Abstract
Analysis of complex mixtures is a common challenge in natural products research. Quantitative nuclear magnetic resonance spectroscopy (qNMR) offers analysis of complex mixtures at early stages and with benefits that are orthogonal to more common methods of quantitation, including ultraviolet absorption spectroscopy (UV) and mass spectrometry (MS). Several experiments were conducted to construct a methodology for use in analysis of extracts of fungal cultures. A broadly applicable method was sought for analysis of both pure and complex samples through use of an externally calibrated method. This method has the benefit of not contaminating valuable samples with the calibrant, and it passed scrutiny for line fitting and reproducibility. The method was implemented to measure the yield of griseofulvin and dechlorogriseofulvin from three fungal isolates. An isolate of Xylaria cubensis (coded MSX48662) was found to biosynthesize griseofulvin in the greatest yield, 149 ± 8 mg per fermentation, and was selected for further supply experiments.
Keywords: NMR, 1H NMR, qNMR, Griseofulvin, Fungi, Secondary metabolites
Introduction
When using fungi as a source for drug discovery, pure compounds are often isolated in quantities of 1.0 mg or less. Although structure elucidation via NMR can be accomplished on this scale, this amount is quickly consumed through biological testing and other experimentation. This valuable compound may then require resupply through the fermentation of new batches of the fungal culture. In order to expedite future restocking, several organisms and media optimization studies may be tested to find the most productive organism and conditions for the resupply of the analyte of interest.[1–5]
The key challenge then falls to analysis of the samples. There are many ways that one could employ to analyze the various fermentations. Historically, analysis of resupply conditions re-isolated the compound of interest, so as to quantitate the yield under new conditions.[2] However, this process can be time intensive, impeding further research on promising leads. Recent innovations in mass spectrometry, particularly ambient ionization techniques like LAESI,[6] DESI,[7] and MALDI,[8] or surface sampling probes like droplet–liquid microjunction–surface sampling probe,[9, 10] continuous flow–liquid microjunction–surface sampling probe,[11] and liquid extraction surface analysis[12] have drastically reduced the analysis time. These techniques can provide an in situ snapshot of the metabolite profile of a culture’s surface. However, quantitative information is difficult to derive from these techniques without further study. They are also incumbent upon the ionization properties of the metabolites. LC-MS analysis of the extract can be used to quantify the results,[13] but this requires the use of a reference standard of the analyte for calibration of ionization efficiency and creation of a standard curve. This is often not feasible if the residual pure sample after early experimentation is extremely low. Without calibration, LC-MS can only be used to give relative production of compounds of interest, and while that may provide a picture of the relative biosynthesis, it can be useful to acquire quantitative information to further evaluate the productivity of the fermentation conditions.
qNMR is a validated method[14] that can be used to quantify and analyze secondary metabolites upstream in the isolation and purification process. Aside from its non-destructive nature, qNMR offers several benefits over LC-MS. Quantitation does not necessitate a purified standard of the analyte to calculate a standard curve.[15] Additionally, NMR sepectroscopy inherently contains some separation of constituent signals,[15] such that complex samples can be analyzed upstream of relatively pure samples in a way that is orthogonal to LC-MS or LC-UV quantitation. With these benefits, qNMR can be applied early in an isolation process, providing quantitative measurements with which to compare differing culture conditions. Moreover, since NMR is frequently incorporated into natural products research schemes, [16–24] this process does not necessitate acquisition of new equipment or severe deviation in protocols. The end result is the selection of an efficient fungal strain and/or specific fermentation conditions to resupply valuable compounds extrapolated from the quantitative information.
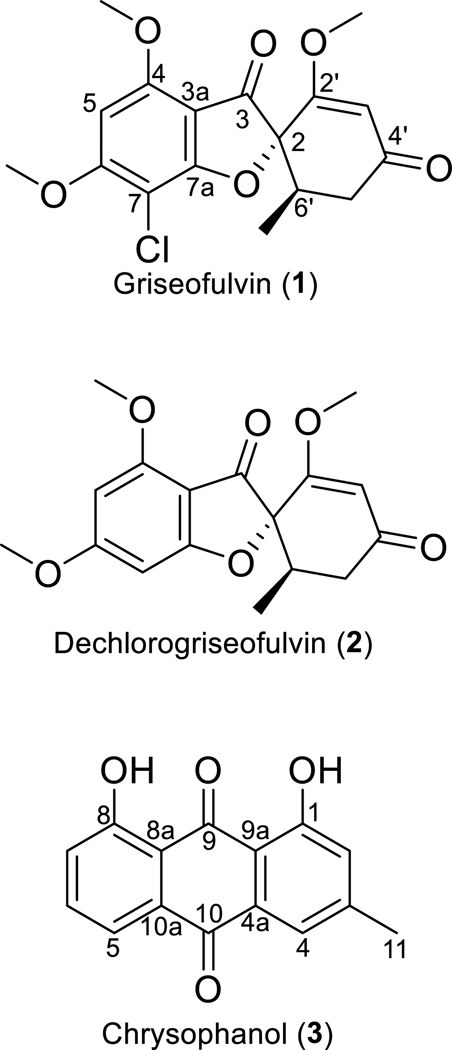
Griseofulvin
Three fungal isolates were evaluated for the biosynthesis of secondary metabolites, with the goal of determining the best culture for their large-scale production. Griseofulvin (1) and dechlorogriseofulvin (2) (Figs. 1, S1, S2, and S4) were observed in extracts of three separate isolates of Xylaria cubensis, which were coded MSX54665, MSX48662, and G536.[25] Originally isolated from a filamentous fungus in 1939,[26] compound 1 was one of the first antifungal compounds isolated from a natural product source and has been in the market for the treatment of several dermatological fungal infections in animals and humans.[27–29] The recent literature on 1 for activity against cancer and suppression of hepatitis C virus replication, in conjunction with the influx of patents for analogues of 1, indicate the expanding interest in this class of compounds.[30–33] Production of 1 is well established via industrial fermentation processes.[34–36] Our interest was to examine cultures that biosynthesize 1 for two complementary reasons. As described herein, it made a good test case for the use of qNMR for profiling biosynthesis. Moreover, as will be reported in the future, in the context of a program to discover new anticancer drug leads from fungal cultures,[37] we needed to enhance the supply of 1 and related analogues for a series of semisynthetic experiments.
