Abstract
Background:
Triple-negative breast cancer (TNBC) is a heterogeneous group of tumors characterized by their aggressive nature and poor associated survival. MicroRNAs (miRs) have been found to play an important role in the occurrence and development of human cancers, but their role in the prognosis of TNBC patients remains unclear. We performed a meta-analysis to explore the prognostic value of miRs in TNBC.
Methods:
We systematically searched the PubMed, Embase, and Web of Science databases to identify eligible studies. A meta-analysis was performed to estimate the pooled hazard ratios (HRs) and their corresponding 95% confidence intervals (CIs) for the associations between levels of miR expression (predictive factors) and overall survival (OS) and disease-free survival (DFS) (outcomes) in patients with TNBC.
Results:
After performing the literature search and review, 21 relevant studies including 2510 subjects were identified. Six miRs (miR-155, miR-21, miR-27a/b, miR-374a/b, miR-210, and miR-454) were assessed in the meta-analysis. Decreased expression of miR-155 was associated with reduced OS (adjusted HR = 0.58, 95% CI: 0.34–0.99; crude HR = 0.67, 95% CI: 0.58–0.79). High miR-21 expression was also predictive of reduced OS (crude HR = 2.50, 95% CI: 1.56–4.01). We found that elevated levels of miR-27a/b, miR-210, and miR-454 expression were associated with shorter OS, while the levels of miR-454 and miR-374a/b expression were associated with DFS.
Conclusions:
Specific miRs could serve as potential prognostic biomarkers in TNBC. Due to the limited research available, the clinical application of these findings has yet to be verified.
Keywords: biomarker, meta-analysis, microRNA, prognosis, triple-negative breast cancer
1. Introduction
Breast cancer is the most commonly diagnosed malignancy and the leading cause of cancer-related mortality among women worldwide, with an estimated 1.7 million new cases and 521,900 deaths in 2012.[1] Of the breast cancer cases, approximately 10% to 20% have been reported to be triple-negative breast cancer (TNBC).[2] TNBC is a heterogeneous group of tumors characterized by the absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2/Neu (HER2), and this malignancy has been found to be often, but not always, a basal-like breast cancer.[3] Because it cannot be treated with current endocrine therapies and exhibits an aggressive nature, TNBC has been regarded as being associated with one of the worst prognoses of all breast cancer subtypes.[2,4]
Advances in in-depth research on genetic biomarkers, such as miRs, in TNBC have promoted the utility of biomarkers in the diagnosis, treatment, and prognosis of the disease. MiRs are a class of small noncoding RNA molecules that are 19 to 25 nucleotides in length, can modulate gene expression, and are easily accessible and quantifiable.[5] A growing body of evidence indicates that aberrant expression of miRs may be linked with the development and progression of human cancers,[6] including renal cell carcinoma,[7] pancreatic ductal adenocarcinoma,[8] and brain tumors.[9] However, until now, no systematic review has been performed to explore the role of particular miRs in the survival of patients with TNBC.
In this study, we systematically reviewed relevant studies on the prognostic value of miRs in TNBC and pooled the effect estimates reported in these studies to provide a better understanding of associations between specific miRs and prognosis in TNBC and provide a rationale for miR-based therapeutics.
2. Materials and methods
2.1. Search strategy
We followed the guidelines of the Meta-analysis of Observational Studies in Epidemiology (MOOSE) group and Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement.[10] We searched the PubMed, Embase, and Web of Science databases to identify relevant studies that assessed the utility of candidate miRs as prognostic factors in TNBC. The databases were searched to identify articles published from January 1990 to December 2016 using the following search strategy: (microRNA OR miRNA OR miR) AND (triple-negative breast cancer) AND (prognosis OR prognostic OR survival OR outcome OR mortality). The searches were limited to articles published in English. Two investigators (LL and XM) reviewed the titles and abstracts of studies identified in the initial search to determine the relevance of these publications. Then, the full texts of the remaining articles were obtained and carefully reviewed. We also manually screened the reference lists of retrieved articles to identify other potentially relevant studies.
2.2. Eligibility criteria
Articles were considered eligible if they met all the following initial inclusion criteria: focused on patients undergoing treatment for TNBC; measured miR expression levels in tumor or blood samples; clearly defined the utilized miR cut-off points; clearly described the utilized miR detection methods; analyzed the correlations between survival outcomes and miR expression; and clearly described the follow-up duration. Articles were excluded if they were case reports, letters, commentaries, conference records or reviews; had a sample size less than 30 cases; calculated HRs based on a combination of multiple miRs; lacked sufficient data for estimating HRs and 95% CIs; or used survival data that originated from the TCGA, PROGmiR, METABRIC, or BreastMark dataset. Data were extracted from articles fulfilling all the aforementioned selection criteria. Two individual investigators (LL and XM) independently assessed the eligibility of the retrieved articles. Discrepancies were resolved by consensus or consultation with a third investigator (PS).
2.3. Quality assessment
The quality of the included studies was assessed according to the following checklist, which was developed based on the criteria proposed by the MOOSE group:[10] clearly defined study design; clearly described study population (country); sufficiently large sample size (N > 30); clearly described the outcomes (OS or DFS); clearly defined the method of miR measurement (quantitative real-time polymerase chain reaction (qRT-PCR), in situ hybridization (ISH), etc.); clear defined the utilized cut-off values; measured miR expression in tumor or blood samples; and had a sufficiently long follow-up duration (>60 months). To assure the quality of this meta-analysis, studies were excluded if they did not meet these criteria.
