Abstract
Information embedded in different ubiquitin chains is transduced by proteins with ubiquitin-binding domains (UBDs) and erased by a set of hydrolytic enzymes referred to as deubiquitinases (DUBs). Understanding the selectivity of UBDs and DUBs is necessary for decoding the functions of different ubiquitin chains. Critical to these efforts is the access to chemically defined ubiquitin chains bearing site-specific fluorescent labels. One approach toward constructing such molecules involves peptide ligation using sortase (SrtA), a bacterial transpeptidase responsible for covalently attaching cell surface proteins to the cell wall. Here, we demonstrate the utility of SrtA in modifying individual subunits of ubiquitin chains. Using ubiquitin derivatives in which an N-terminal glycine is unveiled after protease-mediated digestion, we synthesized ubiquitin dimers, trimers, and tetramers with different isopeptide linkages. SrtA was then used in combination with fluorescent depsipeptide substrates to effect the modification of each subunit in a chain. By constructing branched ubiquitin chains with individual subunits tagged with a fluorophore, we provide evidence that the ubiquitin specific protease USP15 prefers Ub trimers, but has little preference for a particular isopeptide linkage. Our results emphasize the importance of subunit-specific labeling of Ub chains when studying how DUBs process Ub chains.
Keywords: sortase, ubiquitin, ubiquitin chains, deubiquitinases, fluorophore-labeling, N-terminal modification
Table of Contents
We synthesize branched ubiquitin trimers modified with fluorophores at individual subunits using the bacterial transpeptidase, sortase. Using these novel fluorescent probes, we have created a first-of-its-kind fluorescence based assay for monitoring the linkage selectivity of deubiquitinases when hydrolyzing branched ubiquitin chains. Using this assay we show that the deubiquitinase, USP15 may have a preference for branched ubiquitin chains over their linear counterparts.
INTRODUCTION
The human genome encodes ~80 active deubiquitinating enzymes that remove ubiquitin (Ub) modifications and disassemble Ub chains.[1] The precise timing of their action is critical, as aberrant activity is associated with several human diseases, including many cancers and neurological disorders.[2] While the importance of DUBs in disease pathology has been well documented, molecular details underlying the activity of these enzymes remain poorly understood. Central to the challenge in studying DUBs is the diversity of potential substrates. Proteomics studies have identified over ~19,000 ubiquitination sites on 5,000 human proteins. Moreover, there is tremendous complexity just within Ub chains. Each of the eight amino groups of Ub (Met1, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) can be used to extend a Ub chain from a particular substrate.[3] As a result, chains of varying length can be generated bearing a single linkage (homotypic) or a mixture of linkages (heterotypic) in a linear or branched configuration.
Given the diversity in both substrate proteins and Ub chains, DUBs have the ability to regulate Ub-dependent pathways on several levels.[1b, 1c, 4] For instance, a DUB may directly recognize a specific ubiquitinated substrate and remove mono-Ub. In the context of Ub chains, DUBs have been shown to discriminate between the eight different linkages and selectively act on the end (exo) or in the middle (endo) of a chain. Many of these discoveries have been made using a limited set of defined Ub conjugates. A comprehensive biochemical characterization of DUBs requires access to a large array of substrates.
To address this issue a number of research groups, including ours, have been interested in developing chemical methods to synthesize Ub chains.[5] Recently we discovered that by replacing a lysine of interest with cysteine and installing an alkene at the C-terminus of Ub, free-radical thiol-ene coupling (TEC) could be used to effect isopeptide bond formation between Ub molecules.[5a, 6] This method is straightforward to carry out, as it requires standard recombinant proteins, minimal synthetic manipulations, and can be conducted under non-denaturing conditions. Most importantly, thiol-ene coupling provides access to large quantities of all linkage types and topologies that serve as functional surrogates of native oligomers.
In this study, we sought to expand the utility of Ub chains derived from TEC chemistry by installing fluorophores on individual subunits in a site-specific manner. Fluorescent labeling of Ub chains facilitates efforts to better understand the selectivity of DUBs. Current approaches to the site-specific installation of fluorophores employ cysteine reactive dyes.[5e, 7] Although these methods have been successfully applied to the study of homotypic chains, this strategy is not compatible with TEC chemistry. The bacterial transpeptidase, sortase (SrtA), is a powerful tool for site-specific labeling of proteins and is orthogonal to TEC chemistry.[8] We demonstrate that SrtA can be used to install fluorophores on specific subunits within a large variety of Ub chains. Our work has led to the discovery that the ubiquitin specific protease USP15 prefers branched trimers over dimers, but does not have a preference for a particular isopeptide linkage.
