Abstract
Background:
Candida parapsilosis is one of the five common strains of yeasts involved in invasive candidiasis. The expression analysis of sterol biosynthesis pathway genes, which are associated with resistance, can assist the better understanding of antifungal resistance mechanisms.
Methods:
The antifungal susceptibility of 120 clinical C. parapsilosis isolates was examined. The changes in the gene expression related to resistance were analyzed.
Results:
Eight strains were resistant to fluconazole (FLC), itraconazole (ITC), and amphotericin B (AMB). The regulation variations included increased mRNA levels of ERG3, ERG6, and ERG11 and decreased mRNA levels of ERG3 and ERG6 in response to FLC. ERG11 mRNA level increases in response to ITC and AMB.
Conclusion:
The mechanism of resistance to azoles in C. parapsilosis is very similar to C. Albicans. This feature may help to design new treatment strategy for candidiasis
Keywords: Candida parapsilosis, Gene expression, Ergosterol biosynthesis
INTRODUCTION
The rate of candidiasis among patients has increased largely in recent years. One of the five frequent yeast strains involved in invasive candidiasis is Candida parapsilosis that is mainly common in neonates and in catheter-associated candidemia[1].
Lanosterol 14-demethylase is a member of cytochrome P450 enzyme family that is required for the synthesis of ergosterol. This enzyme is encoded by the ERG11 gene, and it is a target for azoles. Azoles, especially fluconazole (FLC), are the most common drugs used for the treatment of candidiasis[2].
Long-term treatment of candidiasis and the emergence of resistance to azole and polyene drugs usually result in treatment failure[3]. However, the pathway for fungal sterol biosynthesis is still a known and confirmed target for antifungal drug development. In ergosterol biosynthesis pathway, there are other genes, i.e. ERG3 and ERG6, that have not yet been characterized completely.
The 14-methyl fecosterol accumulation is caused by mutations in the ERG3 gene. C. albicans erg3 mutants are able to resist both polyene and azole treatment [4]. Sterol content analysis of erg3 mutants shows an accumulation of sterol intermediates, i.e. 14-methyl fecosterol, which led to an impairment in the final steps of the ergosterol pathway[5].
Sequencing of ERG6 gene has indicated a specific missense mutation in ERG6 where cysteine is replaced with phenylalanine[5].
Resistance to polyenes is less common than azoles, but it has been recently reported in Candida species[6,7].
In the present study, eight resistant strains of C. parapsilosis were isolated from clinical samples. Their resistance to specific antifungal agents was validated by in vitro susceptibility assay. Using real-time PCR method, we made an attempt to investigate the possible alterations in expression profile of some ergosterol biosynthetic genes such as ERG3, ERG6, and ERG11 in resistant species[8].
MATERIALS AND METHODS
Clinical isolates
In total, 120 clinical C. parapsilosis isolates, obtained from a collection of clinical isolates, were recovered during an epidemiological study in three provinces of Iran (Tehran, Mazandaran, and Isfahan), between June 2009 and June 2010[9].
Antifungal agents
FLC (Tehran Daru, Iran), amphotericin B (AMB, Sigma-Aldrich, USA) and itraconazole (ITC, Tehran Daru, Iran) were selected as drugs to be used in susceptibility tests. The stock solutions of FLC were prepared in distilled water. However, for AMB and ITC, we used DMSO. The solutions were then kept frozen at -70°C until use. Dilution of antifungal drugs was performed with RPMI 1640 medium (Invitrogen, USA) and buffered to pH 7.0 with 0.165 M morpholine propane sulfonic acid buffer as described previously (Sigma, USA)[10].
Antifungal susceptibility testing
Reference antifungal susceptibility testing of the isolates was performed by the broth microdilution method described in Clinical and Laboratory Standards Institute (CLSI) guidelines, document M27-S3[11]. C. parapsilosis ATCC 22019 type strain from the American type culture collection was used as a control for antifungal susceptibility testing[11]. According to the guidelines of CLSI, concentration ranges were 0.125-64 µg/ml for FLC and 0.03-16 µg/ml for AMB and ITC[11]. The assay was carried out in 96-well round-bottom microtiter plates. Cell suspensions were prepared in RPMI 1640 medium and were adjusted to give a final inoculum concentration of about 0.5×103-2.5×103 cells/ml. The plates were then incubated at 35°C and read after 48 h[12]. The minimum inhibitory concentrations (MICs) were then determined from the readings and compared with a drug-free control. All tests were performed in duplicate. The MIC results were read according to the M27-S3 supplement of the CLSI Guide.
