Supplemental Digital Content is available in the text
Keywords: complementary sequence analysis, differentially expressed miRNAs, epilepsy, interaction network, rno-miR-187-3p
Abstract
This study aimed to explore the role of aberrant miRNA expression in epilepsy and to identify more potential genes associated with epileptogenesis.
The miRNA expression profile of GSE49850, which included 20 samples from the rat epileptic dentate gyrus at 7, 14, 30, and 90 days after electrical stimulation and 20 additional samples from sham time-matched controls, was downloaded from the Gene Expression Omnibus database. The significantly differentially expressed miRNAs were identified in stimulated samples at each time point compared to time-matched controls, respectively. The target genes of consistently differentially expressed miRNAs were screened from miRDB and microRNA.org databases, followed by Gene Ontology (GO) and pathway enrichment analysis and regulatory network construction. The overlapping target genes for consistently differentially expressed miRNAs were also identified from these 2 databases. Furthermore, the potential binding sites of miRNAs and their target genes were analyzed.
Rno-miR-187-3p was consistently downregulated in stimulated groups compared with time-matched controls. The predicted target genes of rno-miR-187-3p were enriched in different GO terms and pathways. In addition, 7 overlapping target genes of rno-miR-187-3p were identified, including NFS1, PAQR4, CAND1, DCLK1, PRKAR2A, AKAP3, and KCNK10. These 7 overlapping target genes were determined to have a different number of matched binding sites with rno-miR-187-3p.
Our study suggests that miR-187-3p may play an important role in epilepsy development and progression via regulating numerous target genes, such as NFS1, CAND1, DCLK1, AKAP3, and KCNK10. Determining the underlying mechanism of the role of miR-187-3p in epilepsy may make it a potential therapeutic option.
1. Introduction
Epilepsy is a serious neurological disorder characterized by spontaneously recurring seizures which are attributed to abnormal and synchronous firing of neurons in the brain.[1] Epilepsy affects approximately 65 million individuals of all ages worldwide, making it second to stroke as one of the most common serious brain disorders.[2] The recent developments in treatment approaches such as surgery,[3] antiepileptic drug therapy,[4] and brain stimulation,[5] have made great advances in the control of seizures. However, almost one-third of all patients with epilepsy still have intractable seizures or adverse effects.[6] One compelling challenge for the treatment of epilepsy is to elucidate the molecular mechanisms of disease development and progression.
MicroRNAs (miRNAs) are a class of endogenous, small, non-coding RNA molecules, which post-transcriptionally fine-tune the expression of their target protein-encoding genes by binding to conserved sequences within their target genes, mainly within the 3′-untranslated region (3′-UTR).[7] There is increasing evidence to support that miRNA changes are involved in the pathophysiology of epilepsy.[8] Using either rodent epilepsy models or human tissue from patients with epilepsy, a number of previous studies have demonstrated that miRNA expression is altered in epilepsy.[9] For instance, Song et al[10] found that 23 miRNAs were expressed differentially in temporal lobe epilepsy (TLE) rats, including 18 upregulated miRNAs (miR-23a/b included) and 5 downregulated miRNAs (let-7e included). Moreover, Hu et al[9] demonstrated the pattern of increased expression of the pro-apoptotic miR-34a in the hippocampus in post-status epilepticus rats. Additionally, 1 previous study reported that miR-146a, an inflammation-associated miRNA, was highly expressed in astrocytes in the hippocampus in a rat model of TLE, as well as in hippocampal tissue from TLE patients.[11] Recently, Henshall[8]found that miRNAs might play critical roles in the control of cell death, inflammation, synaptic structure, and the immune response in epilepsy. Thus, research on miRNAs has the potential to deliver diagnostics and therapeutics that link directly to how the regulatory molecules influence the pathogenesis of epilepsy.[8] However, the functional roles of the aberrantly expressed miRNAs and how miRNAs are involved in the process of epileptogenesis via regulating gene expression remain to be explored.
