Abstract
Hemobartonellosis is caused by Mycoplasma haemofelis, previously known as Haemobartonella felis. Cats infected with this organism typically develop regenerative anemia. The related species Mycoplasma haemominutum may also cause anemia. The purposes of this study were to use polymerase chain reaction technology to determine if both organisms exist in naturally infected cats from Saskatchewan and Alberta, and to determine if disease manifestation corresponds to mycoplasma species. Thirteen of 18 cats with regenerative anemia were infected, 12 with M. haemofelis and 1 with M. haemominutum. Eight of 22 cats with nonregenerative anemia were infected, 4 with M. haemofelis and 4 with M. haemominutum. Two of 20 cats with normal complete blood (cell) counts were infected with M. haemominutum. Although both mycoplasma species were identified, ill cats were more often infected with M. haemofelis.
Abstract
Résumé — Détection de Mycoplasma haemofelis et de Mycoplasma haemominutum par amplification en chaîne par polymérase chez des chats de Saskatchewan et d’Alberta. L’hémobartonellose est causé par Mycoplasma haemofelis, anciennement Haemobartonella felis. Il est typique que les chats infectés par cet organisme développent une anémie régénérative. Les espèces apparentées M. haemofelis et Mycoplasma haemominutum peuvent aussi causer une anémie. Le but de cette étude était d’utiliser la technologie d’amplification en chaîne par polymérase pour déterminer si les 2 organismes sont présents chez des chats naturellement infectés de Saskatchewan et d’Alberta et de déterminer si les manifestations de la maladie correspondent aux espèces de mycoplasmes. Treize des 18 chats atteints d’anémie régénérative étaient infectés, 12 par M. haemofelis et 1 par M. haemominutum. Huit des 22 chats présentant une anémie non régénérative étaient infectés, 4 par M. haemofelis at 4 par M. haemominutum. Bien que les 2 espèces de mycoplasmes aient été identifiées, les chats malades étaient le plus souvent infectés par M. haemofelis.
(Traduit par Docteur André Blouin)
Introduction
The causative agent of hemobartonellosis, a transmissible form of anemia affecting cats, has long been known as Haemobartonella felis. This extracellular pleomorphic bacterium attaches to the surface of feline erythrocytes (1–7), causing hemolytic anemia through extravascular destruction of erythrocytes by the mononuclear phagocyte system and intravascular lysis by direct damage to the cell membrane, increased osmotic fragility, or both (7,8). Following recovery from infection, many cats appear to harbor the organism in a latent carrier state (2,3,5,9).
This organism is fastidious, and attempts to culture it in the laboratory have been unsuccessful. Based on this characteristic and morphology (lack of outer cell membrane), the agent was originally classified into genus Haemobartonella, family Anaplasmataceae, order Rickettsiales. However, recent advances in molecular techniques have demonstrated, based on nucleotide sequences of the 16S rRNA gene, that there are 2 closely related species that cluster phylogenetically with organisms of the genus Mycoplasma (10–13). While these details were emerging, the organisms underwent several name changes in the corresponding reports. Thus, H. felis “Ohio/Florida strain,” “large form,” or “Hflg” has been renamed Mycoplasma haemofelis and H. felis “California strain,” “small form,” or “Hfsm” has been renamed Mycoplasma haemominutum. The revised terminology will be used throughout the remainder of this paper. Also, the term “hemoplasma” is now used to describe mycoplasmas that infect erythrocytes (14–16). However, renaming and reclassification of the organisms have not yet led to the renaming of the disease state. Therefore, the traditional term “hemobartonellosis” will be used here to refer to the clinical syndrome of regenerative anemia caused by hemoplasmas in cats.
Hematologic findings, including blood smear examination, often lead to the initial suspicion of hemobartonellosis in cats. However, organisms are not consistently visualized on Romanowsky-stained smears (1,3,9). Given the serious nature of hemobartonellosis, specific treatment requirements, and possible development of a carrier state, improved detection of the organisms is desirable. Polymerase chain reaction (PCR) technology provides a useful adjunct for the diagnosis of hemoplasma infection in cats.
Messick et al (11) developed a PCR assay with primers designed to amplify sequences from the 16S rRNA gene (16S rDNA) of M. haemofelis. By using this technology, these researchers (12) and others (13) were able to demonstrate the presence of M. haemofelis and M. haemominutum DNA in cats after experimental inoculation with the organisms and development of hemobartonellosis. Following experimental inoculation, M. haemofelis produced severe anemia (9,13,17), whereas M. haemominutum appeared to be less pathogenic, producing mild or no clinical signs and no hematological abnormalities (13,17).
