Abstract
Single-stranded regions (gaps) in nucleosomal DNA interfere with action of the RSC chromatin-remodeling complex, monitored by exposure of restriction endonuclease cutting sites. Single-strand breaks (nicks) in the DNA, by contrast, have no effect. Gaps on one side of the cutting site are inhibitory, but gaps on the other side are not. A gap >100 bp from the cutting site is as effective as a gap <20 bp from the site. These findings suggest a remodeling process involving bending, but not twisting, of the DNA and further point to the propagation of a bent region (loop or bulge) from one end of the nucleosome to the other.
Keywords: nucleosome, restriction endonuclease, RSC
Chromatin-remodeling complexes typified by the SWI/SNF and RSC complexes of yeast perturb the structure of nucleosomes in an ATP-dependent manner (1, 2). The perturbation is revealed by exposure of the nucleosomal DNA to interacting proteins, such as restriction enzymes and nonspecific nucleases (1, 3, 4). The perturbation is transient: after removal of the remodeling complex or of ATP, the DNA is again protected from nuclease attack, although the location of the nucleosome may have changed as a result of sliding (movement of the histone octamer along the DNA) (5, 6) or translocation (movement of the histone octamer to another DNA molecule) (7).
Two types of perturbation have been considered, DNA twisting and DNA bending. In either case, the perturbation is believed to be local and to propagate through the nucleosome from one end of the DNA to the other. The possibility of local twisting was demonstrated by the crystal structure of a nucleosome, revealing 12 bp of overwound DNA within an otherwise almost uniform superhelix (8). The SWI/SNF complex alters the local twist of a naked DNA fragment, as shown by conversion of a duplex containing a palindromic sequence to a cruciform structure (9). Evidence for a role of twisting in chromatin remodeling has come from studies of nucleosomes containing nicked DNA. Nicks will impede the propagation of torsional strain in DNA, because they enable the swiveling of one duplex region with respect to another. In one experiment, nicks caused a 2- to 3-fold reduction in the rate of restriction enzyme cleavage of nucleosomal DNA, depending on RSC and ATP (10). In another case, nicks caused a 20-30% reduction in the extent of restriction enzyme cleavage, depending on the SWI/SNF complex and ATP (11). These findings have led to the conclusion that DNA twisting is not required for chromatin remodeling by RSC and SWI/SNF but can enhance its efficiency.
The alternative view of chromatin remodeling, involving DNA bending, originated in studies of exonuclease III digestion of nucleosomes and the transcription of nucleosomal templates. Exonuclease III degrades a strand in a duplex from the 3′ end and was shown to invade the core particle of the nucleosome, with pauses every 10 residues, corresponding to the periodicity of the double helix (12). This pattern of digestion was attributed to the occasional dissociation of a turn of the double helix from an end (entry/exit site) of the nucleosome, followed by the rapid digestion of the dissociated DNA. RNA polymerases were also shown to invade a nucleosome, and this capacity was similarly attributed to the dissociation of DNA from the ends, with the enzyme advancing along the dissociated region (13). After polymerase passage, the location of the nucleosome was shifted on the DNA in a direction opposite to that of transcription (14). This rearward shift was attributed to binding of DNA from the region behind the nucleosome to the surface of the histone octamer exposed by transcription; the resulting loop or bulge of DNA, protruding from the nucleosome and containing the polymerase, would be translocated by transcription. An analogous mechanism was suggested to explain the exposure of DNA by chromatin-remodeling complexes. It was imagined that a remodeling complex might produce a bulge of exposed DNA at one end of the nucleosome that travels, either spontaneously or by translocation of the remodeling complex, to the other end.
Several observations are consistent with a bulge propagation mechanism. RSC interacts with the ends of the DNA in a nucleosome and releases 60-80 base pairs from the histone octamer surface (6). The product of SWI/SNF action on a nucleosome may contain a stable loop, due to sliding of the octamer beyond one end of the DNA and binding of the other end of the DNA back to the exposed octamer surface (15). Theoretical considerations support a bulge propagation mechanism as well (16).
Despite these findings, the occurrence of a transient DNA bulge during remodeling remains hypothetical. To put the idea to a test, we sought to introduce an impediment to bulge propagation. We report here on the use of DNA with a gap in one strand for this purpose. A degree of stiffness is required for the propagation of bending energy. A bulge traversing a DNA molecule would be expected to collapse at a gap, due to the flexibility of the single-stranded region.