2.4. Data extraction
Data were extracted independently by 2 investigators (LL and XM), who used a predefined sheet to retrieve information from all studies qualifying for final inclusion. The data sheet was designed based on previous studies focusing on similar topics and the PRISMA guidelines.[11] The following data were extracted: title; first author; publication year; study design; study population; participant number; sample types; miRs; miR expression assessment methods; cut-off values; follow-up duration; and HRs for OS or DFS and their corresponding 95% CIs and P values. If HRs (95% CIs) and P values could not be extracted from the original article, we estimated these values using the available data or the Kaplan–Meier curves presented in the articles using the methods described by Parmar et al[12] and Tierney et al.[13]
2.5. Statistical analysis
OS was defined as the interval from the date of primary surgery to the date of all-cause mortality. DFS was defined as the interval from the date of primary surgery to the date of disease relapse or all-cause mortality.[14] We pooled the HRs (95% CIs) extracted from the studies using the Stata 13.0 software (StatCorp, College Station, TX). Heterogeneity was assessed using the Cochran Q test and Higgins I-squared statistic. P values less than .1 for the Q test and I2 value >40% indicated the presence of significant heterogeneity across studies. The fixed-effects model was applied in the absence of between-study heterogeneity, while the random-effects model was applied when heterogeneity was observed. An observed HR > 1 indicated worse prognosis in the group with elevated miR expression. Conversely, an observed HR < 1 indicated worse prognosis in the group with a decreased miR expression.[15] Egger test was used to assess publication bias.
2.6. Ethical consideration
Ethical approval was not required for this study.
3. Results
3.1. Selection of studies
A flow diagram of the study selection process is shown in Fig. 1. A total of 370 publications were identified in the initial search. After reviewing the titles and abstracts of these articles, we identified 51 articles evaluating the use of prognostic miR biomarkers in TNBC. We then carefully reviewed the full texts of these articles and excluded an additional 32 articles. Two articles described independent cohorts that were analyzed separately.[16,17] Thus, we regarded these 2 articles as 4 studies. In total, 19 articles (21 studies) were eligible for inclusion in this meta-analysis.
Figure 1.
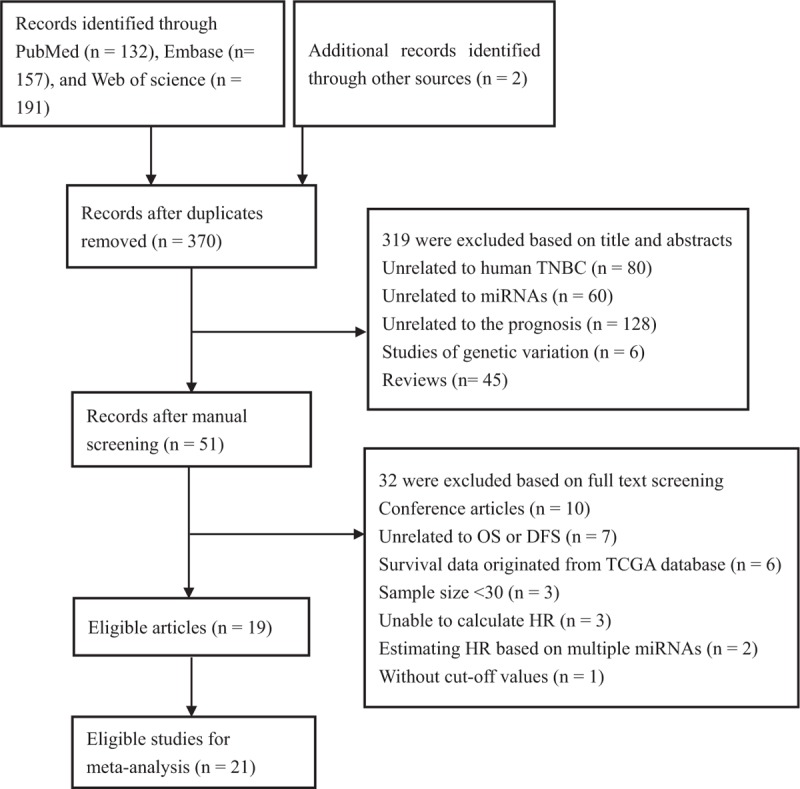
Flow diagram of the study selection procedure. DFS = disease-free survival, OS = overall survival, TCGA = The Cancer Genome Atlas.
3.2. Characteristics of the included studies
A total of 2510 TNBC patients were assessed in the 19 included articles, with a median sample size of 82 patients (range, 39–456 patients). These studies reported the prognostic values of 24 different miRs. The levels of miR expression were mainly detected in tumor tissues. Two studies used serum samples. Five studies did not directly report HR data. Thus, we estimated the HRs using the methods described above. Twelve studies reported adjusted HRs; the models described in these studies included covariates such as age, tumor site, grade, or disease stage (Table 1). In the included articles, increased expression of miR-27a/b,[15,16,18,19] miR-34b,[20] miR-210,[14] miR-125b,[21] miR-655,[21] miR-21,[22,23] miR-18b,[24] miR-103,[24] miR-107,[24] miR-652,[24] miR-301a,[25] miR-30e,[15] miR-214[26] and miR-454[17] decreased expression of miR-155,[15,21,27,28] miR-16,[21] miR-374a/b,[19,21] miR-497,[15,29] miR-493,[15] miR-185,[30] miR-26a,[31] miR-126-3p,[19] miR-218-5p[19] and miR-361-5p[32] were associated with poor prognosis in TNBC. Among these miRs, 6 (miR-155, miR-21, miR-27a/b, miR-374a/b, miR-210, and miR-454) were reported by at least 2 studies (Table 2). Thus, we performed this meta-analysis to summarize the effect of these 6 miRs.
Table 1.
Main characteristics of the eligible studies.
