Abstract
Notch receptor activation initiates cell fate decisions, and is distinctive in its reliance on mechanical force and protein glycosylation. The 2.5 angstrom crystal structure of the extracellular interacting regions of Notch1 complexed with an engineered, high-affinity variant of Jagged1 (Jag1) reveals a binding interface that extends ∼120 angstroms along five consecutive domains of each protein. O-Linked fucose modifications on Notch1 EGF domains 8 and 12 engage the EGF3 and C2 domains of Jag1, respectively, and different Notch domains are favored in binding to Jag1 compared to those that bind to the Delta-like 4 ligand. Jag1 undergoes conformational changes upon Notch binding, exhibiting catch bond behavior that prolongs interactions in the range of forces required for Notch activation. This mechanism enables cellular forces to regulate binding, discriminate amongst Notch ligands and potentiate Notch signaling.
The Notch pathway specifies cell fate decisions in all meta-zoans. In mammals, Notch signals are transmitted when transmembrane ligands Jagged1 (Jag1), Jagged2 (Jag2), Delta-like 1 (DLL1) or Delta-like 4 (DLL4) stimulate Notch receptor paralogs (Notch1-4) on the surface of adjacent cells. The prevailing model for canonical Notch activation includes forces that pull on the Notch-ligand complex as a result of receptor or ligand endocytosis (1, 2). A few piconewtons (pN) of molecular tension activate Notch (3–5) by unfolding its negative regulatory-region (NRR) to expose a cryptic S2 proteolytic site (6), cleavage of which releases the Notch ICD (NICD) to translocate to the nucleus and serve as a transcriptional co-factor (7–9). It is presently unclear whether applied forces, such as those encountered under conditions of shear between cells, induce structural changes in Notch or its ligands, and how these forces propagate from the ligand binding site of Notch to the membrane proximal NRR.
Notch, Jag and DLL proteins encode large extracellular domains (ECDs) with modular architectures. Notch ECDs comprise up to 36 epidermal growth factor-like (EGF) domains followed by an autoinhibitory NRR that protects the S2 cleavage site (Fig. 1A) (6, 10, 11). Several Notch EGF domains are modified with O-linked glycans that influence receptor sensitivity to ligand stimulation (12–16). Jag1 and DLL ECDs consist of an N-terminal C2 domain that has been implicated in lipid binding (17), a Delta-Serrate-Lag-2 (DSL) domain, 6 to 16 EGF domains, and a cysteine-rich domain (CRD) present only in Jag1 and Jag2 (Fig. 1A) (10, 18). Jag and DLL bind Notch1 primarily through EGF domains 11 and 12 (19), and co-crystal structures of Notch1 and DLL4 revealed that the N-terminal DSL and C2 domains of DLL4 engage Notch EGF11 and EGF12, respectively (20). However, additional regions of Notch may contribute to differential recognition of DLL and Jag ligands (16, 21, 22).
Fig. 1. Engineering of high-affinity Jag1 proteins and crystal structure of the Notch1-Jag1JV1 complex.