Materials and Methods
DNA and Nucleosome Preparation. Unnicked 32P-labeled 160-bp DNA was made by PCR from the 160-bp core DNA fragment of the Xenopus laevis 5S rRNA gene (6, 17), with the primers 5′-CCCGGGTATGCTGCTTGACTTCGGTGATCG-3′ (160 end1) and 5′-GAATTCCAACGAATAACTTCCAGGGATTTA-3′ (160 end2).
An unnicked 32P-labeled 160-residue strand (lower strand in Fig. 1) was made by PCR with a 1,000-fold excess of the 160 end2 primer over the 160 end1 primer. The single strand was separated by electrophoresis in a 5% polyacrylamide gel; it migrated slightly more slowly than the 160-bp double-stranded fragment and was recovered from the gel by electroelution.
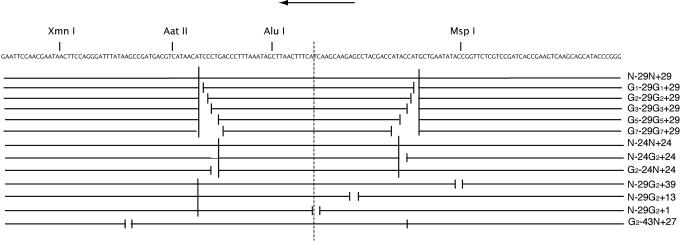
Fig. 1.
Nicked and gapped nucleosomal DNAs. The locations of nicks and gaps in the 160-bp DNAs used in this work are indicated schematically beneath the sequence of the DNA. The DNAs are named according to the presence of a nick (N) or gap (G), followed by the location (distance from the dyad axis of the nucleosome, indicated by a vertical dashed line). Nicks and gaps are on the strand whose sequence is shown, and the size of the gap is indicated by a subscript. An arrow at the top indicates the downstream direction (the inferred direction of chromatin remodeling by RSC in this case).
Nicked and gapped DNAs were made by annealing oligonucleotides, as indicated (Fig. 1), to the unnicked 32P-labeled 160-residue lower strand. All DNAs were equimolar in the annealing reaction, and the product was purified with the use of a Minielute PCR clean-up kit (Qiagen, Valencia, CA).
Nucleosomes were prepared with the use of rat-liver histones, as described (17). Peak maltose gradient fractions of nucleosome monomer typically contained ≈0.2 ng of DNA/μl and were concentrated where necessary by centrifugation in a Microcon 30 (Amicon).
Chromatin Remodeling and Restriction Endonuclease Digestion. Reaction mixtures contained 0.1 ng of nucleosomes, 0.5 μg of RSC prepared as described (2, 17), and 20 u restriction endonuclease. For AluI and AatII, the mixtures contained 15 mM Hepes, pH 7.5, 50 mM potassium acetate, 6 mM MgCl2, 75 μg/ml BSA, 1 mM DTT, with ATP added or not, and reactions were for 10, 20, 40, and 60 min at 30°C. For XmnI, the mixtures contained 15 mM Hepes, pH 7.5, 20 mM potassium acetate, 3 mM MgCl2, 75 μg/ml BSA, with ATP added or not, and reactions were for 5, 10, 15, and 20 min at 30°C. Reactions were stopped by the addition of SDS and proteinase K, followed by incubation for 30 min at 42°C, extraction of the DNA, and electrophoresis in a 7.5% polyacrylamide gel in TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3). Detection and quantitation were performed with a PhosphorImager, and data were fit to linear or simple exponential curves with the data analysis package of excel. Only experiments yielding fits with R2 values of 0.9 or better were included in Results.
Results
Nucleosomes were formed on a series of DNAs with the nucleosome-positioning sequence of the X. laevis 5S rRNA gene (1). In all cases, one strand was 160 residues in length, whereas the opposite strand contained two nicks, a nick and a gap, two gaps, or no nicks or gaps (Fig. 1). Nicks and gaps had no discernable effect on the assembly of nucleosomes (as judged from the relative amounts of nucleosomal and free DNA, revealed by gradient sedimentation; data not shown) or on nucleosome positioning (revealed by the pattern of DNase I digestion of end-labeled DNA; data not shown). In measurements of bending flexibility by ligation of a 68-residue DNA to a circular form, nicks had little effect, whereas a two-residue gap enhanced the rate >2-fold (data not shown).