Recently, Bot et al[12] observed that complex changes in miRNA and their predicted target genes in the dentate gyrus could participate in several molecular events, especially the immune response and neuronal plasticity in epilepsy. Moreover, Meng et al[13] demonstrated that neuronal calcium signaling pathways were associated with the development of epilepsy. However, they did not focus on the miRNAs that were consistently up- or down-regulated at all time points following stimulation in the epileptic dentate gyrus of rats, which could be used as biomarkers. In this study, we downloaded the same miRNA expression profile of GSE49850 used by Bot et al[12] and Meng et al[14] from a public database, and reanalyzed the data using different methods and tools. Microarray analysis was performed to identify the differentially expressed miRNAs in the rat epileptic dentate gyrus at 7, 14, 30, and 90 days after electrical stimulation compared with those from sham-operated time-matched controls. The miRNAs that were consistently down- or upregulated from 7 days to 90 days were our focus. In addition, target genes of the consistently differentially expressed miRNAs were screened, and subjected to functional enrichment analysis and construction of regulatory network. Furthermore, conserved complementary sites between the miRNAs and their target genes were identified. This research aimed to explore the role of aberrant miRNA expression in epilepsy and to identify more potential genes or pathways associated with epilepsy progression based on rat miRNA expression profiles.
2. Methods
2.1. Microarray data
The Gene Expression Omnibus (GEO) repository at National Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/geo/) is currently the largest fully public gene expression resource, which facilitates the submission, storage and retrieval of microarray and other forms of high-throughput data generated by the scientific community.[15] In this study, the miRNA expression profile of GSE49850 was downloaded from the GEO database, which was deposited by Bot et al.[12] The platform is GPL17566 Exiqon miRCURY LNA microRNA array; 7th generation; batch 208500–2, 208510; lot 35009-hsa, mmu &rno (miRBase 19.0). As previously described in the original study by Bot et al[12] the status epilepticus was induced by electrical stimulation of the amygdala in adult male Sprague-Dawley rats, which was used as a model of TLE. Time-matched control animals had electrodes implanted but did not receive electrical stimulation. The dentate gyrus was collected at 7, 14, 30, and 90 days after stimulation. The dataset consisted of 40 samples, including 20 samples of electrically stimulated dentate gyrus, at a range of time points (7, 14, 30, and 90 days) after stimulation and 20 additional samples of sham-operated time-matched controls. Each time point included 5 replicates from each group. This study just re-analyzed the microarry data downloaded from public database and performed bioinformatics analysis. The authors declare that no experiments were performed on humans or animals for this investigation. Thus, ethics approval or consent to participate was not applicable.
2.2. Data preprocessing and differentially expressed miRNA screening
The raw data and the probe annotation file were downloaded. Median normalization was performed on the raw data using the robust multiarray average (RMA) algorithm [16] in R.[17] The missing values were imputed. The t-test in the multtest package [18] in R was used to identify the significantly differentially expressed miRNAs in the stimulated groups at 4 time points, which were compared with the respective time-matched controls. Only the miRNAs with P< .05 were considered significantly differentially expressed. The miRNAs that were consistently up- or downregulated at these 4 time points compared with their respective time-matched controls would be our focus and were thus used for the follow-up analysis.
2.3. Prediction of target genes of the focused differentially expressed miRNAs
A growing number of miRNA-related database systems have been developed to provide further insights into miRNAs and their target genes.[19] For example, miRDB (http://mirdb.org/miRDB/) is an online database for the retrieval of computationally predicted miRNA targets and for miRNA functional annotations with a wiki editing interface.[4] microRNA.org (http://www.microrna.org/microrna/home.do/) is a comprehensive resource of miRNA target predictions, experimentally observed expression patterns, and target down-regulation scores.[20]
In the present study, to identify potential target genes of the consistently aberrant miRNAs, we used two databases: miRDB and microRNA.org. The target genes associated with miRNAs were identified from the 2 databases. The common (overlapping) target genes identified from these 2 databases were deemed to be more reliable.