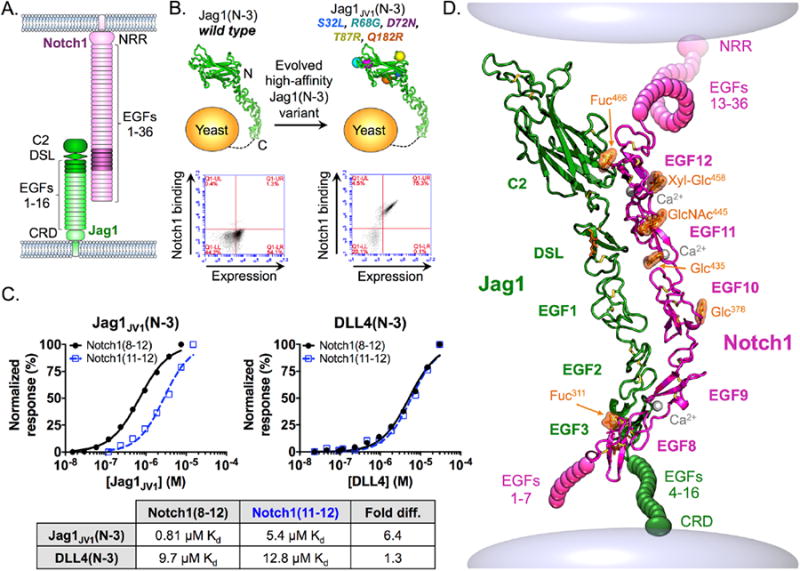
(A) Schematic of Notch1 and Jag1 ECD architecture. Shaded domains represent constructs used for co-crystallization. (B) In vitro evolution of Jag1(N-3) led to the isolation of the high-affinity Jag1JV1 variant containing S32L, R68G, D72N, T87R, and Q182R mutations. Flow cytometry dot plots depict the increased binding of yeast displayed Jag1JV1(N-3) relative to that of WT Jag1(N-3) upon staining with Notch1(8-12) tetramers. Surface expression was detected with an antibody to the c-Myc tag on displayed constructs. (C) Binding isotherms were obtained by surface plasmon resonance. Curves were fitted to a 1:1 binding model to obtain Kd values indicated in the table. (D) Crystal structure of Notch1(8-12) (magenta) bound to Jag1JV1(N-3) (green) represented as the interaction may occur on the surface of two cells. Fuc: fucose; Glc: glucose; GlcNAc: N-acetylglucosamine.
Here, we report the structure of a Notch1-Jag1 complex encompassing a newly visualized interaction site. Molecular force measurements show that Jag1 and DLL4 have different tension thresholds for receptor activation, and both form ‘catch bonds’, or bonds whose lifetimes are prolonged upon the application of tensile forces (23). Notch1-ligand catch bond behavior stands in contrast to ‘slip bond’ behavior (bond lifetimes diminished by tensile forces) and may potentiate signaling by low affinity Notch-ligand interactions in response to intercellular forces.
Structural studies of Notch-ligand interactions are impeded by the low affinities between Notch and Jag or DLL ECDs (10, 18, 20, 22). We therefore used in vitro evolution to engineer a high-affinity Jag1 variant to stabilize Notch1-Jag1 complexes. Jagged variant 1 (Jag1JV1) was generated using an unbiased error-prone mutant library of Jag1 spanning from the N terminus to EGF3 [Jag1(N-3)] on the surface of yeast cells, followed by enrichment of Notch1 binders over several rounds of selection. Sequencing individual clones revealed S32L, R68G, D72N, T87R and Q182R mutations in the C2 domain of Jag1JV1 (Fig. 1B and fig. S1). Enhanced binding of Notch1 to Jag1JV1 relative to that of wild-type (WT) Jag1 was confirmed in a flow cytometry-based assay (Fig. 1B). To investigate the role of each mutation, we reverted mutated residues to the WT sequence and performed a binding assay. Only two of the five mutations, S32L and T87R, strongly affected the affinity of Jag1JV1 (fig. S2).
We determined that Jag1JV1(N-3) bound to Notch1 EGF domains 8 to 12 [Notch1(8-12)] with a KD of 0.81μM and to Notch1 EGF domains 11 and 12 [Notch1(11-12)] with a KD of 5.4μM (Fig. 1C), a 6.4-fold difference in affinity, indicating that EGF8-10 contributes to Jag1JV1-Notch1 binding energetics (Fig. 1C). On the other hand, Notch1(8-12) and Notch1(11-12) bound to DLL4(N-3) with KD values of 9.7μM and 12.8μM, respectively, indicating that EGF8-10 had a minimal role in Notch1-DLL4 engagement (Fig. 1C). However, Notch1 domains 8 through 10 are required for maximal DLL4-mediated Notch1 activation in cellular assays (22). Jag and DLL ligands thus appear to have different energetic requirements for various Notch1 domains.
We determined the 2.5Å x-ray crystal structure of the complex of Notch1(8-12) and Jag1JV1(N-3) proteins, revealing an elongated binding surface that extends for ∼120Å across all five domains of each construct (Fig. 1D and table S1). The Notch1 EGF domains formed a curved structure modified with O-linked fucose, glucose, and N-acetylglucosamine moieties, and calcium ions were bound by EGF domains 9, 11, and 12 (Fig. 1D and fig. S3). Notch1 EGF domains 12, 11, and 10 interacted with the C2, DSL, and EGF1 domains, and the Notch1 EGF9 and EGF8 domains bound to Jag1 EGF2 and EGF3 (Figs. 1D and 2A).
