Abstract
MicroRNAs (miRNAs) constitute a class of small noncoding RNAs that plays an important role in the post-transcriptional regulation of gene expression. Much evidence has demonstrated that miRNAs are involved in regulating the human and mouse pluripotency. Nevertheless, to our knowledge, miRNAs in the pluripotent stem cells of one of the most commonly used model organisms – the Rattus norvegicus have not been studied. In the present study, we performed deep sequencing of small RNA molecules in the embryonic fibroblasts, embryonic stem cells, and induced pluripotent stem cells of laboratory rats. Bioinformatics analysis revealed 674 known miRNAs and 394 novel miRNA candidates in all of the samples. Expression of known pluripotency-associated miRNAs, such as the miR-290–295 and miR-183-96-182 clusters as well as members of the miR-200 family, was detected in rat pluripotent stem cells. Analysis of the targets of differentially expressed known and novel miRNAs showed their involvement in the regulation of pluripotency and the reprogramming process in rats. Bioinformatics and systems biology approaches identified potential pathways that are regulated by these miRNAs. This study contributes to our understanding of miRNAs in the regulation of pluripotency and cell reprogramming in the laboratory rat.
Introduction
Pluripotent stem cells (PSCs) are a unique model for studying early development, disease modelling, and use in regenerative medicine. PSCs are capable of self-renewing and differentiating into all cell types of the adult organism. Two types of pluripotent cells exist – embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). ESCs are derived from the inner cell mass of blastocysts, and iPSCs are generated from somatic cells by the overexpression of Yamanaka factors (Oct4, Sox2, Klf4 and c-Myc)1–5. PSC fate is regulated by a complex network including transcription factors, signalling pathways, epigenetic modifications, and non-coding RNAs6–8. The regulatory pathways of self-renewal and pluripotency are not completely understood, especially in rats. The laboratory rat is widely used in various biomedical research fields such as disease modelling, pharmacology and toxicology9. However, the absence of rat PSCs has, for a long time, limited its further application in biomedicine. Rat pluripotent cells were first derived in 2008 using special serum-free conditions, leukaemia inhibitory factor, and inhibitors of the MEK1/2 and GSK3 kinases10, 11. It is known that PSCs have species-specific properties and that microRNAs (miRNAs) play a substantial role in the regulation of pluripotency12–15.
miRNAs are small 18–23-nt non-coding RNAs that regulate gene expression by mRNA degradation or translation inhibition16, 17. miRNAs are critical for the regulation of self-renewal and pluripotency, as was shown by knockout studies of Dicer and DGCR8 – enzymes that are involved in miRNAs processing18–21. Some miRNAs, such as miR-291, miR-294 and miR-295, can enhance reprogramming that is induced by OSK factors22. Moreover, overexpression of a miR-302-367 cluster was sufficient to reprogramme human and mouse fibroblasts without overexpressing the Yamanaka factors23.
Previous genome-wide sequencing studies of rat miRNAs have been conducted using adult tissues. Rat miRNAs were sequenced and analysed from several neural compartments and cell lines, such as thyroid FRTL-5 cells and their differentiated derivatives24 as well as six different tissues, including spleen, liver, brain, testis, heart, kidneys25. Recently, miRNA analysis was performed on the 21 and 23 organs of male and female Sprague Dawley rats, respectively26. However, miRNA expression in rat PSCs has not been studied yet. We recently derived rat ESCs and iPSCs and characterized their transcriptional profile27. All of our derived ESC and iPSC lines were AP-positive, expressed key pluripotency markers, and were able to differentiate into derivatives of all three germ layers in vitro (embryoid bodies) and in vivo (teratoma test). Female iPSC lines are characterized by two active X-chromosomes. The rat ESCs and iPSCs were also able to contribute to the tissue development of chimeric organisms in the xenogenic rat-mouse model. In this study, we performed next generation sequencing of small RNA libraries from rat ESCs, iPSCs and embryonic fibroblasts (EFs). We analysed miRNA expression in these cell types and identified novel rat miRNAs that were expressed in PSCs and EFs. Using the KEGG signalling pathways and Gene Ontology (GO) enrichment analyses of known and novel miRNA targets, we revealed the involvement of miRNAs in the regulation of pluripotency and cell reprogramming. These findings provide new insights into pluripotency network regulation in rats.
Materials and Methods
Cell cultures
Rat EFs, ESCs and iPSCs were previously obtained and characterized in our lab27. EFs were isolated from E12 rat embryos (WAG and Brattleboro strains). ESCs were derived from the inner cell mass of E4.5 blastocysts as described previously10, 11. iPSCs were produced from rat EFs by the overexpression of Oct4, Sox2, Klf4 and Myc using a lentiviral doxycycline-inducible system28. Cells were cultivated as described27. EFs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) mixed with Ham’s F12 medium (1:1), containing 10% foetal bovine serum, GlutaMAXTM, 5,000 U/mL penicillin/streptomycin, nonessential amino acids, sodium pyruvate, and 0.1 mM 2-mercaptoethanol. ESCs and iPSCs were cultivated in N2B27 medium containing GlutaMAXTM, 5,000 U/mL penicillin/streptomycin, 0.1 mM 2-mercaptoethanol, 1,000 U/mL mouse LIF (StemRD), 1 µM PD0325901 (StemRD), and 3 µM CHIR99021 (StemRD). ESC and iPSC differentiation was performed for 24 days on tissue culture surfaces coated with gelatine according to a previously published protocol27. Cells were disaggregated with collagenase IV or TrypLE and were seeded on a gelatine-treated surface at a density of 104 cells/cm2 in medium containing a 1:1 mixture of N2B27 (without LIF, PD0325901 or CHIR99021) and DMEM/F12 media supplemented with 10% foetal bovine serum, nonessential amino acids, GlutaMAX, 10 ng/mL basic fibroblast growth factor, and 10 mM Y27632 (R&D).
Small RNA extraction, sequencing and qRT-PCR analysis
Cells were resuspended in RNAlater and used for small RNA extraction with the mirVana™ miRNA Isolation Kit (Ambion, #AM1560). RNA quality was analysed using an Agilent Bioanalyzer. A 50-ng quantity of small RNA was used to prepare a library, which was then sequenced on the Illumina GAIIX. Raw data were deposited into the SRA database with the accession numbers SRX395433-395442.
Small RNA for cDNA synthesis were extracted from cells and tissues with the mirVana™ miRNA Isolation Kit. cDNA was synthesized by the NCode™ VILO™ miRNA cDNA synthesis Kit (Invitrogen) from 1 µg of total RNA. Real-time PCR was performed using the HS-qPCR SYBR Blue mix (Biolabmix) with a custom miRNA-specific primer and a universal primer from Invitrogen on the LightCycler480 System (Roche). Reaction conditions were 95 °C for 5 min followed by 40 cycles of 95 °C for 15 secs and 60 °C for 30 sec. Reaction specificity was verified by melting curve analysis. Reactions were performed in triplicate. Sequences of the miRNA-specific primers are provided in Supplementary Table S1. Data were normalized to Rnu1a expression.