RNA purification
Total RNA was extracted from C. parapsilosis cells using a commercial kit (Fermentas, EU). Yeast cells were harvested at the exponential phase of growth. C. parapsilosis culture was grown in sabouraud media (without antifungal drugs) at 32°C for 48 h and grown to an optical density of approximately 0.5-1.0 at 600 nm. Total RNA was extracted according to the manufacturer’s instruction.
RT-PCR
First-strand cDNA synthesis
First-strand cDNA was synthesized from 0.1 ng to 5 µg of total RNA in a 20 µl reaction volume using a commercial kit (Fermentas, EU) according to the manufacturer’s instructions. Primers were designed using the Oligo Explorer (version 15) software and were listed in Table 1. The obtained PCR fragment was estimated to be 150–200 bp.
Table 1.
Primers used in quantitative real-time PCR analysis
| Gene | Primer | Sequence |
|---|---|---|
| ERG11 | Forward | 5’ CAG AAA AGT GGC GTT GTT GA 3’ |
| Reverse | 5’ GCA GCA TCA CGT TTC CAA TA 3’ | |
| ERG3 | Forward | 5’ AGT GGG TGC AGT GAT ACA GT 3’ |
| Reverse | 5’ TGC GGG TAA GAA GGT TGG TT 3’ | |
| ERG6 | Forward | 5’ AGC TAC CGT TCA TGC TCC AG 3’ |
| Reverse | 5’ GTT CGG CAA CTT CAC GAC TG 3’ |
Real-time PCR
Real-time PCR was performed in a Step One Plus real-time PCR system (Applied Biosystems, Foster City, CA), and SYBR Premix Ex Taq II was used as a reagent specifically designed for intercalator-based real-time PCR. PCR reaction mixtures contained 2 µl of first strand cDNA, 10 µl SYBR green, 0.8 µl of each primer, and 6.4 µl dH2O to make a final volume of 20 μL. PCR was performed on a Rotor-Gene 3000 system (Corbett Life Sciences, Sydney, Australia) with a preliminary hold at 94°C for 30 s as initial denaturation step, followed by the 45 cycles PCR step consisting of 95°C for 50 s, 58°C for 20 s and 72°C for 30 s. Final holding was performed at 72°C for 1 min, and melting step was performed at 65-99°C.
To quantify the possible changes in ERG3, ERG6, and ERG11 genes expression levels in C. Parapsilosis, RT PCR was performed. ERG3, ERG6, ERG11 genes expression were normalized to the housekeeping gene, ACT1, and analyzed by using REST© software (2008, v. 2.0.7). The software uses the comparative Ct method (ΔΔCt) to analyse the data. A sensitive strain (positive control) of C. parapsilosis was included in each run of the experiment as a positive control. Experiments under each condition were performed in duplicate, and each experiment was repeated twice on two different days to assess the reproducibility[13].
RESULTS
Determination of MIC
Evaluation of the antifungal susceptibility tests showed that three (2.5%) isolates of C. Parapsilosis strains were resistant to FLC (FLCR1, FLCR2, FLCR3: MIC≥8 µg/ml). In addition, three (2.5%) and two (1.66%) isolates indicated resistance to ITC (ITCR1, ITCR2, ITCR3: MIC≥1 µg/ml) and AMB (AMBR1, AMBR2: MIC≥1µg/ml), respectively. There was no cross-resistance to drugs between the eight strains.
Expression analysis of ERG3, ERG6, and ERG11 genes using ΔΔCt method
ERG3, ERG6, ERG11, and ACT1 (housekeeping gene) mRNA levels were examined in all resistant strains (FLCR1, FLCR2, FLCR3, ITRR1, ITRR2, ITRR3, AMBR1, and AMBR2). The output of REST© software (2008, v. 2.0.7) was calculated for indication of ERG3, ERG6, and ERG11 gene expression in the treated cells after the normalization of their expression to the housekeeping gene in all strains. Table 2 shows the results of data analysis using REST© software (2008, v. 2.0.7). Figure 1 indicates the relative gene expression level of ERG3, ERG6, and ERG11.
