Abstract
To better understand the function role of the melon CmLOX18 gene in the biosynthesis of C6 volatiles during fruit ripening, we biochemically characterized CmLOX18 and identified its subcellular localization in transgenic tomato plants. Heterologous expression in yeast cells showed that the molecular weight of the CmLOX18 protein was identical to that predicted, and that this enzyme possesseed lipoxygenase activity. Linoleic acid was demonstrated to be the preferred substrate for the purified recombinant CmLOX18 protein, which exhibited optimal catalytic activity at pH 4.5 and 30 °C. Chromatogram analysis of the reaction product indicated that the CmLOX18 protein exhibited positional specificity, as evidenced by its release of only a C-13 oxidized product. Subcellular localization analysis by transient expression in Arabidopsis protoplasts showed that CmLOX18 was localized to non-chloroplast organelles. When the CmLOX18 gene was transgenically expressed in tomato via Agrobacterium tumefaciens-mediated transformation, it was shown to enhance expression levels of the tomato hydroperoxide lyase gene LeHPL, whereas the expression levels of six TomLox genes were little changed. Furthermore, transgenic tomato fruits exhibited increases in the content of the C6 volatiles, namely hexanal, (Z)-3-hexanal, and (Z)-3-hexen-1-ol, indicating that CmLOX18 probably plays an important role in the synthesis of C6 compounds in fruits.
Introduction
Aroma volatiles are vital characteristic that determine the quality and commercial value of fruits. Different fruits have different suites of volatile compounds, which contribute to their unique aromas1. For example, in apple, strawberry, and melon, esters are the main flavour compounds that impart a fruity flavour2–4. Previous studies have indicated that degradation of linoleic and linolenic acids contribute to the synthesis of esters, containing 2, 4, and 6 carbon straight chains5, 6. Lipoxygenase (LOX) enzymes might contribute to the breakdown of long-chain fatty acids to C6 aldehydes by hydroperoxide lyases (HPL), which were further converted to alcohols by aldehyde dehydrogenase (ADH), followed by the conversion of alcohols to esters by alcohol acetyl transferase (AAT)7–9.
Plant LOX enzymes, which are encoded by a multi-gene family, are 95–100 kDa monomeric proteins containing an N-terminal β-barrel domain (25–30 kDa) that is probably involved in membrane or protein interactions, and a C-terminal α-helix-rich domain (55–65 kDa) containing the catalytic site10. On the basis of their positional specificity, LOX enzymes can be classified into three types: 9-LOXs, 13-LOXs, and 9/13-LOXs11. 9-LOX and 13-LOX enzymes can produce 9- and 13-hydroperoxy products, respectively, whereas 9/13-LOXs can produce both 9- and 13-hydroperoxy products12, 13. Heterologous expression of plant LOXs in Escherichia coli or yeast has enabled elucidation of the positional specificity for substrate oxygenation. A few of the 9/13-LOXs, such as maize ZmLOX1 14, pea PsLOXN2 15, rice OsLOX1 16, and tea CsLOX1 17, have been shown to confer dual positional specificity since they release both C-9 and C-13 oxidized products in equal proportions.
Different LOX isoforms initiated the synthesis of functionally diverse products, and thus LOXs have been regarded to play different roles in plant development and ripening18. 9-LOXs were implicated in diverse physiological processes in plants, such as growth and development -19, 20, defence21, 22, and resistance to pathogens23. In contrast, 13-LOXs have been reported to produce the ‘green note’ compounds in plant tissues24, and also played an important role in wound- and herbivore-induced jasmonate (JA) accumulation25–27. Furthermore, the subcellular location of a protein is very important because it is closely related to that proteins biological function28. LOX enzymes have been found in various organelles of plants, which could determine the specific function of the different LOXs29. For example, tomato 13-lipoxygenase TomloxC was localized in the chloroplast, where it was specifically involved in the generation of fatty acid-derived straight-chain five- (C5) and six- (C6) carbon flavour compounds (aldehydes and alcohols) in both fruit and leaves30, 31. In potato, LOX H1 and LOX H3 were localized in both the chloroplast stroma and thylakoids, and both of these enzymes were involved in the synthesis of JA and C-6 aldehydes32, 33. Other LOX proteins, for example, cucumber LOX, were found in microsomal membranes34, whereas soybean LOX was found in both the cytosol and vacuole35, 36.