Mapping and filtration of reads
Raw reads of 11 samples were processed with the FASTX Toolkit v0.0.14 in two steps. First, the data were trimmed with the fastq_quality_trimmer tool, leaving only reads with a tail quality of at least 30. Then, an adapter sequence was clipped with the fastx_clipper program, leaving only 15-nt reads or longer.
Filtered reads were mapped to the R. norvegicus reference genome (version rn5) from the UCSC Genome Browser29–32 combined with the ribosomal RNA reference for R. norvegicus from the Silva database33. The BWA v0.7.10-r806 software was utilized for gapped/ungapped alignment of single-ended reads. If a read could be mapped to several places in the reference, only one random place was chosen. The portion of reads that mapped solely to chr1-20,X varied from 94.2% to 97.4% in different samples.
To assess the quality of reads, a set of 486 genomic intervals of known premature miRNAs for R. norvegicus was downloaded from miRBase v21 (ftp://mirbase.org/pub/mirbase/CURRENT/genomes/rno.gff3)34. The absolute majority (99%) of reads that mapped to the genomic intervals of pre-miRNA had a length of 18–27 nt. To keep the focus of this study on miRNA, we filtered out reads that were shorter than 18 and longer than 27 nt because they likely represented short RNA species, which are different from the miRNA class. The rest of the reads were further filtered from the ones mapped onto parts of the genome that is annotated with rmsk repeats (http://hgdownload.soe.ucsc.edu/goldenPath/rn5/database/rmsk.txt.gz)29 of class “RNA” (including rRNA, scRNA, snRNA, tRNA, srpRNA, RNA, and MARNA). This filtration did not influence any reads that were mapped to known miRNAs. A final set of mapped and filtered reads was called informative reads. A summary is presented in Table 1.
Table 1.
Amount of informative reads and known miRNA reads in all samples.
| Cell type | Sample | informative reads | known miRNA reads | Portion of known miRNA reads |
|---|---|---|---|---|
| iPSC | SU3.1.1 | 2652510 | 945963 | 35,66% |
| iPSC | SU3.1.2 | 430889 | 154478 | 35,85% |
| iPSC | SU3.2 | 413721 | 185124 | 44,75% |
| iPSC | SU3.3 | 209961 | 97608 | 46,49% |
| iPSC | NF13 | 253724 | 132544 | 52,24% |
| iPSC | NF21 | 272430 | 135334 | 49,68% |
| iPSC | QV8 | 604002 | 268954 | 44,53% |
| ESC | RES27 | 788002 | 388023 | 49,24% |
| ESC | dB50 | 184668 | 56953 | 30,84% |
| EF | RNFM1 | 1482905 | 1212691 | 81,78% |
| EF | RNFF1 | 1568042 | 1170241 | 74,63% |
Novel miRNA discovery with the custom peaks2mirna method
Wig files were created from the genome maps of the informative reads for two separate groups of samples: PSCs and EFs. The wig files contained RPM (reads per million) expression values for each position on each chromosome. RPM is calculated as the number of reads overlapping a position in the genome divided by the total number of informative reads in a group of samples. Then, wig files were inspected for genome intervals with specific double-peak signals, as explained in Supplementary Figure S1, with a custom Perl script using the following parameters: (i) peak height of 0.9 RPM or higher; (ii) peak length of 18–50 nt; (iii) distance between peaks of 8–30 nt; and (iv) no peaks within 50 nt vicinity. These parameters were selected to maximize the number of detectable, expressed, and known rat miRBase pre-miRNAs (245 of 403, or 60.7%). Then, the following additional filters were applied to the 718 newly identified potential pre-miRNAs to retain only the high-quality candidates: (1) no overlap with the 486 known rat miRBase miRNAs, (2) no overlap with the RNA repeats from the UCSC rn5 rmsk.txt (http://hgdownload.soe.ucsc.edu/goldenPath/rn5/database/rmsk.txt.gz)29, and (3) RNAFold program35 predicted a hairpin structure. As a result, 73 potential pre-miRNAs remained. Following further analyses of sequence homology, some of them were found to belong to the snoRNA class and had to be analysed separately.
Novel miRNA discovery with the miRanalyzer tool
The miRanalyzer program36 was applied to each sample and entire dataset of 11 samples. The R. norvegicus genome (assembly version rn4) was used. The default parameters were used except for the “Fast detection of novel microRNAs”, which was unmarked. For each sample and separately for entire dataset of 11 samples, potential miRNAs that were named “perfect Dicer” were selected from the predictions. Then, only “perfect Dicers” that did not intersect with genome elements (empty “genome elements” column) were selected. Potential miRNA coordinates in rn4 were converted to rn5 using the UCSC LiftOver tool and then intersected with refSeq genes (“refGene.txt” downloaded from UCSC on 14.04.15). Only the entries that did not overlap the refSeq genes in rn4 and rn5 were kept. If a potential miRNA was found in at least two samples, it was labelled as a “cluster” miRNA. The longest or the most strongly expressed isoform was selected as a representative of the cluster. Potential novel miRNAs that were found by miRanalyzer and were predicted35 to have a hairpin structure were filtered against known rat miRNAs (ftp://mirbase.org/pub/mirbase/CURRENT/genomes/rno.gff3)34 and RNA repeats (http://hgdownload.soe.ucsc.edu/goldenPath/rn5/database/rmsk.txt.gz)29. As a result, 13 potential pre-miRNAs remained.
Novel miRNA discovery with the miRDeep2 tool
MiRDeep2 v2.0.0.737 was launched on the collapsed genome (rn5) maps of informative reads. A training set of known mature miRNAs included R. norvegicus (ftp://mirbase.org/pub/mirbase/CURRENT/genomes/rno.gff3)34 and Mus musculus (ftp://mirbase.org/pub/mirbase/CURRENT/genomes/mmu.gff3)34 miRNA sequences acquired from miRBase v21. Additionally, a set of miRNA hairpin sequences was taken from miRBase v21 (ftp://mirbase.org/pub/mirbase/CURRENT/hairpin.fa.gz)34. Potential novel miRNAs found by mirDeep2 were filtered against known rat miRNAs (ftp://mirbase.org/pub/mirbase/CURRENT/genomes/rno.gff3)34 and overlapping RNA repeats (http://hgdownload.soe.ucsc.edu/goldenPath/rn5/database/rmsk.txt.gz)29, which resulted in a list of 315 potential pre-miRNAs.
Search for homologs of novel miRNAs
We took advantage of the UCSC genome browser tracks of rat and other species’ genes, including miRNA, that were mapped to the rn6 genome. To find homologs of novel miRNAs, we intersected their genomic intervals with exons and introns of the ensGene track (last updated 14 Sept, 2015) and the xenoRefGene track (last updated June 19, 2016) (http://hgdownload.soe.ucsc.edu/goldenPath/rn6/database/)29, with consideration of the DNA strand.
