Abstract
The adaptive immune system of jawed vertebrates relies on V(D)J recombination as one of the main processes to generate the diverse array of receptors necessary for the recognition of a wide range of pathogens. The DNA cleavage reaction necessary for the assembly of the antigen receptor genes from an array of potential gene segments is mediated by the Recombination Activating Gene proteins RAG1 and RAG2. The RAG proteins have been proposed to originate from a transposable element (TE) as they share mechanistic and structural similarities with several families of transposases and are themselves capable of mediating transposition. A number of RAG-like proteins and TEs with sequence similarity to RAG1 and RAG2 have been identified, but only recently has their function begun to be characterized, revealing mechanistic links to the vertebrate RAGs. Of particular significance is the discovery of ProtoRAG, a transposon superfamily found in the genome of the basal chordate amphioxus. ProtoRAG has many of the sequence and mechanistic features predicted for the ancestral RAG transposon and is likely to be an evolutionary relative of RAG1 and RAG2. In addition, early observations suggesting that RAG1 is able to mediate V(D)J recombination in the absence of RAG2 have been confirmed, implying independent evolutionary origins for the two RAG genes. Here, recent progress in identifying and characterizing RAG-like proteins and the TEs that encode them is summarized and a refined model for the evolution of V(D)J recombination and the RAG proteins is presented.
Keywords: V(D)J recombination, RAG1, RAG2, Transposition, Transib, ProtoRAG, RAG-like proteins, Evolution
Graphical abstract
V(D)J recombination is the process that assembles and generates diversity in lymphocyte antigen receptor genes and underpins the adaptive immune system in jawed vertebrates. This process is mediated by the Recombination Activating Gene (RAG) proteins. In this review, we highlight the known features and the latest progress on the emergence and divergence of the RAG proteins and the evolution of V(D)J recombination.
Introduction
The antigen receptor genes that give rise to the T cell receptor, B cell receptor, and secreted immunoglobulins are comprised of arrays of gene segments. The assembly of these gene segments in a variety of combinations provides a unique source of diversity to the antigen recognition sites of these proteins and is key to adaptive immunity in jawed vertebrate species ranging from cartilaginous fish to primates. V(D)J recombination is the process by which the gene segments are brought together, and the Recombination Activating Gene (RAG) proteins 1 and 2 mediate the site specific DNA double stranded breaks necessary for this process [1, 2].
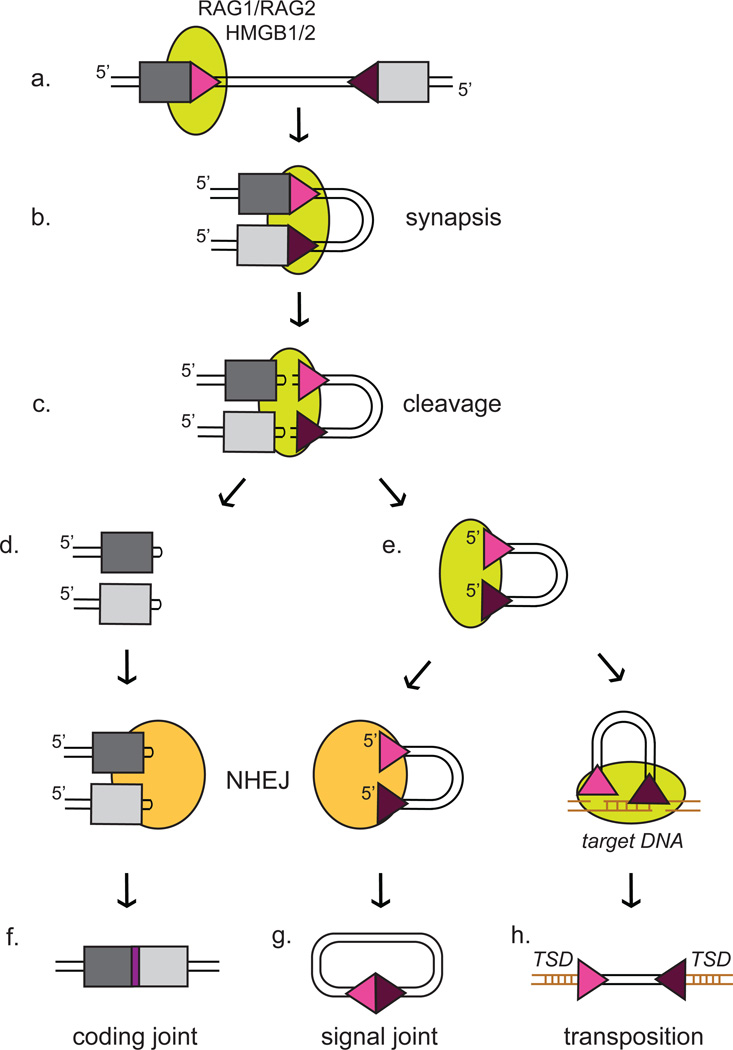
Much work has been done to characterize the RAG proteins and delineate their biochemical reaction. Key features relevant to the evolution of these proteins will be highlighted here, but the reader is directed to additional reviews and papers for a more detailed summary of RAG protein structure and activity [3–6]. V(D)J recombination begins with RAG binding to the recombination signal sequences (RSSs) that demarcate the gene segments of the antigen receptor loci (Fig. 1a). These sequences consist of two conserved motifs, the heptamer and the nonamer, interrupted by a less well conserved spacer sequence of 12 or 23 base pairs [7] (Fig. 2B). The length of the spacer defines the RSS as either a 12RSS or a 23RSS, and efficient recombination occurs only when the RAGs bind one 12RSS and one 23RSS (the "12/23 rule"). Binding of a RAG1/RAG2 heterotetramer together with the high mobility group protein B1 or B2 (HMGB1 or HMGB2) to one RSS and synapsis with a partner RSS allows for the DNA cleavage reaction to proceed (Fig. 1b, c). Nicking of the top strand of DNA just 5’ of the heptamer leaves a free 3’ hydroxyl group that attacks the bottom strand through a direct transesterification reaction, leading to the formation of a DNA hairpin intermediate [8, 9] (Fig. 1c). This generates two types of ends, the hairpin coding ends, which contain the gene segments (Fig. 1d), and the signal ends, which contain the RSSs (Fig. 1e). In vivo, coding and signal ends are processed by the NHEJ pathway [10, 11] to yield coding (Fig. 1f) and signal (Fig. 1g) joints, respectively. In vitro, however, the “signal end complex” (consisting of the RAG proteins bound to the signal ends) (Fig. 1e) can efficiently attack double stranded “target” DNA with a characteristic 5 bp offset, or stagger, on the two strands. This RAG mediated transposition reaction integrates the signal ends and intervening DNA into the target with the staggered attack generating a target site duplication (TSD) that is typically 5 bp in length (Fig. 1h) [12, 13].
Figure 1. Outline of RAG DNA cleavage reaction and products.
RAG mediated cleavage begins with the binding of RAG1/RAG2 along with HMGB1/2 (green oval) to a single RSS (triangle) (a). Synapsis with a partner RSS (b) allows for the cleavage reaction to proceed (c), specifically enabling the hairpin formation step of the nick-hairpin cleavage mechanism. Note that the reaction is most efficient with one 12RSS (pink triangle) and one 23RSS (purple triangle), a restriction known as the 12/23 rule. After cleavage, two types of products are generated. The hairpins are present on the DNA ends containing the gene segments, known as the coding ends (d). They are opened and processed by the NHEJ pathway (orange oval) to generate the coding joint (f). The blunt DNA ends containing the RSSs, known as the signal ends (e), are also usually handed over to the NHEJ pathway to be ligated together, forming the signal joint (g). See [3] for a more detailed summary of this reaction. However, the signal ends also have the potential to undergo a transposition reaction in which the free 3’ hydroxyl groups attack a target piece of DNA (brown) with a stagger of 5 base pairs between the top and bottom strands, generating the TSD and integrating the RSSs and intervening DNA into the target DNA (h). This transposition reaction can lead to genome instability and is very rare in vivo.