In oriental melon, the genes CmLOX01, CmLOX03, and CmLOX18 were expressed during fruit ripening37. The 13-LOXs CmLOX03 and CmLOX18 were potentially important candidate genes involved in straight-chain ester production in melon; however, further studies are needed to verify this supposition38. In the present study, we transformed the 13-lipoxygenase CmLOX18 gene into tomato and analysed its enzymatic characteristics by using the recombinant protein expressed in yeast. Our results showed that CmLOX18 was a member of the 13-LOX family and was localized in non-chloroplast organelles. Transgenic tomato plants over-expressing CmLOX18 exhibited a significant increase in the biosynthesis of C6 compounds; however, the biosynthesis of C5 compounds were not changed. In addition, the expression levels of members of the Tomlox gene family were not altered in the transgenic plants. Therefore, we concluded that a critical role for CmLOX18 was the formation of C6 compounds, such as hexanal, (Z)-3-hexenal, and (Z)-3-hexenol, in fruit, and that it might be involved in straight-chain ester production in melon.
Results
Biochemical activity and characterization of recombinant yeast CmLOX18
The ORF of CmLOX18 was cloned into a vector pYES2.1 for expression in yeast to characterize the enzymatic activity of the encoded protein. The yeast containing the recombinant plasmid grew in selective medium (SC-U) with 2% galactose as inducer of the recombinant protein expression. The recombinant lipoxygenase encoded by CmLOX18 was expressed and purified. Heterologously expressed CmLOX18 was identified by western blot analysis using an anti-His-tag antibody. Western blotting and SDS–PAGE analysis revealed one unique band of approximately 100 kDa protein and were in good agreement with the predicted molecular mass of CmLOX18 (approximately 102.1 kDa) (Fig. 1).
Figure 1.
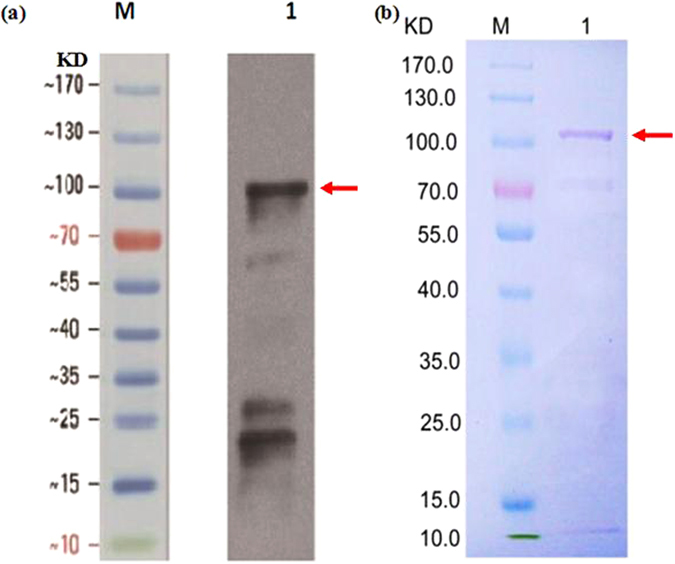
Analysis of recombinant CmLOX18 (a) Western blot analysis. Total proteins (20 μg) were isolated from yeast cells expressing CmLXO18 which was harvested at 24 hours induction and separated on 12% SDS/PAGE gel, then transferred to a PVD membrane and LOX detected with anti-His antibody. (b) SDS-PAGE analysis of the recombinant CmLOX18. The proteins were stained with Coomassie brilliant blue R-250. M: Positions of marker protein. 1: SDS-PAGE of the purified CmLOX18 extracted from pYES2/lox transformed yeast cultures (20 μg). The expressed CmLOX18 was indicated by red arrows.
Biochemical activity of the purified recombinant CmLOX18 was investigated by measuring the increase in A234 using linoleic acid as substrates. To optimum pH was determined by varying in the pH values of the reaction buffers from 3.0 to 9.0. At 25 °C, the optimum pH turned out to be 4.5 since CmLOX18 showed the highest activity at that pH. By contrast, CmLOX18 activity decreased by 32% at pH 3.0 and by 82% at pH 7.0, respectively, and was almost completely deactivated at pH 8.0–9.0 (Fig. 2a). To determine the optimum temperature for recombinant CmLOX18, enzyme activity was measured over a range of temperatures (20 °C–45 °C). Maximal activities for recombinant protein were observed at 30 °C, while the activity of the protein exhibited the lowest activity at 20 °C (Fig. 2b).
Figure 2.