Calculation of digital gene expression (DGE) of known and novel miRNA
Coordinates in the rn5 genome of 832 mature miRNAs were downloaded from miRBase v21 (ftp://mirbase.org/pub/mirbase/CURRENT/genomes/rno.gff3)34. The number of All mapped reads overlapping each miRNA genome interval, with consideration of DNA strand, was calculated in each sample. If a read overlapped the border of an interval, it was counted as 1. The sum of overlapping reads served as a raw digital expression level for each miRNA. Alternatively, raw expression was calculated for the informative reads (a subset of All reads that were 18–27 nt in length without RNA-type repeats). The raw expression of known miRNAs calculated by an alternative method was only 1% less than the expression that was calculated based on All mapped reads. To be less stringent, we took the expression level that was based on All reads for downstream analyses. In order to account for the different initial sequencing depths of samples, the RPM normalization procedure was applied to the raw expression values:
| 1 |
where i = 1, …, 11 samples; j = 1, …, 832 miRNAs; – raw expression; – RPM expression; and Informative_readsi – number of informative reads in sample i (Supplementary Table S2). The expression values of novel miRNAs, based on their genome coordinates, were calculated in a similar way as in the known miRNAs with three exceptions (Supplementary Tables S3 and S4). First, primary sequences of novel pre-miRNAs were mapped to the rn6 genome with the blastn program38. If genome intervals of the blastn hits of two different pre-miRNAs overlap, their expression values were summed together. It was done to account for the random locations in the genome for multimapped reads. Second, novel pre-miRNA intervals longer than 50 nt were split into two equal subintervals to mimic possible mature miRNAs in the 5′ and 3′ shoulders. Third, alternative DGE levels were computed using just informative reads to calculate the in formula (1). Both types of DGE based on All and informative reads were used to further classify novel miRNAs to Tier 1 and Tier 2. Tier 1 contained the novel miRNA species that were most likely to be real.
Classification of novel miRNAs into tiers
By default, all potential pre-miRNAs were located in Tier 1. For each item, the following quality scores were calculated in order to sort them into two different tiers. First, the ratio of the number of informative reads to the number of All mapped reads was used to assess the portion of 18–27 nt reads. If this ratio was less than 0.5, the novel RNA molecule was moved to Tier 2. If the ratio was between 0.5 and 0.85 and the difference between the counts of All and informative reads was higher than 3, the pre-miRNA was also moved to Tier 2. The second classification parameter was calculated for novel pre-miRNA candidates in which the genomic interval was 50 nt or longer. Such intervals were divided into left and right parts of equal length. The parameter was used to assess the level of expression of the stem loop. It was calculated as the ratio m/(l + r), where l and r are the number of reads overlapping the left and right parts of the interval, respectively (they share reads overlapping the internal border), and m is the number of reads overlapping the border between the left and right parts. If the ratio was higher than 0.05, the potential pre-miRNA was moved to Tier 2.
Principal component analysis (PCA) of miRNA expression levels
RPM expression data of 832 known miRNAs, excluding 158 zero-expression entries, were used. Additionally, 304 novel miRNAs with unique sequences and not homologous to known miRNAs were used independently. Using the R statistical computing program (http://www.R-project.org/)39, data were centred around the mean expression values, scaled to unit-variance and then processed using PCA40.
Differential expression analysis of miRNAs
ESC and iPSC groups were combined together (PSCs) and compared with EFs. P-values were calculated with the edgeR software41 for each of the 674 expressed rat miRNAs from miRBase, assuming an overdispersed Poisson model of raw (not RPM normalized) expression counts. Parameters of overdispersion were fitted by the software, taking into account the common dispersion, trended dispersion and tagwise dispersion of the miRNA expression values. A set of 227 miRNAs satisfied a p-value cut-off of 10−4, which corresponded to the 3.0 × 10−4 false discovery rate (Benjamini-Hochberg). Alternatively, ANOVA was also performed using the ‘aov’ function in R after replacing zero values with uniform random values from interval (0 to 0.1) and log2 transformation (Supplementary Table S2). However, p-values based on just two samples in one of the groups (EFs) might have little credit. To get over this problem, differentially expressed miRNAs were required to satisfy very strong fold-change conditions without considering the p-value. The ratio between the group averages was required to be at least 3. Additionally, miRNAs upregulated in the PSC group must have a minimum RPM expression in PSC samples that is 2 times (or 10 times in a parallel analysis) higher than its maximum level in the EFs samples. We called this the extremum fold change method. In the same way, miRNAs were defined as upregulated in EFs if their minimum RPM in EFs was 2 times (or 10 times) higher than its maximum level in PSCs, and additionally, they had to have at least three mapped reads in each of the EF samples. Only 219 miRNAs passed this procedure of selection, and they had an edgeR p-value of less than 0.01. Differentially expressed novel miRNAs were defined in the same way.
Clustering according to miRNA expression levels
Expression values of zero were replaced with 0.25, which is below the minimum positive value. After that, log2 transformation was applied. The clustering of miRNAs and samples based on expression values was performed using the ‘pheatmap’ function in R using the default parameters.
Search for target genes of differentially expressed miRBase miRNAs
Known rat, mouse and human miRNA-target interactions were downloaded from the MiRTarBase database42 (http://mirtarbase.mbc.nctu.edu.tw/php/download.php; Catalogued by Species: hsa_MTI.xlsx, mmu_MTI.xls, rno_MTI.xls; accessed on 26/07/2016). Lists of the targets of differentially expressed known miRNAs were obtained taking the miRNA names as the key. They were additionally filtered using gene expression data obtained from sequencing experiments that were performed on the same biological samples27. Upregulated targets were kept if the corresponding miRNA was downregulated, and downregulated targets were kept if the corresponding miRNA was upregulated (Supplementary Table S5).
Search for the target genes of novel miRNAs
Mature sequences of novel miRNAs were analysed for targets in the 3′ UTRs of rat refSeq transcripts using the TargetSpy v1.143, miRanda v3.3a44 and TargetScan v7.045 tools. To reduce the false positive rate, only targets that were predicted by all three programs were accepted (Supplementary Table S6).
KEGG and GO enrichment analysis
Lists of genes found to be targeted by miRNAs were subjected to KEGG and GO enrichment analysis with the DAVID 6.8 Beta online tool46 using the default parameters. All rat genes were used as the background, and alternatively, only differentially expressed genes27 were used.
Results
Small RNA sequencing
Small RNA libraries from two EF (RNFM1, RNFF1), two ESC (RES27, dB50), and seven iPSC samples (NF13, NF21, QV8, SU3, one technical, and two biological replicates) were generated and sequenced on the Illumina platform. Reads were filtered as described in the Materials and Methods section. A total of 32.9 million raw reads were generated by the sequencing of eleven libraries followed by trimming and adapter clipping. An absolute majority (31.9 million raw reads) mapped onto the reference genome, including 31.4 million reads that mapped on the chr1-20, X (All reads). After filtration against RNA-type repeats and after keeping only 18–27 nt sequences, a set of 8,860,854 informative reads was obtained. The exact amount of informative reads for each library is presented in Table 1. The length distribution of the informative reads was similar among different samples. Examples are given in Fig. 1a and b. Most of the informative reads were between 21–25 nt. To identify known miRNAs, coordinates of All reads that were mapped to the genome were compared with known miRNA coordinates from miRBase v2134. The portion of known miRNA reads (Table 1) varied from 30.84% (dB50) to 81.78% (RNFM1). In total, 674 known miRNAs were identified among all samples.