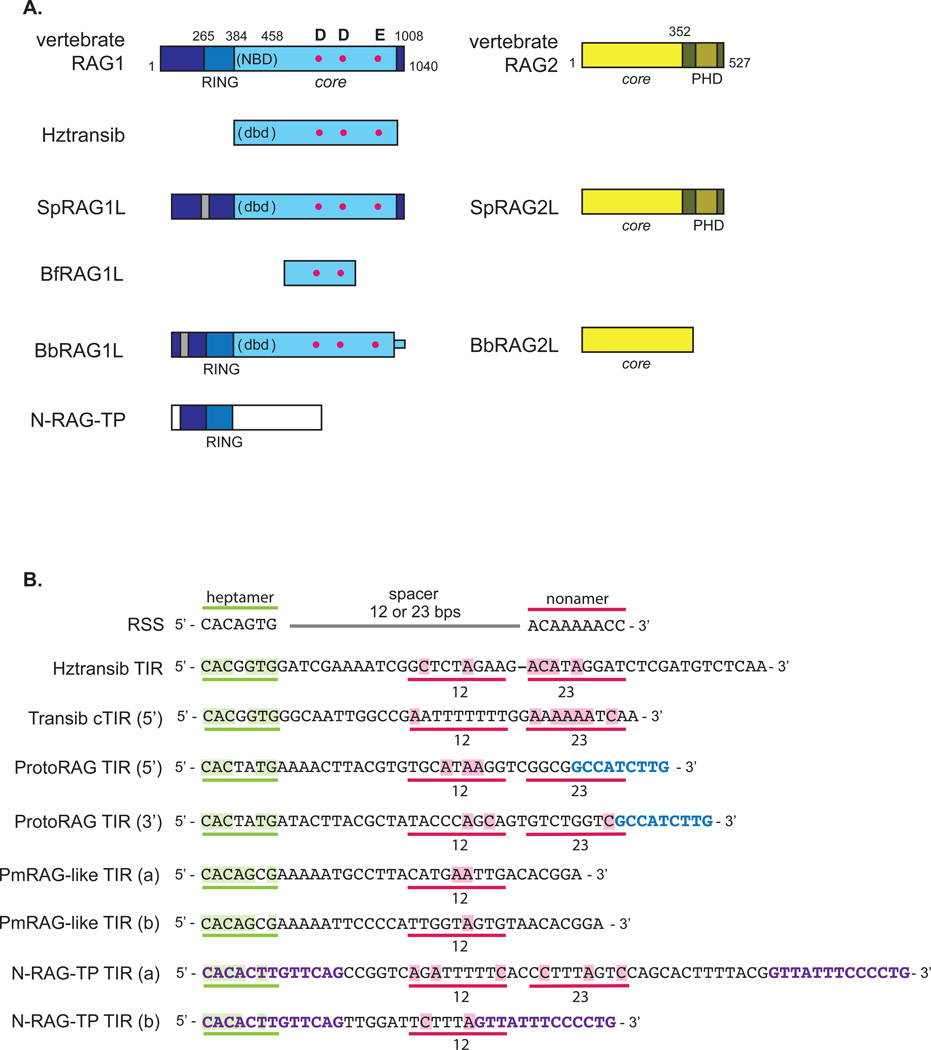
Figure 2. Comparison of RAG and RAG-like proteins: domain structure and DNA binding sequences.
A. The vertebrate RAG1 protein can be truncated to a catalytically active core region (amino acids 384 to 1008 in the mouse protein) (light blue). This contains DNA binding regions, such as the nonamer binding domain (NBD), as well as the three residues that make up the catalytic triad (D600, D708, and E962 in the mouse protein) (red dots). The N-terminal noncore region of RAG1 contains a RING/Zinc finger (RING) that coordinates four zinc atoms [26, 27]. Transib contains sequence similarity only to the core of RAG1, and while it may contain a DNA binding domain (dbd), it but does not have strong sequence similarity to the RAG1 NBD [48]. BfRAG1L contains sequence similarity to only a central portion of RAG1 core, and is therefore missing the last residue of the catalytic triad [54]. BbRAG1L contains sequence similarity that extends into the N-terminal noncore region of RAG1, spanning almost all of the RING/Zinc finger region, but similarity is higher in the core [57], while SpRAG1L contains only limited sequence similarity to the RAG1 RING/Zinc finger region [51]. These two proteins might contain a dbd positioned similarly to the NBD, but do not have sequence similarity to the RAG1 NBD. While HzTransib transposase also lacks sequence similarity to the NBD, some Transib transposases contain NBD-like sequences [53]. Both SpRAG1L and BbRAG1L also contain a series of repeats (gray) but in different locations in their N-terminal regions. Note that the core region of BbRAG1L extends to the C-terminus of the protein, unlike vertebrate RAG1. N-RAG-TP contains sequence similarity solely to the RAG1 N-terminal noncore region. The vertebrate RAG2 protein can be truncated to the core region (amino acids 1 to 352 in the mouse protein) (light yellow), consisting of six kelch repeats which fold into a six-bladed beta propeller, and the noncore PHD (dark yellow). Purple sea urchin and amphioxus contain RAG2-like proteins, but BbRAG2L is missing the PHD finger. The RAG-like proteins from the green sea urchin and bat starfish are not included as their sequences are not well established [53]. No RAG2-like protein has been reported in Aplysia.
B. The RSS consists of a well conserved heptamer (green), a spacer that is either 12 or 23 base pairs, and a well conserved nonamer (pink). The only invariant residues in the RSS are the first three (5’-CAC) of the heptamer. The consensus sequence of the Hztransib TIRs [49], the Transib family TIRs [48], and the 5’ and 3’ TIRs reported for ProtoRAG [57], the bat star (PmRAG-like) [53], and N-RAG-TP from Aplysia are aligned. The corresponding location of the heptamer is underlined in green, and the corresponding location of the nonamer given either a 12 or 23 base pair spacer is underlined in pink. ProtoRAG TIRs contain a 9 base pair sequence (blue) that is well conserved and is separated by either 27 or 31 base pairs from the heptamer while N-RAG-TP has two conserved, distinct tridecamer sequences (purple) separated by either 39 or 13 base pairs. Note that all TIRs of RAG-like TEs have strong conservation of residues in the heptamer (residues highlighted in green), and that alignment of the Hztransib TIR has been shifted by 1 base pair for better alignment of the nonamer. Residues with identity to the nonamer are highlighted in pink.
Both RAG proteins can be truncated to an enzymatically active core portion [14] (Fig. 2A), and functional and structural domains in both the core and non-core regions have been defined. For RAG1, the core region (amino acids 384 to 1008 in the mouse protein) contains several DNA binding domains. The region that makes contacts with the RSS nonamer, the nonamer binding domain (NBD), extends away from the rest of the core on a flexible hinge [4, 5, 15], while heptamer contacts are made by residues located in multiple, more C-terminal portions of the RAG1 core [5]. The RAG1 core also contains three critical acidic residues (D600, D708, E962 in the mouse protein) that coordinate magnesium ions in the active site of the enzyme [16–18] and harbors a structural zinc atom that is coordinated by amino acids located in two regions that are widely separated in the primary amino acid sequence [4, 5, 19]. The noncore portions of RAG1 enhance V(D)J recombination activity [20–25], have E3 ubiquitin ligase activity (contained in an unusual RING/Zinc finger domain) [26, 27], and have been suggested to play a role in protein stability [26], RAG1 dimerization [28, 29], and in mediating interactions with a ubiquitin ligase complex [30] and components of the nonhomologous end joining (NHEJ) pathway [31, 32]. The core region of RAG2 (amino acids 1 to 352 in the mouse protein) (Fig. 2A) is made up of a single domain consisting of a six bladed beta propeller that interacts with RAG1 and with the coding segment DNA that flanks the heptamer of the RSS [4, 5, 33]. The non-core region of RAG2 consists of a flexible, acidic linker followed by a plant homeodomain (PHD) that meditates the interaction between RAG2 and open chromatin through direct binding of the N-terminal tail of histone 3 when lysine 4 is trimethylated (H3K4me3) [34, 35]. The distal C-terminal portion of RAG2 contains a nuclear localization sequence as well as a residue (T490 in the mouse protein) that when phosphorylated by a cyclin-dependent kinase targets the protein for degradation by the proteasome [36, 37]. It is interesting to note that while RAG1 appears to contain all of the domains necessary to bind and cleave the RSS, substantial levels of activity require both RAG proteins.