Table 2.
Results for relative expression of ERG3, ERG6, and ERG11 genes by use of ΔΔCt method (REST©, 2008, v. 2.0.7)
| Gene | Resistance strain | Type | Expression | P(H1) | Result | |
|---|---|---|---|---|---|---|
| Effect of FLC | ERG3 | FLCR1 | TRG | 3.986 | 0.000 | Up |
| FLCR2 | TRG | 20.393 | 0.000 | Up | ||
| FLCR3 | TRG | 3.031 | 0.175 | - | ||
| ERG6 | FLCR1 | TRG | 2.056 | 0.000 | Up | |
| FLCR2 | TRG | 1.803 | 0.000 | Up | ||
| FLCR3 | TRG | 1.847 | 0.166 | - | ||
| ERG11 | FLCR1 | TRG | 1.729 | 0.170 | - | |
| FLCR2 | TRG | 1.028 | 0.837 | - | ||
| FLCR3 | TRG | 12.951 | 0.000 | Up | ||
| Effect of ITC | ERG3 | ITCR1 | TRG | 0.503 | 0.339 | - |
| ITCR2 | TRG | 0.796 | 0.000 | Down | ||
| ITCR3 | TRG | 1.072 | 1.000 | - | ||
| ERG6 | ITCR1 | TRG | 0.108 | 0.000 | Down | |
| ITCR2 | TRG | 0.109 | 0.182 | - | ||
| ITCR3 | TRG | 0.184 | 0.000 | Down | ||
| ERG11 | ITCR1 | TRG | 3.864 | 0.000 | Up | |
| ITCR2 | TRG | 15.945 | 0.000 | Up | ||
| ITCR3 | TRG | 1.905 | 0.000 | Up | ||
| Effect of AMB | ERG3 | AMBR1 | TRG | 0.064 | 0.000 | Down |
| AMBR2 | TRG | 0.015 | 0.000 | Down | ||
| ERG6 | AMBR1 | TRG | 0.142 | 0.000 | Down | |
| AMBR2 | TRG | 0.135 | 0.000 | Down | ||
| ERG11 | AMBR1 | TRG | 1.270 | 0.328 | - | |
| AMBR2 | TRG | 2.071 | 0.000 | Up | ||
| Beta Act | REF | REF | - | - | REF | |
| Positive. control | - | - | 1.000 | |||
Up-regulation (UP) and down-regulation (Down) for ERG genes. REF, reference gene; TRG, target gene; FLC, fluconazole; ITC, itraconazole; AMB, amphotericin B
Fig. 1.
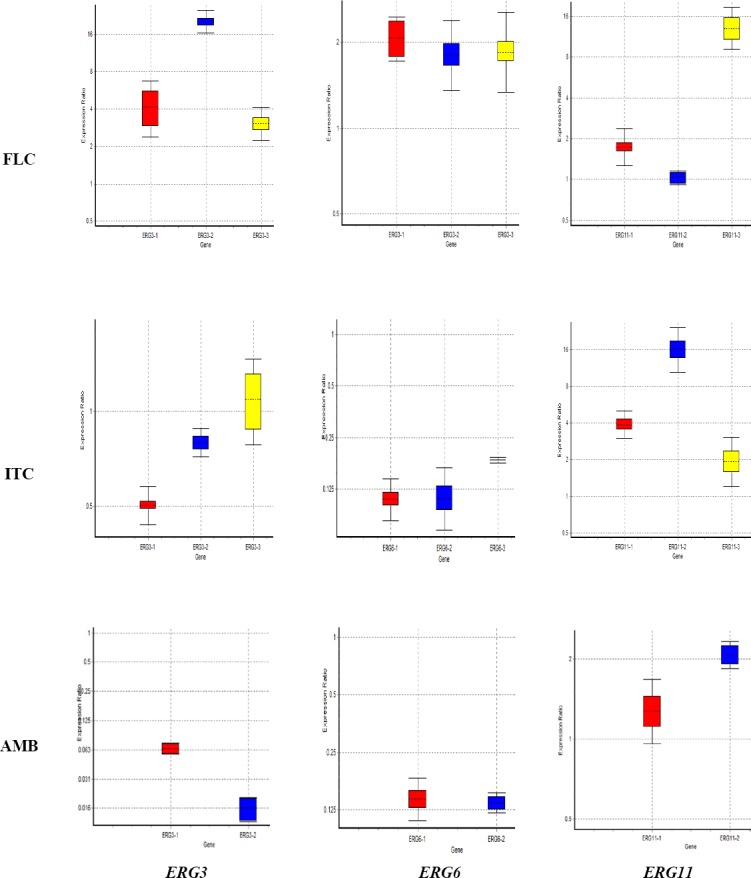
Effect of fluconazole (FLC), itraconazole (ITC), and amphotericin B (AMB) on ERG3, ERG6, and ERG11 genes expression. Colors represent resistant isolates (Red, no. 1; blue, no. 2; yellow, no. 3). Results of relative expression of ERG3, ERG6, and ERG11 genes was calculated using ΔΔCt method (REST©, 2008, v2.0.7. Boxes represent the interquartile range, or the middle 50% of observations. The dotted lines signify the median gene expression.
DISCUSSION