Figure 1.
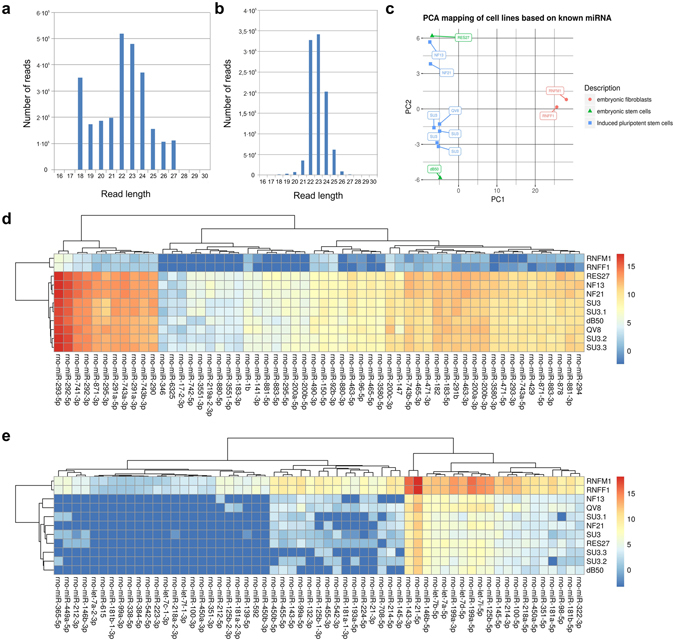
(a) The number of informative reads at different lengths for sample SU3. (b) The number of informative reads at different lengths for sample SU3 that were mapped to known rat miRNAs. (c) Principal component analysis of 11 samples based on RPM expression of 219 differentially expressed miRbase miRNAs. (d and e) Heatmaps of 56 upregulated miRNAs in PSCs (d) and 59 upregulated miRNAs in EFs (e). Known miRNAs with an extremum fold change more than 10, and the log2 transformation.
Novel miRNA discovery
To discover novel miRNA species, the informative reads that were mapped to the rn5 genome were processed by three different methods: the custom peaks2mirna, miRanalyzer36 and miRDeep237. Pre-miRNAs discovered by more than one method were collapsed by taking the maximum of their genomic intervals. Likely false positives, in which the primary sequence was repeated in the genome more than 15 times, were excluded. The number of repeats in the genome was calculated as the number of blastn38 hits, and the threshold was fitted to avoid false negatives. Finally, 388 potential novel pre-miRNAs including genomic duplicates were identified. This number included 39 pre-miRNAs that were homologous to rat or non-rat miRNAs and 40 molecules that were homologous to small nucleolar RNA (snoRNA). Homology analysis was done by overlapping genome intervals with the xenoRefGene and ensGene UCSC genome tracks29. This analysis allows nucleotide gaps, deletions and mismatches between homologs, including variance in miRNA seed sequence. The rest of the 309 candidates were split into Tier 1 and Tier 2 lists according to their quality scores as described in the Materials and Methods section and each had 248 and 61 items, respectively. Tier 1 and novel species that are homologous to known miRNAs are presented in Supplementary Table S3, which contains 394 mature miRNAs including genomic duplicates. Hereafter, this table will be referred to as Tier 1. Tier 2 and snoRNA homologs are presented in Supplementary Table S4, which includes 163 potential mature miRNAs including genomic duplicates. Hereafter, this table will be referred to as Tier 2. The two lists, in total, contain 557 potential novel miRNAs. Serial number in identificators of novel miRNA were assigned arbitrary and their order don’t imply to reflect importance or quality in both Supplementary Tables S3 and S4.
Differential expression of miRNAs in rat pluripotent stem cells and embryonic fibroblasts
To reveal differentially expressed miRNAs, reads per million (RPM) gene expression was calculated for the 832 known and the 557 potential novel miRNAs using All mapped reads as described in the Materials and Methods section. Among the known miRNAs, 77 were found to be upregulated and 142 were found to be downregulated in PSCs compared to EFs, with at least a double extremum fold change (Supplementary Table S7). For novel Tier 1 miRNAs, the corresponding up- and downregulated genes were 10 and 13, respectively (Table 2). In Tier 2, there were 9 and 2, respectively (Supplementary Table S4). The criteria for miRNA up- or downregulation were described in the Materials and Methods section. Most of the miRNAs that were upregulated in PSCs have expression levels that are below the median in adult rat tissues, according to RT-PCR expression profiling with the TaqMan® Rodent miRNA card A v2.0 (GEO accession GSE66195, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66195). On the other hand, most of the downregulated miRNAs have high expression levels (above median) in somatic tissues in the same experiment (data not shown).
Table 2.
Novel miRNA differentially expressed between PSCs and EFs.
| Position (rn5) | Shoulder | PSC or EF | iPSC, RPM | ESC, RPM | EF, RPM | Homolog | miRdeep2 | Custom | Pre-miRNA primary sequence |
|---|---|---|---|---|---|---|---|---|---|
| chr5:+:152026238–152026277 | 3p | EF > PSC | 0 | 0 | 2.98 | 1 | 0 | AAGGGGCAGGTTGCTGATACATTGTGACATAGACTTGGAAACAGTTGGTAATAGTGTACTGTGACCTGTCCTAGGGTGA | |
| chr8:+:67350015–67350048 | 3p | EF > PSC | 0 | 0 | 6.31 | 1 | 0 | CTCAGACATGGTTCCTGTCCTCTTGGGTCTCAGTATTGATGTGGAGTACAGAGCACATGTTTGAGGGG | |
| chr8:+:67395497–67395530 | 3p | EF > PSC | 0 | 0 | 6.31 | 1 | 1 | TCAGACATGGTTCCTGTCCTCTTGGGTCTCAGTATTGATGTGGAGTACAGAGCACATGTTTGAGGGG | |
| chr5:+:83208017–83208048 | 5p | EF > PSC | 0 | 0 | 3.65 | 1 | 1 | TGGTCATGGTTCATGAACTGGAGCGGGTCCATCGTGTGTGTTCCAGTTCAACCTTGTCCCTGTT | |
| chr3:−:124104501–124104531 | 5p | EF > PSC | 0 | 0 | 3.26 | 1 | 0 | CATGTTCCACTCACTCTCAGACGAAAACAGAGAGGCTCTGGGAGAGGTGTGGGACCAGTAC | |
| chr10:−:32125896–32125927 | 5p | EF > PSC | 0 | 0 | 2.64 | 1 | 0 | TCTGTGAGTCTGAAGCCCACCTGGTTTACATATCAAGTTCCAAGTGCGCCAGACCCACATAGTG | |
| chr6:−:137837930−137837960 | 5p | EF > PSC | 2.34 | 0.63 | 15.73 | 1 | 0 | TCTGCTCCTGCTCTTTCTGCTGCTGCTGTTCCTGTTGCCAGCGGGCGAGGAGGAGCGCAGCT | |
| chr2:+:217974496–217974523 | 5p | PSC > EF | 17.74 | 17.01 | 2.66 | 1 | 0 | ATTGCCTGGTTTAGTCTCTGCTACTTGCAAGTACCAGGTCACTAAAACAGGCAGGG | |
| chrX:−:151288045–151288072 | 3p | PSC > EF | 11656.24 | 11023.61 | 3.90 | 1 | 0 | TACTCAGATTAGTGTCACTCCAGGACATAAATATGTATGGCGCTCCTCTGAGTAGAA | |
| chr14:+:26226537–26226565 | 5p | PSC > EF | 61.89 | 72.76 | 0 | 1 | 1 | TGCTTGCCTGGATTACAGTGAAGGTATTCATGTTTCACTGCACTCTAGACAGGCATTA | |
| chrX:−:151288073–151288101 | 5p | PSC > EF | 107.52 | 169.08 | 0 | 1 | 0 | TACTCAGATTAGTGTCACTCCAGGACATAAATATGTATGGCGCTCCTCTGAGTAGAA | |