2.4. Functional enrichment analysis of target genes
To facilitate the functional enrichment analysis, all target genes identified from the miRDB and microRNA.org databases were subjected to functional enrichment analysis using the Database for Annotation Visualization and Integrated Discovery (DAVID, version 6.8).[21] Over-represented Gene Ontology (GO) terms (including biological process [BP], molecular function [MF], and cellular component [CC] categories),[22] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified based on a hypergeometric distribution algorithm. The false discovery rate (FDR) [23] was employed for multiple testing correction using the Benjamini and Hochberg method.[24] The threshold was set as FDR < .05.
2.5. Construction of the interaction network
The Osprey network visualization system is used to design a complex interaction network to enable visualization and manipulation.[25] Osprey represents genes as nodes and interactions of the proteins as edges between nodes.[25] In this study, to annotate functional interactions between the focused differentially expressed miRNAs and their target genes, an interaction network was constructed using the General Repository for Interaction Datasets (GRID) [25] and the Biomolecular Interaction Network Database (BIND),[26] and was generated by the Osprey network visualization system.[25]
2.6. Target gene-miRNA binding site analysis
The miRNA gene sequences were collected from the miRBase sequence database (http://www.mirbase.org/).[27] The sequences of the common target genes found in both databases were identified in the NCBI Gene (http://www.ncbi.nlm.nih.gov/gene). Moreover, the BLAST program in NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) was used to search for the binding sites of the miRNAs and their target genes.
3. Results
3.1. Data preprocessing and screening of differentially expressed miRNAs
After data normalization and screening of differentially expressed miRNAs, we found 51 differentially expressed miRNAs in the simulated group at 7 days following electrical stimulation of the amygdala compared with the control group at 7 days. Additionally, 38, 100, and 62 differentially expressed miRNAs were identified in the other simulated groups (at 14, 30, and 90 days) compared with their respective time-matched controls (Supplementary Table 1). The top 5 upregulated miRNAs and downregulated miRNAs in the stimulated group (ordered according to decreasing |log2 fold change [FC]|, respectively) at each time point are shown in Table 1, such as upregulated rno-miR-212-3p at 7 days, downregulated rno-miR-187-3p at 14 days, upregulated rno-miR-146a-5p at 30 days, and downregulated rno-miR-187-3p at 90 days. However, among all the identified differentially expressed miRNAs, only rno-miR-187-3p was consistently downregulated at each time point from 7 days to 90 days. The decrease in rno-miR-187-3p expression was 0.56-fold, 0.62-fold, 0.73-fold, and 0.56-fold of that in the time-matched controls at 7, 14, 30, and 90 days after stimulation, respectively (P < .01) (Table 1). The expression values of rno-miR-187-3p in each sample at each time point are represented in Fig. 1. Thus, we decided to direct our attention toward rno-miR-187-3p.
Table 1.
The top 5 upregulated miRNAs and downregulated miRNAs between the stimulated and control group at each time point.
Figure 1.
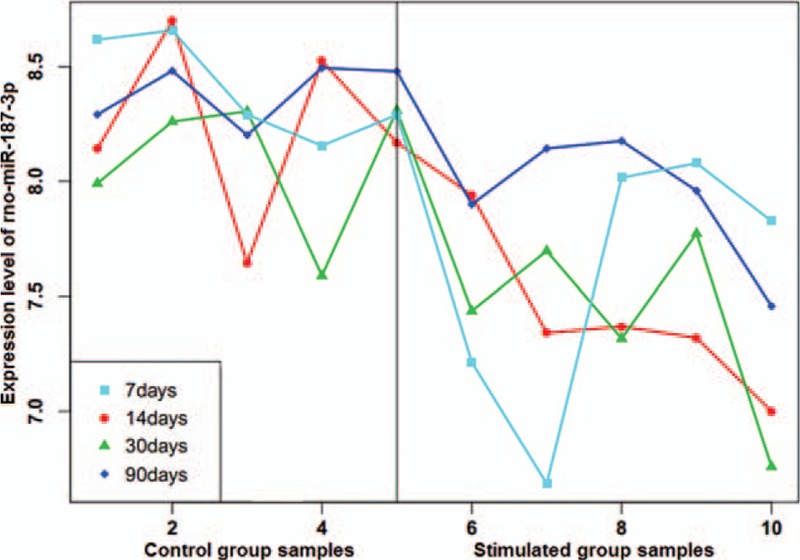
Expression of rno-miR-187-3p at each time point after electrical stimulation of the amygdala and time-matched controls. As each time point had 5 replicates of each group, the left 5 numbers represent control samples and the right 5 numbers represent stimulated samples.