Infections due to C. parapsilosis have been reported in European and Asian countries and Latin America[10]. Azoles, especially FLC, are the most widely used drugs for the treatment of candidiasis. However, due to the high use of azole antifungals, the incidence of resistant strains of Candida has been increased[14-18]. C. parapsilosis is not prone to development of antifungal resistance, but recent reports indicated its increased resistance to azoles[10,19-21]. In the present study, we tried to look into the possible changes in the expression profile of ERG3, ERG6, and ERG11 genes in response to FLC, ITC, and AMB within eight resistance isolates of C. parapsilosis. In three isolates that showed resistance to ITC, we found a significant decrease in ERG3 mRNA level in ITCR2. However, mRNA levels of ERG6 were decreased in ITCR1 and ITCR3 isolates. Surprisingly, ERG11 mRNA levels increased in all mentioned isolates, i.e., ITCR1, ITCR2, and ITCR3.
Further analysis showed different expressions involved in the development of resistance to FLC among three isolates: increased mRNA levels of ERG3 (in FLCR1 and FLCR2), of ERG6 (in FLCR1 and FLCR2), and of ERG11 (only in FLCR3). Morio[22] indicated genetic alterations in ERG3 that may have resulted from f FLC therapy. Berkow et al.[23] found mutations in the sequence of the sterol biosynthesis genes (ERG3 and ERG11). Based on their findings, azole resistance contributes to MDR1 and CDR1 (putative drug transporters). Their findings also demonstrated that among azole-resistant isolates, Y132F substitution in ERG11 is the only substitution. Also, mutation in ERG3 allows the fungal cell to produce toxic intermediate sterols and to become resistant to azoles and AMB[23]. Other investigations have shown that experimental increase in ERG11 level can cause increased azole resistance[24,25]. In addition, drug resistance to antifungals may be regulated by transcription factors[26].
Resistance to the polyenes is rare but could be acquired by the loss-of-function mutations in ERG3, which can inhibit the formation of the drug-lipid complex, prevent osmotic cellular lysis and finally block the production of ergosterol. Mutations in ERG6 led to the accumulation of last sterol intermediates and reduced susceptibility to the polyenes in C. glabrata[5,27].
Based on our results, AMBR1 and AMBR2 isolates showed a decrease in mRNA level of ERG3 and ERG6, but AMBR2 isolate revealed an increase in ERG11 mRNA level. Therefore, we can conclude that the regulation of ERG3 and ERG6 and ERG11 genes could be different in the investigated isolates.