| chr1:−:63523357–63523389 | 3p | PSC > EF | 3395.86 | 3010.64 | 0.97 | MIR372 | 1 | 1 | CACTCAAATGTAGGAGCGCTCTTCTGATCTGGTTTAAGTGGAAAGTGTTGCATCGTTTGGGTGTCAT |
| chr1:−:63523390–63523423 | 5p | PSC > EF | 243.23 | 199.24 | 0.33 | MIR372 | 1 | 1 | CACTCAAATGTAGGAGCGCTCTTCTGATCTGGTTTAAGTGGAAAGTGTTGCATCGTTTGGGTGTCAT |
| chr2:+:251220594–251220625 | 3p | PSC > EF | 45.96 | 47.38 | 0 | MIR302B | 1 | 0 | CCACTTTAACATGGGAATGCTTTCTGTCACATTGAAGAGTAAGTGCTGCCATGTTTTAGTAGA |
| chr2:+:251220844–251220873 | 3p | PSC > EF | 37.22 | 30.67 | 0 | MIR302 | 1 | 1 | TAAACGTGGTTGTCCTTGCTTTGGAACGAAGAAAGTAAGTGCTTCCATGTTTGGGTGAT |
| chr1:+:148620388–148620416 | 5p | PSC > EF | 95.17 | 32.11 | 0.97 | MIR147B | 1 | 0 | GTGGAAACACTTCTGCACAAACTCGATTTTGATGCCAGTGTGCAGAAATGCTTCTGCT |
| chrX:−:140167614–140167645 | 5p | PSC > EF | 18.99 | 31.98 | 0.67 | MIR18B | 0 | 1 | TAAGGTGCATCTAGTGCTGTTAGTGAAGCAGCTTATAATCTACTGCCCTAAATGCCCCTTCTCG |
| chr7:+:70463555–70463594 | 5p | EF > PSC | 2640.99 | 2751.3 | 9939.24 | MIR26 | 0 | 1 | GAGAGCCGGCTGTGGCTGGATTCAAGTAATCCAGGATAGGCTGTTTCCATCTGTGAGGCCTATTCTTGATTACTTGTTTCT |
| chr7:+:70463595–70463635 | 3p | EF > PSC | 2609.2 | 2734.29 | 9854.73 | MIR26 | 0 | 1 | GAGAGCCGGCTGTGGCTGGATTCAAGTAATCCAGGATAGGCTGTTTCCATCTGTGAGGCCTATTCTTGATTACTTGTTTCT |
| chr3:+:16697079–16697110 | 5p | EF > PSC | 8.53 | 7.95 | 2241.76 | MIR199 | 0 | 1 | CCCAGTGTTTAGACTACCTGTTCAGGACTCCCAAATTGTACAGTAGTCTGCACATTGGTTAGGCT |
| chr3:+:16697111–16697143 | 3p | EF > PSC | 81.95 | 73.78 | 14456.4 | MIR199 | 0 | 1 | CCCAGTGTTTAGACTACCTGTTCAGGACTCCCAAATTGTACAGTAGTCTGCACATTGGTTAGGCT |
| chrX:+:105464335–105464360 | 5p | EF > PSC | 0 | 0 | 3.3 | Mir7093 | 1 | 0 | TTTCCATCTGTCACCCTGCAGGTCTGCTGTAGCAGGTGGCACTGCTTGAAGCA |
| chr14:−:30139894–30139924 | 3p | EF > PSC | 1.74 | 0 | 74.28 | MIR365 | 0 | 1 | GGGACTTTTGGGGGCAGATGTGTTTCCATTCCACTATCATAATGCCCCTAAAAATCCTTATTG |
PCA was performed based on the RPM values of 219 differentially expressed known miRNAs. Figure 1c shows the PCA projection of eleven cell lines onto the first two principal components (PC). The first PC perfectly separated EF samples from the others. The second PC divided samples by gender into two groups: NF13, NF21 and RES27 were female and SU3, QV8 and dB50 were male. PC1 explained 81.1% while PC2 explained 6.6% of the total variance. PC1 also perfectly separated PSCs from EFs if considering all (not just differentially expressed) known or novel miRNAs (data not shown). We then performed sample clustering based on differentially expressed miRBase miRNAs (extremum fold change of at least 10) and on all expressed miRNAs. Figure 1d and e show heat maps of the clusters of miRNAs that are overexpressed in PSCs and EFs, respectively. There was no significant difference between the results of the hierarchical clustering of samples performed on all miRNAs and on only differentially expressed miRNAs (data not shown). Both methods, PCA and hierarchical clustering, revealed that there are principal differences in miRNA expression between EFs and PSCs but no major differences between ESCs and iPSCs.
A number of miRNAs that were found to be upregulated in rat PSCs, such as the miR-290-295 cluster and the miR-200 family miRNAs, are involved in pluripotency induction and maintenance in mice21, 47. Several miRNAs, such as members of the miR-183-96-182 cluster, miR-741, miR-881, miR-878, miR-743a, miR-743b, miR-465, miR-871, miR-880, are known to be expressed in mouse ESCs and were also expressed in rat PSCs; however, their function in pluripotency induction and regulation has not been directly shown48–50.
GO and pathway analyses of targets of the differentially expressed miRNAs
Analysis of known miRNA targets
To reveal the function of differentially expressed miRNAs in rat PSCs and EFs, we performed an analysis of the published targets of these miRNAs in humans, mice, rats, and other species. We took known miRNAs with an extremum fold change of more than 10 between EFs and PSCs. Among them, 58 miRNAs were upregulated in the PSCs and 69 were upregulated in the EFs (Supplementary Table S7). Known miRNA targets were found using information from the MiRTarBase database42. Additionally, the targets were required to be downregulated for upregulated miRNAs and upregulated for downregulated miRNAs, which is consistent with the expected miRNA function. By this method, we found 388 published target genes for miRNAs that were upregulated in PSCs and 69 published targets for downregulated miRNAs (Supplementary Table S5). All of the up- and downregulated target genes were analysed with the DAVID46 web application tool to identify the associated KEGG pathways and GO terms. KEGG pathways enriched with target genes, using all rat genes as a background, are presented in Fig. 2a and in Supplementary Table S8. Most of the target genes were associated with cancer pathways, stem cell pluripotency regulatory pathways, and the Hippo signalling pathway. Of note is that the results persist if only differentially expressed genes were used as a background (data not shown). Among the enriched GO terms, stem cell differentiation, response to retinoic acid, somatic stem cell population maintenance, regulation of apoptotic process were present (Fig. 2b). The obtained results suggest that these differentially expressed miRNAs play a substantial role in the regulation of pluripotency and cell reprogramming in rats.