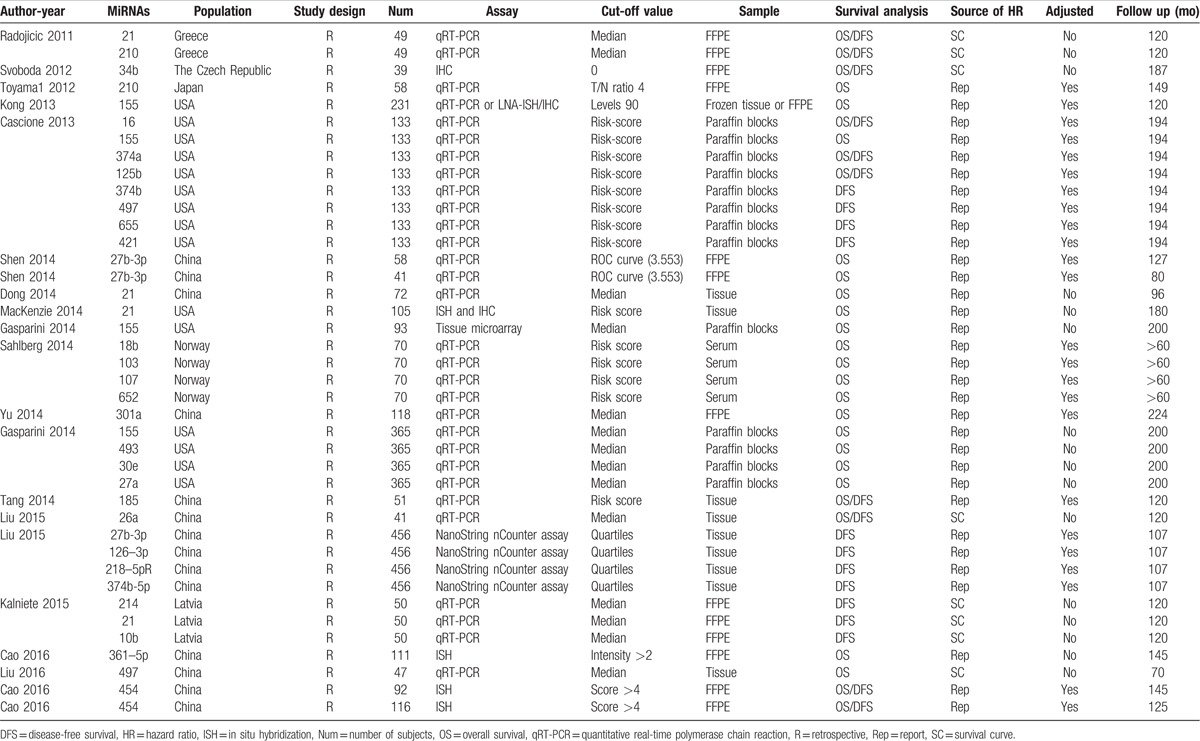
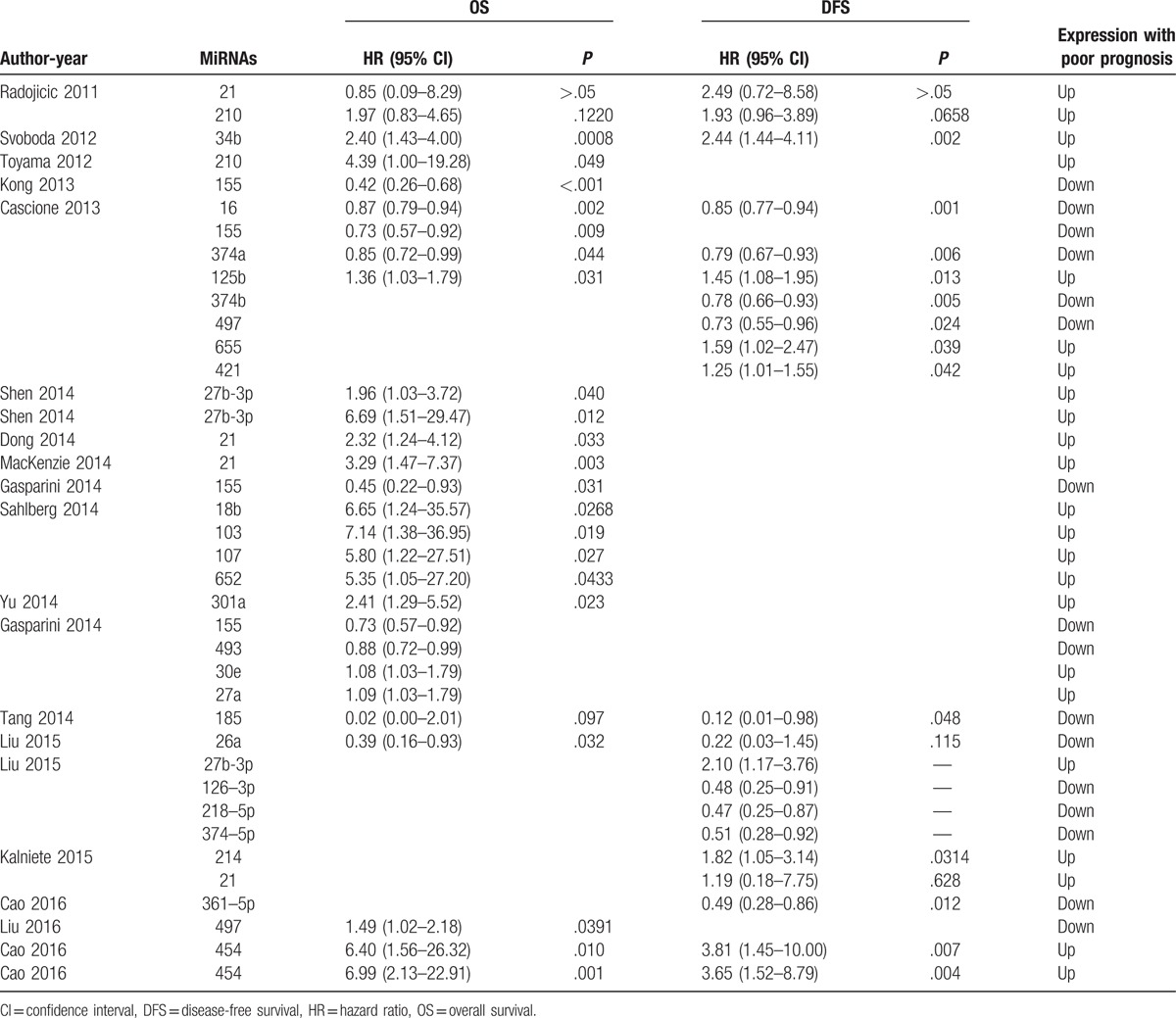
Table 2.