The Transposon/Split Receptor Gene Hypothesis
Since the discovery of V(D)J recombination, the similarities between this process and that of cut-and-paste transposition have been highlighted [38]. The RSSs resemble the terminal inverted repeats (TIRs) that target DNA binding and cleavage by the transposase, and the nick-hairpin DNA cleavage reaction mediated by RAG bears deep mechanistic similarities to that mediated by several families of transposases [9, 39]. Also notable in this regard is the compact structure of the RAG locus, with RAG1 and RAG2 being found adjacent to one other and convergently transcribed (Fig. 3A) in all jawed vertebrate genomes characterized to date, with an intergenic distance that is typically less than 10 kb. Furthermore, in many species including rodents and primates, each RAG open reading frame is contained in a single large exon [1, 2, 40, 41].
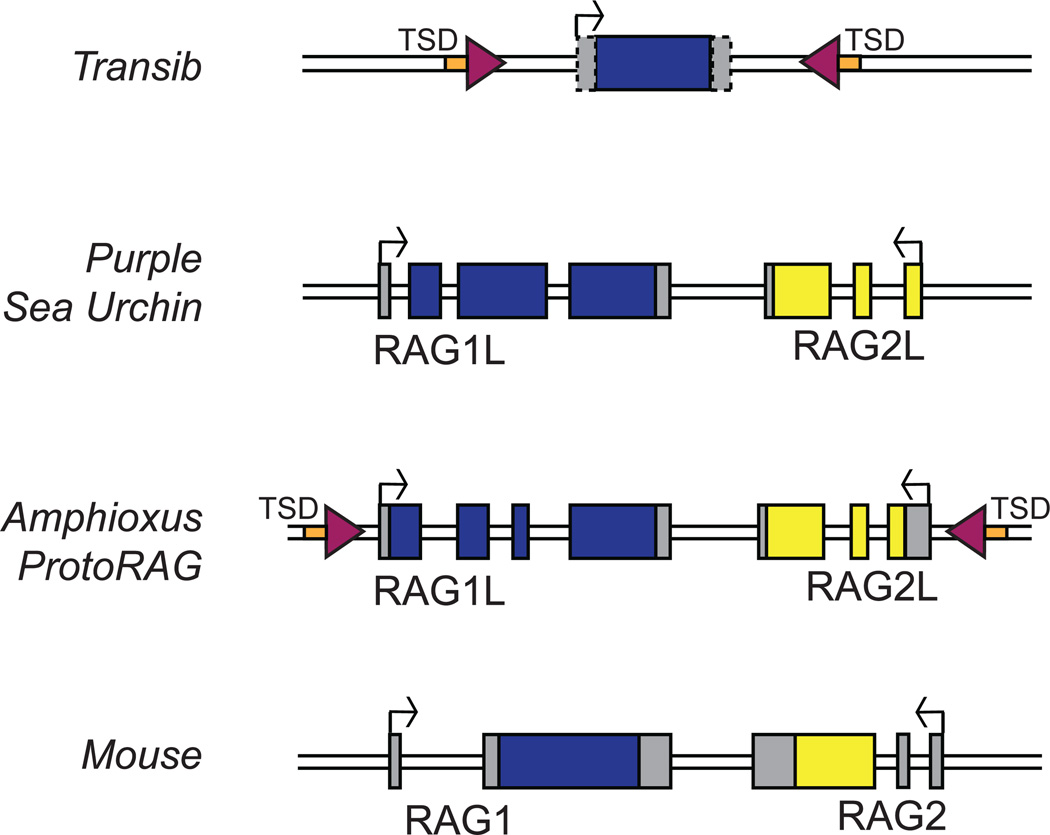
Figure 3. RAG and RAG-like genes.
The structure of the vertebrate RAG locus and of several RAG-like genes or TEs is depicted schematically [51, 57]. Transib and the ProtoRAG element from amphioxus contain TIRs (purple triangles) and 5 base pair TSDs (orange boxes) [49, 57]. Transib shares sequence similarity to RAG1 and does not contain a RAG2-like gene [48]. The coding regions for RAG1/RAG1-like genes and RAG2/RAG2-like genes are in blue and yellow respectively, while gray boxes represent untranslated portions of exons. Start sites of transcription are represented with arrows. The 5’ and 3’ untranslated regions of the Transib transposase gene are not well defined (hatch-marked boxes).
These similarities along with the demonstration of RAG mediated transposition provided support for the transposon/split receptor gene hypothesis for the origins of V(D)J recombination [38, 42, 43]. This hypothesis states that a transposable element (TE) containing RAG1-like and RAG2-like genes and flanked by TIRs that resembled RSSs (hereafter referred to as the RAG transposon), gave rise to the gene segments of the antigen receptor loci and to RAG1 and RAG2. The pivotal evolutionary event is proposed to have been the insertion of a RAG transposon into an exon of a cell surface receptor gene, interrupting the exon with a TIR-flanked fragment and marking the two portions of the exon for reassembly. Expression of the RAG1/RAG2 transposase genes from another site in the genome (perhaps a distinct integration of the RAG transposon) would have provided the necessary enzyme in trans to bind the TIRs and generate the DNA double stranded breaks needed to recreate a functional receptor gene. No additional RAG transposon integration event would have been necessary for generation of the known antigen receptor loci [41], making this an attractive hypothesis that accounts for all of the key building blocks of this system from a single origin.
Identifying ancient relatives of RAG1 and RAG2
While the idea that a RAG transposon gave rise to the antigen receptor loci and the RAG proteins has been a dominant hypothesis for a number of years [39, 42, 43], identifying potential precursor elements and linking them to the modern day proteins proved challenging. Early clues were derived from the comparison of RAG cleavage biochemistry to that used by the cut-and-paste class of TEs. The lack of covalent bond formation between RAG and the DNA during cleavage and the use of three acidic residues in the catalytic triad to coordinate metal cations placed the RAG transposon in the DDE family of TEs [16–18]. Like RAG, these elements cleave their target DNA through nicking and transesterification and some members, such as Tn5, Tn10, and Hermes, also generate a hairpin intermediate [44–46]. However, none of these elements contained significant homology to the vertebrate RAGs. Characterization of RAG mediated transposition provided an additional set of clues in the search for the primordial RAG TE as it revealed the TSD generated by RAG to be predominantly 5 base pairs in length with a preference for GC rich integration sites [12, 13, 47].
Transib
The subsequent identification of the Transib family of TEs finally provided a candidate with sequence similarity to the core region of RAG1, including conservation of the catalytic residues [48] (Fig. 2A). In addition, the TIRs of the Transibs have some sequence similarity with the RSS, with a well conserved heptamer that includes the 5’ CAC residues critical for RAG mediated cleavage (Fig. 2B). Subsequent biochemical characterization of an active member of the Transib family, Hztransib, named after its host organism, Helicoverpa zea (commonly known as corn earworm), demonstrated a similar cleavage mechanism and reaction polarity to that of the RAG reaction, leaving hairpins on the DNA flanking the TE excision site [49]. Like RAG mediated transposition, Hztransib also generates 5 base pair TSDs and has a preference for GC rich integration sites [48, 49].