3.2. Screening rno-miR-187-3p target genes
As miRNAs negatively regulate gene expression by targeting mRNAs at the posttranscriptional level, it is necessary to identify putative target genes for a better understanding of the function of miRNAs. In our study, 15 target genes of rno-miR-187-3p were identified from the miRDB database (there are 2 rat miRNAs found for “miR-187” in the miRDB database, rno-miR-187-3p [previously rno-miR-187] and rno-miR-187-5p [previously rno-miR-187∗]; rno-miR-187-3p targets were exactly what we needed). When we searched for potential target genes of rno-miR-187-3p using microRNA.org database, no results were found using rno-miR-187-3p as the direct miRNA identifier. After sequence alignment, we found that the results of rno-miR-187 (corresponding to rno-miR-187-3p currently; rno-miR-187-5p were not included) in the microRNA.org database were exactly what we needed. Thus, a total of 415 target genes of rno-miR-187-3p were identified from the microRNA.org database. Furthermore, 7 overlapping genes for rno-miR-187-3p were identified from these 2 databases, namely, NFS1 Cysteine Desulfurase (NFS1), Progestin And AdipoQ Receptor Family Member 4 (PAQR4), Cullin Associated And Neddylation Dissociated 1 (CAND1), Doublecortin Like Kinase 1 (DCLK1), Protein Kinase CAMP-Dependent Type II Regulatory Subunit Alpha (PRKAR2A), A-Kinase Anchoring Protein 3 (AKAP3), and Potassium Two Pore Domain Channel Subfamily K Member 10 (KCNK10).
3.3. Functional enrichment analysis of all target genes of rno-miR-187-3p
After pathway and GO analyses, we found that the target genes of rno-miR-187-3p were enriched in different GO terms and pathways. The top 5 GO terms in each category and all the pathways enriched by all target genes are shown in Table 2. From these results, we found that the target genes of rno-miR-187-3p were related to positive regulation of cell proliferation, protein binding, and kinase activity. Moreover, the target genes of rno-miR-187-3p were involved in different pathways such as the hypoxia-inducible factor 1 (HIF-1) signaling pathway, long-term depression, and the phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway.
Table 2.
Top 5 GO terms in each category and the pathways enriched by all putative target genes of rno-miR-187-3p.

3.4. Interaction network construction of rno-miR-187–3p and all target genes
Interactions between rno-miR-187-3p and all of its potential target genes, and interactions between these target genes, are presented in the network as shown in Fig. 2. rno-miR-187-3p was the center of the network, and we found that rno-miR-187-3p could regulate 430 genes. Additionally, from this interaction network, 80 interaction pairs among the target genes could also be identified. Therefore, one of the overlapping target genes, PRKAR2A, could interact with A-Kinase Anchoring Protein 1 (AKAP1), a member of the AKAP family.
Figure 2.