Figure 1.
Structures of compounds 1–3.
Experimental Section
Fungal strains and identification
Fungal strain MSX48662 was isolated from cedar wood collected in Little Rock, Arkansas, in May of 1990, while MSX54665 was isolated from leaf litter obtained in Wilson County, Tennessee, in April of 1991. Fungal strain G536 was isolated from surface sterilized twigs of pawpaw (Asimina triloba (L.) Dunal, Annonaceae) collected from Pfafftown, North Carolina, USA as described in detail previously.[25]
The fungal strains were identified via morphological and molecular methods, and the identification of strain G536 has been described in detail;[25] the sequence data for G536 were deposited in the GenBank (KU560914, KU560915, KU560916). Fungal strains MSX48662 and MSX54665 were characterized using molecular sequence data from nuclear ribosomal internal transcribed spacers and 5.8S gene (nuc ITS) and approximately 600 base pairs of the adjacent D1/D2 regions of the nuclear ribosomal large subunit (nuc LSU) were amplified using methods described earlier.[38–42] Molecular characterization of strain G536 suggests that the fungus belongs to Xylaria cubensis.[25] BLAST search via GenBank showed high coverage and percent identity values with several sequences of Xylaria cubensis, Sordariomycetes, Ascomycota.[43, 44] All sequences with high similarity to the BLAST search were downloaded from GenBank and incorporated into a multiple sequence alignment and subjected to a maximum likelihood (ML) phylogenetic analysis[45] using RAxML. Results of the ML analysis showed that both MSX strains are nested with numerous Xylaria cubensis strains, including authentic sequences GU991523 and AB625440, which have been utilized in molecular phylogenetics of Xylaria spp.[46] (Fig. S5). The sequence data were deposited in GenBank (MSX48662: KX229783; MSX54665:KX229784).
Extraction
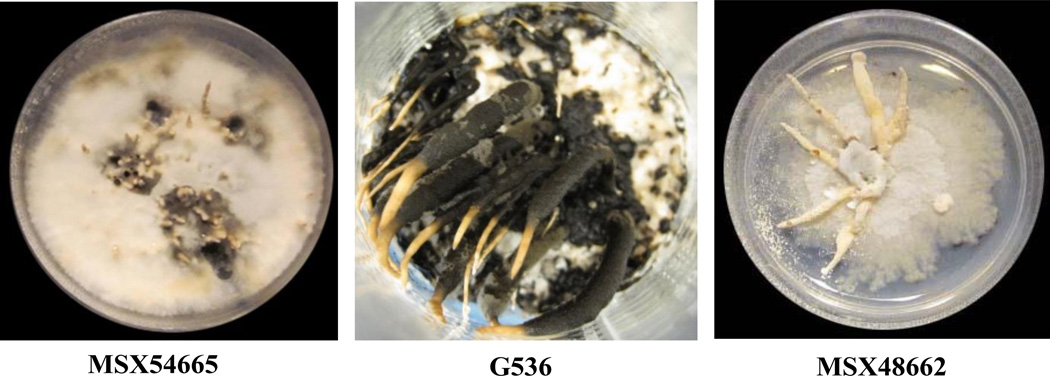
Separately, solid-substrate fermentations of the three fungi (Fig. 2) were chopped with a spatula and shaken for 16 h at 100 rpm with 500 mL MeOH/CHCl3 in a 1:1 mixture. The supernatants were collected via vacuum filtration, and solid substrates were washed with 100 mL of 1:1 MeOH/CHCl3. To the filtrates, 900 mL CHCl3 and 1500 mL H2O were added followed by 2 h of stirring. The mixtures were transferred to separatory funnels, and the two layers were drawn off into independent flasks. The bottom layers were evaporated to dryness under vacuum and reconstituted in 300 mL of 1:1 MeOH/CH3CN and 300 mL of hexanes. These solutions were transferred back to seperatory funnels and shaken vigorously. The MeOH/CH3CN layers (hereafter referred to as organic extract) were evaporated to dryness under vacuum.
Figure 2.
Solid substrate fermentations of MSX54665, G536, and MSX48662.
Sample preparation
Organic extracts were reconstituted in DMSO-d6 at 2.0 mg/mL for MSX48662, 5.0 mg/mL for G536, and 10 mg/mL for MSX54655. The samples were weighed on a micro-analytical balance (XS105, Mettler Toledo), with precision of ± 0.01 mg. DMSO-d6 99.9% (Lot #: PR-26893/10075DM1) was purchased from Cambridge Isotope Laboratories. Chrysophanol (3) (Figs. 1 and S3) (99.3 % Lot No. 870622; Madaus, Germany) was used as a calibrant and was reconstituted using DMSO-d6 at 0.50 mg/mL. All reconstituted fungal extracts were prepared in single stocks and then transferred in triplicate aliquots of 0.50 mL into standard 5 mm NMR tubes.