Fig. 2. Molecular determinants of the Notch1-Jag1JV1 interface.
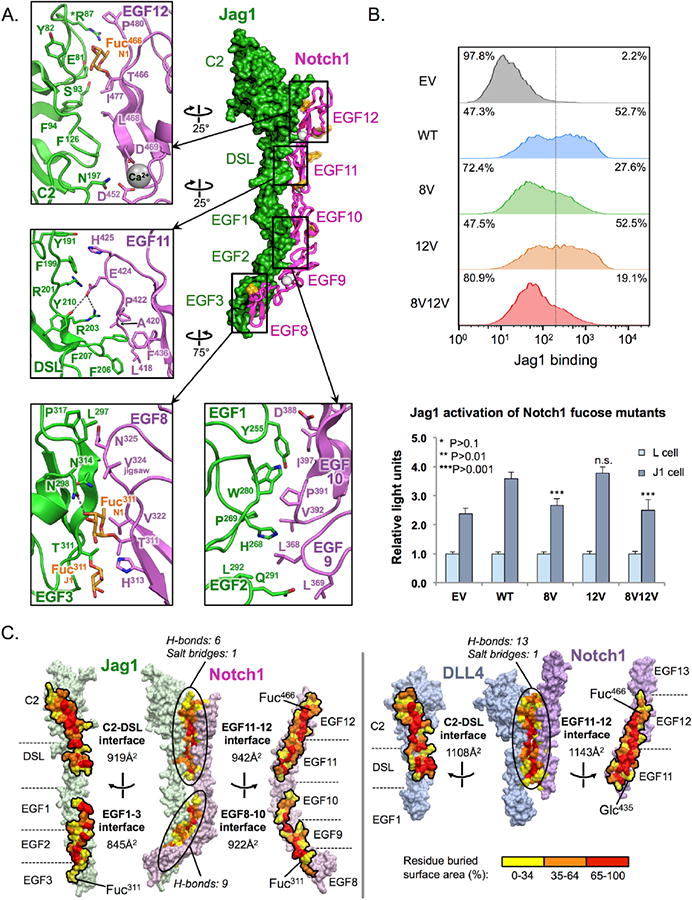
(A) Structure of the Notch1(8-12)-Jag1JV1(N-3) complex with selected interface contacts visualized in zoom windows. Polar contacts are depicted as grey dotted lines. The affinity-enhancing Jag1JV1 T87R mutation is marked with an asterisk. (B) Jag1-Notch1 binding and signaling assays were performed using WT Notch1 expressing cells, or cells expressing Notch1 proteins lacking interface O-fucose modifications in EGF8 (8V: Notch1 T311V), EGF12 (Notch1 T466V) or EGF8/EGF12 (8V12V: Notch1 T311V/T466V). Flow cytometry histograms depict the binding of Notch1 and Notch1 mutants to Jag1-Fc that was pre-clustered using a secondary antibody to the Fc region of human IgG. Bar graphs show the reporter activity of Notch1 or Notch1 mutant reporter cells stimulated by co-culture with L cells, or with L cells expressing Jag1 (J1 cells). Statistical significance of WT Notch1 versus each mutant was determined using one-way ANOVA. Bar graph shows mean +/- SD; two independent experiments n = 6 were analyzed. ***P < 0.0001; **P < 0.001; *P < 0.01; EV: empty vector. (C) Notch1-Jag1JV1 and Notch1-DLL4SLP complexes are depicted in an “open book” view, with residues colored according to percent buried surface area.