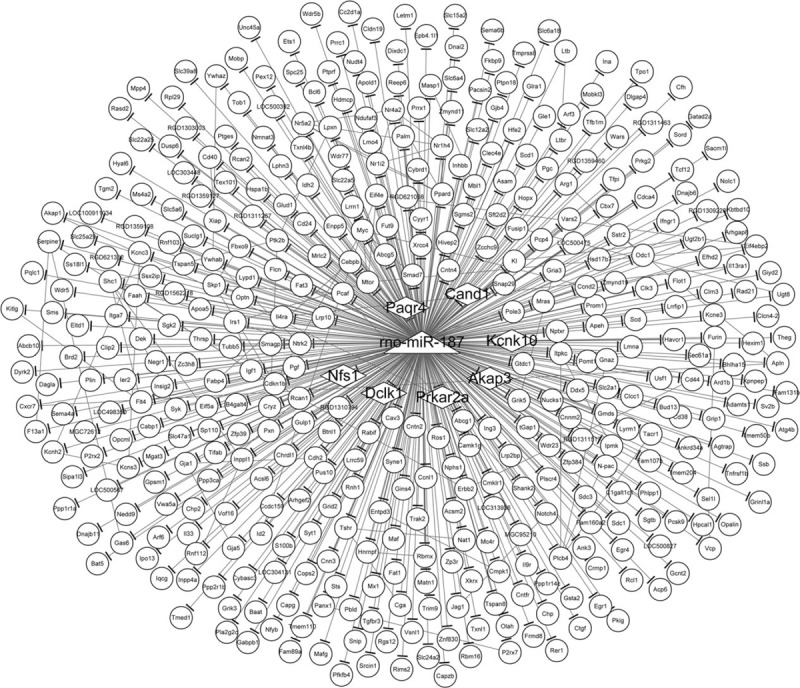
Interaction network of rno-miR-187-3p and its target genes. The triangle node indicates rno-miR-187-3p. The oval nodes indicate the target genes of rno-miR-187-3p. The diamond nodes represent the overlapping target genes of rno-miR-187-3p that were identified in both the miRDB and microRNA.org databases. The edge with a short thick line at the end indicates interaction between rno-miR-187-3p and its target gene. The edge among 2 nodes without thick line at the end indicates the interaction among 2 target genes.
3.5. Analysis of rno-miR-187-3p and its target gene binding sites
To understand the mechanism of the miR-187-3p-target gene interactions, the miR-187-3p sequence and the sequence of the overlapping target genes were subjected to sequence alignment. From these results, we found that the overlapping predicted target genes NFS1, PAQR4, CAND1, DCLK1, PRKAR2A, AKAP3, and KCNK10 had a different number of matched binding sites responsive to rno-miR-187-3p. In detail, NFS1, PAQR4, CAND1, DCLK1, PRKAR2A, AKAP3, and KCNK10 had 1, 1, 1, 3, 2, 1, and 5 matched binding sites responsive to rno-miR-187–3p, respectively.
4. Discussion
Epilepsy is a chronic disease and places both a health and social burden on society.[28] Recently, dysregulation of miRNAs dysregulation was found to be a key regulator of many biological processes in epilepsy.[8] In this study, rno-miR-187-3p was determined to be consistently down-regulated in stimulated groups at 4 time points compared with time-matched controls. The predicted target genes of rno-miR-187-3p were enriched in different GO terms and pathways. In addition, 7 overlapping target genes of rno-miR-187-3p were identified from 2 databases, including NFS1, PAQR4, CAND1, DCLK1, PRKAR2A, AKAP3, and KCNK10. These 7 overlapping target genes were determined to have a different number of matched binding sites for rno-miR-187-3p.
In this study, miRNA alterations were identified in the electrically stimulated dentate gyrus of rats compared with the time-matched controls, indicating that status epilepticus led to an aberrant miRNA expression pattern, consistent with previous studies.[29,30] In special, we found that rno-miR-187-3p was consistently downregulated at all time points. miR-187 has been reported to be related to certain types of cancer and other diseases.[31,32] For instance, Zhang et al. showed that decreased miR-187 expression could induce retinal ganglion cell apoptosis through upregulating SMAD Family Member 7 (SMAD7) in glaucoma.[32] In accordance with this study, we determined that the target genes of miR-187-3p could be enriched in the positive regulation of cell proliferation, suggesting that miR-187-3p may play a critical role in regulating cell proliferation and apoptosis associated with the progression of epilepsy. Moreover, 1 previous study reported that miR-187 expression was significantly downregulated in patients with TLE and demonstrated a critical role for miR-187 in the physiological regulation of IL-10 anti-inflammatory responses in the pathogenesis of TLE.[33] In this study, we concluded that rno-miR-187-3p was consistently down-regulated in the electrically-stimulated dentate gyrus of rats. Additionally, Wang et al[29] demonstrated that deregulated miR-106b-5p had significant diagnostic value for epilepsy with higher sensitivity and specificity, suggesting that miR-106b-5p may serve as a novel, noninvasive biomarker to improve the current diagnosis of epilepsy. Moreover, miRNA-based treatments could be employed as anti-epileptogenic or disease-modifying treatments.[8] Taken together, modulation of the miR-187-3p axis may serve as a novel therapeutic approach for epilepsy. However, more investigations are necessary to verify the role of miR-187-3p in the development and progression of epilepsy.