RESULTS & DISCUSSION
SrtA recognizes the small pentapeptide motif LPXTG (X is any amino acid) and cleaves the scissile bond between Thr and Gly.[9] The resulting acyl-enzyme intermediate is intercepted by an N-terminal oligoglycine motif to furnish a new bioconjugate. Utilizing SrtA to catalyze fluorophore-labeling of Ub chains therefore requires a Ub monomer carrying an N-terminal oligo-Gly appendage. To prepare chains for SrtA-mediated modification, we introduced a tobacco etch virus (TEV) protease recognition sequence (ENLYFQ) at the N-terminus of Ub followed by a pentaglycine (5xGly) motif. We also synthesized depsipeptides I and II using Fmoc solid phase peptide synthesis (SPPS) (Scheme 1). Recent studies have shown that peptides containing a depsipeptide linkage between the scissile Thr-Gly bond greatly enhance the efficiency of N-terminal SrtA ligation by rendering the reaction irreversible.[8b, 10]
Scheme 1.
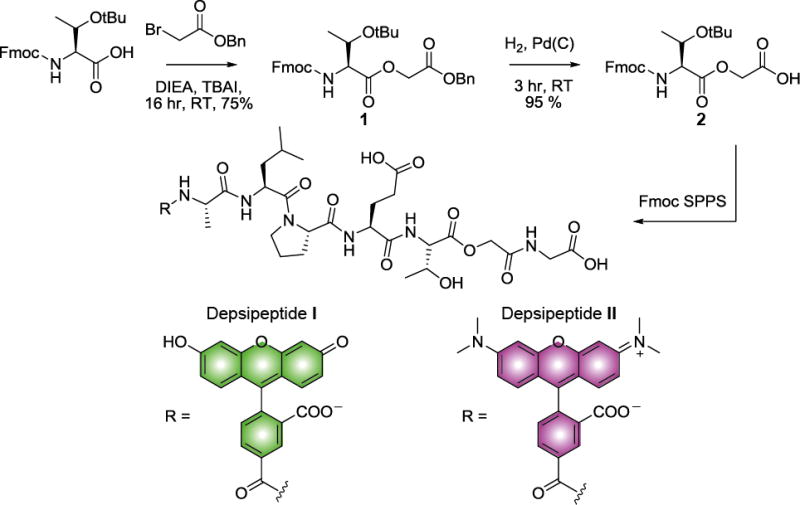
Synthesis of depsipeptides I and II. DIEA; diisopropylethylamine, TBAI; tetrabutylammonium iodide, rt; room temperature, SPPS; solid phase peptide synthesis.
With building blocks in hand, we explored the SrtA-catalyzed fluorophore-labeling of Lys63-linked Ub dimers. After the dimers were synthesized using TEC chemistry, TEV protease was used to unmask the 5xGly motif. Labeling experiments were then performed using 0.5 and 10 equivalents of SrtA and depsipeptide I, respectively. Under these conditions, we observe two new bands by Coomassie and fluorescence imaging at early time points (Figures 1B and 1C). The lower molecular weight band corresponds to a singly labeled species, whereas the higher molecule weight band represents a doubly labeled dimer. By ninety minutes the unlabeled and singly labeled dimers are completely converted to fully labeled dimers. To confirm the SDS-PAGE results, we obtained the intact mass of the products by MALDI-TOF mass spectrometry (Figure S1). The results of this analysis indicate that both subunits are indeed conjugated to a fluorescein-labeled peptide. Encouraged by these findings, we synthesized Lys6-, Lys11-, and Lys48-linked dimers using TEC chemistry, and subjected them to our optimized labeling conditions (Figure S1). As judged by SDS-PAGE and MALDI-TOF analysis, both subunits of the dimers are efficiently labeled with fluorophores; only trace amounts of the singly labeled species are detected.
Figure 1.
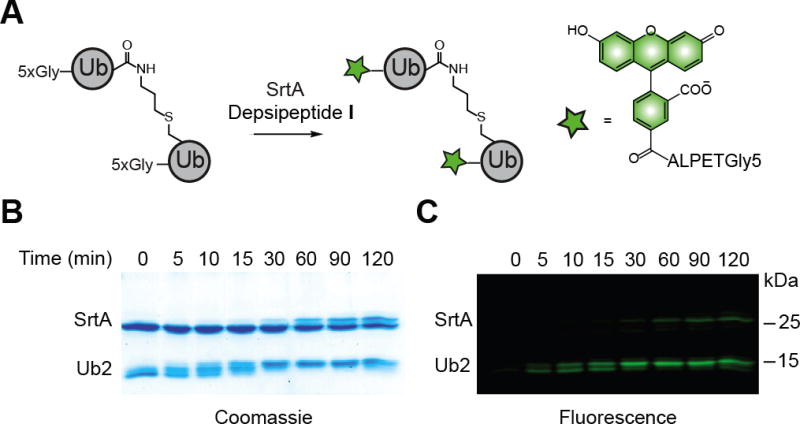
Site-specific labeling of Ub dimers using SrtA. A) Labeling schematic. B) Coomassie-stained 15 % SDS-PAGE gel showing the transition from unlabeled to singly labeled to fully labeled Ub dimer. C) Fluorescence image of the same gel. Fluorescence was detected using the blue laser (473 nm) on a Typhoon FLA 9500 imager (GE Healthcare) equipped with a 515–545 nm filter.