Action of RSC on nucleosomes was assessed by the rate of digestion with a set of restriction endonucleases, with cleavage sites at distances from the center (dyad) of the nucleosomal DNA, as follows (Fig. 1): AluI, -10 bp; AatII, -36 bp; and XmnI, -65 bp. The rate of cleavage in the absence of RSC increased with distance from the dyad (Table 1), consistent with previous studies showing a greater accessibility to restriction enzymes closer to the ends of nucleosomal DNA (18). There was, however, a pronounced asymmetry with respect to the dyad, because cutting by MspIat +39 was 14 times faster than cutting by XmnI at -65. None of the nicks or gaps had a measurable effect on cutting rates in the absence of RSC, except when located within the actual restriction enzyme recognition site (gap at +39 for MspI).
Table 1. Apparent first-order rate constants for restriction enzyme cleavage of nucleosomal DNA.
| Enzyme | Cutting rate constant, k (% DNA cut per min) |
|---|---|
| XmnI | 1.68 |
| AatII | 0.0125 |
| AluI | 0.0074 |
| MspI | 24 |
Nicks Have No Effect on Chromatin Remodeling by RSC. Rates of restriction enzyme digestion in the presence of RSC and ATP were determined under conditions of saturating RSC and limiting restriction enzyme concentration. First-order rate constants were derived from initial rates of appearance of cut fragments (<5% complete digestion) for AluI (Fig. 2) and AatII and from the exponential decay of the uncut DNA for XmnI (up to 80% digestion). All measurements were made at least twice, and in some cases, such as the comparison of AluI digestion rates of nicked and unnicked nucleosomal DNAs, four or more measurements were made on multiple nucleosome preparations.
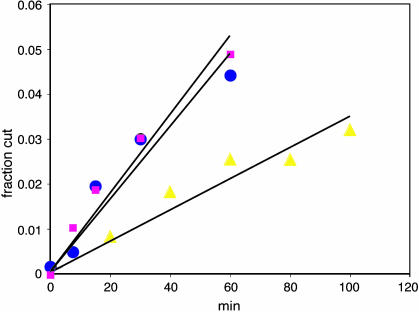
Fig. 2.
Time course of AluI digestion of unnicked, nicked, and gapped DNAs. The extent of AluI digestion, determined from the amount of 70-bp fragment in a gel, is plotted as a function of time. The DNAs were unnicked (blue circles), nicked (N-29N + 29, magenta squares) and gapped (N-29G2 + 13, yellow triangles).
RSC and ATP increased AluI digestion rate 4- to 5-fold (Fig. 2). The increase in cutting rate constant (difference between rate constants measured in the presence and absence of ATP), denoted Δk, was the same for nicked and unnicked DNAs (Table 2). Any small differences in Δk values between these DNAs or between DNAs with nicks in different locations were less than the SDs of the measurements (Table 2). The nicks were located on both sides of the AluI site, insulating it from twisting strain introduced at either end of the nucleosome. Chromatin remodeling by RSC is therefore unlikely to be due to twisting strain originating at or near the ends and propagating through the nucleosome.
Table 2. Increase in rate of AluI cutting of nicked and unnicked nucleosomal DNAs due to RSC and ATP.
| Position of nicks | Increase in cutting rate constant, Δk (% DNA cut per min) | No. of observations | SD |
|---|---|---|---|
| None | 0.027 | 3 | 0.007 |
| -24, +24 | 0.031 | 4 | 0.004 |
| -29, +29 | 0.031 | 3 | 0.004 |
Gaps Interfere with Chromatin Remodeling by RSC. Gaps of one, two, three, and five residues were introduced on both sides of the AluI site (Fig. 1). A one-residue gap had little effect on enhancement of the digestion rate by RSC and ATP (an increase in cutting-rate constant, Δk, of 0.026% min-1 for G1-29G1 + 29, compared with a Δk of 0.031 for the nicked control N-29N + 29). A two-residue gap had a significant interfering effect (Δk of only 0.010 for G2-29G2 + 29). Increasing the size of the gap to three, five, or seven residues did not increase the magnitude of the interfering effect (data not shown).
To our surprise, a single gap at +24 (N-24G2 + 24, Δk = 0.015) was as inhibitory as a pair of gaps at +/-24 (G2-24G2 + 24, Δk = 0.017, compared with a Δk of 0.031 for the nicked control N-24N + 24). This suggested that the gap at -24 was inconsequential, and indeed, a single gap in this location had no effect (G2-24N + 24, Δk = 0.036). Experiments with nucleosomes containing single gaps at other locations (Fig. 3A) led to two conclusions. First, only gaps on the upstream side of the AluI site (to the right of the site in Fig. 1) diminished the cutting rate in the presence of RSC and ATP; gaps on the downstream side did not. Second, all upstream gaps were equally effective, regardless of distance from the AluI site. These findings are most consistent with a remodeling process that traverses the nucleosome from the upstream end of the DNA in the downstream direction.