Jensen et al (18) applied PCR technology in natural infections in the United States, using primers that differentiated the 2 hemoplasma species. In cats with clinical signs suggesting hemobartonellosis, infection with M. haemofelis and M. haemominutum occurred in 12% and 11%, respectively, and 4.9% were infected with both species (18). It was also shown that 13.8% of control animals (not suspected to have hemobartonellosis) were infected with M. haemominutum, and 0.7% were infected with both hemoplasmas (18).
The objectives of this study were to apply a PCR assay that differentiates the 2 mycoplasma organisms to determine if these species are present in naturally infected cats from Saskatchewan and Alberta, and to relate the species to disease manifestation.
Materials and methods
Specimen collection
Feline blood was retrieved for PCR analysis from ethylene-diaminetetraacetic acid (EDTA) samples submitted for complete blood (cell) counts (CBC) to Prairie Diagnostic Services (Saskatoon, Saskatchewan). The groups of cats sampled were as follows: group 1: 18 cats suspected of having hemobartonellosis (regenerative anemia, defined as hematocrit < 0.29 L/L, and either a reticulocyte count > 2.5%, mycoplasma organisms noted on blood smears, or both); group 2: 22 cats with nonregenerative anemia (NRA) that was suspected to be due to other illness (hematocrit < 0.29 L/L, and reticulocyte count < 2.5%, with no mycoplasma organisms apparent on blood smears); and group 3: 20 cats with normal CBCs, no significant biochemical abnormalities, and no, or minimal, nonspecific, clinical signs. Two cats were tested that did not fit the criteria of any group; they had been diagnosed previously with hemobartonellosis but were not currently anemic: 1 had been diagnosed 2 wk previously and was receiving antibiotic therapy; the other had been diagnosed and treated 5 mo previously but was not receiving antibiotics at the time of retesting. When available, the retrovirus status of cats was recorded. Four samples came from cats residing in Alberta; the remaining samples were from cats residing in Saskatchewan.
Specimen preparation and DNA extraction
Extraction of DNA from whole blood was performed by using a DNA extraction protocol (Qiagen DNeasy Tissue Kit; Qiagen, Mississauga, Ontario) according to the manufacturer’s directions, except that the final elution step was performed with 100 μL of ultra pure water and that incubation occurred at 70°C for 5 min before centrifugation.
Amplification of DNA
Primers were used that target the 16S rRNA gene, producing a 170 base pair (bp) product from M. haemofelis and a 193 bp amplicon from M. haemominutum (forward primer, 5’- ACG AAA GTC TGA TGG AGC AAT A-3’ and reverse primer 5’- ACG CCC AAT AAA TCC GRA TAA T-3’) (18). The PCR assay was optimized, yielding the following reaction conditions: initial denaturation for 2 min at 94°C, followed by 45 cycles of 1-min denaturation at 94°C, 1-min primer annealing at 60°C, and 30-s extension at 72°C. Template DNA (2 μL) was added to 48 μL of reaction mixture containing 29.5 μL of sterile ultra-pure water, 5.0 μL of 10 × PCR buffer, 7.0 μL of MgCl2 (25 mM), 1.25 μL of deoxynucleotide triphosphates (dNTPs) (10 mM), 2.5 μL of each primer (25 pmol/μL) and 0.25 μL of Taq polymerase (5 U/μL, Invitrogen, Burlington, Ontario). Mycoplasma haemofelis and M. haemominutum DNA (gifts from Dr. Messick, University of Illinois, Urbana-Champaign) were used as positive controls. A reagent negative control (reagents with no DNA) was included in each PCR run to monitor for contamination. Reaction products (10 μL) were electrophoresed through 2.5% agarose gels, stained with ethidium bromide, and photographed. Pictures were stored digitally (AlphaImager 2000; Alpha Innotech, San Leandro, California, USA). Identification of the DNA as M. haemofelis or M. haemominutum was made by comparing the size of the PCR product to the size of known positive control DNA and a 100 bp DNA ladder (Invitrogen, Burlington, Ontario).
Nucleotide sequencing
In order to confirm the specificity of the PCR product, 13 randomly selected amplicons were sent to a reference laboratory (DNA Services Laboratory, National Research Council, Saskatoon, Saskatchewan) for nucleotide sequencing. Sequencing was performed in both directions using the same primers used for PCR amplification. Sequences were aligned with software (MegAlign; DNA Star, Madison, Wisconsin, USA) and compared with M. haemofelis and M. haemominutum DNA sequences by using GenBank nucleotide data (accessions #AF178677, #U88563, and #U95297 for M. haemofelis; accessions #AF271154 and #U88564 for M. haemominutum).