Quantitative NMR
Quantitative NMR measurements were conducted on a JEOL ECA-500, operating at 500 MHz for 1H and 125 MHz for 13C using parameters recommended in literature.[15] For each set of 6 to 12 samples, auto-tuning was employed to optimize the probe. The autogain program routine was then run on the first sample in a preliminary experiment to establish an optimal gain value. Each sample was then set to 90 % of the optimal gain value so as to maximize signal while avoiding clipping. A 60 s relaxation delay was incorporated to ensure relaxation of most protons. Two dummy scans were applied to achieve steady state for each sample, and those were followed by 8 scans. A 90° pulse was applied to give maximum detector response. Acquisition time was set to 3 s, and a 20 ppm spectral width was used, centered on 6.5 ppm. Sample temperature was maintained at 25 °C.
NMR files were processed using MestReNova software (Mestrelab Research, S.L.). Exponential apodization was applied using a value of 0.40 Hz, followed by phase correction. The baseline of each spectrum was corrected using the Whittaker Smoother routine included in the MestReNova software. The chemical shift was then adjusted to the DMSO-d5 peak, which was set to 2.500 ppm. Analyte peaks used in quantitation were selected based on high intensity and relative isolation from neighboring peaks. Based on a close inspection of peak shape, peaks were selected if the majority of peak area was due to the analyte signal, rather than neighboring peaks. Peaks that did not pass this scrutiny were also excluded from use for quantitation. Integration was then applied in a manner to exclude neighboring peaks. In the standard, and where possible in the analyte samples, integration was taken for a spectral width of 30 Hz. The solvent peak was integrated identically between the standard and analyte samples to ensure consistent integration and then was normalized to an arbitrary large value (i.e. 10,000.00). By setting the solvent peak to a large value, the relatively small analyte and standard integrals were comparable, so as to give a sense of variation of the concentrations in the samples. 13C satelites were visible for the solvent peak but were not included in integration or purity calculations. Complete spectra and the assignment of peaks for 1 and 2 are included in the supplementary data (Figs. S1, S2, and S4) and have been reported.[47]
Results and discussion
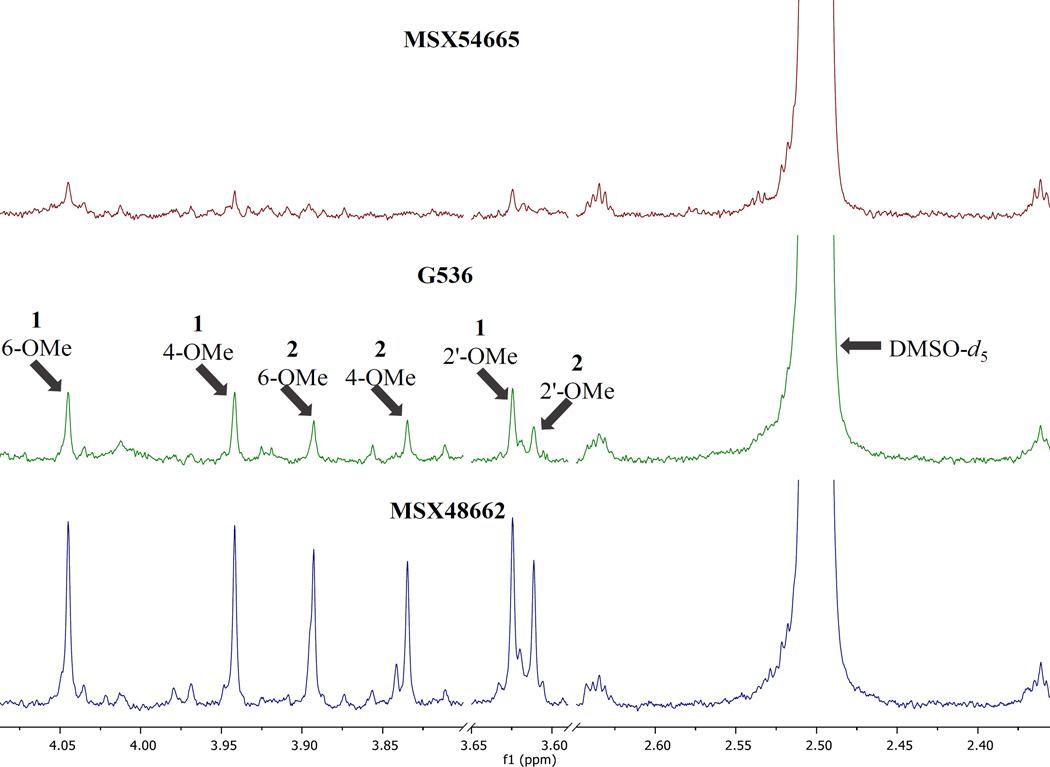
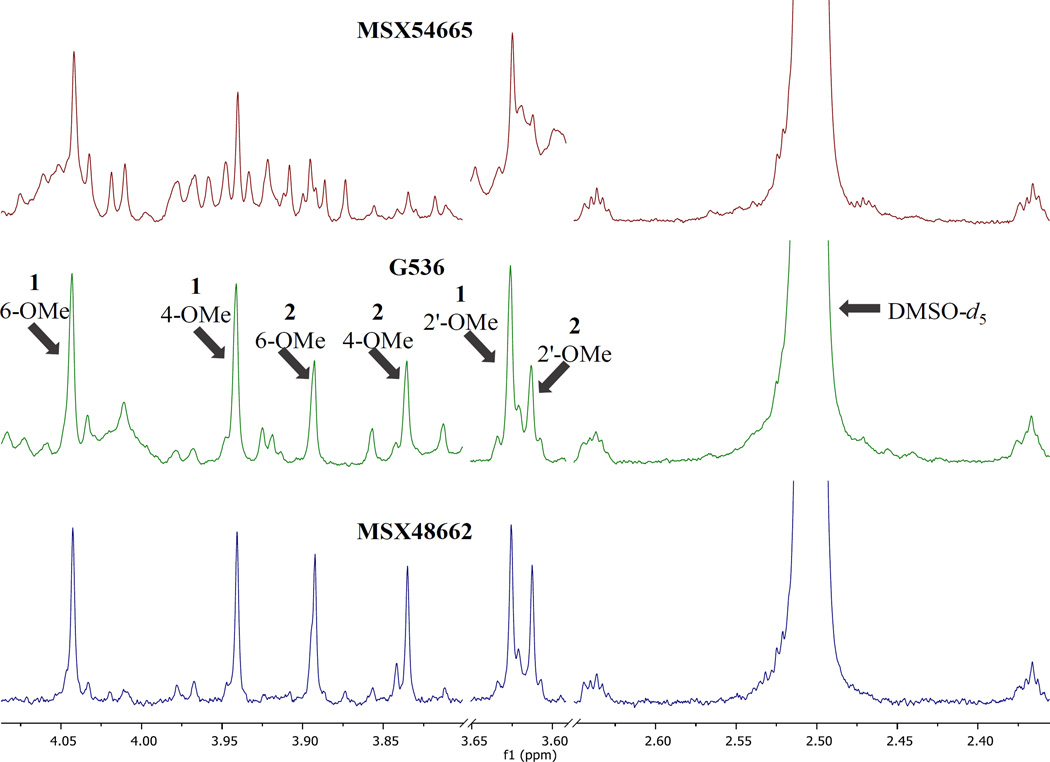
Three separate fungal cultures were observed to biosynthesize griseofulvin (1) and dechlorogriseofulvin (2). The goal of this study was to determine which isolate produced compounds 1 and 2 in the greatest yield. The intensity of the analyte signals for MSX48662 were high enough to generate meaningful quantitative results at the initial concentration of 2.0 mg/mL. However, the organic extracts of MSX54665 and G536 dissolved at 2.0 mg/mL had low signal intensity of 1 and 2 with S/N ratios ranging from 40–80 (Fig. 3), which were not ideal for quantitation.[14, 15, 48] In general, low S/N can hinder the ability to detect small impurities in the baseline surrounding analyte peaks.