The interface between the Jag1 C2 domain and Notch1 EGF12 contained a functionally important Notch1 Thr466 O-fucose modification (Fig. 2A) (24), which formed a hydrogen bond (H-bond) with the backbone carbonyl of Tyr82 of the Jag1 C2 domain, and formed Van der Waals (VDW) interactions with the side chains of Glu81 and Tyr82, and with main chain atoms of Tyr82 and Gln84 (Fig. 2A and table S2). Elongation of the Thr466 O-fucose to a disaccharide by Fringe glycosyltransferase enzymes is known to enhance ligand binding to Notch1(11-13) (25). The engineered T87R mutation, present at the C2-EGF12 interface, may enhance affinity by creating a VDW contact between the extended Arg87 side chain and Pro480 of Notch1. At the Jag1 DSL-Notch1 EGF11 interface, prominent contacts included an H-bond and salt bridge formed between Glu424 of Notch1 and Tyr210 and Arg203 of Jag1, and a hydrophobic peg generated by insertion of Jag1 Phe206 and Phe207 side chains into a pocket in Jag1 (Fig. 2A and table S2). The interface formed between Notch1 EGFs 10, and 9 and Jag1 EGFs 1and 2, respectively, is anchored by Jag1 residues Tyr255, His268, Pro269, Trp280 and Leu292, which protrude outward from the bent “elbow” between EGF domains 1 and 2 to fill a cavity along Notch1 EGF domains 9- and 10 (Fig. 2A). Tyr255 of Jag1 is not conserved in DLL1 and DLL4, and thus could function as the selectivity filter that facilitates the biased engagement of this surface by Jag1 (fig. S3).
An O-linked fucose modification on Thr311 of Notch1 EGF8 contacted EGF3 of Jag1 by forming an H-bond with the side chain of Jag1 Asn298 (Fig. 2A and table S2), raising the possibility that distinct glycan signatures can exert combinatorial effects on ligand responsiveness (16, 26). On the opposing side of the interface, the O-fucose modification of Thr311 of Jag1 EGF3 formed a minor VDW contact with His313 of Notch1 EGF8 (Fig. 2A). Also present at the Notch1 EGF8-Jag1 EGF3 interface was a conserved Val324 residue that, when mutated to a methionine (termed the “jigsaw” mutation), selectively ablates Notch1-Jag1 interactions (Fig. 2A and fig. S3) (21).
To elucidate the relative functional contributions of Notch1 interface O-fucose moieties, we measured binding and receptor activation in cells that expressed either WT or Notch receptors lacking O-fucose modifications. Reporter cells were generated in which fucose-modified Notch1 residues EGF8 Thr311, EGF12 Thr466, or EGF8 Thr311 and EGF12 Thr466 were mutated from threonine to valine (fig. S4). Cells lacking both EGF 8 and EGF12 fucose moieties exhibited a pronounced decrease in Jag1 binding but a similar decrease in activation to that seen in cells lacking only the EGF8 fu-cose (P<0.0001) (Fig. 2B). Thus, the O-fucose of Notch1 EGF8 appears to be of greater functional importance than the EGF12 modification in the context of Jag1, and both O-glycans appear to be required for optimal receptor engagement.
When comparing the binding interfaces of the Notch1(11-13)-DLL4SLP(N-1) and Notch1(8-12)-Jag1JV1(N-3) complexes, the former buries ∼200 Å2 more surface area than the analogous interface on Jag1 (Fig. 2C). The smaller Notch EGF11 and 12 binding surface of Jag1 is compensated by additional contacts with Notch1 EGF8 through 10 (Fig. 2C and table S2).
We examined the mechanical requirements for Notch activation with a tension gauge tether (TGT) assay. Cells were exposed to ligands tethered to a surface through rupturable tethers (3, 5) with estimated tension tolerances of 4 pN, 12 pN and 54 pN (Fig. 3). WT Jag1 activated Notch when it was linked to the surface through the 12 pN and 54 pN TGTs but not through the 4 pN TGT. Thus, Notch activation through WT Jag1, like that through DLL1 (5), appears to require tension larger than 4 pN but smaller than 12 pN. These data are consistent with the requirement of 5 pN force for NRR conformational change and cleavage (4). In contrast, Jag1JV1 activated Notch even with the 4 pN TGT (Fig. 3). We attribute the differential tension requirements to the difference in the frequency of events that expose the NRR cleavage site. The probability that a Notch receptor is engaged with a lig-and during the period of cellular tension application to the receptor will be higher if the affinity is higher. Therefore, JagJV1 would allow a higher frequency of pulling events than would WT Jag1, and even through the 4 pN TGTs, a sufficient number of NRRs may experience the ∼5 pN tension needed for their cleavage and signaling activation (Fig. 3). In support of this model, we observed a differential tension requirement when we changed ligand density instead of affinity (fig. S5).