Figure 2.
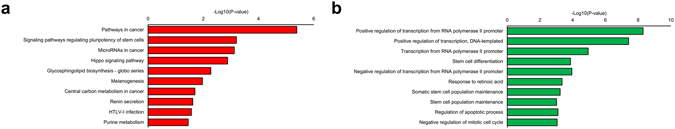
KEGG signalling pathway (a) and GO (b) enrichment analyses of the published targets of differentially expressed known miRNAs. The top 10 KEGG signalling pathways and 10 GO terms with the most significant p-value.
Analysis of novel miRNA putative targets
Targets of 304 novel miRNAs with unique sequences that are not homologous to known miRNAs were predicted by TargetSpy v1.143, miRanda v3.3a44, and TargetScan v7.045 as described in the Materials and Methods section. In total 8,437 transcript targets were identified for novel miRNAs (Supplementary Table S6). GO enrichment and KEGG signalling pathway analyses were performed using the DAVID tool46 on the targets of the top 10 highly expressed novel miRNAs in the PSC group and the top 10 highly expressed novel miRNAs in the EF group, including all rat genes as a background. The results are presented in Fig. 3 and Supplementary Table S9. According to the GO annotations, the predicted target genes of novel miRNAs expressed in PSCs are related to processes of embryonic development, cell migration, signal transduction and gene expression regulation. According to KEGG annotation, putative targets are involved in endocytosis, pathways in cancer and the Hippo, FoxO, Wnt, and TGF-beta signalling pathways (Fig. 3a,b). The two latter networks are known to be involved in cell reprogramming and the regulation of pluripotency in rats12, 47.
Figure 3.
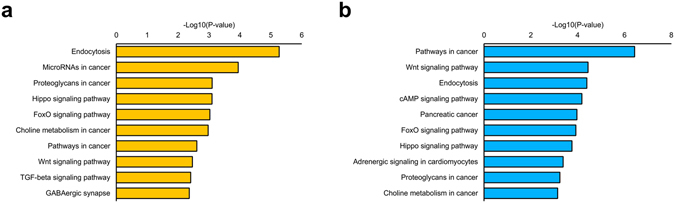
KEGG signalling pathway analysis of putative targets of (a) the top 10 highly expressed novel miRNAs in the PSC group and (b) the top 10 highly expressed novel miRNAs in the EF group.
Validation of novel miRNAs by qRT-PCR
To validate some of the novel miRNAs, we performed qRT-PCR analysis using the NCode™ system in the EF and PSC lines. Four novel miRNAs with the following coordinates: chrX:−:151288045–151288101 (rno-miR-novel-8); chr7:+:70463555–70463594 (rno-homolog-miR-26); chr3:+:16697111–16697143 (rno-homolog-miR-199); and chr18:−:69422790–69422857 (rno-miR-sno-57) were selected for the validation. rno-miR-novel-8, rno-homolog-miR-26, and rno-homolog-miR-199 miRNAs were selected from Tier 1, and rno-miR-sno-57 miRNA was selected from Tier 2. In addition, we analysed the expression of miR-741-3p and miR-743a-3p and found that, in accordance with sequencing data, they were highly expressed in rat PSCs. A pluripotency-associated miRNA, miR-295-3p, was used as a positive control. Expression of miR-295-3p was observed in rat PSCs and absent in rat EFs, which was consistent with its expression pattern in the mouse51 (Fig. 4a). qRT-PCR analysis demonstrated that the rno-miR-novel-8 and rno-miR-sno-57 miRNAs as well as the known miRNAs miR-741-3p and miR-743a-3p were expressed in rat PSCs and absent in rat EFs (Fig. 4a). rno-homolog-miR-26 and rno-homolog-miR-199 miRNAs were expressed in EFs, ESCs and iPSCs, which is consistent with the data obtained from sequencing. We also analysed the expression of the rno-miR-novel-8 miRNA, rno-miR-sno-57 miRNA, miR-741-3p, and miR-743a-3p, which are upregulated in PSCs during differentiation. We induced spontaneous differentiation of rat ESCs and iPSCs in a monolayer and analysed the expression of the miRNAs at various time points. We found that rat miR-295-3p, miR-741-3p and miR-743a-3p expression was significantly decreased during ESC and iPSC differentiation (Fig. 4b). For novel miRNAs, the expression of rno-miR-novel-8 miRNA was absent at the first time point (day 4) and subsequent time points of differentiation, whereas the expression of rno-miR-sno-57 miRNA increased at the first stages of differentiation and then decreased to levels lower than that in PSCs (Fig. 4b). We also assessed the expression of rno-miR-novel-8 miRNA, rno-miR-sno-57 miRNA, and the known miRNAs in different rat adult organs, including the heart, lungs, liver, kidneys, ovaries, testes, eyes and spleen. We found that miR-741-3p, miR-743a-3p, and rno-miR-novel-8 miRNA were expressed in the testis at levels comparable to that in PSCs. miR-741-3p was also expressed in the lungs, ovaries and spleen but at lower levels (Fig. 4c).
Figure 4.
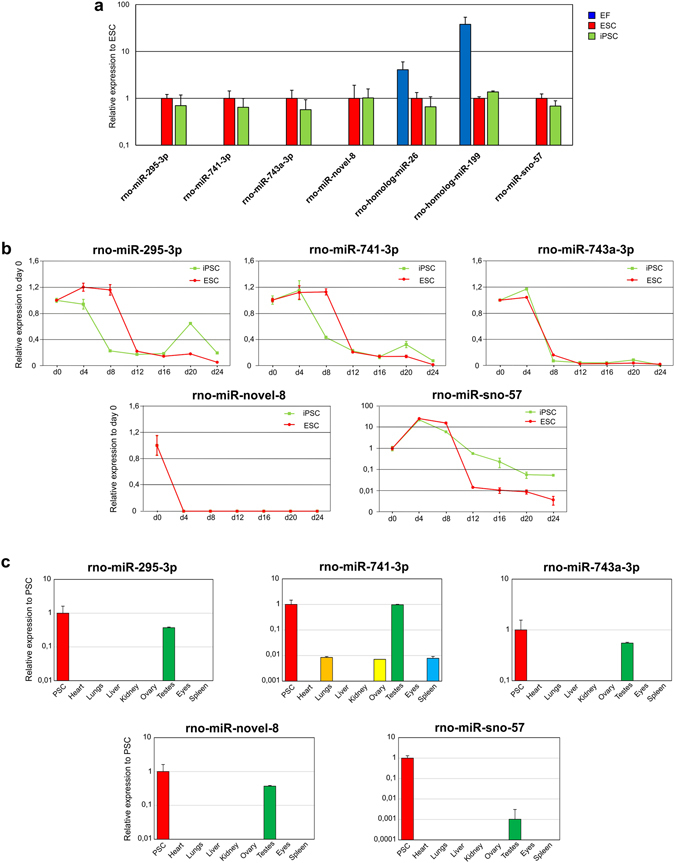
qRT-PCR analysis of known and novel miRNAs in (a) iPSCs, ESCs and EFs (b) during the differentiation of ESCs and iPSCs and (c) in rat adult organs.
