experiments were performed in accordance with relevant guidelines and regulations. We enrolled a patient in whom we observed cardiac structure, leading to diagnosis of TOF and CLP by transthoracic echocardiogram.
DNA Extraction
Genomic DNA was extracted from peripheral blood lymphocytes of the patient. Genomic DNA was prepared for testing by DNeasy Blood and TissueKit (Qiagen, Valencia, CA) on the QIAcube automated DNA-extraction robot (Qiagen, Hilden, Germany), as previously described.4 The quality and quantity of the DNA sample were measured by the use of the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc, Waltham, MA), from which 3 μg of DNA of each sample was used for analysis.
Targeted Next-Generation Sequencing
Targeted, next-generation sequencing (NGS), including library construction, capture, and sequencing, was carried out at Oxford Gene Technologies (Oxford, UK). Enrichment of target regions and library preparation were performed by the use of a SureSelectXT2 Custom kit (1–499 kb, 16) in accordance with SureSelect protocol (version 1.2; Agilent Technologies, Changsha, China). Library concentrations were determined using Agilent's QPCR NGS Library Quantification Kit (G4880A), with each sample at a final concentration of 10 nmol/L. A HiSeq2000 sequencer ordered the samples using TruSeq chemistry and protocols (version 3, Illumina Inc, San Diego, CA). The TOF with CLP patient was analyzed separately using all CHD or CLP patients grouped into 1 enrichment kit and sequencing run.3
Data Analysis and Filtering
The preliminary data analyses, including read alignment, variant calling, and annotation, were carried out by Oxford Gene Technologies. All variants that affect protein-coding regions for each sample were categorized into “novel” and “known” categories according to their presence in Single-Nucleotide Polymorphism Database (dbSNP) 137. Minor allele frequencies of all known variants were reported according to Exome Sequencing Project (ESP) or Yan Huang (YH), 1000 Genomes Project (TGP) if not present in dbSNP137. All variants were subjected to in silico analysis, which included prediction programs, such as SIFT, PolyPhen-2, and Mutation Taster.
Variant Validation
Variants warranting further investigation included novel variants, which were predicted to be “probably damaging,” “disease causing,” or “damaging,” according to PolyPhen-2, mutation tasting, and SIFT predictions, or these variants were known to be “probably damaging” and possessed minor allele frequencies <0.1%. Variants were verified by Sanger sequencing. To confirm the related disease genes, we surveyed the relevant literature on PubMed (https://www.ncbi.nlm.nih.gov/pubmed).
Polymerase Chain Reaction
Entire exons and exon–intron junctions of MTHFR (Refseq:NM_005957) were amplified by polymerase chain reaction (PCR, primer sequences will be provided upon request). Sequences of the PCR products were determined using an ABI 3100Genetic Analyzer. The primer sequences are as follows:
Forward primer: 5′ ATAGGTGACCAGTGGGAAGA 3′,
Reverse primer: 5′ CTGATCACTGTGTCCTGAACC 3′.
RESULTS
In our study, there were 1390 SNPs and 130 indels mutations (a total of 1520 mutations) that successfully passed the filtering criteria as identified by targeted, next-generation sequencing. In addition, there were 6 gene mutations (ZNF528, PVRL2, methylenetetrahydrofolate reductase [MTHFR], EVC2, DAND5, consensus coding sequence [CCDC39]) not found in the databases used, such as dbSNP, YH, ESP, and TGP (Table 1).
TABLE 1.
The Results of Targeted Sequencing
| Category | |
| The total number of mutations | 1520 |
| The number of SNP | 1390 |
| The number of missing mutations | 82 |
| The number of insertion mutations | 48 |
| Database of dbSNP | 515 |
| Database of TGP | 173 |
| Database of YH | 46 |
| Database of ESP | 6 |
dbSNP, Single Nucleotide Polymorphism Database; ESP, Exome Sequencing Project; TGP, 1000 Genomes Project; YH, Yan Huang.
Four genes (ZNF528, PVRL2, EVC2, CCDC39) were all predicted to be “tolerated,” “benign,” and/or “polymorphic” by SIFT, PolyPhen-2, and Mutation Taster. The DAND5 gene was predicted to be “possibly damaging” and “disease causing” respectively by PolyPhen-2 and Mutation Taster, but the SIFT program predicted this mutation to be “tolerated.” The MTHFR gene mutation was predicted to be “damaging,” “possibly damaging,” and “disease causing” respectively by SIFT, PolyPhen-2, and Mutation Taster (Table 2). At last, we verified the variant by Sanger sequencing (Fig. 1). There is no relevant report about MTHFR gene mutation (c.G586A, p.G196S) on PubMed.