Fig. 3. Different molecular tension thresholds for receptor activation by various Notch ligands.
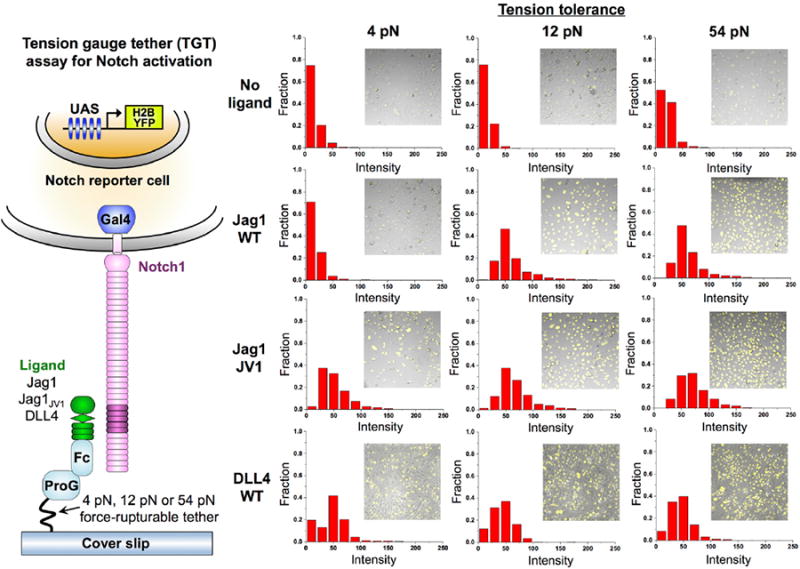
Molecular tension requirements for Notch activation were determined using a TGT assay (3, 5). Fc-tagged Jag1(N-3), Jag1JV1(N-3) and DLL4(N-3) were attached to surfaces using tethers calibrated to rupture upon application of 4, 12 or 54 pN of tension. Binding to surfaces was mediated by Fc-capture with protein G (ProG). Cells expressing Notch1 with the NICD replaced by a Gal4 activator and a H2B-YFP-reporter controlled by an upstream activator sequence (UAS) (30) were cultured on the surfaces and reporter activity was monitored. Each panel shows an example composite image of micrographs of adhered cells with YTF fluorescence and a YFP intensity histogram.
Notch-ligand affinity is determined by association (kon) and dissociation (koff) rate constants, and the latter may also be a function of tension. We therefore used biomembrane force probe (BFP) force-clamp spectroscopy to measure lifetimes of single Notch1(8-12)-Jag1(N-3) bonds in a range of tensile forces (27). At “low” tension values below 9 pN (the tension range relevant to Notch activation according to the TGT analysis), the lifetime of the bound state was longer for JagJV1 than for Jag1, suggesting that much of the higher affinity of JagJV1 at zero force may reflect a lower koff (Fig. 4A). The bond lifetime became comparable between Jag1 and Jag1JV1 at forces of 9 pN or greater, probably because of tension-induced changes in lifetimes, which increased for Jag1 (a ‘catch bond’) but decreased for JagJV1 (a ‘slip bond’) (Fig. 4A). Therefore, JagJV1 appears to adopt a high affinity conformation at low forces, whereas WT Jag1 adopts the high affinity conformation only under tension, which is consistent with a conformational switch we observe in the Notch1-JagJV1 structure (see below).
Fig. 4. Conformational changes in Jag1 domains implicate a catch bond in Notch1 engagement.