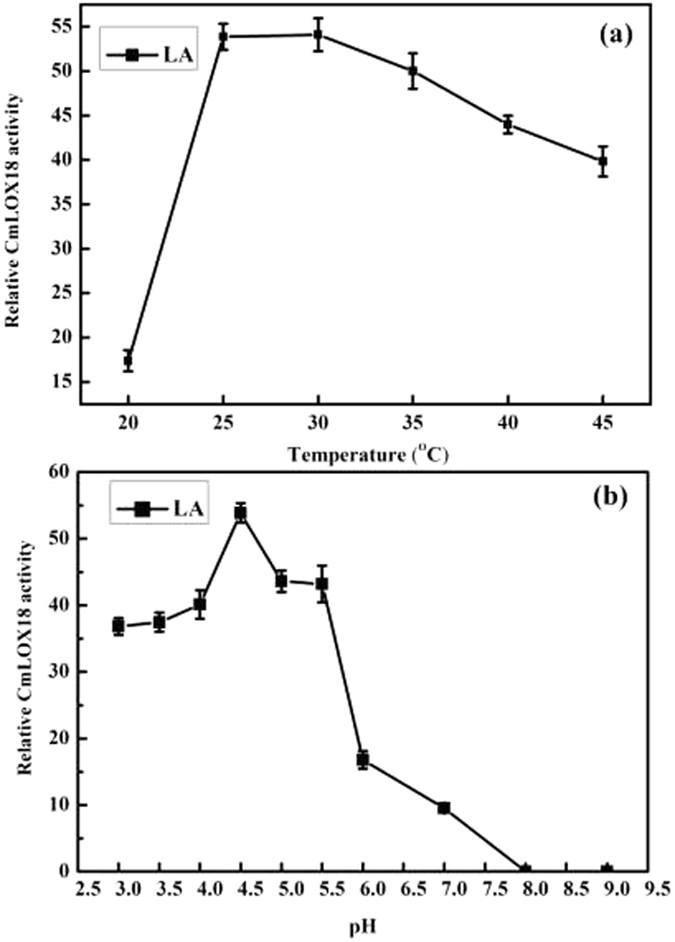
The optimum temperature (a) and Ph (b) for recombinant CmLOX18. The effects of pH and temperature on the enzymatic activity of recombinant CmLOX18 were determined using linoleic acid (LA) as substrates. The recombinant CmLOX18 proteins displayed the highest catalytic activity at pH 4.5 (a) respectively. The optimum temperatures for CmLOX18 were observed at 30 °C (b), respectively. The maximum activity was estimated as 100%. Means ± SD were obtained from three independent measurements.
The kinetic parameters were analysed using linoleic and linolenic acids as substrates (Fig. S2). The recombinant CmLOX18 showed 3.12-fold higher K m value for linoleic acid (126.9 μM) than for linolenic acid (40.72 μM). Comparison of the V max values showed that CmLOX18 oxidized linoleic acid approximately 4-fold faster than linolenic acid. The k cat/K m values of recombinant CmLOX18 for linoleic acid and linolenic acid were 34.54 and 26.96 s−1 μM−1, respectively (Table 1). These results indicated that linoleic acid was clearly the preferred substrate for the recombinant CmLOX18.
Table 1.
Kinetic parameters of purified recombinant CmLOX18.
| Substrate | Km (μM) | Vmax (μmol s−1) | Kcat (s−1) | Kcat/Km (s−1 μM−1) |
|---|---|---|---|---|
| Linoleic acid | 126.9 ± 18.6 | 32.87 ± 3.1 | 4383 ± 295 | 34.54 ± 2.7 |
| Linolenic acid | 40.72 ± 2.65 | 8.19 ± 0.8 | 1098 ± 78 | 26.96 ± 2.2 |
Kinetic parameters were calculated using a Michaelis-Menten plot analysis program. Values represent the mean ± SD of 3 independent replicates. Kcat is defined to equal Vma/Et. Et is total enzyme concentration is molar.
In order to study the positional-specificity of recombinant CmLOX18, we separated the reaction products by SP-HPLC. The retention time of CmLOX18 product was consistent with authentic standards of 13-HPOD (Fig. 3a), which indicated that CmLOX18 was 13-LOX. The stereochemistry of the reaction products was analysed chiral-phase HPLC.13-HPOD was predominantly in the S configuration, indicating that it had been derived from the activity of a specific enzyme (Fig. 3b). The result was consistent with the positional specificity rule of plant LOXs, in which S/CF motif at the active site indicated the LOXs were 13-LOXs.
Figure 3.