[15] The low S/N was seen to affect baseline correction and integration of analyte peaks, thereby introducing error into quantitation. Additionally, a high noise level can cause overcompensation of automatic baseline correction routines, which can skew integration downward. Minor impurities were likely to be hidden by noise if their low concentration does not permit their detection, due to the complexity of the spectral region of interest. To compensate for the low signal, the samples of MSX54665 and G536 were concentrated to 10 mg/mL and 5 mg/mL, respectively, to yield higher peak intensity. Subsequently, the NMR experiments were repeated to yield S/N ratio ranging from 120–240 for analyte peaks (Fig. 4).
Figure 3.
Selected sections of the 1H NMR spectra of MSX54665, G536 and MSX48662 at 2.0 mg/mL in DMSO-d6. Given are the sections useful for quantitation of the methoxy groups. The DMSO-d5 peak is also included for comparison. Spectra were collected on a JEOL ECA-500 operating at 500 MHz with 2 dummy scans followed by 8 scans.
Figure 4.
Selected sections of the 1H NMR spectra of MSX54665, G536 and MSX48662 at 10, 5.0, and 2.0 mg/mL, respectively, in DMSO-d6. Given are the sections useful for quantitation of the methoxy groups. The DMSO-d5 peak is also included for comparison. Peak height of spectra were normalized to the DMSO-d5 peak. Spectra were collected on a JEOL ECA-500, operating at 500 MHz with 2 dummy scans followed by 8 scans.
Purity Calculation
To calculate purity and yield of 1 and 2 from the various fungal cultures, the following formula was applied, where mx is the calculated mass of analyte, 1 or 2, inside of the sample mixture, mcal is the measured mass of calibrant in the prepared calibrant sample, Sx is the normalized integral of the anlyte, Scal is the normalized integral of the calibrant, Ncal is the number of protons represented by calibrant peak, Nx is the number of protons represented by the peak of the analyte, Mx is the molar mass of the analyte and Mcal is the molar mass of the calibrant.
| (1) |
In this project, the contents of 1 and 2 within the three fungal extracts were measured and 3 was used as a calibrant. Simple analyte integrals were normalized to the solvent signal, so as to compensate for variation in integrals due to any dilution effect from contaminants (i.e. water), any signal variation between NMR tubes, or sample shimming in the magnet. As noted by Krunic and Orjala,[49] using a single batch of DMSO-d6 gave the best consistency of DMSO-d5 concentration and corresponding consistency of raw integrals of the DMSO-d5 peaks.
Sample mass was then used to calculate purity of the sample using the following equation:
| (2) |
where mx is the sample mass as in equation 1, Vsmp is the total volume used in the NMR experiment, and Csmp is the concentration of sample inside the tube (Table 1 and Fig. S6). Purity was then extrapolated to the extract to yield a calculated mass of analyte (mtot x) (Table 2) that was produced by the fungal strain using the equation:
| (3) |
where mtot corresponds the mass of the organic extract.
Table 1.