RAG1-like and RAG2-like proteins
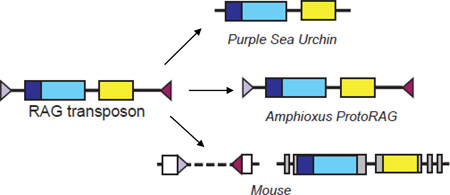
While the discovery of the Transibs provided a candidate for a primordial RAG TE, ascertaining whether a transposon containing RAG1-like and RAG2-like genes ever existed, and if so, piecing together its evolutionary history, has been difficult. Members of the Transib family contain only a RAG1-like gene (Fig. 3) [48, 50] leaving the origins of RAG2 uncertain and hinting at independent origins for the two RAG genes. The picture was further complicated with the identification of adjacent RAG1-like and RAG2-like genes in the purple sea urchin (Fig. 3) [51]. Like Transib, the purple sea urchin RAG1-like (SpRAG1L) protein shares sequence similarity with the core of RAG1 including the catalytic residues, but unlike Transib, the similarity extended to include portions of the N-terminal RING domain. While the purple sea urchin RAG2-like (SpRAG2L) protein lacks extended sequence similarity to RAG2, it is predicted to contain an N-terminal six bladed beta propeller domain and a C-terminal PHD finger, as is the case for vertebrate RAG2 [43, 51] (Fig 2A). It was subsequently found that while the RAG2 PHD binds H3K4 trimethyl, the SpRAG2L PHD binds H3K4 dimethyl [52]. When co-expressed, the purple sea urchin RAG-like proteins were able to interact with each other, and SpRAG1L was also capable of interacting with shark RAG2 [51], highlighting potential similarities between the purple sea urchin proteins and the vertebrate RAGs. In addition, SpRAG1L and SpRAG2L are coordinately expressed in embryonic and adult sea urchin cells [51], similar to the coordinate expression of vertebrate RAG1 and RAG2. However, no endogenous function or DNA cleavage, recombination, or transposition activity had been attributed to these proteins, leaving their relationship to the vertebrate RAGs uncertain. The lack of TIRs flanking the SpRAG1L–SpRAG2L locus also made their relationship to the Transibs and the putative RAG transposon difficult to ascertain.
More clues to the existence of the RAG transposon and the evolutionary history of RAG were recently provided by the identification of additional RAG-like genes or gene fragments in the genomes of chordates such as the green sea urchin, amphioxus, and starfish. The green sea urchin RAG1-like (LvRAG1L) gene, though harboring multiple inactivating mutations, is predicted to encode protein fragments with 50% sequence identity to SpRAG1L [53]. The green sea urchin RAG2-like (LvRAG2L) gene, which lies adjacent to LvRAG1L in convergent transcriptional orientation, is potentially intact and can encode a protein with 48% identity to SpRAG2L [53]. Hence, these two sea urchin species contain tandem RAG-like genes that are strongly related. Interestingly, the green and purple sea urchin RAG-like loci are located in non-syntenic regions of the genomes of the two species. This is consistent with the possibility that the two loci derived from two independent transposon insertion events, although there is no evidence of TIRs or TSDs in either locus [53]. The bat star genome also appears to contain adjacent, convergently oriented RAG1-like (PmRAG1L) and RAG2-like (PmRAG2L) genes that encode predicted proteins or protein fragments with substantial sequence similarity to the SpRAG-like proteins [53]. Although the bat star genome assembly is incomplete, comparison of sequences flanking one copy of PmRAG1L to the (as yet incomplete) bat star genome assembly revealed low copy number repeat sequences with well defined boundaries, the ends of which contain 13 base pair identical sequences with clear resemblance to the RSS heptamer (Fig. 2B). Hence, PmRAG1L–PmRAG2L might be flanked by TIRs that resemble the RSS, and additional analysis raised the possibility of a 5 base pair TSD in one set of adjacent contigs [53]. Finally, an early study of the amphioxus genome identified a region encoding a RAG1-like protein fragment (BfRAG1L) with sequence similarity solely to a central portion of vertebrate RAG1 [54] (Fig 2A). Notably, this fragment exhibited endonuclease activity, and when reconstituted with the missing portions of the RAG1 core, the resulting chimeric protein was capable of mediating V(D)J recombination when paired with mouse RAG2 [54]. Identification of these RAG-like proteins in various organisms expanded the evidence in favor of the existence of a RAG transposon that was more ancient and widespread than previously thought. Currently, nothing is known about the function or biochemical properties of the RAG-like proteins from the green sea urchin and the bat star.
Functional links between RAG and its ancient relatives
Some progress was made towards piecing together the relationship between RAG and various RAG-like proteins by our recent study that utilized a highly sensitive assay for V(D)J recombination in mouse fibroblasts. These experiments revealed that both Hztransib and SpRAG1L could mediate V(D)J recombination when paired with mouse RAG2 [55], providing the first functional link between these proposed ancient relatives and vertebrate RAG1. Like SpRAG1L, Hztransib was shown to be able to interact physically with shark RAG2 and mouse RAG2, suggesting functional similarities between Hztransib and RAG and RAG-like proteins. This study also demonstrated, as had first been suggested during the original discovery of RAG1 [1], that RAG1 has low levels of V(D)J recombination activity in the absence of RAG2 [55], a result also obtained in RAG2-deficient mice [56]. Together, these result illustrated striking parallels between RAG1 and Transib: both proteins have the capacity to function in the absence of RAG2, and yet both also have the capacity to interact physically and functionally with RAG2. We therefore favor the hypothesis that a Transib element was the original source of RAG1. These experiments also revealed that recombination by RAG1 in the absence of RAG2 was not substantially more efficient on a 12/23 RSS pair than on a 12/12 RSS pair, in contrast to recombination mediated by RAG1 with RAG2, which strongly favors the 12/23 pair. This hints at a potential role for RAG2 in helping to establish the 12/23 rule during the evolution of V(D)J recombination.
In this same study, biochemical assays documented the ability of RAG2 to stimulate transposition activity by Hztransib on substrates containing Transib TIRs and on substrates with a pair of 23RSSs or a 12/23RSS pair [55]. Curiously, Hztransib transposase was capable of mediating transposition of a substrate containing a pair of 12RSSs in the absence of RAG2, and this activity was heptamer dependent but nonamer independent, consistent with heptamer (but not nonamer) conservation in the Hztransib TIR and the lack of a recognizable nonamer binding domain in Hztransib transposase. These results reveal the ability of mouse RAG2 to alter the biochemical activity of Hztransib transposase and lead us to suggest that collaboration between a RAG2-like protein and an early RAG1-like Transib would have led to enhanced transposition activity and perhaps altered TIR substrate preferences, with implications for how to integrate the various RAG-like proteins into the evolutionary history of the vertebrate RAGs (see model below).
ProtoRAG: the missing link
Prior to the identification of RAG-like genes in the purple sea urchin, it had been presumed that the RAG transposon was introduced into the genome of a common jawed vertebrate ancestor by horizontal transfer from another species [13, 42]. After this discovery, however, a different possibility was raised [43, 51]: that linked RAG1-like and RAG2-like genes existed in an early chordate ancestor and were passed down to vertebrates and echinoderms by the more conventional vertical transmission process (discussed further below). What remained unresolved on this alternative model was whether the vertically transmitted element was a RAG transposon, or was instead simply linked RAG1-like and RAG2-like genes that lacked TIRs and transposon function. The primary reason for uncertainty was that no active TE fulfilling the criteria for a RAG transposon had been identified in any species, leading to doubts as to whether the RAG transposon ever actually existed. The discovery of ProtoRAG [57] puts these doubts to rest.
The ProtoRAG superfamily of cut-and-paste transposons was identified in amphioxus (also known as the lancelet), a cephalochordate with a genome that exhibits extreme polymorphism and a remarkably high diversity of TEs [58]. In all identified copies of ProtoRAG, the open reading frames were closely flanked by extended inverted repeat sequences that terminate in identical seven base pair sequences. Both the sequence analysis and functional data clearly indicate that these inverted repeats constitute the TIRs of ProtoRAG. Like Transib, ProtoRAG TIRs have sequence similarity to the RSS heptamer (Fig. 2B) and are flanked by 5 base pair TSDs [57]. Unlike Transib, however, ProtoRAG contains both a RAG1-like (BbRAG1L) and a RAG2-like (BbRAG2L) gene, oriented in a tail-to-tail, convergently transcribed orientation (Fig. 3). BbRAG1L displays sequence similarity to vertebrate RAG1 throughout most of the core, including conservation of active site residues and zinc coordinating ligands, but lacks similarity to the NBD of RAG1 (Fig. 2A), and correspondingly, ProtoRAG lacks sequence similarity in its TIRs to the RSS nonamer (Fig. 2B). BbRAG1L also exhibits sequence similarity to SpRAG1L, and curiously, both proteins contain a block of highly repeated amino acid sequence in their N-terminal regions, although the repeats differ in sequence and length and are inserted at different locations in the two proteins. In addition, BbRAG1L shares sequence similarity to the noncore N-terminal RING/Zinc finger of RAG1, with conservation of nearly all of the zinc coordinating residues of this domain. BbRAG2L, like SpRAG2L, shares low sequence similarity to RAG2 but contains residues predicted to allow the protein to adopt a beta propeller structure. Indeed, computer modeling predicts that BbRAG1L and BbRAG2L contain secondary structure elements very similar to those found in the RAG1 and RAG2 core regions, respectively, and have the potential to fold into tertiary structures similar to those adopted by the RAG cores (M. Surleac and A. Petrescu, personal communication). Notably, however, BbRAG2L is missing the entire RAG2 C-terminal region including the PHD finger present in SpRAG2L and RAG2 [57].