Lees et al.[28] found that ERG11 (lanosterol demethylase) is essential for aerobic growth but is suppressed by mutations in the ERG3 gene, which is in accordance with our obtained results. Silva et al.[14] reported that C. parapsilosis, like C. albicans, acquires resistance to azoles either through increased expression of the sterol biosynthetic pathway genes or via the up-regulation of the MDR1 multidrug transporter family. In Silva’s study[14], the expression of ERG3 and ERG11 was reduced in FLCR (-4.86 and -2.69fold), whereas in our study, the expression of ERG3, ERG6, and ERG11 was increased or remained unchanged in FLCRS (from +1.028 to + 20.39). Morio et al.[22] suggested more extensive investigations on other genes, such as ERG3 and ERG6, which are involved in the ergosterol biosynthesis pathway, when azole resistance is suspected. Vandeputte et al.[7] showed that a nonsense mutation detected in the ERG6 gene led to a decrease in ergosterol content in C. glabrata isolates. Expression of ERG11 and ERG3 genes was decreased upon exposure to AMB. Liu et al.[8] observed that ketoconazole increases the expression of genes involved in sterol metabolism, lipids, and fatty acid, including ERG3 and ERG11. Similar to those findings, our results revealed that ERG3 and ERG6 genes were down-regulated due to exposure to AMB.
In summary, we can conclude that the mechanisms of resistance to azole drugs in C. parapsilosis and in C. albicans are the same. In addition, this finding may help in designing new strategies for antifungal therapy in Candida infections. However, further analysis is needed to determine the process by which mRNA levels for ERG3 and ERG6, as well as ERG11 are altered in these isolates.
ACKNOWLEDGMENTS
This study was supported by the School of Public Health, Tehran University of Medical Sciences, Tehran (Iran) (Grant Number 26956).
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Mirhendi H, Bruun B, Schoenheyder HC, Christensen JJ, Fuursted K, Gahrn-Hansen B, Johansen HK, Nielsen L, Knudsen JD, Arendrup MC. Molecular screening for Candida orthopsilosis and Candida metapsilosis among Danish Candida parapsilosis group blood culture isolates:proposal of a new RFLP profile for differentiation. Journal of medical microbiology. 2010;59(Pt 4):414–420. doi: 10.1099/jmm.0.017293-0. [DOI] [PubMed] [Google Scholar]
- 2.Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhäuser J, Rogers PD. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryotic cell. 2008;7(7):1180–1190. doi: 10.1128/EC.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morschhauser J. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biochimica et biophysica acta. 2002;1587(2-3):240–248. doi: 10.1016/s0925-4439(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 4.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Rolley N, Kelly DE, Kelly SL. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrobial agents and chemotherapy. 2010;54(11):4527–4533. doi: 10.1128/AAC.00348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandeputte P, Tronchin G, Bergès T, Hennequin C, Chabasse D, Bouchara J-P. Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrobial agents and chemotherapy. 2007;51(3):982–990. doi: 10.1128/AAC.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodero L, Mellado E, Rodriguez AC, Salve A, Guelfand L, Cahn P, Manuel Cuenca-Estrella, Graciela Davel, Juan L. Rodriguez-Tudela. G484S amino acid substitution in lanosterol 14-αdemethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrobial agents and chemotherapy. 2003;47(11):3653–3656. doi: 10.1128/AAC.47.11.3653-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandeputte P, Tronchin G, Larcher G, Ernoult E, Bergès T, Chabasse D, Bouchara JP. A nonsense mutation in the ERG6 gene leads to reduced susceptibility to polyenes in a clinical isolate of Candida glabrata. Antimicrobial agents and chemotherapy. 2008;52(10):3701–3709. doi: 10.1128/AAC.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu TT, Lee RE, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrobial agents and chemotherapy. 2005;49(6):2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammadi R, Mirhendi H, Rezaei-Matehko-laei A, Ghahri M, Shidfar MR, Jalalizand N, Makimura K. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Medical mycology. 2013;51(6):657–663. doi: 10.3109/13693786.2013.770603. [DOI] [PubMed] [Google Scholar]