Purity of 1 and 2 in Fungal Extracts
| Compound 1 | Compound 2 | |||||
|---|---|---|---|---|---|---|
| Culture | Purity | SD | CV | Purity | SD | CV |
| MSX54665 | 0.0055 | 0.0003 | 5.8% | N/D | N/A | N/A |
| G536 | 0.0164 | 0.0005 | 3.3% | 0.0095 | 0.0007 | 7.3% |
| MSX48662 | 0.076 | 0.004 | 5.0% | 0.052 | 0.001 | 2.7% |
Included are the results from one qNMR experiment. Each extract sample was weighed and aliquoted into triplicate NMR tubes and compared to a single triplicate set of standard. Purity represents a proportion of analyte in the extract;
SD = Standard Deviation; CV = Coefficient of Variation.
Table 2.
Mass of 1 and 2 in Fungal Extracts
| Compound 1 | Compound 2 | |||||
|---|---|---|---|---|---|---|
| Culture | Mass (mg) |
SD (mg) |
CV | Mass (mg) |
SD (mg) |
CV |
| MSX54665 | 5.4 | 0.3 | 5.8% | N/D | N/A | N/A |
| G536 | 4.6 | 0.1 | 3.3% | 2.6 | 0.2 | 7.3% |
| MSX48662 | 149 | 8 | 5.0% | 102 | 2 | 2.7% |
Total mass of 1 and 2 in each organic extract are shown. Signals of 2 were not readily identifiable in the extract of MSX54665 and thereby mass of 2 was not calculated.
Result of Extract Comparison
Culture MSX48662 had the highest total yield of 1 and 2 (149 ± 8 mg per fermentation and 102 ± 2 mg per fermentation, respectively) (Table 2 and Fig. S6). In contrast, culture MSX54665 yielded 5.4 ± 0.3 mg of 1 per fermentation and G536 yielded 4.6 ± 0.1 mg of 1 and 2.6 ± 0.2 mg of 2 per fermentation. These results suggested that MSX48662 would be the best of the three candidate fungal cultures for the resupply of compounds 1 and 2.
Challenges in Analysis
qNMR of complex mixtures suffers from signal overlap and requires careful consideration of solvent and instrumentation to give the best possible isolation of quantifiable peaks. During analysis, MSX54665 displayed numerous signals between 3.8 and 4.1 ppm, which had a similar shape to methoxy signals observed for both 1 and 2. While these signals could indicate a number of griseofulvin analogues, which could be interesting from a research standpoint, the signals did not have baseline resolution between neighboring peaks. At the 10 mg/mL concentration of the organic extract of MSX54665, the methoxy peaks of compound 2 were not readily identifiable within this population of signals and was therefore not calculated (Tables 1 and 2, Figs. 4 and S6).
In the organic extracts of MSX48662 and G536, the 6-OMe signal of 1 and 2 had subtle shoulders that indicated peaks with little separation from the analyte peaks. Although the 4-OMe had similar contaminant signals, the peaks were better resolved. To determine the effect of the neighboring signals on the 4-OMe signals used for quantitation, contaminant and analyte peaks were fitted using line fitting procedures included in the MestReNova software. Peaks were fitted using the automated “fit” routine in combination with manual adjustments to yield even residual noise, once all differentiable peaks were modeled. The side peaks were labeled as impurities and integrals were recalculated using the EditedSum integration routine, which subtracts modeled peak areas of impurities from the measured integrals. Thereby, the analyte peaks were well represented by the resulting integral. The peak fitting integration (Fig. S7) for the extracts of MSX54665 and MSX48662 reflected the original calculations well and yielded notably improved standard deviations. For the extract of G536, overlapping contaminant peaks formed a faux baseline around the analyte peaks, and these interfered with the software’s fitting routines. As the routines would try to explain a multitude of poorly defined contaminant peaks, peak models were “over-fitted” to the data. Since the peak fittings of the better resolved spectra correlated well with the raw integrations and purity calculations, the previous method was deemed acceptable for the calculation of analyte purity. However, a peak fitting method may be preferable, and close inspection of data is required to determine the benefit of peak modeling.
2D qNMR has been used to deconvolute complex samples as well. These 2D methods can require advanced digital or mathematical manipulation of peaks to ensure accurate quantitation. 2D methods have seen improvements on long acquisition times through optimization techniques, such as J-resolved and DQF-COSY,[50, 51] which have reduced the impact of acquisition time on the accuracy of their measurements. Although such methods may help in the deconvolution of the complex peaks, the high sensitivity, short acquisition time and the relative ease of chromatographic sample purification give good reasons to use 1D 1H NMR as the basis for a general method for analysis of small molecules, at least with respect to natural products research.