TABLE 2.
The Results of Prediction
| Genes | Chromosome | SIFT | PolyPhen 2 | Mutation Taster | CCDS | Amino Acid |
| ZNF528 | Chr19 | Tolerated | Benign | Polymorphism | A598G | S200G |
| PVRL2 | Chr19 | Tolerated | Benign | Polymorphism | C214G | R72G |
| MTHFR | Chr1 | Damaging | Probably damaging | Disease causing | G586A | G196S |
| EVC2 | Chr4 | Tolerated | Benign | Polymorphism | G2623A | V875I |
| DAND5 | Chr19 | Tolerated | Possibly damaging | Disease causing | G515A | R172Q |
| CCDC39 | Chr3 | Tolerated | Benign | Polymorphism | G1035T | R345S |
CCDS, the consensus coding sequence; MTHFR, methylenetetrahydrofolate reductase.
FIGURE 1.
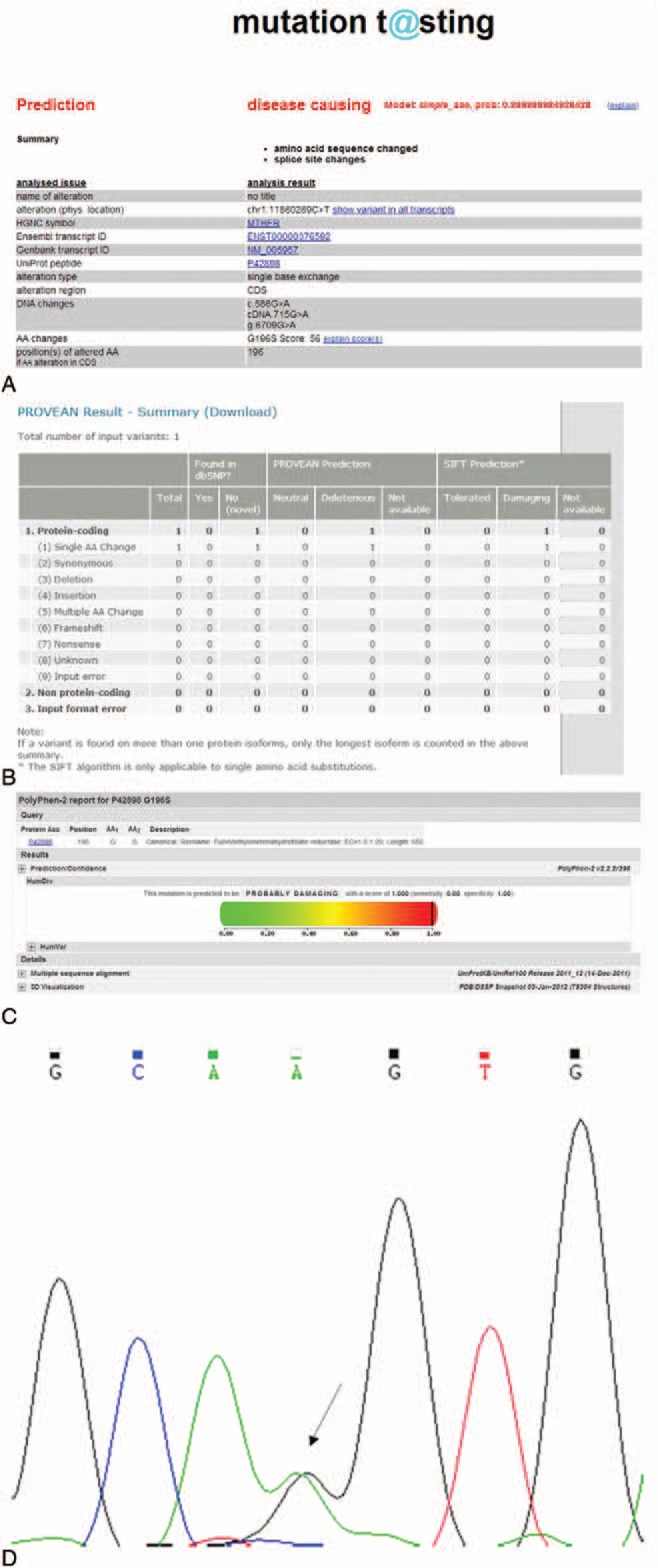
The results of prediction programs. (A) Mutation Taster prediction. (B) SIFT prediction. (C) Polyphen-2 prediction. (D) The results of Sanger sequencing.