Together, BbRAG1L/2L have TIR-dependent DNA cleavage activity in vitro that, like RAG, is supported by Mg2+ but not Ca2+ cations, is stimulated by human HMGB1, and proceeds through a nick-hairpin mechanism [57]. BbRAG1L/2L are also capable of mediating transposition of target sequences flanked by ProtoRAG TIRs, and like RAG, they generate predominantly a 5 base pair TSD and have a preference for GC rich integration sites. When expressed in human cells, BbRAG1L/2L can mediate TIR-dependent DNA cleavage of episomal substrates, and the cleaved TIR DNA ends can then undergo transposition or be joined to yield structures resembling the signal joints formed during V(D)J recombination (Fig. 1g, h). In addition, the non-TIR ends (containing the flanking donor DNA) generated by BbRAG1L/2L cleavage can be joined to form structures closely resembling V(D)J coding joints (Fig. 1f). This extensive array of parallels between the sequences and activities of RAG1/2 and the ProtoRAG proteins strongly argues for an evolutionary relationship between the two systems. ProtoRAG therefore represents the first description of a functional transposable element containing both RAG1-like and RAG2-like genes.
Model for the evolution of the RAG proteins
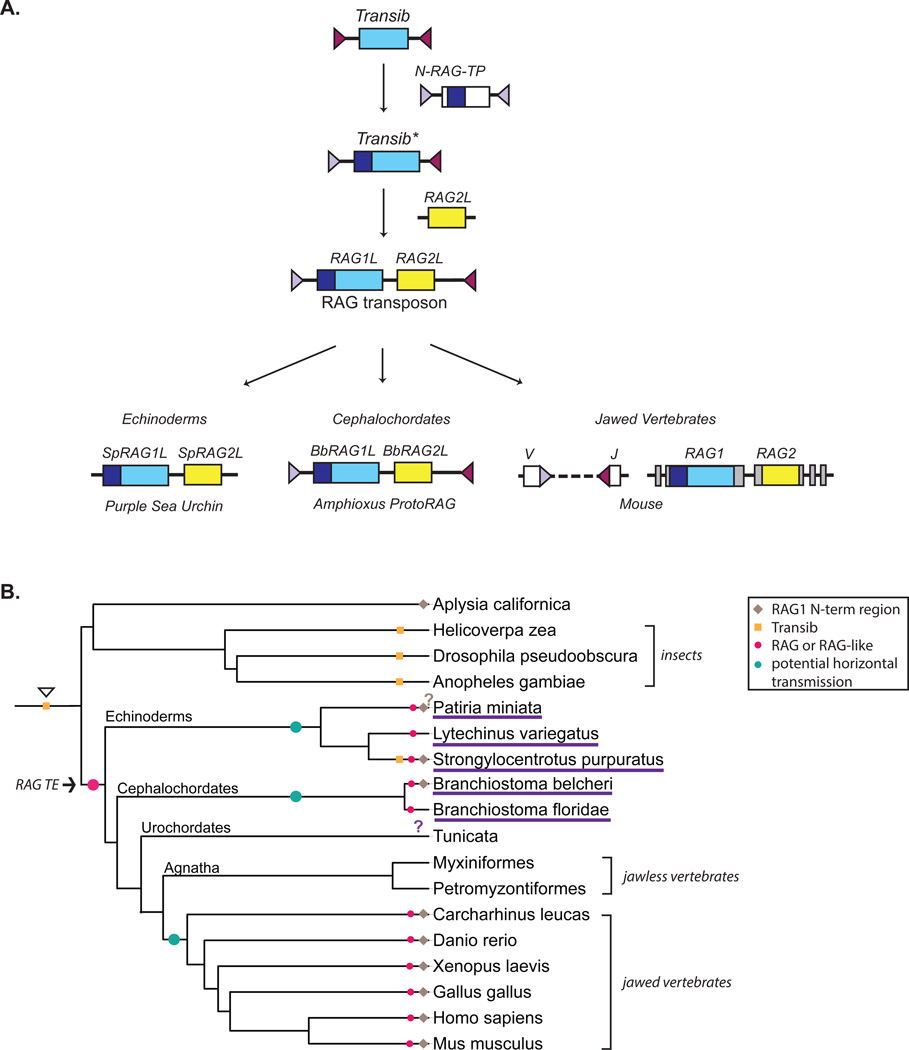
The recent work identifying and characterizing various RAG-like proteins has allowed for the formulation of a more complete and evidence based model for the evolution of RAG1 and RAG2 and the proteins they encode, and argues against an alternative proposal of a viral origin for RAG1 [59]. The similarities between Transib transposase and RAG1 and the ability of Hztransib transposase to mediate recombination when paired with mouse RAG2 provides evidence for a relationship between these proteins [55]. Given that Transib is found in the genomes of organisms whose ancestors branched off during eukaryote evolution much earlier than amphioxus, it is likely that Transib is an older superfamily of TEs than ProtoRAG, and that a Transib TE was the evolutionary precursor of RAG1 and RAG1-like genes (Fig. 4A) [48, 50]. We refer to this hypothetical element as Transib*. The Transib* transposase would have been capable of mediating transposition on targets containing its own TIRs, and it is likely that these TIRs contained an RSS heptamer-like sequence necessary for cleavage.
Figure 4. Model for the evolution of the RAG proteins and transmission of the ancestral RAG transposon.
A. We propose that RAG1 originated from an ancient relative of the Transib TEs. A recombination event between this ancient element and a relative of N-RAG-TP could have generated a new element, Transib*, containing an open reading frame containing both the noncore N-terminal domain and core region of the RAG1 precursor, as well as one new TIR. Subsequent acquisition of a RAG2-like gene by Transib* led to the emergence of the RAG transposon that gave rise to the RAG and RAG-like genes found in echinoderms, cephalochordates, and jawed vertebrates. In the sea urchin, the TIRs of the RAG-like element appear to have been lost while in amphioxus, the TIRs have been maintained and the ProtoRAG element is active in vitro and may retain activity in vivo. In jawed vertebrates, the TIRs went on to become the RSSs after being inserted into a gene (the ancestral split receptor gene) that gave rise into the antigen receptor loci as predicted by the transposon/split receptor gene hypothesis [38, 42, 43]. RAG1/RAG1-like and RAG2/RAG2-like genes are in blue and yellow respectively, and the N-terminal noncore region of RAG1 is in dark blue. TIRs and RSSs are purple triangles. No attempt has been made to depict the origins of RAG2 C-terminal region containing the PHD, although the most parisomonious hypothesis is that it was present in RAG2L of the RAG transposon.
B. We propose that the RAG transposon emerged in the genome of a common deuterostome ancestor (arrow and large pink dot), leading to the existence of RAG-like genes in numerous lineages (small pink dots). The widespread presence of these genes can be accounted for through vertical transmission of the RAG TE and loss of RAG-like genes in the jawless fish and at least some tunicates. A model involving horizontal transmission is also possible, and in this case, three independent integration events (teal dots) of a RAG TE would have been required to explain the presence of RAG-like genes in various lineages. Orange squares mark the presence of Transib TEs [48] and the presumed presence of ancient Transib TEs (open arrow head) prior to the divergence of protostomes and deuterostomes. Organisms in which RAG1-like genes have been reported are underlined in purple. The presence or absence of RAG1-like genes in tunicates has not been definitively determined (purple question mark). The N-terminal noncore domain may have originated from a relative of the N-RAG-TP element reported in Aplysia californica. Species with a RAG1-like protein containing the N-terminal domain are marked with a brown diamond. It is not clear whether the N-terminal region of the bat star RAG1-like has homology to RAG1 (brown question mark).