Reproducibility
Chrysophanol (3) was chosen as a calibrant due to its availability and to maximize reproducibility of the experiments. As 3 is a solid at room temperature, the concentration of calibrant should remain constant relative to the slowly evaporating DMSO solvent, even over long periods of time. The volatility of 3 is contrast to calibrants used in internal calibration methods, like dichloromethane, which readily evaporate and is thereby advantageous for the simplicity of analyte recovery but may not remain constant over a long period of time. Although the stability was only tested over a period of three days, using a nonvolatile calibrant served to remove variability due to volatility from the measurements. This could allow for repeated measurements of these samples over extended periods of time, in this case likely being limited by signal contamination from water introduction to the DMSO. Other calibrants suggested by Pauli and Rundlöf include DMSO, caffeine, and 3-sulfolene as well as other compounds with a range of resonances to suit various applications.[15, 52]
The reproducibility of the measurements was established through three replicate analyses of the purity of MSX48662. Triplicate aliquots were made of a sample of 3 (Figs. S3 and S4) at 0.50 mg/mL and the extract of MSX48662 at 2.0 mg/mL. The latter aliquots were analyzed using the above method for content of 1. The calculations were averaged to give purity and standard deviation. To determine the precision of the measurement, this process was completed twice more, each on separate days to account for multi-day variability, including weighing and dissolving new batches of sample extract and calibrant (Table 3, Fig. S6). The average purity calculated from three days of the experiment was 7.4 ± 0.5 % (Table 3, Fig. S6). The standard deviation was less on several of the individual days than for the average of all days. To estimate multi-day variability of the NMR spectrometer and post-collection processes, a single set of triplicate standards and analytes from MSX48662 were processed through the above qNMR analysis over three days (Table 4, Fig. S6). The average purity from the one set of triplicates was 7.7 ± 0.3 %. The coefficient of variation dropped from 7.1 % variation among the complete replicates to 4.0 % among the replicate measurements of a single set of replicates.
Table 3.
Complete Repetition of Method on MSX48662 Extract
| Compound 1 | Compound 2 | |||||
|---|---|---|---|---|---|---|
| Repetition | Purity | SD | CV | Purity | SD | CV |
| 1 | 0.076 | 0.004 | 5.0% | 0.052 | 0.001 | 2.7% |
| 2 | 0.078 | 0.006 | 7.6% | 0.055 | 0.005 | 8.2% |
| 3 | 0.069 | 0.002 | 3.5% | 0.048 | 0.002 | 4.4% |
| Tot. | 0.074 | 0.005 | 7.1% | 0.051 | 0.004 | 7.5% |
Each repetition was made from a separate weighing of MSX48662, separated into 3 aliquots. Each repetition represents a complete set of sample triplicates and calibrant triplicates. Each repetition was completed on a separate day to include multi-day variation.
Table 4.
Analysis of Single MSX48662 Extract Triplicate
| Compound 1 | Compound 2 | |||||
|---|---|---|---|---|---|---|
| Day | Purity | SD | CV | Purity | SD | CV |
| 1 | 0.076 | 0.004 | 5.0% | 0.052 | 0.001 | 2.7% |
| 2 | 0.076 | 0.002 | 2.0% | 0.054 | 0.002 | 3.3% |
| 3 | 0.081 | 0.001 | 1.5% | 0.056 | 0.001 | 1.3% |
| Tot. | 0.077 | 0.003 | 4.0% | 0.054 | 0.002 | 4.0% |
A single weighing of MSX48662 extract was separated into three aliquots. This set of triplicate samples was measured over 3 days. The results imply that variation due to the day-to-day drift of the NMR spectrometer is small compared to other factors.
Previous literature has described the use of a solvent signal that is calibrated externally for quantification.[15] Using this method allows the sample to remain untainted from introduction of a calibrant into a valuable sample, but it requires careful weighing and pipetting. The smaller variability between days using the same sample, and the higher variability between separate samples on separate days, was taken to indicate that the majority of the error involved in the analysis occurs in the weighing and pipetting.
Applicability
Use of this method gave clear indication that MSX48662 biosynthesized the largest amounts of 1 and 2 per fermentation. This technique could be used to analyze separate growth conditions as well, since it is well documented that different growth conditions produce varying metabolite profiles.[1] Additionally, because we have chosen to use a method that does not require any internal calibrant, the contamination of the analyte is not a concern. Thereby, this method also lends itself to use for purified samples, as it is unnecessary to complete further purification steps to recover the measured material. In particular, this method should be useful for novel compounds for which analytical reference standards are not available. This method could also be used to verify reference standards before their use for quantitation in other methods. Applications could include forensic science of novel designer drugs, where new illicit drugs often have no standards available.[53] As a starting point, this general method allows for the analysis of small molecules produced from natural products research. Techniques like 2D NMR, spectral deconvolution, 13C decoupling, or chromatographic methods can be employed subsequently to overcome the predominant drawback of this method, which may be its inability to overcome extensive signal overlap. Somewhat redeeming is that a single, relatively pure signal from nonexchangeable proton is sufficient to quantify a compound using the described method.[15]
Although the method outlined above utilizes common instrumentation and equipment, supplemental NMR and chromatographic instrumentation and techniques were observed to further assist in the quantitation procedures. Flash chromatography of extracts can enhance the purity of the analytes 1 and 2, and simplify the spectrographic complexity within a few hours of work (Fig. S8). Additionally, S/N could be improved with use of a greater number of scans,[15] with reduced volume NMR tubes,[49] or with the use of a modern cryoprobe (Fig. S8).[48, 49] Moreover, minor impurities could be revealed if a higher magnetic field was used to increase spectral resolution, thereby separating closely shifted peaks.[48] For this study, an increase in resolution by using a higher magnetic field, or use of deconvolution methods, could help to elucidate the signals of 2 in the extract from MSX54665.[54] In efforts to make the method generally applicable, however, standard instrumentation and a shorter timeframe was emphasized. Therefore, concentrations of the two extracts with low analyte signal were increased to give the desired S/N, and the method yielded comparative results with 8 scans, requiring 10 minutes of collection per sample.
Conclusion
In natural products research, the world’s supply of a promising compound can be rapidly consumed in follow-up bioassay studies. Analysis of the experiments designed to efficiently resupply these compounds can be challenging without a well characterized reference sample. NMR has the ability to quantitate any proton, even in the absence of a reference standard. Thereby, nearly any secondary metabolite can be measured using this method. In this study, MSX48662 yielded the most of the target compounds and was therefore the best of the three Xylaria cubensis isolates to supply griseofulvin (1) and dechlorogriseofulvin (2) for further experimentation. The use of a qNMR method afforded an orthogonal measurement to LC-MS of 1 and 2 within extracts.