DISCUSSION
With the rapid development of technology, many methods had been used to find the disease-causing genes, such as Array-SNP, CNVs, targeted, next-generation sequencing, and whole exome sequencing. Targeted, next-generation sequencing offers opportunities for genetic testing and can analyze large amounts of genetic information rapidly. We consider targeted, next-generation sequencing to be more clinically useful than whole exome sequencing, due to speeder turnaround time (reduced sequencing volume and associated data analysis), higher and more reliable coverage, plus the ability to avoid incidental findings.3 In the last few years, many researchers have published reports on gene mutations, discovered by targeted sequencing, for many genetic diseases, including CHD and CL/P.
In this study, a total of 1520 mutations were successfully found in the patient via the targeted, next-generation sequencing. However, there were 6 genes (ZNF528, PVRL2, MTHFR, EVC2, DAND5, CCDC39) with mutations not found in existing databases, such as dbSNP, YH, ESP, and TGP. We predicted character of the named gene mutations through PolyPhen-2, SIFT, and Mutation Taster programs. The ZNF528, PVRL2, EVC2, CCDC39 genes were all predicted to be “tolerated,” “benign,” and “polymorphic” respectively by SIFT, PolyPhen-2, and Mutation Taster. Thus, we can eliminate those genes mutations. The DAND5 gene was predicted to be “possibly damaging” and “disease causing,” respectively by PolyPhen-2 and Mutation Taster, while the SIFT program predicted DAND5 to be “tolerated.” However, there is a close relationship between the said gene and spiradenoma according to our survey of published reports. The MTHFR gene mutation (c.G586A, p.G196S) was predicted to be “damaging,” “possibly damaging,” and “disease causing” in turn, by SIFT, PolyPhen-2, and Mutation Taster. These predictions support the notion that the MTHFR gene variant may contribute to TOF and CLP pathogenesis. We also studied the subject's parents using Sanger sequencing, but the results were meaningless. The result should be further verified through a model organism, such as zebra fish or mouse.
The human MTHFR gene, consisting of 11 exons, catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a cosubstrate for the remethylation of homocysteine to methionine, which plays a key role in neural tube and vascular defects during embryogenesis.5 According to the Human Gene Mutation Database, 74 mutations on the MTHFR gene have been reported in several congenital heart diseases. So far, more than 8 genes (JAG1, NKX2–5, GATA4, MTHFR, ZFPM2, GDF1, TBX1, GATA6) have been implicated in TOF.6–11 Moreover, more than 12 genes (MSX1, MTHFR, IRF6, PVRL1, SUMO1, BMP4 in addition to the previous list) have been reported in CLP.12–15 Among these genes, MTHFR is a disease gene with high-penetrance mutations.
Over the last few years, impaired MTHFR has been widely investigated to establish its potential role as a risk factor or marker of cardiovascular disease, neural tube defect, maxillofacial malformation, cognitive disorders, and cancer.16–21 Abnormal folate metabolism has been previously described as a possible risk factor for TOF.6 It has been hypothesized that genetic polymorphisms in folate-metabolizing enzymes affect global DNA methylation as well as changes the synthesis and repair of DNA. Furthermore, animal experiments have shown that disruption of the MTHFR gene results in decreased methylation capacity.22 There are, however, no reports about mutations on the MTHFR gene in TOF with CLP.
By contrast, this study found a novel MTHFR mutation (c.G586A, p.G196S) located in the functional region that may affect the enzyme activities. For the first time, levels of homocysteine in a patient with TOF and CLP were shown to be affected by the mutation using targeted, next-generation sequencing, which mutation had not been described in any previous reports or databases. The physiological function of MTHFR gene is changed by the mutation, a defect that leads to methionine deficiency and over-accumulation of homocysteine, in turn which leads to lowered MTHFR enzymatic activity or lowered folate levels. As the result of reduced enzymatic activity and elevated plasma homocysteine levels, the patient may exhibit congenital malformations (tetralogy of Fallot, plus cleft lip, and palate). Our study broadens the mutation spectrum of the MTHFR gene and suggests that this approach will facilitate etiological elucidation of this congenital malformation, which causes TOF and CLP, by effective identification of the causative genetic mutations.
In spite of this study has found a new mutation by the new research method, but there are still some shortcomings in this study. On the one hand, the sample size is not enough. On the other hand, this research has not carried out further functional studies on the new mutation found such as zebra fish or mouse.