Next, we sought to label longer chains. A series of Lys63-linked chains were generated by TEC chemistry, ranging in length between two and five subunits. After incubating with TEV protease, the chains were labeled at their individual N-termini using depsipeptide I (Figure 2B). It is important to note the amounts of SrtA and depsipeptide I had to be adjusted to account for the total concentration of Ub monomer in each reaction. As with the Ub dimers, we detect robust labeling of chains of different length. Fluorescence imaging shows single bands for each of the discrete oligomers. To measure the extent of labeling, we then purified the chains and measured the dye to protein ratio. This required removal of excess depsipeptide, TEV, and SrtA and TEV (Figure S2 and S3). Ratios of dye to protein indicate that individual subunits are conjugated to the fluorophore (Figure 2C). In this case, the resulting products are similar to those obtained using N-terminal fluorophore-labeled mono-Ub and Ub conjugating enzymes.[11] However, since thiol-ene coupling is more versatile than enzymatic methods, the site-specific installation of fluorophores can be applied toward the synthesis of fluorescent chains bearing different linkages and topologies.
Figure 2.
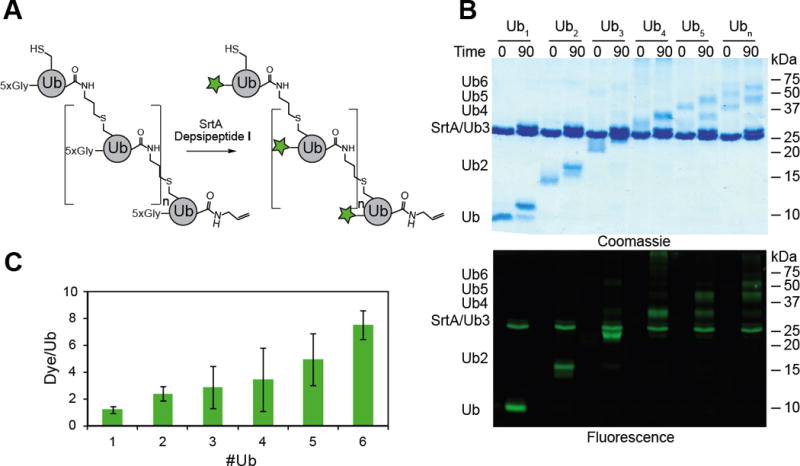
Site specific labeling of Ubiquitin chains of discrete length. A) General scheme for the labeling of Ub chains of different length. B) SDS-PAGE analysis of labeling reactions. Ub oligomers (40 μM adjusted for the relative concentration of monoUb) were incubated with TEV protease (2 μM) for 2 hours at room temperature prior to the addition of SrtA (20 μM) and I (400 μM). Time points were taken at the indicated times by quenching an aliquot with 6× Laemmli loading buffer and separated on a 15 % SDS-PAGE gel. The top gel is the Coomassie-stained SDS-PAGE gel of labeled Ub chains before and after 90 min incubation with SrtA and depsipeptide I. In each reaction a discrete band can be seen for the labeled oligomer with a mass shift consistent with that of the added peptides. The bottom image is the fluorescence scan of the same gel. C) Quantification of labeled Ub chains. Chains were separated from SrtA, TEV protease, and excess peptide I using a two step purification strategy employing Ni2+ affinity chromatography to remove His-tagged enzymes and a desalting column to remove excess I. The ratio of dye to protein is plotted for each oligomer of different length. Error bars show the replicates of three measurements.
A topology of particular interest is a branched Ub chain. Branched Ub chains in which a single Ub has been modified at positions Lys11 and Lys48 have been shown to advance the cell cycle through enhanced degradation by the proteasome.[12] The mechanisms by which Lys11/Lys48 branched chains trigger degradation are, however, unclear. Recent data suggests that single linkage (homotypic) Lys11 chains bind to Ub receptors on the proteasome with low affinity, whereas mixed linkage (heterotypic) Lys11 chains strongly interact with the proteasome.[12] These results suggest that in contrast to the prevailing view that branched Ub chains may adopt distinct conformations relative to their linear counterparts.[13] Access to fluorescent, branched chains is therefore important for testing this hypothesis. We constructed branched Lys6/Lys48 and Lys11/Lys48 Ub trimers containing a single oligoglycine motif at the base of the chain. Introduction of fluorescent labels at the base proceeds efficiently (Figure S4), demonstrating that SrtA-mediated modification enables unprecedented access to fluorophore-labeled branched oligomers.