The proteins encoded by NFS1 supply inorganic sulfur to iron-sulfur (Fe-S) clusters by removing the sulfur from cysteine, creating alanine in the process.[34] Lipoic acid synthetase (LIAS) depends on Fe-S cluster prosthetic groups, and a deficiency in LIAS was demonstrated to cause neonatal-onset epilepsy.[35]CAND1 acts as a key assembly factor of SCF (SKP1-CUL1-F-box protein) E3 ubiquitin ligase complexes. Studies have shown that E3 ubiquitin ligase restricts the activity of Ca2+- and voltage-activated K+ (BK) channels, and prevents epileptogenesis .[36,37]DCLK1 encodes a member of the protein kinase superfamily and the doublecortin family.[38] Kerjan et al[38] reported that mice lacking doublecortin and doublecortin-like kinase 2 displayed altered hippocampal neuronal maturation and spontaneous seizures. AKAP3 encodes a member of AKAPs, a family of functionally related proteins that target protein kinase A to discrete locations within the cell.[39] It was demonstrated that AKAP3 synthesis was mediated by RNA binding proteins and PKA signaling.[40] Additionally, a previous study showed that N-methyl-D-aspartate preconditioning could protect against quinolinic acid-induced seizures via PKA signaling pathways.[41]KCNK10, also known as TREK-2, belongs to the family of potassium channel proteins containing two pore-forming P domains.[42] Numerous studies have indicated a role for potassium channel regulation during epilepsy.[43] On this basis, we postulate that genes NFS1, CAND1, DCLK1, AKAP3, and KCNK10 may play essential roles in the development of epilepsy. Nevertheless, there are few articles concerning the function of PAQR4 and PRKAR2A. In this study, NFS1, PAQR4, CAND1, DCLK1, PRKAR2A, AKAP3, and KCNK10 were identified to be overlapping target genes of rno-miR-187-3p using 2 databases. On the other hand, evidence demonstrates that the number of binding sites is an important factor that influence miRNA repression .[44] Genes are more repressed as the number of binding sites increases.[44,45] In this study, we found the overlapping genes had different number of binding sites responsive to rno-miR-187-3p. Overall, downregulation of miR-187-3p may be significantly associated with epilepsy progression via regulating these genes (NFS1, CAND1, DCLK1, AKAP3, and KCNK10). However, further experiments and investigations are necessary to confirm our findings.
However, this study had several limitations. First, the sample size was small. Future studies with a larger sample size are needed. Second, we tried to find additional microarray datasets to validate the findings in our study. However, due to the animals used, specific tissues used, and the status epilepticus induction by means of electrical stimulation of the amygdala, we could not find appropriate datasets that could be used for validation. More data analyses are warranted to confirm our findings. Third, we plan to perform experimental validations, such as the luciferase assay, to validate our findings in our future studies.
In conclusion, our study showed that rno-miR-187-3p, which may represent a potential molecular target in epileptogenesis, was consistently downregulated in stimulated groups compared with time-matched controls. Additionally, our analysis identified several target genes of miR-187-3p (NFS1, CAND1, DCLK1, AKAP3, and KCNK10), which might play crucial roles in epilepsy development and progression. The increase in knowledge about the miRNA expression changes and genetic changes in epilepsy may promote miRNAs as diagnostic and therapeutic options.