The acquisition of a RAG2-like gene by Transib* would have been the pivotal step in the evolution of the RAG transposon (Fig. 4A). The RAG2-like protein might have enhanced and modulated the transposition activity of the element, consistent with our findings [55], thereby providing a selective advantage for maintaining both proteins. It is very attractive to think that this initial two-gene TE (RAG transposon) was the evolutionary precursor of modern day ProtoRAG, which retains transposase function, of the RAG1L/RAG2L gene pairs observed in echinoderm genomes (whose functions and activities are unknown), and of the RAG transposon presumed to have been present in early vertebrate ancestors (Fig. 4A).
From here, it is easy to imagine how the generation of a split receptor gene early in jawed vertebrate evolution could have provided the necessary selective pressure for the development of a more highly active recombinase enzyme in the jawed vertebrate lineage (Fig. 4A). If the receptor gene contributed to organism fitness, for example by encoding a receptor involved in immune recognition, it would have been advantageous for the organism to reassemble the receptor gene by TE excision, a function provided by the RAG1/RAG2 proteins encoded either by the inserted TE itself or another copy of the TE present elsewhere in the genome. Mutations in the RAG proteins that enhanced their recombination activity while reducing or regulating their transposition activity would then have been positively selected. In this regard, it is noteworthy that BbRAG2L does not contain a PHD finger, as this domain reduces the transposition activity of the RAGs [60–62] but plays an important role in helping the RAG complex localize to sites of open chromatin [34, 35, 63–65]. Retention of regions such as the PHD could have led to a less active transposase, while additional changes could have enhanced recombination activity allowing for efficient reassembly of the receptor gene. Conversely, loss of the PHD from BbRAG2L might have been important for enhancing transposition of ProtoRAG TEs.
In the echinoderm lineage, the RAG transposon might have remained active for an extended period of time. This is suggested by the fact that, as noted above, putative RAG transposon descendants are found in different locations in the green sea urchin and purple sea urchin genomes (Fig. 4A) [53]. Subsequent loss of the TIRs would have immobilized the genes in their current locations and eliminated the selective pressure on the open reading frames to maintain transposase activity. The fact that both SpRAG1L and SpRAG2L retain intact open reading frames suggests that these proteins have been “domesticated” to carry out new functions in the purple sea urchin [43, 51]. It also appears that ProtoRAG elements have been active quite recently during lancelet evolution, as members of the family are found in different genome locations in different individuals [57]. It is unknown if the ProtoRAG-encoded proteins have a function in the lancelet apart from that of mediating transposition. It will be of great interest to address this and related questions regarding putative functions of RAG1L/RAG2L gene pairs in echinoderms.
Unanswered Questions, Old and New
The origins of RAG2
As depicted in the model (Fig. 4A), the acquisition of a RAG2-like gene by a Transib transposon was likely a key step in the evolution of the RAG transposon. The origins of RAG2, however, remain elusive. Vertebrate RAG2 is composed of a six-bladed beta propeller core linked to a C-terminal noncore PHD finger. Searches for these two domains in predicted protein structures from organisms predating jawed vertebrates did not reveal candidate proteins other than SpRAG2L [51]. Hence, there is currently no strong candidate for the precursor of RAG2 that is not already linked to a RAG1-like gene. The search for a RAG2 predecessor is complicated by several factors. First, while it is likely that the RAG2 predecessor contained a PHD finger (because this domain is found in both SpRAG2 and vertebrate RAG2), this is not certain given the lack of a PHD in BbRAG2L (Fig. 2A) [51, 52]. Second, while it is possible to identify many proteins with a propeller structure similar to that of RAG2, it is difficult to narrow down this list of candidates. And third, the sequence similarity between RAG2 and BbRAG2L or SpRAG2 is considerably lower than that between RAG1 and BbRAG1L, SpRAG1, or Transib, further complicating a comparative genomics approach to identifying RAG2 related sequences [48, 51]. Perhaps new genome sequences or biochemical approaches will allow identification of candidates for the predecessor of RAG2.
It has been postulated [66] that the ancestral RAG2 protein was a host factor in the organism that contained Transib*. The ability of current day RAG2 to interact with Transib transposase lends support to the hypothesis that the ancestral RAG2 protein could also have interacted with the Transib* transposase, potentially enhancing its activity. Therefore, the ancestral RAG2 protein could have influenced the activity of Transib* transposase prior to its incorporation into the RAG transposon, and could have provided benefits that would have helped maintain it as part of the RAG transposon.
TIR asymmetry in Transib and the RAG transposon
12RSS/23RSS asymmetry and the 12/23 rule provide an important level of regulation of RAG function and V(D)J recombination. When did such asymmetry first arise during evolution of the system? While most Transib TIRs resemble the 23RSS, there is a subset of Transibs with asymmetric TIRs that resemble a 12RSS/23RSS pair [48]. Hence, the asymmetry could have arisen as early as Transib* and hence been a feature of the earliest RAG transposon (see discussion below regarding the RAG1 N-terminal region for speculation on how TIR asymmetry arose). Alternatively, asymmetry might have arisen well after the genesis of the RAG transposon. In either case, the finding that RAG2 strengthens adherence to the 12/23 rule on the part of RAG1 [55] argues that acquisition of a RAG2-like gene during formation of the RAG transposon could have strengthened or enabled a preference for asymmetry. Curiously, vertebrate RAG2 specifically facilitates Hztransib-mediated transposition with 23RSS but not 12RSS substrates [55], despite the fact that Hztransib does not contain a domain with detectable sequence similarity to the NBD of RAG1. The mechanism of this selective stimulation is not known, but it suggests that if Transib* contained one or two 23RSS-like TIRs, the Transib* transposase might have realized a significant boost in activity when present in cells also expressing the RAG2 precursor.
ProtoRAG TIRs do not contain a sequence similar to that of the RSS nonamer but do contain highly conserved 7 bp and 9 bp sequences separated by less well conserved regions of either 27 bp or 31 bp [57] (Fig. 2B). Hence, ProtoRAG elements exhibit TIR asymmetry, with a “27-TIR” upstream of the BbRAG1L gene and a “31-TIR” upstream of the BbRAG2L gene, and initial functional assays indicate a preference for 27/31 over 27/27 and 31/31 [57]. How strong this preference is, the extent to which it regulates ProtoRAG mobilization, and whether BbRAG1L/BbRAG2L cleave DNA in a synaptic complex comparable to the paired complex formed by RAG1/RAG2 and a 12/23RSS pair remain to be determined.
Given the presence of an RSS-like nonamer sequence in many Transib TIRs (and a corresponding NBD-like sequence in some Transib transposases [48]), it seems probable that the TIRs of the initial RAG transposon more closely resembled modern day RSSs than ProtoRAG TIRs. The lack of an RSS-like nonamer sequence in ProtoRAG TIRs is mirrored by the lack of a recognizable NBD in BbRAG1L; a similar situation pertains for Hztransib TIRs and transposase. This likely explains why BbRAG1L/BbRAG2L and Hztransib transposase lack substantial activity on RSS substrates [55, 57] and suggests that both ProtoRAG and Hztransib represent evolutionary branches on which the nonamer sequence and its binding domain co-evolved away from the nonamer and NBD, respectively.