Having established a method for the synthesis and labeling of Ub chains, we evaluated whether DUBs accept the modified dimers as substrates. We performed a series of assays comparing the hydrolysis of labeled dimers to that of unmodified dimers (Figures 3A and B). For these experiments, we chose two different members of the ubiquitin-specific protease (USP) family of DUBs, USP7 and USP15. The USP DUBs have previously been shown to display little selectivity toward different Ub dimers, making them ideal candidates for testing whether a fluorophore label on different dimers perturbs activity.[14] With both USP7 and USP15, we observe robust chain hydrolysis similar to that of the unmodified dimers. These results indicate that the introduction of a fluorophore does not impede the ability to cleave dimers.
Figure 3.
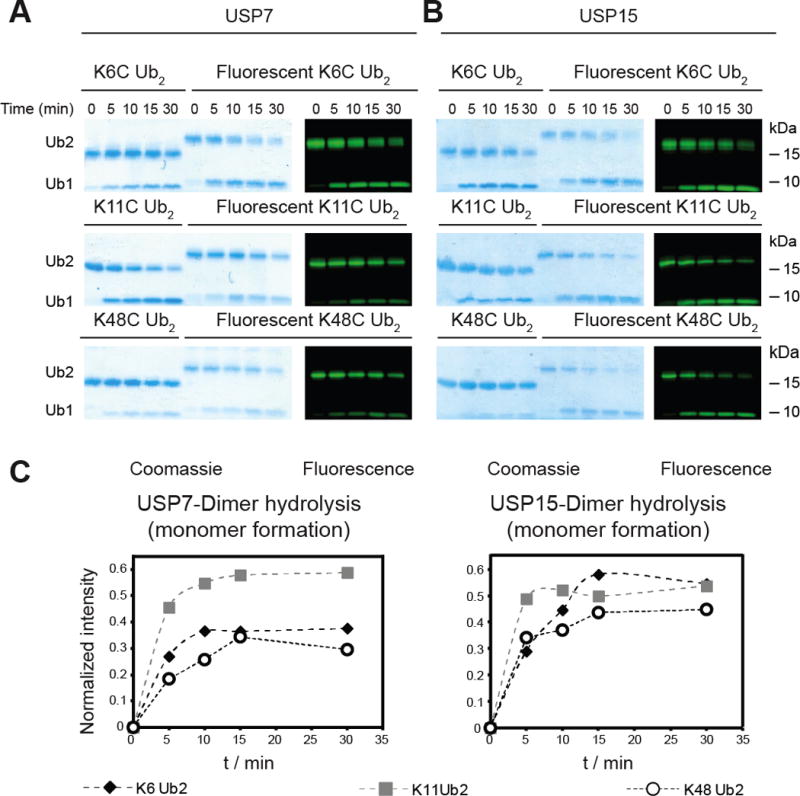
Ub dimers labeled with SrtA are suitable substrates for DUBs. A) USP7-catalyzed hydrolysis of Lys6-, Lys11-, and Lys48-linked unlabeled and fluorescently labeled Ub dimers. B) USP15-catalyzed hydrolysis of Lys6-, Lys11-, and Lys48-linked unlabeled and fluorescently labeled Ub dimers. For both sets of DUB assays, Ub dimer (10 μM) was incubated with the indicated DUB (100 nM) at 37 °C. Time points were taken at the indicated time by quenching an aliquot of reaction mixture with 6x Laemmli loading buffer. Samples were then separated on a 12 % Bis-Tris NuPAGE gel (Invitrogen). C) Densitometric analysis of fluorescent gels in panel B following the appearance of fluorescent monoUb over time. The intensity of the monomer band was normalized against the intensity of the fluorescent dimer at t = 0. The dotted lines do not represent a fit to a mathematical expression. Densitometry was performed on the fluorescence image using ImageJ (NIH) to quantify the relative amounts of monoUb.
We then tested branched trimers as substrates. As with the fluorescently labeled dimers, the fluorophore-labeled trimers were hydrolyzed by USP7 and USP15 with similar efficiency as the unlabeled oligomers (Figures 4B and C). With USP15, the fluorescent gels show the TEC-derived Lys6/Lys48 branched trimer is rapidly converted from trimer to dimer, however, hydrolysis of the resulting dimer occurs at a much slower rate. Densitometric analysis confirmed these observations, showing fast appearance of fluorescent dimer and a significant a delay before the appearance of fluorescent monomer in the gels (Figure 4C). Considering the fluorophore is tethered to the base of the trimer, complete hydrolysis from trimer to monomer is necessary for the appearance of fluorescent monomer. The delay in monomer formation was also observed when the TEC-derived Lys11/Lys48 branched trimer was tested against USP15, but to a lesser extent than the Lys6/Lys48 trimer. Selective hydrolysis of branched chains was not observed with USP7. The results with USP15 were unexpected, as this DUB displays little selectivity toward homotypic chains,[14c] but here it appears that cleavage of a branched chain is preferred over that of the dimer product.