Supplementary Material
Footnotes
Abbreviations: AKAP1 = A-Kinase Anchoring Protein 1, AKAP3 = A-Kinase Anchoring Protein 3, BIND = Biomolecular Interaction Network Database, BP = biological process, CAND1 = Cullin Associated And Neddylation Dissociated 1, CC = Cellular component, DCLK1 = Doublecortin Like Kinase 1, FC = Fold change, FDR = false discovery rate, GEO = gene expression omnibus, GO = gene ontology, GRID = general repository for interaction datasets, KCNK10 = Potassium Two Pore Domain Channel Subfamily K Member 10, KEGG = Kyoto Encyclopedia of Genes and Genomes, LIAS = lipoic acid synthetase, MF = molecular function, miRNAs = microRNAs, NCBI = National Center of Biotechnology Information, NFS1 = NFS1 cysteine desulfurase, PAQR4 = Progestin And AdipoQ Receptor Family Member 4, PI3K = Phosphatidylinositol 3-kinase, PRKAR2A = Protein Kinase CAMP-Dependent Type II Regulatory Subunit Alpha, RMA = robust multiarray average, SMAD7 = SMAD Family Member 7.
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Yin YH, Ahmad N, Makmorbakry M. Pathogenesis of epilepsy: challenges in animal models. Iranian J Basic Med Sci 2013;16:1119–32. [PMC free article] [PubMed] [Google Scholar]
- [2].Mooney C, Becker BA, Raoof R, et al. EpimiRBase: a comprehensive database of microRNA-epilepsy associations. Bioinformatics 2016;32:1436–8. [DOI] [PubMed] [Google Scholar]
- [3].Costa-Ferro ZS, Vitola AS, Pedroso MF, et al. Prevention of seizures and reorganization of hippocampal functions by transplantation of bone marrow cells in the acute phase of experimental epilepsy. Seizure 2010;19:84–92. [DOI] [PubMed] [Google Scholar]
- [4].Hwang H, Kim KJ. New antiepileptic drugs in pediatric epilepsy. Brain Dev 2008;30:549–55. [DOI] [PubMed] [Google Scholar]
- [5].Fridley J, Thomas JG, Navarro JC, et al. Brain stimulation for the treatment of epilepsy. Neurosurg Focus 2012;32:E13. [DOI] [PubMed] [Google Scholar]
- [6].Amini E, Rezaei M, Mohamed Ibrahim N, et al. A molecular approach to epilepsy management: from current therapeutic methods to preconditioning efforts. Mol Neurobiol 2014;52:492–513. [DOI] [PubMed] [Google Scholar]
- [7].Haenisch S, Zhao Y, Chhibber A, et al. SOX11 identified by target gene evaluation of miRNAs differentially expressed in focal and non-focal brain tissue of therapy-resistant epilepsy patients. Neurobiol Dis 2015;77:127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Henshall DC. MicroRNA and epilepsy: profiling, functions and potential clinical applications. Curr Opin Neurol 2014;27:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hu K, Xie Y-Y, Zhang C, et al. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci 2012;13:115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [10].Song YJ, Tian XB, Zhang S, et al. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res 2011;1387:134–40. [DOI] [PubMed] [Google Scholar]
- [11].Aronica E, Fluiter K, Iyer A, et al. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci 2010;31:1100–7. [DOI] [PubMed] [Google Scholar]
- [12].Bot AM, Dębski KJ, Lukasiuk K. Alterations in miRNA levels in the dentate gyrus in epileptic rats. PLoS One 2013;8:e76051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Meng F, You Y, Liu Z, et al. Neuronal calcium signaling pathways are associated with the development of epilepsy. Mol Med Rep 2015;11:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Meng F, You Y, Liu Z, et al. Neuronal calcium signaling pathways are associated with the development of epilepsy. Mol Med Rep 2014;11:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].López-Romero P, González MA, Callejas S, et al. Processing of Agilent microRNA array data. BMC Res Notes 2010;3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat 1996;5:299–314. [Google Scholar]
- [18].Springer, Pollard KS, Dudoit S, van der Laan MJ. Multiple testing procedures: the multtest package and applications to genomics. Bioinformatics and Computational Biology Solutions Using R and Bioconductor 2005;249–271. [Google Scholar]