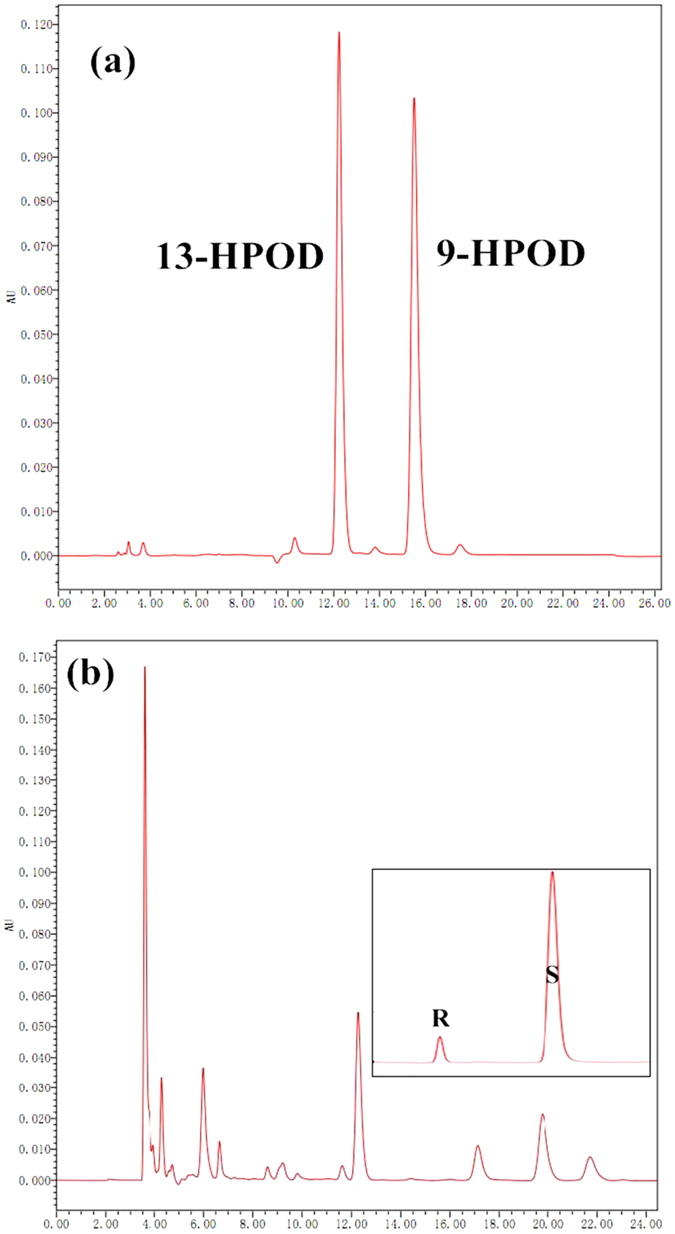
Determination of positional specificity of the recombinant CmLOX18. (a) The retention time of the isomers produced by soybean LOX1 compared with authentic standards of 13-HPOD and 9-HPOD. (b) SP-HPLC analysis of the reaction mixture catalysed by recombinant CmLOX18. Boxes: chiral-phase HPLC showing the enantiomer composition of 13-HPOD.
Subcellular localization of CmLOX18 protein
To confirm the subcellular location of CmLOX18, a green fluorescent protein (GFP) reporter gene was fused in-frame to the N-terminus of CmLOX18 and transiently transformed into Arabidopsis protoplasts under the control of the 35 S promoter. As shown in Fig. 4, CmLOX18 was not observed in chloroplasts, which was not in accord with the result of the bioinformatics analysis, and the results showed that CmLOX18 was localized in non-chloroplast organelles.
Figure 4.
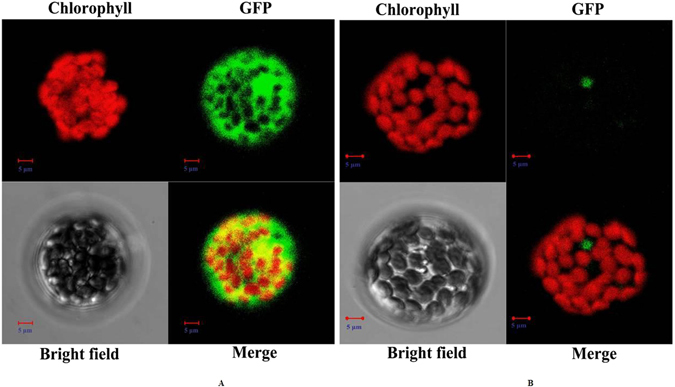
Subcellular localization of oriental melon CmLOX18 in Arabidopsis protoplasts. Arabidopsis protoplasts were transformed by the vectors containing the 35S-GFP (A) and 35S-CmLOX18-GFP (B), respectively. Chlorophyll and GFP fluorescence were examined using Zeiss LSM510 confocal microscope excited with a 488-nm Ar laser line. Merge is the computed overlay of the two fluorescence images. Reference bar is 5 μm.