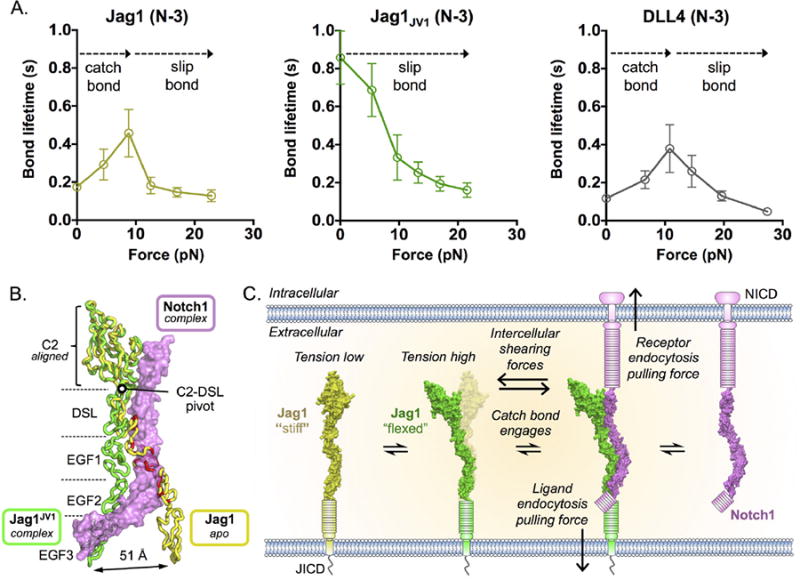
(A) BFP force-clamp spectroscopy was used to measure the average bond lifetimes between Notch1(8-12) and WT Jag1(N-3), Jag1JV1(N-3) or DLL4(N-3) as a function of applied force. Lifetimes were binned into different force groups and represented as the average lifetime in each bin associated with the standard error of the mean of the lifetimes in that bin. (B) The C2 domains of Jag1JV1(N-3) (green) in complex with Notch1(8-12) (pink) and unbound Jag1(N-3) (yellow, PDB ID: 4CC0) were aligned to highlight hinge-like motions that occur upon Notch1 binding (17). Steric clashes predicted to occur between the apo conformation of Jag1 and Notch1 are colored red. (C) A possible model for a catch bond in Notch-ligand engagement. Unbound, “stiff” Jag1 incurs molecular force, either due to endocytosis or intercellular shearing, and then becomes “flexed” so that it may optimally engage Notch1. The catch bond may then extend the Notch1-Jag1 bond lifetime such that the force may be permeated to the NRR to drive activation.
DLL4 binds Notch with higher affinity than does Jag1 (Fig. 1C and fig. S6), and may even activate Notch at tensions as low as 1 pN (4). TGT experiments with DLL4 indicated that Notch was activated even with the 4 pN TGT, in contrast to Jag1, which required tension above 4pN (Fig. 3), suggesting that mechanical tuning through affinity modulation may also occur in natural systems. BFP experiments revealed that DLL4 had a similar bond lifetime vs. tension profile to that of Jag1, indicating that the higher affinity of DLL4 results from its higher kon (Fig. 4A).
A hallmark of catch-bond forming proteins is a hingelike motion of domains involved in ligand binding (28, 29). In the Notch1-Jag1 complex, several Jag1JV1 domains undergo hinge-like movements, the most dramatic of which is a 32° pivot about the C2-DSL linker that shifts the position of EGF3 51Å between the bound and unbound structures (Fig. 4B and fig. S7). The S32L substitution in the Jag1JV1 linker region (fig. S1) may enhance affinity by favoring the “flexed” conformation observed in the Notch1-Jag1JV1 structure (Fig. 4C and fig. S7).
The formation of a catch bonds has broader repercussions within the context of cellular Notch signaling. For Notch to become activated, the initial, low affinity ligand interaction must persist long enough not only to propagate cellular forces to the NRR, but also to maintain the NRR in an unfolded state until its S2 site may be cleaved by an ADAM protease. We found applied force to extend Notch-ligand bond lifetimes (Fig. 4A), which would increase the probability of satisfying the above requirements such that a receptor can become activated (Fig. 4C). The apparently different force requirements needed to activate Notch signaling by different ligands may be a mechanism, perhaps in concert with lipid-binding (17), for Notch-expressing cells to tune their sensitivity and discriminate between the multitude of Notch ligands.