Supplementary Material
Acknowledgments
This research was supported in part by program project grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA.
Footnotes
Supporting Information
Supporting information includes illustrations of the data in Tables 1–4, a phylogram of Xylaria cubensis isolates, the NMR data and spectra for compounds 1–3, and a NMR spectrum of MSX48662 with line fitting applied and is available along with the digital version of this article.
References
- 1.Frisvad JC. In: Fungal Secondary Metabolism: Methods and Protocols. Keller NP, Turner G, editors. New York: Springer; 2012. pp. 47–77. [Google Scholar]
- 2.Xu F, Tao W, Cheng L, Guo L. Biochem. Eng. J. 2006;31:67–73. [Google Scholar]
- 3.Cai M-H, Zhou X-S, Sun X-Q, Tao K-J, Zhang Y-X. J. Ind. Microbiol. Biotechnol. 2009;36:381–389. doi: 10.1007/s10295-008-0507-6. [DOI] [PubMed] [Google Scholar]
- 4.Revankar MS, Lele SS. Process. Biochem. 2006;41:581–588. [Google Scholar]
- 5.Deswal D, Khasa YP, Kuhad RC. Bioresour. Technol. 2011;102:6065–6072. doi: 10.1016/j.biortech.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 6.Nemes P, Vertes A. Anal. Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 7.Leuthold LA, Mandscheff J-F, Fathi M, Giroud C, Augsburger M, Varesio E, Hopfgartner G. Rapid Commun. Mass Spectrom. 2005;20:103–110. doi: 10.1002/rcm.2280. [DOI] [PubMed] [Google Scholar]
- 8.Stump MJ, Fleming RC, Gong W-H, Jaber AJ, Jones JJ, Surber CW, Wilkins CL. Appl. Spectrosc. Rev. 2002;37:275–303. [Google Scholar]
- 9.Kertesz V, Van Berkel GJ. Anal. Chem. 2010;82:5917–5921. doi: 10.1021/ac100954p. [DOI] [PubMed] [Google Scholar]
- 10.Sica VP, Raja HA, El-Elimat T, Kertesz V, Van Berkel GJ, Pearce CJ, Oberlies NH. J. Nat. Prod. 2015;78:1926–1936. doi: 10.1021/acs.jnatprod.5b00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Berkel GJ, Kertesz V, King RC. Anal. Chem. 2009;81:7096–7101. doi: 10.1021/ac901098d. [DOI] [PubMed] [Google Scholar]
- 12.Walworth MJ, ElNaggar MS, Stankovich JJ, Witkowski C, Norris JL, Van Berkel GJ. Rapid Commun. Mass Spectrom. 2011;25:2389–2396. doi: 10.1002/rcm.5132. [DOI] [PubMed] [Google Scholar]
- 13.VanderMolen KM, Raja HA, El-Elimat T, Oberlies NH. AMB Express. 2013;3:1–7. doi: 10.1186/2191-0855-3-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniara G, Rajamoorthi K, Rajan S, Stockton GW. Anal. Chem. 1998;70:4921–4928. doi: 10.1021/ac980573i. [DOI] [PubMed] [Google Scholar]
- 15.Pauli GF, Chen S-N, Simmler C, Lankin DC, Gödecke T, Jaki BU, Friesen JB, McAlpine JB, Napolitano JG. J. Med. Chem. 2014;57:9220–9231. doi: 10.1021/jm500734a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alali F, El-Elimat T, Albataineh H, Al-Balas Q, Al-Gharaibeh M, Falkinham JO, Chen W-L, Swanson SM, Oberlies NH. J. Nat. Prod. 2015;78:1708–1715. doi: 10.1021/acs.jnatprod.5b00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sy-Cordero AA, Figueroa M, Raja HA, Meza Aviña ME, Croatt MP, Adcock AF, Kroll DJ, Wani MC, Pearce CJ, Oberlies NH. Tetrahedron. 2015;71:8899–8904. doi: 10.1016/j.tet.2015.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur A, Raja HA, Darveaux BA, Chen W-L, Swanson SM, Pearce CJ, Oberlies NH. Magn. Reson. Chem. 2015;53:616–619. doi: 10.1002/mrc.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd DA, Gulledge TV, Britton ER, Oberhofer M, Leyte-Lugo M, Moody AN, Shymanovich T, Grubbs LF, Juzumaite M, Graf TN, Oberlies NH, Faeth SH, Laster SM, Cech NB. PLoS ONE. 2015;10:e0124276. doi: 10.1371/journal.pone.0124276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Elimat T, Raja HA, Figueroa M, Swanson SM, Falkinham Iii JO, Lucas DM, Grever MR, Wani MC, Pearce CJ, Oberlies NH. J. Antibiot. 2015;68:191–196. doi: 10.1038/ja.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Elimat T, Figueroa M, Raja HA, Graf TN, Swanson SM, Falkinham JO, Wani MC, Pearce CJ, Oberlies NH. Eur. J. Org. Chem. 2015;2015:109–121. doi: 10.1002/ejoc.201402984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaroszewski JW. Planta Med. 2005;71:691–700. doi: 10.1055/s-2005-871298. [DOI] [PubMed] [Google Scholar]
- 23.Keifer PA. Drugs Future. 1998;23:301–317. [Google Scholar]
- 24.Molinski TF. Nat. Prod. Rep. 2010;27:321–329. doi: 10.1039/b920545b. [DOI] [PubMed] [Google Scholar]