Discussion
R. norvegicus is one of the most promising models for disease modelling and translational biomedicine. Pluripotent stem cells are currently widely used to generate various disease models52. An understanding of pluripotency and self-renewal regulation is required to improve the approaches to PSCs generation and cultivation, which will help realize the full potential of their application. Although much progress has been made in the understanding of the pluripotency regulatory network, it has not been fully investigated yet, especially in rats. miRNAs are known to play substantial roles in the regulation of pluripotency and self-renewal14. To study miRNA expression in rat PSCs and to find novel rat miRNAs, we performed genome-wide sequencing of small RNA libraries from rat ESCs, iPSCs, and EFs. Analysis of the sequencing data revealed the expression of 674 known miRNAs across all samples. Differential expression analysis of known miRNAs demonstrated that 77 miRNAs were upregulated and 142 miRNAs were downregulated in rat PSCs compared to EFs. Members of the miR-290-295 cluster (miR-290, miR-291a, miR-292, miR-291b, miR-293, miR-294 and miR-295-1) were the most abundant among known miRNAs that were upregulated in rat PSCs. It is known that mouse miR-290-295 and its human homologues in the miR-372 clusters are ESC specific and regulate cell cycle progression20, 21. Among the miRNAs upregulated in rat PSCs, we also identified miR-205 and members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429), which promote mesenchymal to epithelial transition (MET) in mouse cells, a key step in fibroblast reprogramming47. Members of the miR-200 family are known to suppress the Zeb2 transcription factor, and its inhibition promotes MET53. Another miRNA cluster, miR-183-96-182, is known to be expressed in mouse PSCs49 and is also expressed in rat PSCs. The microRNA-183-96-182 cluster inhibits epithelial to mesenchymal transition (EMT) in cancer cells, and thus, the miRNA cluster may facilitate the reprogramming process by promoting MET54–57. Members of the miR-106-363 and miR-106b-25 clusters, which were shown to be upregulated during fibroblast reprogramming in mice58, were also upregulated in rat PSCs. Most of the miRNAs upregulated in rat PSCs are also expressed in mouse ESCs or iPSCs48. In addition, we found several rat miRNAs (including miR-3580, miR-3551, and miR-6325) whose expression had not been established in PSCs before.
A cluster of miRNAs, comprised of miR-463, miR-465, miR-471, miR-741, miR-743a, miR-743b, miR-871, miR-878, miR-880, miR-881, and miR-883, was located on the X chromosome and was expressed at a high level in rat PSCs according to the sequencing data. These miRNAs are known to be expressed in mouse ESCs and testes48, 50, 59. These miRNAs are testis specific in rats, according to the miRNA sequencing data of 21 male rat organs26. Using qPCR, we have shown that members of the miR-741-3p and miR-743a-3p clusters were expressed at high levels in rat ESCs and iPSCs and were downregulated during differentiation. These miRNAs are also expressed in some adult rat tissues, according to our data and RT-PCR expression profiling with the TaqMan® Rodent miRNA card A v2.0 (GEO accession GSE66195, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66195).
The number of known rat miRNAs is much lower compared to that in humans and mice. In this study, 394 novel mature miRNAs were identified using three methods – mirDeep2, miRanalyzer, and custom peaks2mirna. Most of the novel miRNAs were identified by the mirDeep2 method. In rat PSCs and EFs, 23 novel miRNAs were differentially expressed. Among them, 10 novel miRNAs were upregulated and 13 were downregulated in PSCs. This list includes homologues of human miR-302a and miR-302b. We found that these miRNAs were upregulated in rat PSCs. We also found a homologue of human miR-367. The human and mouse miR-302-367 cluster has been demonstrated to play an important role in the regulation of pluripotency. Moreover, overexpression of the miRNA cluster was sufficient to reprogramme fibroblasts to iPSCs21, 23, 60. However, expression levels of miR-302a and miR-302b were low in rat PSCs compared to the ESC specific miR-290-295 cluster. This is in agreement with the fact that high expression levels of miR-302 cluster are typical of mouse epiblast stem cells48. We also found a homologue of human mir-372 located inside the rat miR-290-295 cluster. miR-372 in humans is known to be connected to fibroblasts reprogramming, particularly by promoting MET60.
Four novel miRNAs were validated using qRT-PCR analysis. Three miRNAs were selected from Tier 1, and one was selected from Tier 2. Expression of the two validated novel miRNAs was observed in rat PSCs and was absent in EFs. Expression of rno-miR-novel-8 miRNA decreased during differentiation and was not observed in the analysed adult organs, except in the testes. The pattern is similar to ESC-specific miRNAs, such as members of the miR-290-295 cluster51. In addition, we found that the rno-miR-novel-8 miRNA was located less than 10 kb from miR-871, mir-3580 and miR-465, which were upregulated in rat PSCs. This may be a cluster of miRNAs that is involved in the regulation of pluripotency, and further investigations are required to shed light on its function.
miRNAs implement their functions through repressing target gene expression. In human and mouse PSCs, clusters of pluripotency-associated miRNAs control cell proliferation and prevent differentiation58, 61. To better understand the functions of known rat miRNAs that are differentially expressed between PSCs and EFs, we analysed their target genes that have been validated in rats, humans or mice. An additional criterion was that the change in the PSC and EF target gene expression levels had to be opposite of that for miRNAs. This criterion helped to improve the analysis of miRNA functions in the rat. In total, 388 downregulated targets were found for the upregulated miRNAs in PSCs, and 69 upregulated targets were observed for miRNAs that were downregulated in PSCs. Then, we performed KEGG and GO enrichment analyses of the target genes. The most common GO terms were related to transcriptional regulation, which indicated that a substantial part of the target genes functioned as transcriptional factors. Many GO terms, such as stem cell differentiation and response to retinoic acid, were related to cell differentiation, which was consistent with the function of miRNAs in suppressing PSCs differentiation61, 62. Self-renewal and differentiation processes are balanced in PSCs, and miRNAs are involved in the regulation of this process58, 61. We also found a stem cell population maintenance category that was relevant for targets of miRNAs that were differentially expressed in PSCs. Regulation of the apoptotic process was also an important category for pluripotent cells, because inhibition of apoptosis promotes pluripotency63 and was demonstrated for the ESC-specific miR-290-295 cluster64. We found enrichment of differentially expressed miRNAs and their targets in the KEGG signalling pathway, the stem cell pluripotency regulatory pathway, pathways in cancer, and the Hippo signalling pathway. Pathways in cancer are relevant for miRNAs that are upregulated in PSCs. It is known that miRNAs that are expressed in PSCs are frequently related to different types of cancer. The human miR-372-373 cluster promotes tumorigenesis, for example65. The Hippo signalling pathway represents a barrier for reprogramming, and its inhibition by LATS2 knockdown increases the efficiency of iPSC generation66. Analysis of the putative target genes of novel rat miRNAs has shown that the miRNAs may take part in the regulation of development and differentiation. In addition, KEGG signalling pathway analysis suggests the involvement of miRNAs in the regulation of the Wnt signalling pathway, which regulates rat PSC maintenance, and the TGF-beta pathway, which participates in the regulation of cell reprogramming12, 47. Based on the results, we suggest that the rat miRNAs identified in this study are involved in the regulation of pluripotency and reprogramming in rats.