The identification of transgenic tomato plants over-expressing CmLOX18
To examine the physiological role of CmLOX18, the full-length cDNA of CmLOX18 was cloned into pB7FWG2.0 under the control of the 35 S promoter (Fig. S1) and transformed into ‘Zhongshu 6’ using the Agrobacterium method. The construct, encoding a carboxy-terminal fusion of CmLOX18 onto EGFP under the control of a cauliflower mosaic virus 35 S promoter, contained bialaphos acetyltransferase gene (Bar) and enhanced green fluorescent protein gene (Egfp). Southern blot, PCR, Western blot, and fluorescence imaging technique indicated that three T0 transgenic plants were obtained (Fig. 5). Three independent transgenic lines (CM18-01, 03, and 04) which contained one copy identified by Southern blot were selected.
Figure 5.
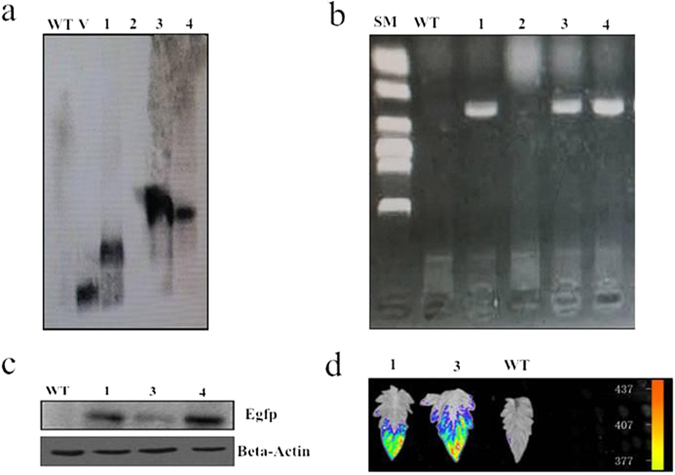
Molecular analysis of T0 transegenic tomato liners expressing CmLOX18. (a) Southern blot of wild-type control and four T0 transgenic tomato lines expression CmLOX18. Genomic DNA was prepared from young leaf material from wild-type control and transgenic plants: 1, 2, and 3 and V (35S-overexpression CmLOX18 construct). The genomic DNA (10 μg/lane) was digested with HindШ and separated in a 0.8% (w/v) agarose gel. Blotted DNA was hybridized to a probe prepared from the bar gene. (b) Detection of CmLOX18 cDNA by PCR analysis. Genomic DNA was extracted from transgenic lines. (c) Western blot. Proteins were extracted and Western blot was carried according to standard procedures using anti-e GFP Mouse Monoclonal antibody. (d) Images of wild-type control and twoT0 transgenic tomato leaves.
In comparison with “Zhongshu 6” tomato, as control plant, which have no expected bands by Southern and PCR, transgenic tomato plants indeed contained the CmLOX18 gene (Fig. 5a and b). Furthermore, western blot (Fig. 5c) and image (Fig. 5d) analysis indicated that Egfp protein was high expressed in lines (CM18-01, 03, and 04). These results indicated that the CmLOX18 gene was successfully transferred into tomato plant.
Real-time qPCR analysis of LOX gene from ripening fruit of wild-type and transgenic plants
Real-time PCR analyses indicated that CmLOX18 transcription were only present in three lines of 35 S::CmLOX18 (CM18-01,03 and 04), which was not detected in the wild-type. The expression level of CmLOX18 mRNA in the fruits of three transgenic lines at the B + 7 stage were about 2-fold higher than that at the breaker stage (Fig. 6a). To find out whether the CmLOX18 gene was involved in catalyzing chain cleavage of hydroperoxides to release C5 and C6 volatiles, real-time quantitative RT-PCR was carried out. Transcript analysis of LeHPL was significantly higher in transgenic fruits than that in wild-type fruits. However, the expression level of LeHPL at the B + 7 stage was lower than that at the breaker in both transgenic and wild-type fruits, which was not consistent with the expression of CmLOX18 (Fig. 6b).
Figure 6.