Descriptive characteristics and related data from included studies.
3.3. miR-155 and TNBC prognosis
Four articles (n = 822) suggested that downregulation of miR-155 was associated with poor prognosis in patients with TNBC. Gasparini et al[15,27] calculated the crude HR for miR-155, while Kong et al[28] and Cascione et al[21] performed multivariate analyses. No significant interstudy heterogeneity was observed (I2 = 48.1%, P = .123), and the Egger test results indicated the presence of no significant publication bias (P = .091). The pooled crude HR was 0.67 (95% CI: 0.58–0.79) (Fig. 2A). If excluding studies only reporting crude HRs, the observed interstudy heterogeneity was significant (I2 = 78.4%, P = .044). The random-effects model revealed that miR-155 expression was consistently associated with OS in TNBC patients (HR: 0.58, 95% CI: 0.34–0.99) (Fig. 2B).
Figure 2.
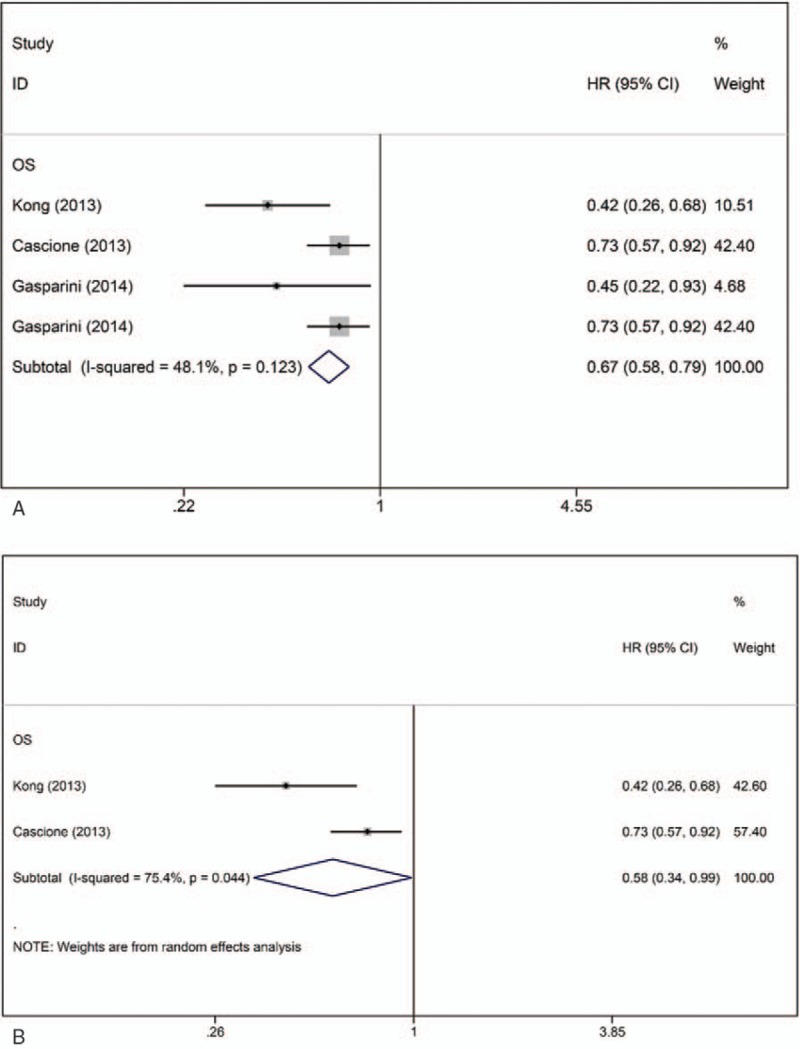
Forest plots of the HRs for the association between miR-155 and TNBC survival. A, Forest plot showing the combined HR based on all studies. B, Forest plot showing the combined HR based on multivariate studies. HR = hazard ratio, OS = overall survival.
3.4. miR-21 and TNBC prognosis
Four articles (n = 276) reported the effect of miR-21 on the prognosis of TNBC patients. Of these studies, 1 reported both OS and DFS data,[33] 2 reported only OS data,[22,23] and 1 reported only DFS data.[26] No significant heterogeneity was observed across studies (OS, I2 = 0.0%, P = .502; DFS, I2 = 0.0%, P = .521). The fixed-effects model revealed that miR-21 expression was inversely associated with OS (crude HR: 2.50; 95% CI: 1.56–4.01) and DFS (crude HR: 1.99; 95% CI: 0.71–5.60) in TNBC patients. There was no significant evidence of publication bias (OS, Egger test, P = .578) (Fig. 3).
Figure 3.