Supplementary Material
Acknowledgments
We thank A. Velasco, K. Jude, B. Bell and S. Fischer for assistance; M. Elowitz for providing Notch reporter cells; the Advanced Photon Source staff for support and access to the 23-ID-B beamline; the staff of the Stanford Synchrotron Radiation Lightsource, which is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract DE-AC02-76SF00515. This work was supported by NIH-1R01-GM097015 and Ludwig Cancer Foundation (K.C.G.); the HHMI (K.C.G., T.H., B.C.K.); an NSF Physics Frontier Center grant (PHY 1430124) (T.H.); an Irvington Postdoctoral Fellowship from the Cancer Research Institute and NIH-K99-CA204738 (V.C.L); NIH-R01-AI044902 (C.Z. and C.G); and NIH-R01-GM061126 (S.K. and R.S.H.). Coordinates and structure factors of the Notch1-Jag1JV1 complex have been deposited in the Protein Data Bank with accession code 5UK5.
Footnotes
References and Notes
- 1.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. Medline. [DOI] [PubMed] [Google Scholar]
- 2.Chapman G, Major JA, Iyer K, James AC, Pursglove SE, Moreau JLM, Dunwoodie SL. Notch1 endocytosis is induced by ligand and is required for signal transduction. Biochim Biophys Acta. 2016;1863:166–177. doi: 10.1016/j.bbamcr.2015.10.021. Medline. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. doi:110.1126/science.1231041. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, Blacklow SC. Mechanical allostery: Evidence for a force requirement in the proteolytic activation of Notch. Dev Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. doi:110.1016/j.devcel.2015.05.004. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury F, Li ITS, Ngo TTM, Leslie BJ, Kim BC, Sokoloski JE, Weiland E, Wang X, Chemla YR, Lohman TM, Ha T. Defining single molecular forces required for Notch activation using nano yoyo. Nano Lett. 2016;16:3892–3897. doi: 10.1021/acs.nanolett.6b01403. doi:110.1021/acs.nanolett.6b01403. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. doi:110.1038/nsmb1227. Medline. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. doi:110.1038/19083. Medline. [DOI] [PubMed] [Google Scholar]
- 8.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. Medline. [DOI] [PubMed] [Google Scholar]
- 9.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. doi:110.1016/S1097-27650080417-7. Medline. [DOI] [PubMed] [Google Scholar]
- 10.Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, Redfield C, Baron M, Lea SM, Handford PA. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. doi:110.1038/nsmb.1457. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisshuhn PC, Sheppard D, Taylor P, Whiteman P, Lea SM, Handford PA, Redfield C. Non-linear and flexible regions of the human Notch1 extracellular domain revealed by high-resolution structural studies. Structure. 2016;24:555–566. doi: 10.1016/j.str.2016.02.010. doi:110.1016/j.str.2016.02.010. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. doi:110.1038/35019553. Medline. [DOI] [PubMed] [Google Scholar]
- 13.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. doi:110.1074/jbc.275.13.9604. Medline. [DOI] [PubMed] [Google Scholar]
- 14.Okajima T, Irvine KD. Regulation of Notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. doi:110.1016/S0092-86740201114-5. Medline. [DOI] [PubMed] [Google Scholar]
- 15.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. doi:110.1073/pnas.0831126100. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakuda S, Haltiwanger RS. Deciphering the fringe-mediated Notch code: Identification of activating and inhibiting sites allowing discrimination between ligands. Dev Cell. 2017;40:193–201. doi: 10.1016/j.devcel.2016.12.013. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chillakuri CR, Sheppard D, Ilagan MXG, Holt LR, Abbott F, Liang S, Kopan R, Handford PA, Lea SM. Structural analysis uncovers lipid-binding properties of Notch ligands. Cell Rep. 2013;5:861–867. doi: 10.1016/j.celrep.2013.10.029. doi:110.1016/j.celrep.2013.10.029. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kershaw NJ, Church NL, Griffin MDW, Luo CS, Adams TE, Burgess AW. Notch ligand delta-like1: X-ray crystal structure and binding affinity. Biochem J. 2015;468:159–166. doi: 10.1042/BJ20150010. doi:110.1042/BJ20150010. Medline. [DOI] [PubMed] [Google Scholar]
- 19.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. doi:110.1016/0092-86749190064-6. Medline. [DOI] [PubMed] [Google Scholar]