- [19].Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res 2010;39:D163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Betel D, Wilson M, Gabow A, et al. The microRNA. org resource: targets and expression. Nucleic Acids Res 2008;36(suppl 1):D149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [22].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reiner-Benaim A. FDR control by the BH procedure for two-sided correlated tests with implications to gene expression data analysis. Biom J 2007;49:107–26. [DOI] [PubMed] [Google Scholar]
- [24].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing, Journal of the Royal Statistical Society. J R Stat Soc B (Methodological) 1995;57:289–300. [Google Scholar]
- [25].Breitkreutz B-J, Stark C, Tyers M. The GRID: the general repository for interaction datasets. Genome Biol 2003;4:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alfarano C, Andrade CE, Anthony K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res 2005;33:D418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Springer, Griffiths-Jones S. miRBase: the microRNA sequence database. MicroRNA Protocols 2006;129–138. [DOI] [PubMed] [Google Scholar]
- [28].Moshé SL, Perucca E, Ryvlin P, et al. Epilepsy: new advances. Lancet 2015;385:884–98. [DOI] [PubMed] [Google Scholar]
- [29].Wang J, Yu JT, Tan L, et al. Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci Rep 2015;5:9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dogini DB, Avansini SH, Vieira AS, et al. MicroRNA regulation and dysregulation in epilepsy. Front Cell Neurosci 2013;7:172–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huang F, Jin Y, Wei Y. MicroRNA-187 induces diffuse large B-cell lymphoma cell apoptosis via targeting BCL6. Oncol Lett 2016;11:2845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang QL, Wang W, Li J, et al. Decreased miR-187 induces retinal ganglion cell apoptosis through upregulating SMAD7 in glaucoma. Biomed Pharmacother 2015;75:19–25. [DOI] [PubMed] [Google Scholar]
- [33].Alsharafi WA, Xiao B, Abuhamed MM, et al. Correlation between IL-10 and microRNA-187 expression in epileptic rat hippocampus and patients with temporal lobe epilepsy. Front Cell Neurosci 2015;9:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Biederbick A, Stehling O, Rösser R, et al. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol Cell Biol 2006;26:5675–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mayr J, Zimmermann F, Fauth C, et al. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am J Hum Genet 2011;89:792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu J, Ye J, Zou X, et al. CRL4A (CRBN) E3 ubiquitin ligase restricts BK channel activity and prevents epileptogenesis. Nat Commun 2014;5:3924. [DOI] [PubMed] [Google Scholar]
- [37].Leo A, Citraro R, Constanti A, et al. Are big potassium-type Ca (2+)-activated potassium channels a viable target for the treatment of epilepsy? Expert Opin Ther Targets 2015;19:911–26. [DOI] [PubMed] [Google Scholar]
- [38].Kerjan G, Koizumi H, Han EB, et al. Mice lacking doublecortin and doublecortin-like kinase 2 display altered hippocampal neuronal maturation and spontaneous seizures. Proc Natl Acad Sci 2009;106:6766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Song MH, Choi KU, Shin DH, et al. Identification of the cancer/testis antigens AKAP3 and CTp11 by SEREX in hepatocellular carcinoma. Oncol Rep 2012;28:1792–8. [DOI] [PubMed] [Google Scholar]
- [40].Xu K, Yang L, Zhao D, et al. AKAP3 synthesis is mediated by RNA binding proteins and PKA signaling during mouse spermiogenesis. Biol Reprod 2014;90:19. [DOI] [PubMed] [Google Scholar]
- [41].de Araújo Herculano B, Vandresen-Filho S, Martins WC, et al. NMDA preconditioning protects against quinolinic acid-induced seizures via PKA, PI3K and MAPK/ERK signaling pathways. Behav Brain Res 2011;219:92–7. [DOI] [PubMed] [Google Scholar]
- [42].Mcclenaghan C, Carpenter E, Stephen TJ. Mechanisms of TREK-2 potassium channel gating. Biophys J 2015;108:437a. [Google Scholar]
- [43].Köhling R, Wolfart J. Potassium channels in epilepsy. Cold Spring Harb Perspect Med 2016;6:pii: a022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol 2007;8:R166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 2004;10:1518–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