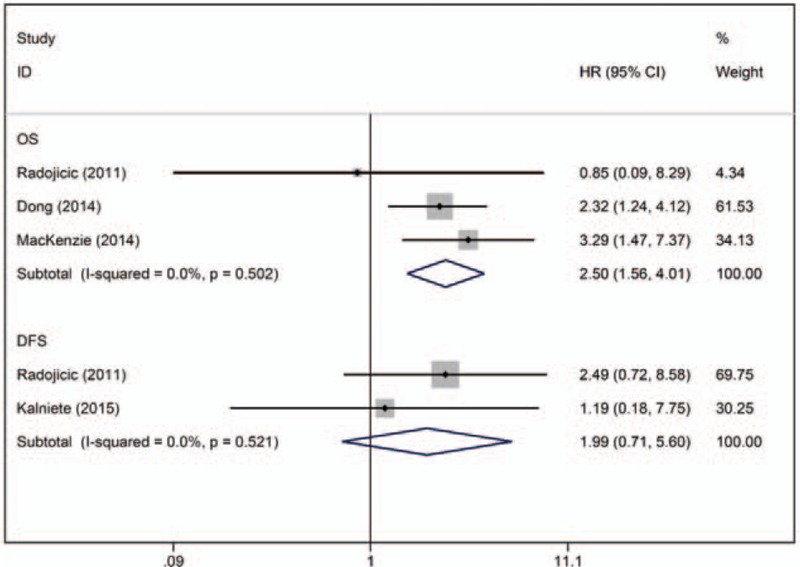
Forest plot of the HRs for the association between miR-21 and TNBC survival. DFS = disease-free survival, HR = hazard ratio, OS = overall survival.
3.5. miR-27a/b and TNBC prognosis
Three articles (4 studies, n = 920) assessed the association between miR-27a/b expression and prognosis in TNBC. Of these studies, 3 provided OS data,[15,16] and 1 provided DFS data.[19] For OS, a univariate HR was calculated in 1 study, while 2 studies performed multivariate analyses. The crude HR for the association between miR-27a/b expression and OS in TNBC was 1.25 (95% CI: 0.98–1.61) (Fig. 4A). After the univariate study conducted by Gasparini et al[15] was excluded, the pooled adjusted HR for OS was 2.38 (95% CI: 1.32–4.29) (Fig. 4B).
Figure 4.
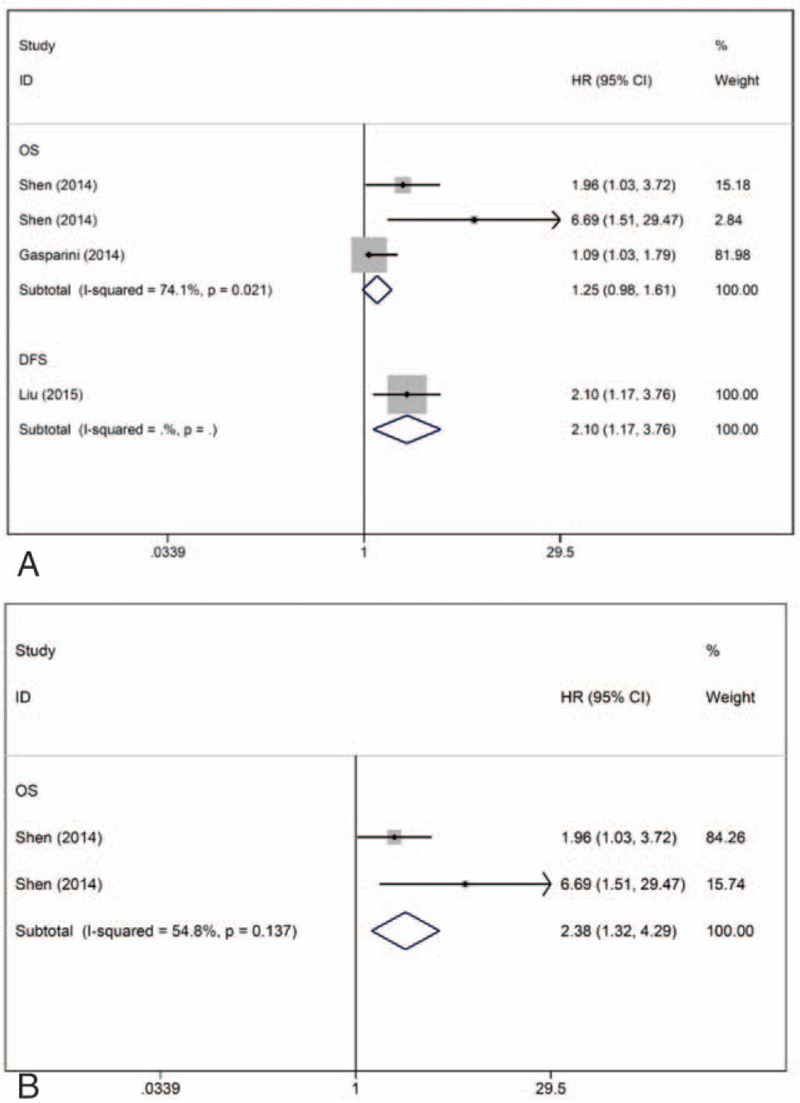
Forest plots of the HRs for the association between miR-27a/b and TNBC survival. A, Forest plot showing the combined HR based on all studies. B, Forest plot showing the combined HR based on multivariate studies. DFS = disease-free survival, HR = hazard ratio, OS = overall survival.
3.6. The miR-374a/b and prognosis of TNBC
Two studies evaluated the association between miR-374a/b expression and the prognosis of TNBC patients (n = 589), of which 1 reported data on DFS[19] and 1 reported data on both OS and DFS.[21] All of these studies provided adjusted HR data for DFS, and no significant heterogeneity was observed (I2 = 48.3%, P = .164). The fixed-effects model revealed that downregulation of miR-374 was associated with shorter DFS (combined adjusted HR: 0.77; 95% CI: 0.65–0.90) (Fig. 5).
Figure 5.