Results
Nucleotide sequencing of 13 randomly selected amplicons confirmed the PCR products as M. haemofelis and M. haemominutum. Results of the PCR analysis of blood from 3 groups of cats are presented in Table 1. In all cases, the 2 organisms were differentiated on the basis of amplicon size following agarose gel electrophoresis (Figure 1). Within the group of cats with suspected hemobartonellosis, the PCR was positive in 13/18 (72%); 12/18 (66%) were infected with M. haemofelis, and 1/18 (6%) with M. haemominutum. Eight of the 22 cats (36%) in the NRA/ other illness group had positive PCR results. In this group, M. haemofelis was diagnosed in 4/22 (18%) and M. haemominutum was identified in the remaining 4. Of the 20 cats with normal CBCs, PCR results were positive in 2 (10%), both with M. haemominutum. A total of 23 of 60 (38%) cats in this study were PCR positive, and 16 of the 23 (70%) had M. haemofelis. Blood from the cat that had been diagnosed with hemobartonellosis 2 wk previously and was currently receiving antibiotics was PCR negative. The cat that had been diagnosed and treated 5 mo previously and was not currently receiving antibiotics was found to be infected with M. haemominutum. The 4 samples obtained from cats from Alberta were negative.
Table 1.
Detection of Mycoplasma haemofelis and M. haemominutum by polymerase chain reaction (PCR) in peripheral blood from 60 cats from Saskatchewan and Alberta
| PCR positive | Anemic, suspicious of hemobartonellosis (n = 18) | Anemic, not suspicious of hemobartonellosis (n = 22) | Normal complete blood (cell) count (n = 20) |
|---|---|---|---|
| M. haemofelis | 12 | 4 | 0 |
| M. haemominutum | 1 | 4 | 2 |
| Total | 13 | 8 | 2 |
Figure 1.
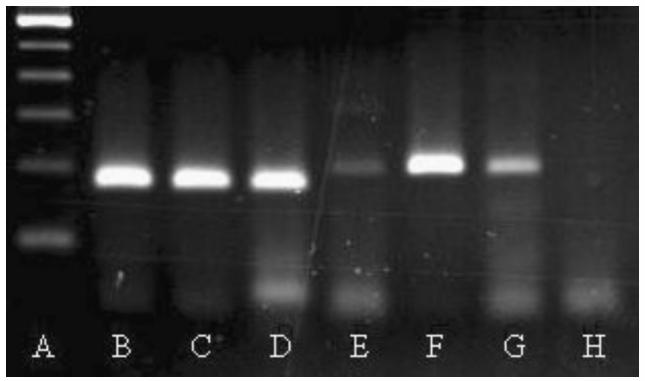
Ethidium bromide-stained agarose gel (2.5%) electrophoretogram demonstrating polymerase chain reaction (PCR) amplified products of mycoplasma 16S rRNA gene from peripheral blood of cats. Lanes are as follows: A — 100 base pairs (bp) ladder (lowest rung is 100 bp); B and C — 2 feline samples identified as Mycoplasma haemofelis; D — Mycoplasma haemofelis positive control (170 bp); E — Mycoplasma haemominutum positive control (193 bp); F and G — 2 feline samples identified as Mycoplasma haemominutum; H — Reagent negative control.
The retroviral status (feline leukemia virus [FeLV], feline immunodeficiency virus [FIV], or both) was known for 12 cats. In the suspect hemobartonellosis group, 1 cat was FeLV negative and 5 cats were both FeLV and FIV negative. In the NRA group, 1 cat was FIV positive, 1 was FeLV positive and FIV negative, and 4 were FeLV and FIV negative.
Discussion
Both species of hemoplasma, M. haemofelis and M. haemominutum, were present in blood from naturally infected cats from Saskatchewan. The majority of cats with suspected hemobartonellosis and confirmed hemoplasma infection were infected with M. haemofelis. Cats with normal CBCs that were PCR positive were infected with M. haemominutum and may have been latent carriers of this organism. These results are consistent with previous reports that M. haemominutum is less pathogenic than M. haemofelis (13,17,18). However, 1 cat with suspected hemobartonellosis in this study was infected with M. haemominutum. This suggests that, under certain circumstances, infection with M. haemominutum can be associated with clinically serious disease.