The evolutionary history of the RAG1 N-terminal region
Transib transposase does not contain sequences similar to the RAG1 N-terminal region, raising the question of the evolutionary origin of this portion of RAG1. A plausible answer came from an unexpected direction with the identification of a TE, named N-RAG-TP, in the sea slug Aplysia californica, that encodes a putative transposase with extended sequence similarity to the RAG1 N-terminal region including the RING/Zinc finger region [67]. The similarity ended abruptly at the RAG1 core, leading to the proposal that the ancestral RAG1 was created by a recombination event between a Transib and an N-RAG-TP element [43]. One possibility is that acquisition of the RAG1 N-terminal region occurred after formation of the RAG transposon, in which case Transib* would have lacked this region. Alternatively, a recent analysis argues that unification of the RAG1 core with the N-terminal region was an early event, preceding even the formation of extant Transib TEs [53]. On this model, Transib* contained the RAG1 N-terminal region, and Transibs (and many other RAG1-like genes) experienced the loss of some or all of this region. It is conceivable that a recombination event between a Transib TE and an N-RAG-TP TE not only unified the RAG1 core and N-terminal regions, but also gave rise to TIR asymmetry, as depicted in Fig. 4A. The finding of a region similar to the RAG1 RING/Zinc finger in the ProtoRAG RAG1-like protein strongly supports the idea that the RING/Zinc finger region was an early component of the RAG1-like gene in the RAG transposon and demonstrates that this region is compatible with both transposase and recombinase activity. The relevant targets and functional significance of the E3 ligase activity associated with the RAG1 RING/Zinc finger region are not well understood.
A curious aspect of the N-terminal region of the ProtoRAG and purple sea urchin RAG1-like proteins is the presence of amino acid repeats not found in vertebrate RAG1. For BbRAG1L, this involves 17 copies of variations on the 12 aa sequence PPTADVRATTSQ inserted N-terminal to the RING/Zinc finger domain, while for SpRAG1L, it involves 10 copies of variants of the 8 amino acid sequence TAPLTPTA inserted within the RING/Zinc finger domain. The two repeats share a proline-rich character and a stretch of 4 residues (PPTA; which occurs in 2 of the 10 repeats in SpRAG1L), raising the possibility that the two inserts are related. The origin and functional significance of the repeat regions is not known.
Transmission of the RAG transposon
The identification of RAG1-like and RAG2-like proteins in animals that predate the jawed vertebrates raises the question of when a RAG TE first integrated into the genome of a deuterostome ancestor that gave rise to the jawed vertebrates. Transibs are widely dispersed, being found in various insect species and even fungi, organisms that diverged much earlier than vertebrates [48, 50]. Adjacent RAG1-like and RAG2-like genes have been found in echinoderms (sea urchin and starfish), cephalochordates (amphioxus), and jawed vertebrates. Perhaps the simplest explanation for this lineage distribution of RAG-like genes is a vertical transmission model that begins with a Transib TE in an ancestral bilaterian (orange square at the base of the tree in Fig. 4B), genesis of the RAG transposon by capture of RAG2L by Transib* in a basal deuterostome (large pink circle, Fig. 4B), and subsequent vertical transmission of the RAG transposon (Fig. 4B). This vertical transmission model accounts for the widespread presence of RAG-like genes in the deuterostomes, requires no horizontal transmission events, but requires a loss event in the agnathan lineage leading to the jawless fish, in which no RAG-like genes have been found [68], as well as a loss event at some level in the tunicate lineage. RAG-like sequences have not be found in the genome of the tunicate Ciona intestinalis (the sea squirt), although the Ciona genome is thought to be under strong selection to remain small and lacks many genes found in vertebrates and echinoderms [69, 70]. The presence or absence of RAG-like sequences in other tunicates has not been well documented.
An alternative to the vertical transmission model is one involving horizontal transfer. This would require three independent integration events of the RAG transposon, one each into an ancestral echinoderm, an ancestral cephalochordate, and an ancestral jawed vertebrate (teal circles, Fig. 4B). While more complex, such a scenario offers one possible explanation for the observation that RAG-like genes appear to have evolved more slowly in amphioxus than in vertebrates or sea urchin [57]. A successful model will have to accommodate the presence of introns in different locations and different phases in the open reading frame of RAG-like genes of amphioxus and sea urchin [57] and the absence of introns in the open reading frame of most vertebrate RAG1 genes [71]. It will also have to accommodate the aforementioned lack of a recognizable nonamer and NBD in ProtoRAG TIRs and the BbRAG1L protein, respectively. Further genome sequencing of invertebrate deuterostome genomes might help to distinguish between the two models and might also help decipher the relationship between the Transibs, ProtoRAG, and the vertebrate RAGs.
If correct, the vertical transmission model indicates that RAG-like genes, and very likely the RAG transposon, existed in the last common ancestor of jawed and jawless vertebrates. The adaptive immune systems of jawed and jawless vertebrates exhibit remarkable parallels including three shared lymphocyte lineages that express diverse, clonally distributed antigen receptors [72, 73]. Antigen receptor gene assembly and diversification rely on RAG (for V(D)J recombination) and activation induced deaminase (AID) (for somatic hypermutation/gene conversion) in jawed vertebrates and are thought to rely on cytidine deaminases related to AID in jawless vertebrates [74]. Hence, an ancestral AID protein might have co-existed with RAG1-like/RAG2-like proteins in the last common vertebrate ancestor. Was one or the other (or both) of these enzyme systems active in immune receptor gene assembly/diversification at this evolutionary stage? If so, what type of receptor was being generated/diversified? Was it composed of immunoglobulin domains (as in jawed vertebrates) or of leucine-rich repeats (as in jawless vertebrates)? If the former, the diversifying locus was subsequently lost in the jawless vertebrate lineage; if the latter, it was lost in the jawed vertebrate lineage. These puzzles cannot be solved readily with current information. It is appealing to think, however, that the last common vertebrate ancestor had substantial raw materials available in the form of RAG, AID, and three functionally distinct lymphocyte-like lineages for the creation of a sophisticated adaptive immune system.
A second implication of the vertical transmission model would be that RAG1-like/RAG2-like gene pairs substantially preceded AID-like genes in evolution, given that the AID/APOBEC family of cytidine deaminases appears to be restricted to vertebrates [75].
Transposase versus recombinase
After DNA cleavage by RAG or BbRAG1L/2L, there are two predominant fates for the excised fragment flanked by RSSs or TIRs (Fig. 1e): joining of the ends to form a signal joint (Fig. 1g) or transposition (Fig. 1h). RAG strongly favors the former outcome. RAG actively directs cleaved signal and coding ends into the NHEJ repair pathway for signal and coding joint formation [76–78], while RAG-mediated transposition is extremely rare in vivo [79–82]. In contrast, BbRAG1L/2L, while allowing some TIR-TIR joints to form, appears to strongly favor transposition [57]. These preferred outcomes make sense given the functional imperatives faced by the two enzyme systems. For RAG, end joining accomplishes the goal of V(D)J recombination while transposition threatens genome integrity; for the ProtoRAG proteins, transposition equates to the survival of the TE and perhaps is beneficial to the highly diverse genome of the lancelet. The mechanisms that suppress RAG-mediated transposition in vivo are poorly understood (see particularly [82]), and similarly, the features of BbRAG1L/2L that favor transposition are not known. While RAG has been shown to interact with DNA repair proteins [31, 32], it is not known if this contributes to efficient NHEJ-mediated end joining and/or suppression of transposition. We anticipate that biochemical and structural analyses of Transib transposase and BbRAG1L/2L are likely to be a rich source of information regarding the functional underpinnings of the evolutionary transition from transposase to recombinase.
Concluding Remarks
The identification of RAG-like proteins and TEs with similarities to the vertebrate RAG genes has provided candidates for functional tests to identify relatives of the long sought RAG transposon, the element hypothesized to have played a key role in the establishment of V(D)J recombination. The discovery of the ProtoRAG superfamily of TEs provides the strongest evidence to date for the existence of the RAG transposon. Together, comparative genomics and functional studies have provided much support for the transposon/split receptor gene hypothesis, and they have begun to shed light on the specific changes and steps this element must have undergone as it evolved from a transposase to a recombinase.
Acknowledgments
We thank Anthony De Tomaso for helpful information regarding tunicate genomes, Sebastian Fugmann for insightful ideas, and Marius Surleac and Andre Petrescu for sharing their models of the protein structures of BbRAG1 and BbRAG2. We also thank Shaochun Yuan and Anlong Xu for helpful comments on the manuscript. Our work on the evolution of the RAG proteins was funded by the Howard Hughes Medical Institute, and L.M.C. received support from a training grant from the National Institutes of Health T32 AI007019.