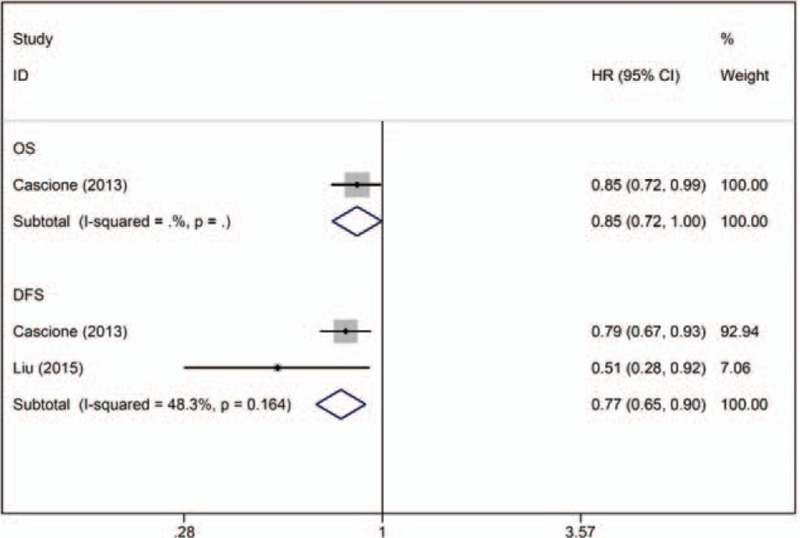
Forest plot of the HRs for the association between miR-374a/b and TNBC survival. DFS = disease-free survival, HR = hazard ratio, OS = overall survival.
3.7. miR-210 and TNBC prognosis
Two studies determined the association between miR-210 expression and prognosis in TNBC (n = 107), of which 1 provided OS data[14] and 1 provided OS and DFS data.[33] For OS, no significant heterogeneity was observed across studies (I2 = 0.0%, P = .359). The fixed-effects model revealed that elevated miR-210 expression was predictive of shorter OS (crude HR: 2.41; 95% CI: 1.15–5.08) (Fig. 6).
Figure 6.
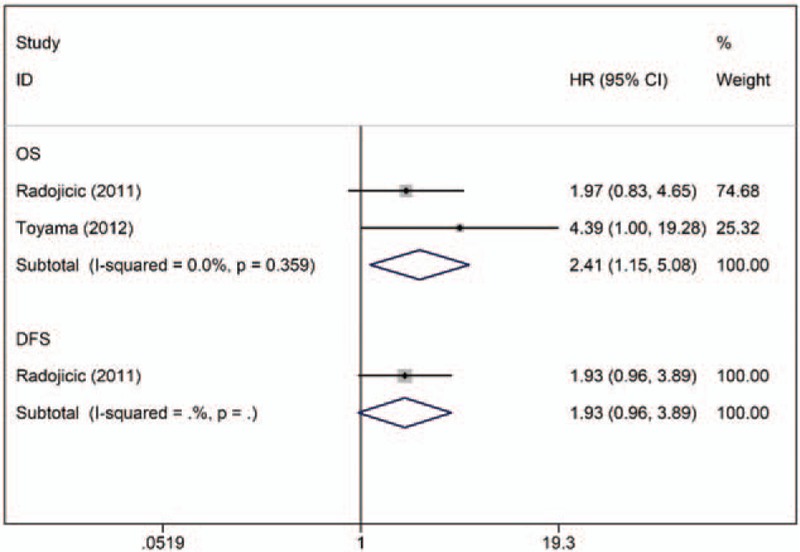
Forest plot of the HRs for the association between miR-210 and TNBC survival. DFS = disease-free survival, HR = hazard ratio, OS = overall survival.
3.8. miR-454 and TNBC prognosis
One article describing 2 studies reported higher miR-454 expression to be a predictive factor for poor OS and DFS in TNBC using multivariate analyses (n = 208).[17] No significant heterogeneity was observed across studies (OS, I2 = 0.00%, P = .925; DFS, I2 = 0.00%, P = .949). The fixed-effects model revealed that miR-454 expression was inversely associated with OS (combined adjusted HR: 6.74; 95% CI: 2.72–16.73) and DFS (combined adjusted HR: 3.72; 95% CI: 1.94–7.12) in TNBC patients (Fig. 7).
Figure 7.
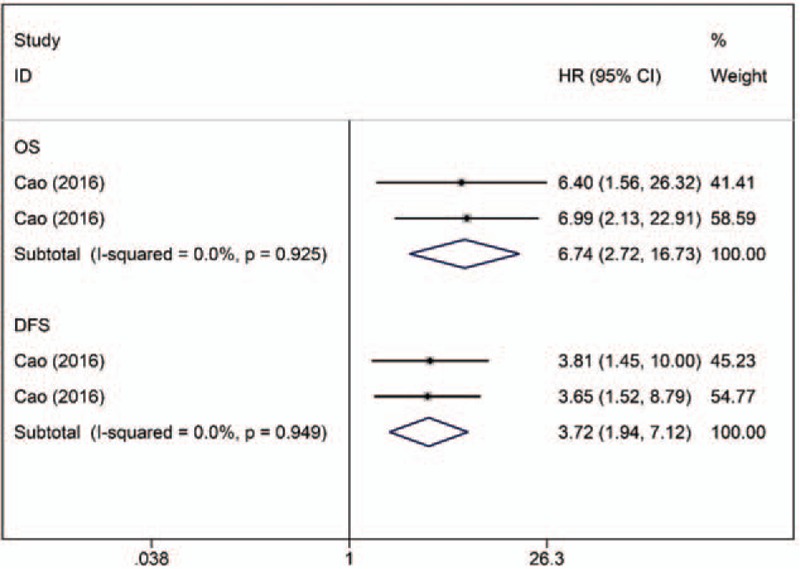
Forest plot of the HRs for the association between miR-454 and TNBC survival. DFS = disease-free survival, HR = hazard ratio, OS = overall survival.
4. Discussion
We conducted a comprehensive systematic literature review to explore the utility of miR biomarkers that can be easily and robustly evaluated in predicting prognosis in TNBC. To our knowledge, this is the first extensive meta-analysis to describe the role of miRs in TNBC prognosis.