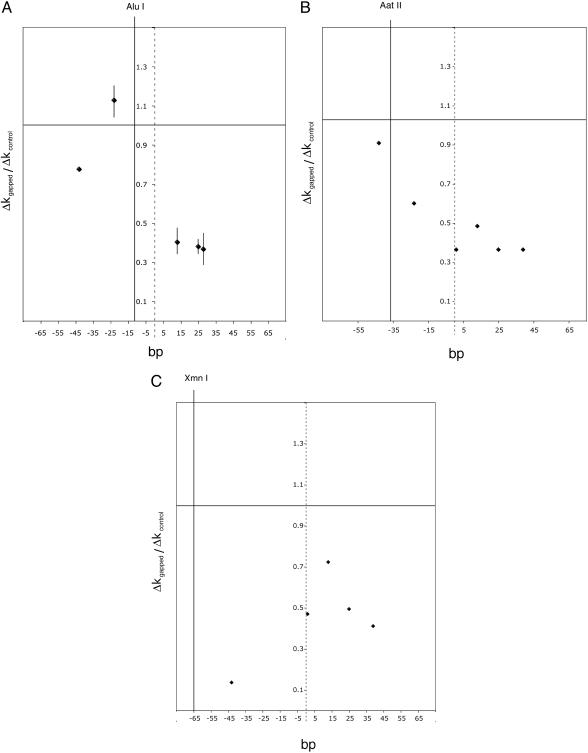
Fig. 3.
Interfering effect of a gap in nucleosomal DNA on remodeling by RSC and ATP, as a function of location of the gap. The increase, Δkgapped, due to RSC and ATP, in the rate constant of cutting gapped nucleosomal DNA (Δk = k+ATP - k-ATP), divided by the Δkcontrol value for unnicked, ungapped DNA, is plotted on the ordinate, and the location of the gap is plotted on the abscissa. (A), AluI; (B) AatII; (C) XmnI. The locations of the cutting site and of the dyad axis of the nucleosome are indicated by solid and dashed vertical lines. All data are averages of at least three independent determinations, and error bars (SDs) are shown in cases where four or more determinations were made.
Range of the Chromatin-Remodeling Process. To assess the generality of the results from AluI digestion and determine the range of the inhibition by a gap, we performed a similar analysis of AatII and XmnI digestion (Fig. 3 B and C). The results were essentially the same: gaps upstream of the restriction site interfered with remodeling, whereas gaps downstream did not. The magnitude of the inhibition by a gap was comparable to that observed for AluI digestion. It is noteworthy that a gap at -24 between the AatII and AluI sites (G2-24N + 24), which did not affect the rate of AluI digestion, did inhibit AatII digestion (Fig. 3B). Similarly, a gap at -43 between the XmnI and AatII sites (G2-43N + 27), which did not affect the rate of AatII digestion, did inhibit XmnI digestion (Fig. 3C). It is furthermore noteworthy that cutting at the XmnIsite at position -65 was inhibited by a gap at position +39 (N-29G2 + 39), a distance 114 bp away (Fig. 3C).
Discussion
The lack of effect of nicks in nucleosomal DNA upon the rate of remodeling by RSC and ATP argues against a primary role for DNA twisting in the remodeling process. RSC might, indeed, cause a degree of untwisting, as reported for SWI/SNF and other remodeling complexes, but it must perturb the structure in other ways as well. Previous studies have shown some diminution in the rate and extent of remodeling of nucleosomes containing nicked DNA (10, 11). Although we cannot explain the discrepancy, we do find a considerable variation in the results of rate measurements (SDs of up to 32% in the data reported here), and our analysis is the most extensive so far made (23 determinations on four nicked and unnicked nucleosome preparations).
Our results on the remodeling of nucleosomes containing gapped DNA not only are consistent with the involvement of bending strain, and thus of bulge formation, but also give evidence of bulge propagation. A gap on one side of the restriction site was inhibitory, whereas a gap on the other side was not, suggesting a remodeling process coming from one side. The location of the gap on that side could be varied, consistent with a remodeling process starting at one end of the DNA and traversing the nucleosome. Finally, the same end of the nucleosomal DNA was more accessible to restriction enzyme attack in the absence of a remodeling complex, possibly due to a greater frequency of dissociation from the nucleosome, which might explain why remodeling tends to start at that end.