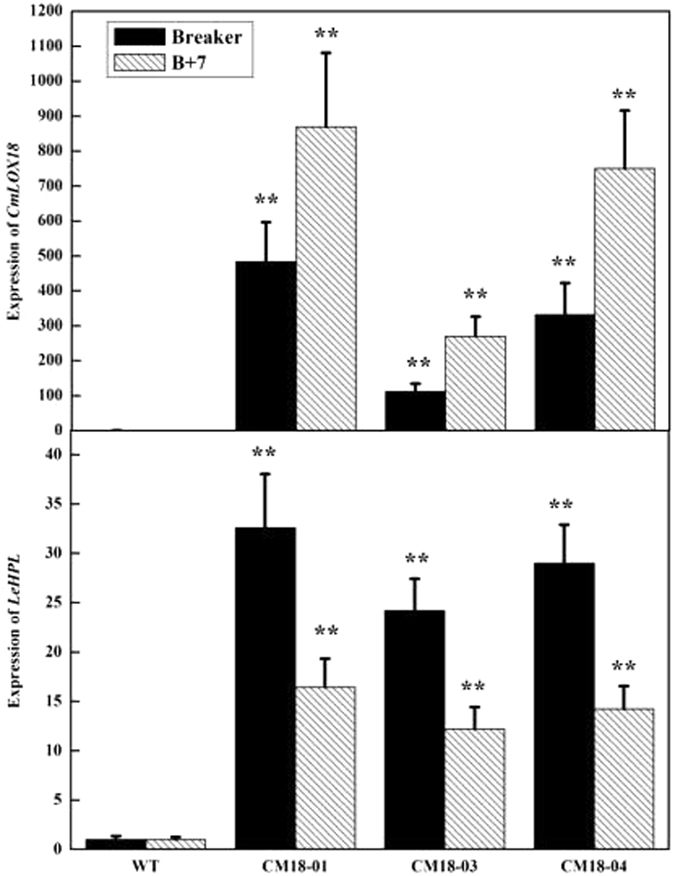
Transcript levels of CmLOX18 and LeHPL in wild type (“Zhongshu 6”) and transgenic (CM18-01, 03 and 04) ripe fruits. Transcripts were quantified by Quantitative PCR. Values are mean ± standard error of three replicates. Significant differences are indicated with asterisks above the bars: *P ≤ 0.05, **P ≤ 0.01.
In order to confirm whether the over-expression of CmLOX18 gene caused significant changes in the mRNA expression levels of six tomato LOXs in the transgenic fruits, we analyzed the expression levels of TomloxA-TomloxF in transgenic and wild-type fruits. The results showed that there was no difference between the expression levels of TomloxA, TomloxB, TomloxC, TomloxD, TomloxE, and TomloxF mRNA in the transgenic and wild-type fruits at the breaker and B + 7. Furthermore, the transcription levels of TomloxA and TomloxC were consistent with the LeHPL in both transgenic and wild-type fruits and higher at the breaker than that at the B + 7. In contrast, transcription levels of TomloxB, TomloxE, and TomloxF were lower at the breaker than at the B + 7, while TomloxD had no difference (Fig. 7). Taken together, the results indicated that the over-expression of CmLOX18 caused significant changes in the expression levels of LeHPL, but there was no change in Tomloxs.
Figure 7.
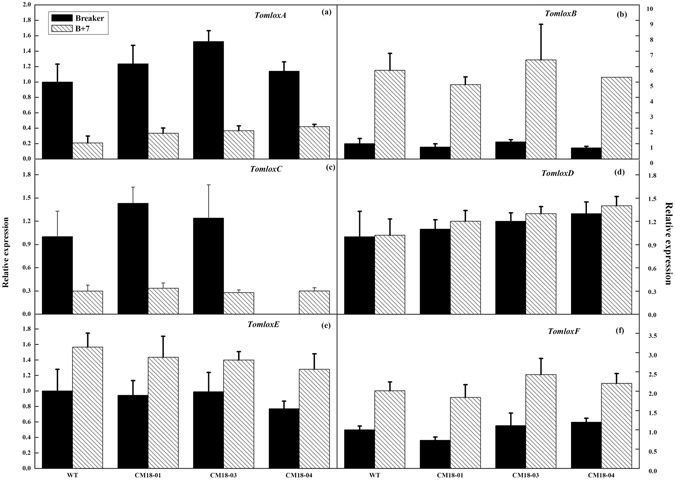
Transcript levels of TomLOXA-F in wild type (“Zhongshu 6”) and transgenic (CM18-01, 03 and 04) ripe fruits. Transcripts were quantified by Quantitative PCR. Values are mean ± standard error of three replicates. Significant differences are indicated with asterisks above the bars: *P ≤ 0.05, **P ≤ 0.01.