- 25.Sica VP, Rees ER, Tchegnon E, Bardsley RH, Raja HA, Oberlies NH. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oxford AE, Raistrick H, Simonart P. Biochem. J. 1939;33:240–248. doi: 10.1042/bj0330240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen AB, Rønnest MH, Larsen TO, Clausen MH. Chem. Rev. 2014;114:12088–12107. doi: 10.1021/cr400368e. [DOI] [PubMed] [Google Scholar]
- 28.Gentles JC. Nature. 1958;182:476–477. doi: 10.1038/182476a0. [DOI] [PubMed] [Google Scholar]
- 29.Williams DI, Marten RH, Sarkany I. Lancet. 1958;2:1212–1213. doi: 10.1016/s0140-6736(58)92363-8. [DOI] [PubMed] [Google Scholar]
- 30.Jin H, Yamashita A, Maekawa S, Yang P, He L, Takayanagi S, Wakita T, Sakamoto N, Enomoto N, Ito M. Hepatol. Res. 2008;38:909–918. doi: 10.1111/j.1872-034X.2008.00352.x. [DOI] [PubMed] [Google Scholar]
- 31.Rathinasamy K, Jindal B, Asthana J, Singh P, Balaji PV, Panda D. BMC Cancer. 2010;10:213. doi: 10.1186/1471-2407-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oda T. J. Antibiot. 2006;59:114–116. doi: 10.1038/ja.2006.17. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y-S, Duh J-S, Jeng J-H, Wang Y-J, Liang Y-C, Lin C-H, Tseng C-J, Yu C-F, Chen R-J, Lin J-K. Int. J. Cancer. 2001;91:393–401. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1070>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 34.Hemming HG, Lehan M, Giles D. 3616247. U.S. Patent. 1971
- 35.John DM, Mitchell ILS, Rule DW, Cecile W. 3069329. U.S. Patent. 1962
- 36.Venkata Dasu V, Panda T. Bioprocess Eng. 1999;21:489–495. [Google Scholar]
- 37.Kinghorn AD, Carache de Blanco EJ, Lucas DM, Rakotondraibe HL, Orjala J, Soejarto DD, Oberlies NH, Pearce CJ, Wani MC, Stockwell BR, Burdette JE, Swanson SM, Fuchs JR, Phelps MA, Xu L, Zhang X, Shen YY. Anticancer Res. 2016;36:5623–5637. doi: 10.21873/anticanres.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Elimat T, Figueroa M, Raja HA, Graf TN, Adcock AF, Kroll DJ, Day CS, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2013;76:382–387. doi: 10.1021/np300749w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raja HA, Shearer CA, Raja HA, Fournier J, Miller AN. Mycoscience. 2012;53:373–380. [Google Scholar]
- 40.El-Elimat T, Figueroa M, Raja HA, Adcock AF, Kroll DJ, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Tetrahedron Lett. 2013;54:4300–4302. doi: 10.1016/j.tetlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raja HA, Oberlies NH, El-Elimat T, Miller AN, Zelski SE, Shearer CA. Mycoscience. 2013;54:353–361. [Google Scholar]
- 42.Raja HA, Oberlies NH, Figueroa M, Tanaka K, Hirayama K, Hashimoto A, Miller AN, Zelski SE, Shearer CA. Mycologia. 2013;105:959–976. doi: 10.3852/12-313. [DOI] [PubMed] [Google Scholar]
- 43.U'Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Am. J. Bot. 2012;99:898–914. doi: 10.3732/ajb.1100459. [DOI] [PubMed] [Google Scholar]
- 44.U’Ren JM, Miadlikowska J, Zimmerman NB, Lutzoni F, Stajich JE, Arnold AE. Mol. Phylogenet. Evol. 2016;98:210–232. doi: 10.1016/j.ympev.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt I, Barker FK. Nat. Prod. Rep. 2009;26:1585–1602. doi: 10.1039/b910458p. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh HM, Lin CR, Fang MJ, Rogers JD, Fournier J, Lechat C, Ju YM. Mol. Phylogenet. Evol. 2010;54:957–969. doi: 10.1016/j.ympev.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 47.Ronnest MH, Harris P, Gotfredsen CH, Larsen TO, Clausen MH. Tetrahedron Lett. 2010;51:5881–5882. [Google Scholar]
- 48.Pauli GF. Phytochem. Anal. 2001;12:28–42. doi: 10.1002/1099-1565(200101/02)12:1<28::AID-PCA549>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 49.Krunic A, Orjala J. Magn. Reson. Chem. 2015;53:1043–1050. doi: 10.1002/mrc.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giraudeau P. Magn. Reson. Chem. 2014;52:ii–ii. doi: 10.1002/mrc.4068. [DOI] [PubMed] [Google Scholar]
- 51.Rance M. Biochem. Biophys. Res. Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 52.Rundlöf T, McEwen I, Johansson M, Arvidsson T. J. Pharm. Biomed. Anal. 2014;93:111–117. doi: 10.1016/j.jpba.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Johansson M, Fransson D, Rundlöf T, Huynh N-H, Arvidsson T. J. Pharm. Biomed. Anal. 2014;100:215–229. doi: 10.1016/j.jpba.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 54.Napolitano JG, Lankin DC, Graf TN, Friesen JB, Chen S-N, McAlpine JB, Oberlies NH, Pauli GF. J. Org. Chem. 2013;78:2827–2839. doi: 10.1021/jo302720h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.