Figure 4.
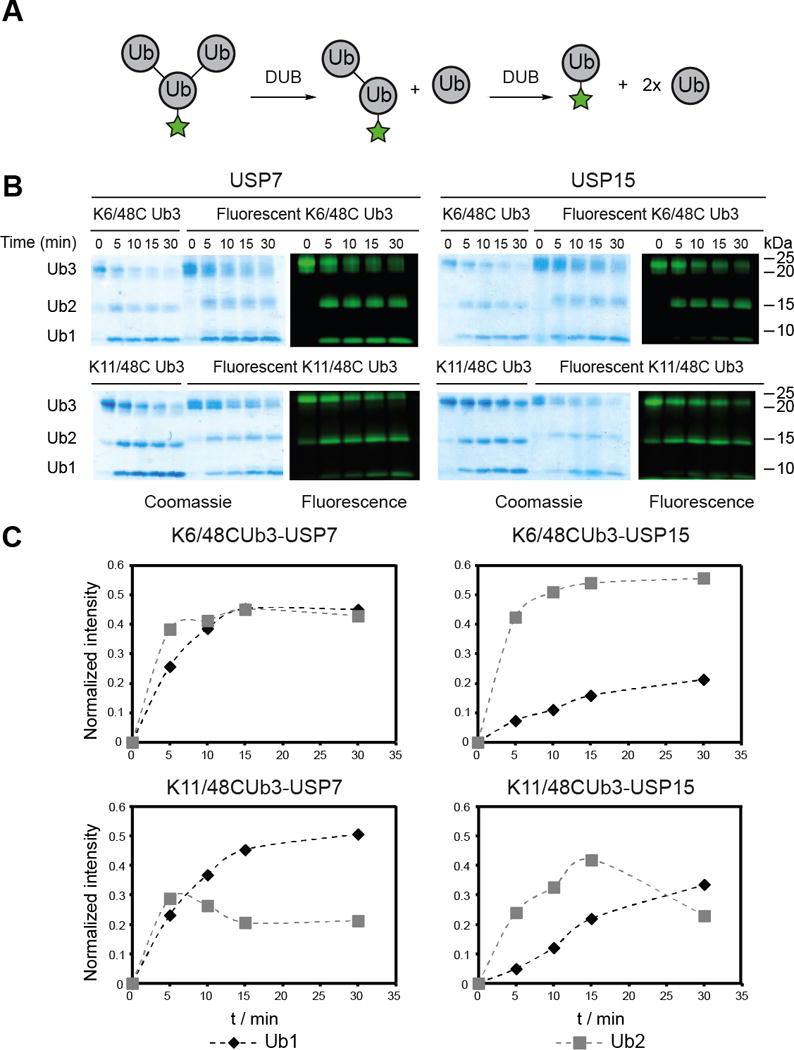
DUB-catalyzed hydrolysis of labeled branched Ub chains. A) General scheme for the DUB-catalyzed hydrolysis of branched Ub labeled with peptide I at the base. B) Time-course analysis of the USP7- and USP15-catalyzed hydrolysis of TEC-derived Lys6/Lys48 and Lys11/Lys48 branched trimers. For both sets of DUB assays, the branched trimer (10 μM) was incubated with the indicated DUB (100 nM) at 37 °C. Aliquots of reaction mixture were quenched by the addition of 6× Laemmli loading buffer at the indicated time points and separated on a 12 % Bis-Tris NuPAGE gel (Invitrogen). C) Densitometric analysis of fluorescent gels in panel B following the appearance of fluorescent mono- and di-Ub over time. The intensities of the bands are normalized to the intensity of the fluorescent branched trimer at t = 0 min.
To confirm our findings with TEC-derived chains and ensure trimer hydrolysis is a consequence of preferential branched chain hydrolysis—and not simply a preference for a specific linkage within the trimer—we synthesized segmentally, fluorophore-labeled native isopeptide-linked trimers. The bacterial E3 ligase NleL was recently shown to build primarily Lys6- linked chains.[15] Thus, we used NleL in conjunction with the Lys48 linkage-specific conjugating enzyme UBE2R1 to synthesize Lys6/Lys48 branched trimers modified with an N-terminal 5xGly motif at specific subunits (Figure 5A). The connectivity of each of the native branched trimers was confirmed using minimal trypsinolysis followed by high-resolution tandem mass spectrometry (MS/MS) on a Fourier-transform ion cyclotron resonance mass spectrometer (FT-ICR) (Figures S5–8).[16] SrtA was then employed to append a peptide carrying either fluorescein (depsipeptide I) or 5-carboxytetramethylrhodamine (TAMRA; depsipeptide II).