C5 and C6 volatiles from the transgenic and wild-type fruits
C5 and C6 volatile compounds in tomato, such as n-hexanal, (Z)-3-hexenal, (E)-2-hexanal, (Z)-3-hexenol, 1-pente-3-ol, 1-pente-3-one, and pentanal which are derived from fatty acids, are known to be formed by 13-TomloxC-HPL pathway30, 31. In order to further identify the role of CmLOX18 in the C5 and C6 volatiles formation, GC-MS was used, and we detected a total of 26 volatile compounds from ripening fruit of wild-type and transgenic tomato at the breaker and B + 7 stages. Furthermore, the contents of C6 volatiles, containing hexanal, (Z)-3-hexenaland (Z)-3-hexen-1-ol were higher in transgenic fruits than that in wild-type fruits at the breaker and B + 7 stages. While the contents of C5 volatiles, including 1-pentanol and 1-pente-3-one, showed similar levels in transgenic and wild-type tomato (Fig. 8 and Table S2).
Figure 8.
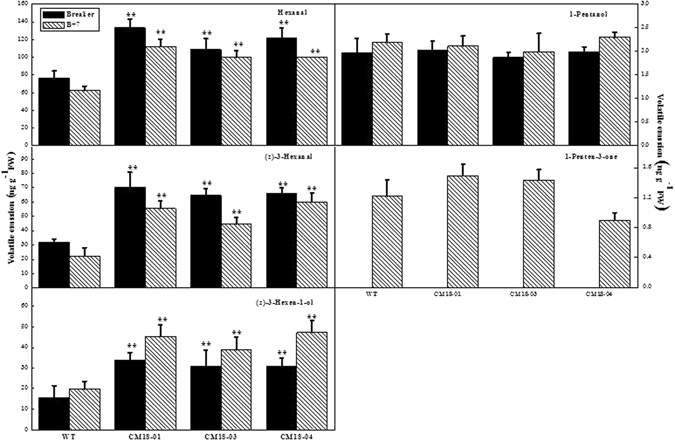
Overexpression CmLOX18 in fruits C6 and C5 volatiles emission in wild type (“Zhongshu 6”) and transgenic (CM18-01, 03 and 04) ripe fruits. Values are mean ± standard error of three replicates. Significant differences are indicated with asterisks above the bars: *P ≤ 0.05, **P ≤ 0.01.
Discussion and Conclusions
On the basis of the position of oxygenation sites in polyunsaturated fatty acids, plant LOXs can be classified as 9- and 13-LOXs. The positional specificity of plant LOXs is dependent on certain conserved sites at which the LOXs containing R/H, R/TF, R/S or R/CF, and R/TV motifs were predicted to be 13-LOX and 9-LOX, respectively11. However, some LOX proteins do not fit the predictive position-specificity models, and can produce both 9- and 13-hydroperoxy products14, 17, 39. In terms of the stereo-specificity of LOXs, the conserved single residue that is important for stereo control is an Ala for S-LOX and a Gly for R-LOX40. In our previous study, phylogenetic analysis grouped CmLOX18 with the characterized members of 13-LOX, which contains an R/CF motif, and was predicted to be a 13S-lipoxygenase37. In the present study, the only detected reaction product of CmLOX18 was a 13-hydroperoxide, which was predominantly in the S configuration, as determined by HPLC analysis. Furthermore, kinetic studies showed that linoleic acid was the preferred substrate for recombinant CmLOX18, which maintained an intact LOX-like activity after being expressed in yeast, and was optimally active at pH 4.5. Similar to CmLOX18, pea PsLOXN2, which was involved in the late mechanisms of host resistance, exhibited normal enzyme activity at pH 4.541. In addition, tea CsLOX1, which was involved in flower development and played a regulatory role in flower senescence, had a lower optimal pH of 3.617. The acidic LOXs from rose and carnation might play important roles in flower senescence via membrane disruption by HPO and free radical action42, 43. In our previous studies, the expression level of CmLOX18 was markedly increased when fruit developed to the climacteric stage, and was up-regulated by ethylene treatment44. These results indicated that the acidic CmLOX18 might be involved in fruit ripening and play a regulatory role in the late development of melon.