- 10.Silva AP, Miranda IM, Lisboa C, Pina-Vaz C, Rodrigues AG. Prevalence, distribution, and antifungal susceptibility profiles of Candida parapsilosis, C. orthopsilosis, and C. metapsilosis in a tertiary care hospital. Journal of clinical microbiology. 2009;47(8):2392–2397. doi: 10.1128/JCM.02379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI. Implementation Guide of POCT for Health Care Providers; Approved Guideline. Wayne, PA; 2007. [Google Scholar]
- 12.CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; 4th Informational Supplement. Wayne, PA: CLSI document M27-S4. Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 13.Mohseni R, Noorbakhsh F, Moazeni M, Nasrollahi Omran A, Rezaie S. Antitoxin Characteristic of Licorice Extract:The Inhibitory Effect on Aflatoxin Production in Aspergillus parasiticus. Journal of food safety. 2014;34(2):119–125. [Google Scholar]
- 14.Silva A, Miranda I, Guida A, Synnott J, Rocha R, Silva R, et al. Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrobial agents and chemotherapy. 2011;55(7):3546–3556. doi: 10.1128/AAC.01127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almirante B, Rodríguez D, Cuenca-Estrella M, Almela M, Sanchez F, Ayats J, Alonso-Tarres C, Rodriguez-Tudela JL, Pahissa A. Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections:case-control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. Journal of clinical microbiology. 2006;44(5):1681–1615. doi: 10.1128/JCM.44.5.1681-1685.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medrano DJA, Brilhante RSN, Cordeiro RdA, Rocha MFG, Rabenhorst SHB, Sidrim JJC. Candidemia in a Brazilian hospital:the importance of Candida parapsilosis. Revista do instituto de medicina tropical de são paulo. 2006;48(1):17–20. doi: 10.1590/s0036-46652006000100004. [DOI] [PubMed] [Google Scholar]
- 17.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis:a persistent public health problem. Clinical Microbiology reviews. 2007;20(1):133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tay ST, Na SL, Chong J. Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. Journal of medical microbiology. 2009;58(2):185–191. doi: 10.1099/jmm.0.004242-0. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn DM, Mukherjee PK, Clark TA, Pujol C, Chandra J, Hajjeh RA, Warnock DW, Soll DR, Ghannoum MA. Candida parapsilosis characterization in an outbreak setting. Emerging infectious diseases. 2004;10(6):1074–1081. doi: 10.3201/eid1006.030873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ng KP, Colombo A, Finquelievich J, Barnes R, Wadula J Global Antifungal Surveillance Group. Geographic and temporal trends in isolation and antifungal susceptibility of Candida parapsilosis:a global assessment from the ARTEMIS DISK Antifungal Surveillance Program 2001 to 2005. Journal of clinical microbiology. 2008;46(3):842–849. doi: 10.1128/JCM.02122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Asbeck E, Clemons KV, Martinez M, Tong A-J, Stevens DA. Significant differences in drug susceptibility among species in the Candida parapsilosis group. Diagnostic microbiology and infectious disease. 2008;62(1):106–9. doi: 10.1016/j.diagmicrobio.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Morio F, Pagniez F, Lacroix C, Miegeville M, Le Pape P. Amino acid substitutions in the Candida albicans sterol Δ5, 6-desaturase (Erg3p) confer azole resistance:characterization of two novel mutants with impaired virulence. Journal of antimicrobial chemotherapy. 2012;67(9):2131–2138. doi: 10.1093/jac/dks186. [DOI] [PubMed] [Google Scholar]
- 23.Berkow EL, Manigaba K, Parker JE, Barker KS, Kelly SL, Rogers PD. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrobial agents and chemotherapy. 2015;59(10):5942–5950. doi: 10.1128/AAC.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman NT, Pham CD, Cleveland AA, Lockhart SR. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a US surveillance system. Antimicrobial agents and chemotherapy. 2015;59(2):1030–1037. doi: 10.1128/AAC.04613-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du W, Coaker M, Sobel JD, Akins RA. Shuttle vectors for Candida albicans:control of plasmid copy number and elevated expression of cloned genes. Current genetics. 2004;45(6):390–398. doi: 10.1007/s00294-004-0499-3. [DOI] [PubMed] [Google Scholar]
- 26.Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryotic Cell. 2004;3(6):1391–1397. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiology and molecular biology reviews. 2011;75(2):213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lees N, Skaggs B, Kirsch D, Bard M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—a review. Lipids. 1995;30(3):221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]