CONCLUSIONS
Using targeted, next-generation sequencing technology, we identified for the first time a mutation (c.G586A, p.G196S) in the MTHFR gene as a possible cause of TOF and CLP in a patient. Due to the complex and long cycle of treatment procedures for CHD and CLP, these diseases can bring a heavy burden to family and society and seriously affect neonatal health. It becomes urgent to reduce the birth rate of children with CHD and CLP by screening of the pathogenic genes and avoid surgical treatment by gene therapy in Chinese rural areas.
Footnotes
LL and HB contributed equally to this work.
The authors report no conflicts of interest.
REFERENCES
- 1.Liang CD, Huang SC, Lai JP. A survey of congenital heart disease in patients with oral clefts. Acta Paediatr Taiwan 1999; 40:414–417. [PubMed] [Google Scholar]
- 2.Barbosa MM, Rocha CM, Katina T, et al. Prevalence of congenital heart diseases in oral cleft patients. Pediatr Cardiol 2003; 24:369–374. [DOI] [PubMed] [Google Scholar]
- 3.Blue GM, Kirk EP, Giannoulatou E, et al. Targeted next-generation sequencing identifies pathogenic variants in familial congenital heart disease. J Am Coll Cardiol 2014; 64:2498–2506. [DOI] [PubMed] [Google Scholar]
- 4.Tan ZP, Xie L, Deng Y, et al. Whole-exome sequencing identifies Y1495X of SCN5A to be associated with familial conduction disease and sudden death. Sci Rep 2014; 4:5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyette P, Pai A, Milos R, et al. Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm Genome 1998; 9:652–656. [DOI] [PubMed] [Google Scholar]
- 6.Benson DW, Silberbach GM, Kavanaugh-McHugh A, et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest 1999; 104:1567–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Mei J, Jiang L, et al. rs1801133 C>T polymorphism is associated with an increased risk of tetralogy of Fallot. Biomed Rep 2014; 2:172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eldadah ZA, Hamosh A, Biery NJ, et al. Familial tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet 2001; 10:163–169. [DOI] [PubMed] [Google Scholar]
- 9.Tomita-Mitchell A, Maslen CL, Morris CD, et al. GATA4 sequence variants in patients with congenital heart disease. J Med Genet 2007; 44:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pizzuti A, Sarkozy A, Newton AL, et al. Mutations of ZFPM2/FOG2 gene in sporadic cases of tetralogy of Fallot. Hum Mutat 2003; 22:372–377. [DOI] [PubMed] [Google Scholar]
- 11.Rauch R, Hofbeck M, Zweier C, et al. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of Fallot. J Med Genet 2010; 47:321–331. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Pan Y, Du Y, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and nonsyndromic orofacial clefts susceptibility in a southern Chinese population. DNA Cell Biol 2011; 30:1063–1068. [DOI] [PubMed] [Google Scholar]
- 13.Jezewski PA, Vieira AR, Nishimura C, et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet 2003; 40:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw GM, Rozen R, Finnell RH, et al. Infant C677T mutation in MTHFR, maternal periconceptional vitamin use, and cleft lip. Am J Med Genet 1998; 80:196–198. [DOI] [PubMed] [Google Scholar]
- 15.Kondo S, Schutte BC, Richardson RJ, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet 2002; 32:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boccia S, Hung R, Ricciardi G, et al. Meta- and pooled analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer risk: a huge-GSEC review. Am J Epidemiol 2008; 167:505–516. [DOI] [PubMed] [Google Scholar]
- 17.Ebadifar A, Ameli N, Khorramkhorshid HR, et al. Incidence assessment of MTHFR C677T and A1298C polymorphisms in Iranian non-syndromic cleft lip and/or palate patients. J Dent Res Dent Clin Dent Prospects 2015; 9:101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan X, Wang P, Yin X, et al. Association between maternal MTHFR polymorphisms and nonsyndromic cleft lip with or without cleft palate in offspring, a meta-analysis based on 15 case-control studies. Int J Fertil Steril 2015; 8:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bezerra JF, Oliveira GH, Soares CD, et al. Genetic and non-genetic factors that increase the risk of non-syndromic cleft lip and/or palate development. Oral Dis 2015; 21:393–399. [DOI] [PubMed] [Google Scholar]
- 20.Xuan C, Li H, Zhao JX, et al. Association between MTHFR polymorphisms and congenital heart disease: a meta-analysis based on 9,329 cases and 15,076 controls. Sci Rep 2014; 4:7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Hou Z, Wang C, et al. Association between 5, 10-methylenetetrahydrofolate reductase (MTHFR) polymorphisms and congenital heart disease: a meta-analysis. Meta Gene 2013; 1:109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Karaplis AC, Ackerman SL, et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet 2001; 10:433–443. [DOI] [PubMed] [Google Scholar]