The involvement of bending strain in remodeling implies a bulge in the DNA but says nothing of the size, mechanisms of formation and translocation, or other aspects of the bulge propagation process. A minimal size in the range of 50-100 bp might be expected from the persistence length of B form DNA and the minimal size for circle formation (19). A bulge of this size would almost certainly suffice for restriction enzyme accessibility and would likely cause nucleosome sliding as well, in a manner analogous to loop propagation in transcription.
ISWI complexes, which form a family distinct from SWI/SNF and RSC, also promote nucleosome sliding but do not cause transient accessibility to restriction enzyme attack. Although remodeling by these complexes had been suggested to involve twisting of the DNA, nicks proved not to impede the remodeling, and a nick immediately adjacent to a nucleosome could even enhance the process (20). It was suggested that a nick could facilitate the formation of a small loop, insufficient in size for restriction enzyme access but able to propagate and promote nucleosome sliding.
Acknowledgments
This research was supported by National Institutes of Health Grant GM36659 (to R.D.K.).
References
- 1.Cote, J., Quinn, J., Workman, J. L. & Peterson, C. L. (1994) Science 265, 53-60. [DOI] [PubMed] [Google Scholar]
- 2.Cairns, B. R., Lorch, Y., Li, Y., Zhang, M., Lacomis, L., Erdjument-Bromage, H., Tempst, P., Du, J., Laurent, B. & Kornberg, R. D. (1996) Cell 87, 1249-1260. [DOI] [PubMed] [Google Scholar]
- 3.Imbalzano, A. N., Kwon, H., Green, M. R. & Kingston, R. E. (1994) Nature 370, 481-485. [DOI] [PubMed] [Google Scholar]
- 4.Lorch, Y., Cairns, B. R., Zhang, M. & Kornberg, R. D. (1998) Cell 94, 29-34. [DOI] [PubMed] [Google Scholar]
- 5.Langst, G., Bonte, E. J., Corona, D. & Becker, P. B. (1999) Cell 97, 843-852. [DOI] [PubMed] [Google Scholar]
- 6.Lorch, Y., Zhang, M. & Kornberg, R. D. (2001) Mol. Cell 7, 89-95. [DOI] [PubMed] [Google Scholar]
- 7.Lorch, Y., Zhang, M. & Kornberg, R. D. (1999) Cell 96, 389-392. [DOI] [PubMed] [Google Scholar]
- 8.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251-260. [DOI] [PubMed] [Google Scholar]
- 9.Havas, K., Flaus, A., Phelan, M., Kingston, R., Wade, P. A., Lilley, D. M. & Owen-Hughes, T. (2000) Cell 103, 1133-1142. [DOI] [PubMed] [Google Scholar]
- 10.Saha, A., Wittmeyer, J. & Cairns, B. R. (2002) Genes Dev. 16, 2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoyagi, S. & Hayes, J. J. (2002) Mol. Cell. Biol. 22, 7484-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prunell, A. & Kornberg, R. D. (1978) Cold Spring Harbor Symp. Quant. Biol. 42, 103-108. [DOI] [PubMed] [Google Scholar]
- 13.Lorch, Y., LaPointe, J. W. & Kornberg, R. D. (1987) Cell 49, 203-210. [DOI] [PubMed] [Google Scholar]
- 14.Studitsky, V. M., Clark, D. J. & Felsenfeld, G. (1994) Cell 76, 371-382. [DOI] [PubMed] [Google Scholar]
- 15.Kassabov, S. R., Zhang, B., Persinger, J. & Bartholomew, B. (2003) Mol. Cell 11, 391-403. [DOI] [PubMed] [Google Scholar]
- 16.Schiessel, H., Widom, J., Bruinsma, R. F. & Gelbart, W. M. (2001) Phys. Rev. Lett. 86, 4414-4417. [DOI] [PubMed] [Google Scholar]
- 17.Lorch, Y. & Kornberg, R. D. (2003) Methods Enzymol. 377, 316-322. [DOI] [PubMed] [Google Scholar]
- 18.Polach, K. J. & Widom, J. (1995) J. Mol. Biol. 254, 130-149. [DOI] [PubMed] [Google Scholar]
- 19.Cloutier, T. E. & Widom, J. (2004) Mol. Cell 14, 355-362. [DOI] [PubMed] [Google Scholar]
- 20.Langst, G. & Becker, P. B. (2001) Mol. Cell 8, 1085-1092. [DOI] [PubMed] [Google Scholar]