Although various miRs were found to be associated with prognosis in TNBC, most of these miRs were assessed in only a single study. Six miRs (miR-155, miR-21, miR-27a/b, miR-374a/b, miR-210, and miR-454) were evaluated in at least 2 studies. We, therefore, performed a meta-analysis of the effect of these 6 miRs on the survival of TNBC patients. The results of this study showed that lower expression of miR-155 predicted worse OS in TNBC patients, while elevated levels of miR-21, miR-27a/b, miR-210, and miR-454 expression were associated with shorter overall survival times. Similarly, lower expression of miR-374a/b and higher expression of miR-454 were associated with shorter DFS.
The miR-155 locus is located within a region known as B-cell integration cluster,[34] and miR-155 is overexpressed in various solid tumors, including breast, lung, colon, pancreatic and thyroid cancers.[35,36] Some studies have reported the pro-oncogenic properties of miR-155 in lung cancer[36] and T-cell leukemia.[37] However, we identified this miR to exhibit opposite behavior, finding that overexpression of miR-155 tended to have a protective effect on survival in TNBC patients. There are a number of molecular mechanisms that could explain this relationship. In TNBC, miR-155 may play a crucial role in DNA damage pathways.[15] miR-155 may regulate DNA repair activity and sensitivity to ionizing radiation by repressing RAD51 recombinase (RAD51),[27] while RAD51 has been identified as a central protein in homologous recombination.
MiR-21 is one of the most extensively studied cancer-related miRs and might play an ever-expanding role in most cancers.[38] miR-21 may serve as a key regulator of oncogenic processes, including tumor growth, migration, and invasion.[39] Elevated miR-21 expression levels have been found to be associated with poor outcomes in cancer patients.[40] miR-21 may target the pro-apoptotic phosphatase and tensin homolog (PTEN) and promote tumor cell proliferation, which, in turn, may inhibit the apoptosis of tumor cells in TNBC cell lines in vitro.[22]
miR-27a/b has been linked to the peroxisome proliferator-activated receptor (PPAR) and PTEN signaling in TNBC cells, acting as a tumor suppressor by regulating the cell division cycle (CDC27) gene.[19] CDC27 has been identified as a core component of the anaphase-promoting complex (APC) and found to be involved in regulating mitotic checkpoints to ensure chromosomal integrity.[41] The results of a pathway analysis showed that miR-374b may regulate critical pathways involved in TNBC tumor development and progression, including the fibroblast growth factor and transforming growth factor pathways.[19] miR-210, a known hypoxia-regulated miR, has been found to be upregulated in many cancers. This miR may serve as a key player in cell response to hypoxia and has been linked to a number of hypoxia-dependent diseases involved in mitochondrial metabolism, angiogenesis, DNA repair, and cell survival.[42] miR-454 has dual functionality, acting as either an oncogenic miR or a tumor suppressor. Previous studies have reported this miR to be downregulated in esophageal cancer[43] and upregulated in colorectal cancer and breast cancer.[44,45] miR-454 has been reported to function as an oncogenic miR by targeting PTEN. Patients with TNBC tumors that lose PTEN expression have poorer survival, as PTEN negatively regulates the PI3K-AKT signaling pathway.[46,47] Further studies are needed to understand the molecular mechanism underlying the effect of miRs in TNBC.
Some limitations must be considered when interpreting the results of the current study. First, the number of studies available was limited. More studies are needed to further assess these associations in the future. Second, marked heterogeneity was observed in some of the analyses, findings that were likely identified due to differences in patient characteristics (ethnicity, nationality, gender, age, tumor stage, and grade) and the use of different assay methods, cut-off values for miR expression levels, sample preparation methods (i.e., paraffin-fixed, formalin-fixed, freshly frozen tumors or serum), follow-up durations, and HR extraction methods. Third, circulating biomarkers are more valuable than tissue biomarkers because they can be assayed before surgery and monitored throughout the lifespan. More studies should be conducted in the future to evaluate the prognostic value of specific miRs in serum in TNBC.
5. Conclusions
Specific miRs may serve as potential prognostic biomarkers in TNBC. Due to the limited research available, the clinical application of these findings has yet to be verified.
Footnotes
Abbreviations: APC = anaphase-promoting complex, CDC27 = cell division cycle 27, CI = confidence interval, DFS = disease-free survival, ER = estrogen receptor, HER2 = human epidermal growth factor receptor-2/Neu, HR = hazard ratio, ISH = in situ hybridization, miR = microRNA, MOOSE = Meta-analysis of Observational Studies in Epidemiology, OS = overall survival, PR = progesterone receptor, PPAR = peroxisome proliferator-activated receptor, PRISMA = Preferred Reporting Items for Systematic Reviews and Meta-analysis, PTEN = pro-apoptotic phosphatase and tensin homolog, qRT-PCR = quantitative real-time polymerase chain reaction, RAD51 = RAD51 recombinase, TNBC = triple-negative breast cancer.
LL and XM contributed equally to this work.