- 20.Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–853. doi: 10.1126/science.1261093. doi:110.1126/science.1261093. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto S, Charng WL, Rana NA, Kakuda S, Jaiswal M, Bayat V, Xiong B, Zhang K, Sandoval H, David G, Wang H, Haltiwanger RS, Bellen HJ. A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science. 2012;338:1229–1232. doi: 10.1126/science.1228745. doi:110.1126/science.1228745. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC, Blacklow SC. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. 2013;288:25477–25489. doi: 10.1074/jbc.M113.454850. doi:110.1074/jbc.M113.454850. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. doi:110.1038/nature01605. Medline. [DOI] [PubMed] [Google Scholar]
- 24.Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. J Biol Chem. 2005;280:32133–32140. doi: 10.1074/jbc.M506104200. doi:110.1074/jbc.M506104200. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS, Handford PA. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc Natl Acad Sci USA. 2014;111:7290–7295. doi: 10.1073/pnas.1319683111. doi:110.1073/pnas.1319683111. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu A, Lei L, Irvine KD. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. J Biol Chem. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. doi:110.1074/jbc.M505569200. Medline. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Lou J, Zhu C. Forcing switch from short- to intermediate- and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. J Biol Chem. 2010;285:35967–35978. doi: 10.1074/jbc.M110.155770. doi:110.1074/jbc.M110.155770. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas W. For catch bonds, it all hinges on the interdomain region. J Cell Biol. 2006;174:911–913. doi: 10.1083/jcb.200609029. doi:110.1083/jcb.200609029. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarti S, Hinczewski M, Thirumalai D. Plasticity of hydrogen bond networks regulates mechanochemistry of cell adhesion complexes. Proc Natl Acad Sci USA. 2014;111:9048–9053. doi: 10.1073/pnas.1405384111. doi:110.1073/pnas.1405384111. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. doi:110.1038/nature08959. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. doi:110.1038/nprot.2006.94. Medline. [DOI] [PubMed] [Google Scholar]
- 32.Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. doi:110.1016/j.str.2006.09.005. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. doi:110.1016/S0076-68799776066-X. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. doi:110.1038/nprot.2009.2. Medline. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. doi:110.1107/S0907444909052925. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. doi:110.1107/S0021889807021206. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. doi:110.1107/S0907444904019158. Medline. [DOI] [PubMed] [Google Scholar]
- 38.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. doi:110.1107/S0907444906005270. Medline. [DOI] [PubMed] [Google Scholar]
- 39.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. doi:110.1107/S0021889805038987. [Google Scholar]
- 40.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. doi:110.1016/j.jmb.2007.05.022. Medline. [DOI] [PubMed] [Google Scholar]
- 41.The PyMOL Molecular Graphics System, v1.5.0.5 (Schrödinger).
- 42.Lee RA, Razaz M, Hayward S. The DynDom database of protein domain motions. Bioinformatics. 2003;19:1290–1291. doi: 10.1093/bioinformatics/btg137. doi:110.1093/bioinformatics/btg137. Medline. [DOI] [PubMed] [Google Scholar]
- 43.Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB, Haltiwanger RS. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. doi:110.1074/jbc.M111.268243. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, Evans EA, McEver RP, Zhu C. Monitoring receptor-ligand interactions between surfaces by thermal fluctuations. Biophys J. 2008;94:694–701. doi: 10.1529/biophysj.107.117895. doi:110.1529/biophysj.107.117895. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans E, Ritchie K, Merkel R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys J. 1995;68:2580–2587. doi: 10.1016/S0006-3495(95)80441-8. doi:110.1016/S0006-34959580441-8. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. doi:110.1016/S0006-34959874074-3. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Rahil Z, Li ITS, Chowdhury F, Leckband DE, Chemla YR, Ha T. Constructing modular and universal single molecule tension sensor using protein G to study mechano-sensitive receptors. Sci Rep. 2016;6:21584. doi: 10.1038/srep21584. doi:110.1038/srep21584. Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