Figure 5.
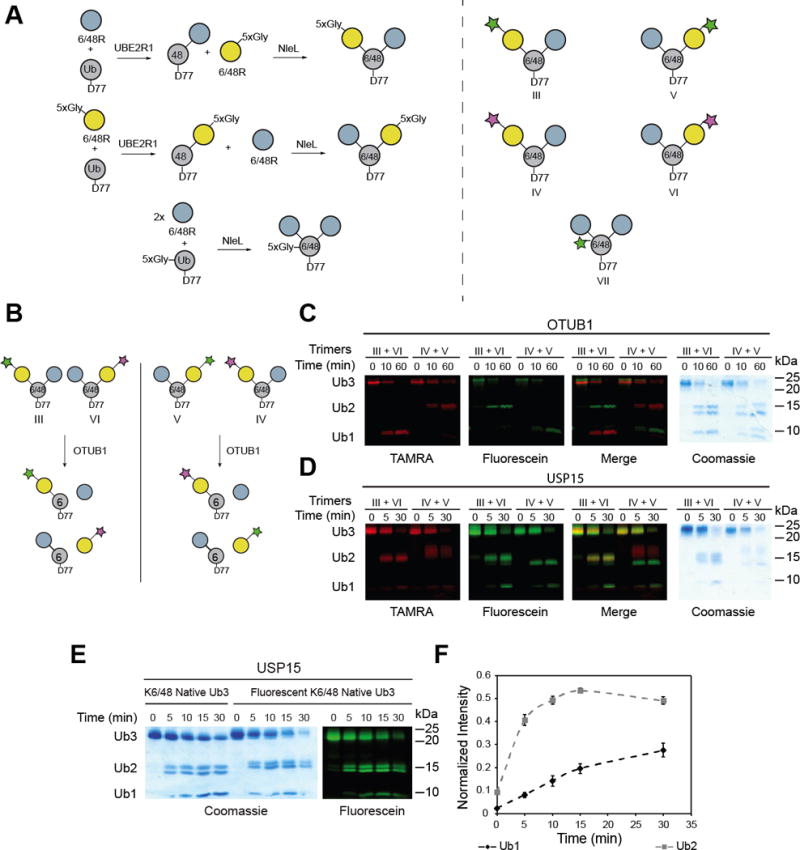
Selectivity of USP15 toward native isopeptide-linked branched trimers. A) Scheme depicting the synthesis of segmentally labeled native isopeptide-linked branched trimers (left). Structures of the segmentally labeled branched trimers used in this study (right). B) Scheme showing how segmental labeling informs on the linkage selectivity of a DUB. The green star denotes Ub labeled with depsipeptide I, the pink start denotes Ub labeled with depsipeptide II. C) Complementary branched trimers labeled with either depsipeptide I or II were mixed at an equal ratio and subjected to hydrolysis by the Lys48 selective DUB OTUB1. Aliquots were taken at indicated time points and quenched by the addition of 6 × Laemmli loading buffer prior to separation on a 12 % Bis-Tris NuPAGE gel (Invitrogen). Gels were then imaged using a Typhoon FLA 9500 imager (GE Healthcare). D) USP15 hydrolysis of complementary labeled branched trimers shows that USP15 does not display linkage selectivity when hydrolyzing Lys6/Lys48 linked branched trimers. D) Time-course analysis of the USP15-catalyzed hydrolysis of native Lys6/Lys48 branched trimer labeled with depsipeptide I at the base (left). E) Densitometric analysis of the fluorescent gel in Figure S9 following the appearance of fluorescent mono- and di-Ub over time. The error bars represent the standard deviation of three replicates.
Since the Ub subuntis are essentially “color-coded” in segmentally labeled native branched trimers we devised a competition based DUB assay to report on the linkage selectivity. Differentially labeled trimers were mixed and subjected to the Lys48 linkage-specific DUB OTUB1 (Figure 5B). When either fluorescein or TAMRA are attached to the Lys48-linked distal Ub subunit, a fluorescent monomer rapidly appears in the presence of OTUB1 (Figure 5C). By contrast, labeling of the Lys6-linked distal Ub subunit results in the formation of only fluorescent dimers, indicative of Lys48 linkage specificity. Using USP15 instead of OTUB1 shows there is little selectivity for the Lys6 or Lys48 isopeptide bond, as fluorescent dimers and monomers are formed regardless of the position of the fluorescent Ub subunit (Figure 5D). Thus, our results demonstrate that segmental labeling can inform on the linkage selectivity of DUBs and USP15 does not have a predilection for a particular linkage within a branched trimer.