Abbreviations
- BbRAG1L
Branchiostoma belcheri (Chinese lancelet) RAG1-like
- BbRAG2L
Branchiostoma belcheri (Chinese lancelet) RAG2-like
- BfRAG1L
Branchiostoma floridae (Florida lancelet) RAG1-like
- H3K4
lysine 4 of histone 3
- H3K4me3
trimethylated lysine 4 of histone 3
- HMGB1/2
high mobility group protein B1 or B2
- Hztransib
Transib element from Helicoverpa zea (corn earworm)
- LvRAG1L
Lytechinus variegatus (green sea urchin) RAG1-like
- LvRAG2L
Lytechinus variegatus (green sea urchin) RAG2-like
- N-RAG-TP
Transposable element from Aplysia californica (sea slug)
- NBD
nonamer binding domain
- NHEJ
nonhomologous end joining
- PHD
plant homeodomain
- PmRAG1L
Patiria minata (bat starfish) RAG1-like
- PmRAG2L
Patiria minata (bat starfish) RAG2-like
- RAG1
Recombination Activating Gene 1
- RAG2
Recombination Activating Gene 2
- RSS
recombination signal sequence
- SpRAG1L
Strongylocentrotus purpuratus (purple sea urchin) RAG1-like
- SpRAG2L
Strongylocentrotus purpuratus (purple sea urchin) RAG2-like
- TE
transposable element
- TIR
terminal invert repeats
- TSD
target site duplication
Footnotes
Author Contributions
LMC and DGS wrote the manuscript.
References
- 1.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 2.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 4.Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little AJ, Matthews AG, Oettinger MA, Roth DB, Schatz DG. The Mechanism of V(D)J Recombination. In: Alt FW, Radbruch THA, Reth M, editors. Molecular Biology of B cells. London, UK: Academic Press/Elsevier Limited; 2015. pp. 13–34. first published online. [Google Scholar]
- 7.Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 9.van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 10.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 12.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 14.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 15.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 17.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwyn LM, Peak MM, De P, Rahman NS, Rodgers KK. A zinc site in the C-terminal domain of RAG1 is essential for DNA cleavage activity. J Mol Biol. 2009;390:863–878. doi: 10.1016/j.jmb.2009.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudley DD, Sekiguchi J, Zhu C, Sadofsky MJ, Whitlow S, DeVido J, Monroe RJ, Bassing CH, Alt FW. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J Exp Med. 2003;198:1439–1450. doi: 10.1084/jem.20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones JM, Simkus C. The roles of the RAG1 and RAG2 “non-core” regions in V(D)J recombination and lymphocyte development. Arch Immunol Ther Exp (Warsz) 2009;57:105–116. doi: 10.1007/s00005-009-0011-3. [DOI] [PubMed] [Google Scholar]
- 23.Roman CA, Cherry SR, Baltimore D. Complementation of V(D)J recombination deficiency in RAG-1(−/−) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- 24.Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 29.Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- 30.Kassmeier MD, Mondal K, Palmer VL, Raval P, Kumar S, Perry GA, Anderson DK, Ciborowski P, Jackson S, Xiong Y, et al. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J. 2012;31:945–958. doi: 10.1038/emboj.2011.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/Ku80 association with full-length RAG1. Nucleic Acids Res. 2008;36:2060–2072. doi: 10.1093/nar/gkn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coster G, Gold A, Chen D, Schatz DG, Goldberg M. A dual interaction between the DNA damage response protein MDC1 and the RAG1 subunit of the V(D)J recombinase. J Biol Chem. 2012;287:36488–36498. doi: 10.1074/jbc.M112.402487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin WC, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci U S A. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 39.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 40.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 41.Hsu E, Lewis SM. The Origin of V(D)J Diversification. In: Alt FW, Radbruch THA, Reth M, editors. Molecular Biology of B Cells. London, UK: Academic Press/Elsevier Limited; 2015. pp. 133–149. first published online. [Google Scholar]
- 42.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 43.Fugmann SD. The origins of the Rag genes--from transposition to V(D)J recombination. Semin Immunol. 2010;22:10–16. doi: 10.1016/j.smim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 45.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 46.Bhasin A, Goryshin IY, Reznikoff WS. Hairpin formation in Tn5 transposition. J Biol Chem. 1999;274:37021–37029. doi: 10.1074/jbc.274.52.37021. [DOI] [PubMed] [Google Scholar]
- 47.Tsai CL, Chatterji M, Schatz DG. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 2003;31:6180–6190. doi: 10.1093/nar/gkg819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hencken CG, Li X, Craig NL. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol. 2012;19:834–836. doi: 10.1038/nsmb.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson DR, Norton DD, Fugmann SD. The PHD domain of the sea urchin RAG2 homolog, SpRAG2L, recognizes dimethylated lysine 4 in histone H3 tails. Dev Comp Immunol. 2008;32:1221–1230. doi: 10.1016/j.dci.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapitonov VV, Koonin EV. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct. 2015;10:20. doi: 10.1186/s13062-015-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Xu K, Deng A, Fu X, Xu A, Liu X. An amphioxus RAG1-like DNA fragment encodes a functional central domain of vertebrate core RAG1. Proc Natl Acad Sci U S A. 2014;111:397–402. doi: 10.1073/pnas.1318843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carmona LM, Fugmann SD, Schatz DG. Collaboration of RAG2 with RAG1-like proteins during the evolution of V(D)J recombination. Genes Dev. 2016;30:909–917. doi: 10.1101/gad.278432.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montaudouin C, Boucontet L, Mailhe-Lembezat MP, Mariotti-Ferrandiz ME, Louise A, Six A, Freitas AA, Garcia S. Endogenous TCR recombination in TCR Tg single RAG-deficient mice uncovered by robust in vivo T cell activation and selection. PLoS One. 2010;5:e10238. doi: 10.1371/journal.pone.0010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S, Chen Z, Yan X, Yu T, Huang G, Yan Q, Pontarotti PA, Zhao H, Li J, Yang P, et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat Commun. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dreyfus DH. Paleo-Immunology: Evidence Consistent with Insertion of a Primordial Herpes Virus-Like Element in the Origins of Acquired Immunity. Plos One. 2009:4. doi: 10.1371/journal.pone.0005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swanson PC, Volkmer D, Wang L. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J Biol Chem. 2004;279:4034–4044. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- 63.Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, et al. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell. 2015;162:751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maman Y, Teng G, Seth R, Kleinstein SH, Schatz DG. RAG1 targeting in the genome is dominated by chromatin interactions mediated by the non-core regions of RAG1 and RAG2. Nucleic Acids Res. 2016;19 doi: 10.1093/nar/gkw633. first published online July. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panchin Y, Moroz LL. Molluscan mobile elements similar to the vertebrate Recombination-Activating Genes. Biochem Biophys Res Commun. 2008;369:818–823. doi: 10.1016/j.bbrc.2008.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–157. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 69.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 70.Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, Marino R, Shida K, Ikeda M, Ikeda M, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- 71.Flajnik MF, Du Pasquier L. Evolution of the Immune System. In: Paul WE, editor. Fundamental Immunology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013. pp. 67–128. first published online. [Google Scholar]
- 72.Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, Cooper MD. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501:435–438. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flajnik MF. Re-evaluation of the immunological Big Bang. Curr Biol. 2014;24:R1060–R1065. doi: 10.1016/j.cub.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 75.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 76.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 77.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 78.Cui X, Meek K. Linking double-stranded DNA breaks to the recombination activating gene complex directs repair to the nonhomologous end-joining pathway. Proc Natl Acad Sci U S A. 2007;104:17046–17051. doi: 10.1073/pnas.0610928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol. 2006;26:1558–1568. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Curry JD, Schulz D, Guidos CJ, Danska JS, Nutter L, Nussenzweig A, Schlissel MS. Chromosomal reinsertion of broken RSS ends during T cell development. J Exp Med. 2007;204:2293–2303. doi: 10.1084/jem.20070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang H, Ross AE, Desiderio S. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J Biol Chem. 2004;279:8478–8486. doi: 10.1074/jbc.M311219200. [DOI] [PubMed] [Google Scholar]