Previous reports have suggested that cats may have more severe anemia and clinical signs with both hemobartonellosis and retroviral infection (FeLV or FIV) than with either disease alone (19–21). Hemobartonellosis typically produces a strongly regenerative anemia (3,13). However, 8 cats with NRA were found to be infected with either M. haemofelis or M. haemominutum. These cats, by definition, were suspected to have other causes for the anemia. Of these 8 animals with NRA, 1 cat was FIV positive and 1 was FeLV positive. In addition to exacerbating the anemia caused by hemoplasmas, concurrent retroviral infection may inhibit erythroid regeneration, either directly or indirectly (22). These findings emphasize the importance of using all available information, in addition to the CBC, to aid in assessing the clinical importance of hemoplasma infection. Additionally, these findings suggest that hemobartonellosis should be a differential diagnosis in cats with both regenerative and nonregenerative anemias.
One cat in this study had been diagnosed with hemobartonellosis 2 wk previously and was receiving doxycycline therapy at the time of PCR testing. No mycoplasma DNA was detected in the peripheral blood of this cat. A second cat that had been diagnosed and treated for hemobartonellosis 5 mo previously, but was not currently receiving antibiotics, was not anemic, and was not exhibiting clinical signs. The blood from this cat was PCR positive for M. haemominutum. These findings are consistent with previous work, wherein the PCR results became negative within 12 h of initiating antibiotic therapy, and PCR positivity returned following cessation of treatment (12,13).
Jensen’s (18) study, using PCR evaluation of naturally infected cats, found that 28% of cats with suspected hemobartonellosis were infected with either one or both species of hemoplasma. An infection rate of 52.5% (21/40 cats) is obtained if the data from our regenerative and NRA groups (all anemic cats) are combined. The disparity between Jensen’s (18) study and ours may reflect differences in group definitions, sampling methods, or sample size. Alternatively, disease prevalence may differ between the geographic areas of the 2 studies. It is less likely that laboratory or technical variables contributed to the discrepancy, as the same primers were used and the PCR conditions were similar. Jensen’s group found a prevalence of 14.5% hemoplasma infection in cats without clinical signs of hemobartonellosis (18), similar to the prevalence in our group with normal CBCs (10%). In another recent study, 9/30 (30%) cats in Spain that had clinical signs of hemobartonellosis were found to be PCR positive (23). Our results cannot be compared easily with those in that study, however, as a control group was not included, and the inclusion criteria were not clear (23).
None of the blood samples in this study were PCR positive for both hemoplasmas. This contrasted to a previous study wherein coinfection was found in both hemobartonellosis suspects and normal control cats (18). Either coinfection was truly not present in our animals, or the PCR may have failed to amplify one species, perhaps due to the relative prevalence of each organism.
Cats in this study resided in Saskatchewan and Alberta. If PCR were used as a tool in a broad survey of cats in Canada, it would help to determine the geographic extent and prevalence of hemoplasma infection, and might help to clarify the role of M. haemominutum in clinical disease, provide insight into latency states, and identify risk factors for infection.
Acknowledgments
The authors thank Ms. B. Trask and the staff of the Prairie Diagnostic Service’s PCR and Clinical Pathology sections for laboratory assistance, and Mr. I. Shirley and Ms. C. Hare for audiovisual assistance. CVJ
Footnotes
Dr. Kewish was supported by a WCVM Interprovincial Graduate Student Fellowship. The study was supported in part by a grant from the WCVM Companion Animal Health Fund.
References
- 1.Flint JC, Moss LC. Infectious anemia in cats. J Am Vet Med Assoc. 1953;122:45–48. [PubMed] [Google Scholar]
- 2.Splitter EJ, Castro ER, Kanawyer WL. Feline infectious anemia. Vet Med. 1956;51:17–22. [Google Scholar]
- 3.Flint JC, Roepke MH, Jensen R. Feline infectious anemia. I Clinical aspects. Am J Vet Res. 1958;19:164–168. [PubMed] [Google Scholar]
- 4.Small E, Ristic M. Morphologic features of Haemobartonella felis. Vet Res. 1967;28:845–851. [PubMed] [Google Scholar]