The present study was supported by the National Natural Science Foundation of China (81673249), Social Development Project in Jiangsu Province (BE2015694), Scientific Research Innovation Project for Graduate Student in Jiangsu Province (KYLX16_1125), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAAD). The funding agencies had no role in the study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Kumar P, Aggarwal R. An overview of triple-negative breast cancer. Arch Gynecol Obstetr 2016;293:247–69. [DOI] [PubMed] [Google Scholar]
- [3].Papa A, Caruso D, Tomao S, et al. Triple-negative breast cancer: investigating potential molecular therapeutic target. Expert Opin Ther Targets 2015;19:55–75. [DOI] [PubMed] [Google Scholar]
- [4].Mathe A, Scott RJ, Avery-Kiejda KA. MiRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. Int J Mol Sci 2015;16:28347–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther 2017;172:34–49. [DOI] [PubMed] [Google Scholar]
- [6].Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gu L, Li H, Chen L, et al. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget 2015;6:32545–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Frampton AE, Krell J, Jamieson NB, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: a meta-analysis. Eur J Cancer 2015;51:1389–404. [DOI] [PubMed] [Google Scholar]
- [9].He XY, Liao YD, Guo XQ, et al. Prognostic role of microRNA-21 expression in brain tumors: a meta-analysis. Mol Neurobiol 2016;53:1856–61. [DOI] [PubMed] [Google Scholar]
- [10].Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008–12. [DOI] [PubMed] [Google Scholar]
- [11].Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Instit 2012;104:528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statist Med 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- [13].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Toyama T, Kondo N, Endo Y, et al. High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol 2012;42:256–63. [DOI] [PubMed] [Google Scholar]
- [15].Gasparini P, Cascione L, Fassan M, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 2014;5:1174–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shen S, Sun Q, Liang Z, et al. A prognostic model of triple-negative breast cancer based on miR-27b-3p and node status. PLoS One 2014;9:e100664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cao ZG, Li JJ, Yao L, et al. High expression of microRNA-454 is associated with poor prognosis in triple-negative breast cancer. Oncotarget 2016;7:64900–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Buffa FM, Camps C, Winchester L, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 2011;71:5635–45. [DOI] [PubMed] [Google Scholar]
- [19].Liu Y, Cai Q, Bao PP, et al. Tumor tissue microRNA expression in association with triple-negative breast cancer outcomes. Breast Cancer Res Treat 2015;152:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Svoboda M, Sana J, Redova M, et al. MiR-34b is associated with clinical outcome in triple-negative breast cancer patients. Diagn Pathol 2012;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cascione L, Gasparini P, Lovat F, et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One 2013;8:e55910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dong G, Liang X, Wang D, et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol 2014;31:57. [DOI] [PubMed] [Google Scholar]
- [23].MacKenzie TA, Schwartz GN, Calderone HM, et al. Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am J Pathol 2014;184:3217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kleivi Sahlberg K, Bottai G, Naume B, et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res 2015;21:1207–14. [DOI] [PubMed] [Google Scholar]
- [25].Yu H, Li H, Qian H, et al. Upregulation of miR-301a correlates with poor prognosis in triple-negative breast cancer. Med Oncol 2014;31:283. [DOI] [PubMed] [Google Scholar]
- [26].Kalniete D, Nakazawa-Miklasevica M, Strumfa I, et al. High expression of miR-214 is associated with a worse disease-specific survival of the triple-negative breast cancer patients. Hered Cancer Clin Pract 2015;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gasparini P, Lovat F, Fassan M, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A 2014;111:4536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kong W, He L, Richards EJ, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 2014;33:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu J, Zhou Y, Shi Z, et al. microRNA-497 modulates breast cancer cell proliferation, invasion, and survival by targeting SMAD7. DNA Cell Biol 2016;35:521–9. [DOI] [PubMed] [Google Scholar]
- [30].Tang H, Liu P, Yang L, et al. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther 2014;13:3185–97. [DOI] [PubMed] [Google Scholar]
- [31].Liu P, Tang H, Chen B, et al. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett 2015;357:384–92. [DOI] [PubMed] [Google Scholar]
- [32].Cao ZG, Huang YN, Yao L, et al. Positive expression of miR-361-5p indicates better prognosis for breast cancer patients. J Thorac Dis 2016;8:1772–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Radojicic J, Zaravinos A, Vrekoussis T, et al. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 2011;10:507–17. [DOI] [PubMed] [Google Scholar]
- [34].Metzler M, Wilda M, Busch K, et al. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 2004;39:167–9. [DOI] [PubMed] [Google Scholar]
- [35].Jiang S, Zhang HW, Lu MH, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 2010;70:3119–27. [DOI] [PubMed] [Google Scholar]
- [36].Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189–98. [DOI] [PubMed] [Google Scholar]
- [37].Ishihara K, Sasaki D, Tsuruda K, et al. Impact of miR-155 and miR-126 as novel biomarkers on the assessment of disease progression and prognosis in adult T-cell leukemia. Cancer Epidemiol 2012;36:560–5. [DOI] [PubMed] [Google Scholar]
- [38].Pfeffer SR, Yang CH, Pfeffer LM. The role of miR-21 in cancer. Drug Dev Res 2015;76:270–7. [DOI] [PubMed] [Google Scholar]
- [39].Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 2009;37(pt 4):918–25. [DOI] [PubMed] [Google Scholar]
- [40].Wang Y, Gao X, Wei F, et al. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene 2014;533:389–97. [DOI] [PubMed] [Google Scholar]
- [41].Ren YQ, Fu F, Han J. MiR-27a modulates radiosensitivity of triple-negative breast cancer (TNBC) cells by targeting CDC27. Med Sci Monit 2015;21:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle 2010;9:1072–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu SG, Qin XG, Zhao BS, et al. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett 2013;5:1639–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liang HL, Hu AP, Li SL, et al. MiR-454 prompts cell proliferation of human colorectal cancer cells by repressing CYLD expression. Asian Pac J Cancer Prev 2015;16:2397–402. [DOI] [PubMed] [Google Scholar]
- [45].Mishra S, Srivastava AK, Suman S, et al. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett 2015;369:67–75. [DOI] [PubMed] [Google Scholar]
- [46].Sun L, Wang Q, Gao X, et al. MicroRNA-454 functions as an oncogene by regulating PTEN in uveal melanoma. FEBS Lett 2015;589(19 pt B):2791–6. [DOI] [PubMed] [Google Scholar]
- [47].Beg S, Siraj AK, Prabhakaran S, et al. Loss of PTEN expression is associated with aggressive behavior and poor prognosis in Middle Eastern triple-negative breast cancer. Breast Cancer Res Treat 2015;151:541–53. [DOI] [PubMed] [Google Scholar]