We then sought to reaffirm that USP15 has a proclivity for branched trimers by shifting our focus on native trimers labeled with a fluorophore at the base of the chain. The experiments are identical to those performed with TEC-derived chains. USP15-catalyzed hydrolysis of the native trimers shows the two possible fluorescent dimers appear simultaneously, again indicating a lack of linkage selectivity (Figure 5E). More importantly, however, the formation of fluorescent mono-Ub is significantly postponed relative to the dimer, similar to the USP15-catalyzed hydrolysis of TEC-derived chains (Figures 5E and 5F). These results further demonstrate that TEC-derived Ub oligomers are a good model for native chains and suggest USP15 prefers branched trimers over Ub dimers. The analysis of Coomassie stained gels does not yield this level of insight, as it is difficult to discern which Ub moiety is represented by the mono-Ub species. Our findings with USP15 therefore underscore the importance of subunit-specific labeling of Ub chains when studying how DUBs process Ub chains.
Our observation that USP15 rapidly cleaves branched trimers leaving behind a dimer that is transformed more slowly into monomers likely has physiological significance. Recent studies have shown that USP15 counteracts the ability of the E3 Ub ligase Parkin to promote the clearance of damaged mitochondria through a degradative process referred to as mitophagy.[17] Parkin synthesizes heterogeneous chains with Lys6, Lys11, Lys48, and Lys63 linkages.[18] Failure to produce these linkages, particularly Lys6 and Lys63, reduces the efficiency of mitophagy.[19] Although the precise topology of the chains Parkin generates is unknown, the finding that USP15 exhibits little linkage selectivity but prefers a branched trimer suggests this DUB might mitigate mitophagy by removing branched conjugates. Another possibility is that USP15 could restrict the length of Ub chains produced by Parkin through an odd-even effect. With an even number of Ub molecules in a chain there is the possibility to form multiple inter-subunit contacts, which may preclude interactions with USP15. With an odd number there will always be a Ub molecule unable to form inter-subunit contacts, making it accessible to USP15. The ramifications of an odd-even effect would be an accumulation of proteins decorated with Ub dimers, which may not be sufficient to drive mitophagy. Future work will focus on how the biochemical activity of USP15 relates to its biological function.
In conclusion, we demonstrate the utility of SrtA in catalyzing the installation of fluorophores on individual subunits of homotypic and heterotypic chains. By combining TEC with SrtA chemistry, we now have the ability to probe the selectivity of a number of DUBs. We highlight this by using SrtA to site-specifically install a fluorophore at each individual subunit of a branched Ub trimer. We further demonstrate the utility of these novel probes by showcasing their ability to reveal insight into the linkage selectivity of DUBs when processing a branched Ub chains using both a linkage selective DUB (OTUB1) and non selective DUB (USP15). Furthermore, we provide evidence that USP15 may have a preference for branched trimers over their linear counterparts.
EXPERIMENTAL
Full details of all experimental methods are provided in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by research grant RO1GM110543 from the National Institutes of Health.
Footnotes
COMPETING INTEREST
The authors declare no competing financial interest.
AUTHOR CONTRIBUTIONS
SOC and ERS designed the experiments and wrote the manuscript. SOC, GHP, JCZ, KKD, and RGG conducted the experiments. YG and ERS supervised the acquisition of data. All authors read and approved the final manuscript.
References
- 1.a) Nijman SMB, Luna-Vargas MPA, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]; b) Komander D, Clague MJ, Urbe S. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]; c) Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbé S. Physiol Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 2.a) Hoeller D, Dikic I. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]; b) Heideker J, Wertz IE. Biochem J. 2015;465:1–26. doi: 10.1042/BJ20140496. [DOI] [PubMed] [Google Scholar]; c) Tai HC, Schuman EM. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]; d) Popovic D, Vucic D, Dikic I. Nat Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 3.a) Komander D, Rape M. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]; b) Husnjak K, Dikic I. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]; c) Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]; d) Clague MJ, Heride C, Urbé S. Trends Cell Biol. 2015;25:417–426. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 4.a) Clague MJ, Coulson JM, Urbé S. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]; b) Reyes-Turcu FE, Ventii KH, Wilkinson KD. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Valkevich EM, Guenette RG, Sanchez NA, Chen Y-c, Ge Y, Strieter ER. J Am Chem Soc. 2012;134:6916–6919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dixon EK, Castañeda CA, Kashyap TR, Wang Y, Fushman D. Bioorg Med Chem. 2013;21:3421–3429. doi: 10.1016/j.bmc.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Castañeda C, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. J Am Chem Soc. 2011;133:17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. J Am Chem Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Nat Chem, Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]; f) Eger S, Scheffner M, Marx A, Rubini M. J Am Chem Soc. 2010;132:16337–16339. doi: 10.1021/ja1072838. [DOI] [PubMed] [Google Scholar]; g) Schneider T, Schneider D, Rösner D, Malhotra S, Mortensen F, Mayer TU, Scheffner M, Marx A. Angew Chem Int Ed. 2014;53:12925–12929. doi: 10.1002/anie.201407192. [DOI] [PubMed] [Google Scholar]; h) Hameed DS, Sapmaz A, Ovaa H. Bioconjugate Chem. 2016 doi: 10.1021/acs.bioconjugatechem.6b00140. [DOI] [PubMed] [Google Scholar]
- 6.Trang VH, Valkevich EM, Minami S, Chen YC, Ge Y, Strieter ER. Angew Chem Int Ed. 2012;51:13085–13088. doi: 10.1002/anie.201207171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Gururaja TL, Pray TR, Lowe R, Dong G, Huang J, Daniel-Issakani S, Payan DG. Methods Enzymol. 2005;399:663–682. doi: 10.1016/S0076-6879(05)99044-7. [DOI] [PubMed] [Google Scholar]; b) Yang LL, Kao M, Chen HL, Lim TS, Fann W, Chen R. Eur Biophys J. 2012;41:189–198. doi: 10.1007/s00249-011-0772-6. [DOI] [PubMed] [Google Scholar]; c) Kao MWP, Yang LL, Lin JCK, Lim TS, Fann W, Chen RPY. Bioconjugate Chem. 2008;19:1124–1126. doi: 10.1021/bc700480j. [DOI] [PubMed] [Google Scholar]; d) Trang VH, Rodgers ML, Boyle KJ, Hoskins AA, Strieter ER. Chembiochem. 2014;15:1563–1568. doi: 10.1002/cbic.201402059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Li YM, Li YT, Pan M, Kong XQ, Huang YC, Hong ZY, Liu L. Angew Chem. 2014;126:2230–2234. doi: 10.1002/anie.201310010. [DOI] [PubMed] [Google Scholar]; b) Williamson DJ, Fascione MA, Webb ME, Turnbull WB. Angew Chem Int Ed. 2012;51:9377–9380. doi: 10.1002/anie.201204538. [DOI] [PubMed] [Google Scholar]; c) Antos JM, Chew GL, Guimaraes CP, Yoder NC, Grotenbreg GM, Popp MWL, Ploegh HL. J Am Chem Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Witte MD, Theile CS, Wu T, Guimaraes CP, Blom AEM, Ploegh HL. Nat Protoc. 2013;8:1808–1819. doi: 10.1038/nprot.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Claessen JHL, Witte MD, Yoder NC, Zhu AY, Spooner E, Ploegh HL. Chembiochem. 2013;14:343–352. doi: 10.1002/cbic.201200701. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Swee LK, Guimaraes CP, Sehrawat S, Spooner E, Barrasa MI, Ploegh HL. Proc Natl Acad Sci USA. 2013;110:1428–1433. doi: 10.1073/pnas.1214994110. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) David Row R, Roark TJ, Philip MC, Perkins LL, Antos JM. Chem Commun. 2015 doi: 10.1039/c5cc04657b. [DOI] [PubMed] [Google Scholar]
- 9.a) Mazmanian SK, Liu G, Ton-That H, Schneewind O. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]; b) Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson DJ, Webb ME, Turnbull WB. Nat Protoc. 2014;9:253–262. doi: 10.1038/nprot.2014.003. [DOI] [PubMed] [Google Scholar]
- 11.a) Lu Y, Lee B-h, King RW, Finley D, Kirschner MW. Science. 2015;348(6231):1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lu Y, Wang W, Kirschner MW. Science. 2015;348(6231):1248737. doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer HJ, Rape M. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Structure. 2013;21:727–740. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Faesen AC, Dirac AMG, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mol Cell. 44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]; b) Faesen AC, Luna-Vargas MPA, Geurink PP, Clerici M, Merkx R, van Dijk WJ, Hameed DS, El Oualid F, Ovaa H, Sixma TK. Chem Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]; c) Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, Kelly SM, Wood NT, Virdee S, Gray NS, Morrice NA, Alessi DR, Trost M. Nat Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hospenthal MK, Freund SMV, Komander D. Nat Struct Mol Biol. 2013;20:555–565. doi: 10.1038/nsmb.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valkevich EM, Sanchez NA, Ge Y, Strieter ER. Biochemistry. 2014;53:4979–4989. doi: 10.1021/bi5006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, Sue C, Gevaert K, De Strooper B, Verstreken P, Vandenberghe W. Hum Mol Genet. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham CN, Baughman JM, Phu L, Tea JS, Yu C, Coons M, Kirkpatrick DS, Bingol B, Corn JE. Nat Cell Biol. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- 19.a) Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, Wells JA, Gygi SP, Schulman BA, Harper JW. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA, Harper JW. Proc Natl Acad Sci USA. 2015;112:6637–6642. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