- 5.Small E, Ristic M. Haemobartonellosis. Vet Clin North Am Small Anim Pract. 1971;1:225–230. doi: 10.1016/s0091-0279(71)50027-2. [DOI] [PubMed] [Google Scholar]
- 6.Demaree RS, Nessmith WB. Ultrastructure of Haemobartonella felis from a naturally infected cat. Am J Vet Res. 1972;33:1303– 1308. [PubMed] [Google Scholar]
- 7.Jain NC, Keeton KS. Scanning electron microscopic features of Haemobartonella felis. Am J Vet Res. 1973;34:697–700. [PubMed] [Google Scholar]
- 8.Maede Y. Sequestration and phagocytosis of Haemobartonella felis in the spleen. Am J Vet Res. 1979;40:691–695. [PubMed] [Google Scholar]
- 9.Harvey JW, Gaskin JM. Experimental feline haemobartonellosis. J Am Anim Hosp Assoc. 1977;13:28–38. [Google Scholar]
- 10.Rikihisa Y, Kawahara M, Wen B, et al. Western immunoblot analysis of Haemobartonella muris and comparison of 16S rRNA gene sequence of H. muris, Hfelis and Eperythrozoon suis. J Clin Microbiol. 1997;35:823–829. doi: 10.1128/jcm.35.4.823-829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messick JB, Berent LM, Cooper SK. Development and evaluation of a PCR-based assay for detection of Haemobartonella felis in cats and differentiation of Hfelis from related bacteria by restriction fragment length polymorphism analysis. J Clin Microbiol. 1998;36:462–466. doi: 10.1128/jcm.36.2.462-466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berent LM, Messick JB, Cooper SK. Detection of Haemobartonella felis in cats with experimentally induced acute and chronic infections, using a polymerase chain reaction assay. Am J Vet Res. 1998;59:1215–1220. [PubMed] [Google Scholar]
- 13.Foley JE, Harrus S, Poland A, Chomel B, Pedersen NC. Molecular, clinical, and pathologic comparison of two distinct strains of Haemobartonella felis in domestic cats. Am J Vet Res. 1998;59:1581–1588. [PubMed] [Google Scholar]
- 14.Neimark H, Johansson KE, Rikihisa Y, Tully JG. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ‘Candidatus Mycoplasma haemofelis’, ‘Candidatus Mycoplasma haemomuris’, ‘Candidatus Mycoplasma haemosuis’, and ‘Candidatus Mycoplasma wenyonii’. Int J Sys Evol Microbiol. 2001;51:891–899. doi: 10.1099/00207713-51-3-891. [DOI] [PubMed] [Google Scholar]
- 15.Neimark H, Johansson KE, Rikihisa Y, Tully JG. Revision of haemotrophic Mycoplasma species names. Int J Sys Evol Microbiol. 2002;52:683. doi: 10.1099/00207713-52-2-683. [DOI] [PubMed] [Google Scholar]
- 16.Foley JE, Pedersen NC. ‘Candidatus Mycoplasma haemominutum’, a low-virulence epierythrocytic parasite of cats. Int J Sys Evol Microbiol. 2001;51:815–817. doi: 10.1099/00207713-51-3-815. [DOI] [PubMed] [Google Scholar]
- 17.Westfall DS, Jensen WA, Reagan WJ, Radecki SV, Lappin MR. Inoculation of two genotypes of Haemobartonella felis (California and Ohio variants) to induce infection in cats and the response to treatment with azithromycin. Am J Vet Res. 2001;62:687–691. doi: 10.2460/ajvr.2001.62.687. [DOI] [PubMed] [Google Scholar]
- 18.Jensen WA, Lappin MR, Kamkar S, Reagan WJ. Use of a polymerase chain reaction assay to detect and differentiate two strains of Haemobartonella felis in naturally infected cats. Am J Vet Res. 2001;62:604–608. doi: 10.2460/ajvr.2001.62.604. [DOI] [PubMed] [Google Scholar]
- 19.Bobade PA, Nash AS, Rogerson P. Feline haemobartonellosis: Clinical, haematological and pathological studies in natural infections and the relationship to infection with feline leukaemia virus. Vet Rec. 1988;122:32–36. doi: 10.1136/vr.122.2.32. [DOI] [PubMed] [Google Scholar]
- 20.Grindem CB, Corbett WT, Tomkins MT. Risk factors for Haemobartonella felis infection in cats. J Am Vet Med Assoc. 1990;196:96–99. [PubMed] [Google Scholar]
- 21.George JW, Rideout BA, Griffey SM, Pedersen NC. Effect of preexisting FeLV infection or FeLV and feline immunodeficiency virus coinfection on pathogenicity of the small variant of Haemobartonella felis in cats. Am J Vet Res. 2002;63:1172–1178. doi: 10.2460/ajvr.2002.63.1172. [DOI] [PubMed] [Google Scholar]
- 22.Shelton GH, Linenberger ML. Hematologic abnormalities associated with retroviral infections in the cat. Semin Vet Med Surg (Small Anim) 1995;10:220–233. [PubMed] [Google Scholar]
- 23.Criado-Fornelio A, Martinez-Marcos A, Buling-Saraña A, Barba-Carretero JC. Presence of Mycoplasma haemofelis, Mycoplasma haemominutum and piroplasmids in cats from southern Europe: a molecular study. Vet Microbiol. 2003;93:307–317. doi: 10.1016/s0378-1135(03)00044-0. [DOI] [PubMed] [Google Scholar]