In conclusion, we first compared the miRNA expression in rat embryonic stem cells and induced pluripotent stem cells with rat embryonic fibroblasts used for reprogramming. The miRNA expression signature in rat PSCs was similar to that in mouse PSCs, and most mouse pluripotency-associated miRNAs were also found in rats. A number of novel rat miRNAs were identified. Overall, our results shed light on the role of miRNAs in the pluripotency regulatory network of the laboratory rat.
Electronic supplementary material
Acknowledgements
This work was supported by the Russian Science Foundation (project №16-14-10084). The authors thank Elena V. Dementyeva for her critical comments and suggestions on the article, and Denis V. Antonets for his help with bioinformatics analysis.
Author Contributions
S.M.Z. and S.P.M. conceived and designed the experiments. E.A.V. cultured the cells. E.A.E., M.T.R., Y.V.V., O.V.S., and D.N.S. performed bioinformatics analyses. S.P.M. and V.V.S. performed the miRNA isolation, cDNA synthesis and PCR analysis. M.T.R., Y.V.V., O.V.S., D.N.S. and V.V.S. prepared the manuscript. All authors read and approved the manuscript. S.M.Z. approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02632-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medvedev SP, et al. Human induced pluripotent stem cells derived from fetal neural stem cells successfully undergo directed differentiation into cartilage. Stem cells and development. 2011;20:1099–1112. doi: 10.1089/scd.2010.0249. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 6.Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell stem cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Huang G, Ye S, Zhou X, Liu D, Ying QL. Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cellular and molecular life sciences: CMLS. 2015;72:1741–1757. doi: 10.1007/s00018-015-1833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaskova EA, Stekleneva AE, Medvedev SP, Zakian SM. “Epigenetic memory” phenomenon in induced pluripotent stem cells. Acta naturae. 2013;5:15–21. [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob HJ, Kwitek AE. Rat genetics: attaching physiology and pharmacology to the genome. Nature reviews. Genetics. 2002;3:33–42. doi: 10.1038/nrg702. [DOI] [PubMed] [Google Scholar]
- 10.Buehr M, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Li P, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Blair K, Smith A. Robust self-renewal of rat embryonic stem cells requires fine-tuning of glycogen synthase kinase-3 inhibition. Stem cell reports. 2013;1:209–217. doi: 10.1016/j.stemcr.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphrey RK, et al. Maintenance of pluripotency in human embryonic stem cells is STAT3 independent. Stem cells. 2004;22:522–530. doi: 10.1634/stemcells.22-4-522. [DOI] [PubMed] [Google Scholar]
- 14.Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nature cell biology. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meek S, et al. Tuning of beta-catenin activity is required to stabilize self-renewal of rat embryonic stem cells. Stem cells. 2013;31:2104–2115. doi: 10.1002/stem.1466. [DOI] [PubMed] [Google Scholar]
- 16.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 17.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. Genetics. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 18.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & development. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi J, et al. microRNAs regulate human embryonic stem cell division. Cell cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nature genetics. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nature biotechnology. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell stem cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsen SE, de Wit E, de Bruijn E, Cuppen E. Small RNA expression and strain specificity in the rat. BMC genomics. 2010;11:249. doi: 10.1186/1471-2164-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith A, et al. The Rat microRNA body atlas; Evaluation of the microRNA content of rat organs through deep sequencing and characterization of pancreas enriched miRNAs as biomarkers of pancreatic toxicity in the rat and dog. BMC genomics. 2016;17:694. doi: 10.1186/s12864-016-2956-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaskova EA, et al. Transcriptome Characteristics and X-Chromosome Inactivation Status in Cultured Rat Pluripotent Stem Cells. Stem cells and development. 2015;24:2912–2924. doi: 10.1089/scd.2015.0204. [DOI] [PubMed] [Google Scholar]
- 28.Carey BW, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent, W. J. et al. The human genome browser at UCSC. Genome research12, 996–1006, 10.1101/gr.229102. Article published online before print in May 2002 (2002). [DOI] [PMC free article] [PubMed]
- 30.Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 31.Havlak P, et al. The Atlas genome assembly system. Genome research. 2004;14:721–732. doi: 10.1101/gr.2264004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic acids research. 2015;43:D670–681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofacker IL. Vienna RNA secondary structure server. Nucleic acids research. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackenberg M, Rodriguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic acids research. 2011;39:W132–138. doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic acids research. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.R: A language and environment for statistical computing (R Foundation for Statistical Computing 2015).
- 40.Jolliffe, I. T. Principal Component Analysis. (Springer-Verlag 1986).
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu SD, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic acids research. 2011;39:D163–169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturm M, Hackenberg M, Langenberger D, Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC bioinformatics. 2010;11:292. doi: 10.1186/1471-2105-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife4, doi:10.7554/eLife.05005 (2015). [DOI] [PMC free article] [PubMed]
- 46.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell stem cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 48.Jouneau A, et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. Rna. 2012;18:253–264. doi: 10.1261/rna.028878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung AK, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nature structural & molecular biology. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao B, et al. Genome-wide mapping of miRNAs expressed in embryonic stem cells and pluripotent stem cells generated by different reprogramming strategies. BMC genomics. 2014;15:488. doi: 10.1186/1471-2164-15-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Developmental cell. 2003;5:351–358. doi: 10.1016/S1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 52.Medvedev SP, Shevchenko AI, Zakian SM. Induced Pluripotent Stem Cells: Problems and Advantages when Applying them in Regenerative Medicine. Acta naturae. 2010;2:18–28. [PMC free article] [PubMed] [Google Scholar]
- 53.Wang G, et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2858–2863. doi: 10.1073/pnas.1212769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 55.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of biological chemistry. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li XL, et al. A p21-ZEB1 complex inhibits epithelial-mesenchymal transition through the microRNA 183-96-182 cluster. Molecular and cellular biology. 2014;34:533–550. doi: 10.1128/MCB.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & development. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greve TS, Judson RL, Blelloch R. microRNA control of mouse and human pluripotent stem cell behavior. Annual review of cell and developmental biology. 2013;29:213–239. doi: 10.1146/annurev-cellbio-101512-122343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes & development. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanyam D, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature biotechnology. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. The EMBO journal. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi E, Choi E, Hwang KC. MicroRNAs as novel regulators of stem cell fate. World journal of stem cells. 2013;5:172–187. doi: 10.4252/wjsc.v5.i4.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng GX, et al. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS genetics. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 66.Qin H, et al. Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming. Human molecular genetics. 2012;21:2054–2067. doi: 10.1093/hmg/dds023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