Differential LOX functions arise from distinct localizations26. The subcellular location of LOXs might determine when and how precursors were fed into the allene oxide synthase (AOS) or hydroperoxide lyase (HPL) branches of the LOX pathway, followed by the synthesis of products with distinct biological properties27. The possible shift in the allocation of LOX-derived substrates to the HPL and AOS pathways and the substantial overlap of green leaf volatile (GLV)- and JA-regulated plant defence responses demonstrated a crosstalk between the two cascades that clearly exceeded the competition for a common substrate: the 13-hydroperoxide of linolenic acid45–47. In Arabidopsis thaliana, tobacco, and potato, there was co-localization of JA- and HPL-associated enzymes in the chloroplast33, 48. However, in this study, GLV-production CmLOX18 was located in non-chloroplast organelles (Fig. 4), which was not consistent with the previous study. Similar results were also obtained for maize ZmLOX10 26. ZmLOX8 49 and ZmLOX10, which were involved in the AOS and HPL pathways, were located in chloroplasts and non-chloroplast organelles, respectively26. The discrepancy between our findings and those of previous studies could be explained by the fact that the GLV and JA pathways in melon and maize were physically separated from each other.
As has been mentioned previously, 13-lipoxygenases was key enzymes in the production of C5 and C6 compounds, and also JA24. For example, grape VvLOXA could be responsible for the formation of C6 compounds under acidic conditions50, whereas in olive, Oep2LOX2 played a role in the biosynthesis of C6 and C5 volatile compounds51. The barley lox2:Hv:3 gene has been proposed to be involved in the channelling of linolenic acid substrate into the HPL-mediated production of C6 volatiles52, 53. In maize, ZmLOX10 and ZmLOX08 were specialized in providing substrates for the GLV and JA biosynthesis pathways, respectively26. In oriental melon fruit, straight-chain esters, such as ethyl acetate, have been demonstrated that they were the most important aroma components and might be derived from the fatty acid LOX–HPL metabolic pathways38, 54. Previous research has indicated that there are at least 18 LOX genes (CmLOX01–CmLOX18) in the melon genome37; however, the roles of these genes in the generation of fruit flavour volatiles have yet to be clarified. CmLOX18 may, however, be an important candidate gene involved in straight-chain ester production in melon37, 38. In the present study, we transferred the melon CmLOX18 gene into the tomato genome, which showed that CmLOX18 might plays an important role in the synthesis of C6 compounds in melon fruits.
Previous research has indicated that the 13-LOX TomloxC is essential for the synthesis of C6 compounds30, 31. TomloxF, showing 13-LOX activity and preferentially useing linolenate as a substrate, was probably involved in the production of C6 volatile compounds55. In the present study, the contents of C6 volatile compounds, including hexanal, (Z)-3-hexenal, and (Z)-3-hexen-1-ol, in the fruits of transgenic tomato plants over-expressing CmLOX18 were higher than those in wild-type fruits (Fig. 8 and Table S2). In addition, the over-expression of CmLOX18 caused a significantly higher expression of LeHPL mRNA, whereas there was no change in the expression levels of TomloxA, TomloxB, TomloxC, TomloxD, TomloxE, and TomloxF mRNA (Fig. 7). These results indicated that the increases in C6 volatiles in transgenic fruits were attributable to the involvement of melon CmLOX18 and tomato LeHPL, and were not related to endogenous tomato LOX. Furthermore, C6 volatiles and C5 compounds might be generated through an additional branch of the LOX pathway. Previous studies have shown that tomato TomloxC and olive Oep2LOX2 are involved in the biosynthesis of C5 compounds31, 41, 56. However, in the present study, we could detect no significant differences between the C5 compounds, including 1-pentanol and 1-penten-3-one, of wild-type and transgenic fruits (Fig. 8). On the basis of the aforementioned results, we suspected that CmLOX18 might be involved in the synthesis of C6 volatiles via its participation in the LOX–13HPL pathway, whereas it was not involved in the synthesis of C5 compounds. Moreover, CmLOX18 and TomloxD shared 77% identity at the amino acid level, whereas the predicted amino acid sequence of CmLOX18 displayed only 41–45% identity to other identified tomato LOXs. In addition to the high similarity in their amino acid sequences, both CmLOX18 and TomloxD might also be involved in JA biosynthesis, given that previous studies have shown that TomloxD is involved in wound-induced JA biosynthesis and plant immunity to insect herbivores27, 57, and that the expression of CmLOX18 mRNA is induced by wounding and methyl jasmonate treatments58.
In conclusion, the purified CmLOX18 recombinant protein over-expressed in yeast showed positional specificity, as evidenced by its release of only a C-13 oxidized product. Linoleic acid was demonstrated to be the preferred substrate for the recombinant CmLOX18 protein, which exhibited optimal catalytic activity at pH 4.5 and 30 °C. CmLOX18, which was localized to non-chloroplast organelles, played an essential role in the synthesis of C6 flavour volatiles in ripe fruits, but was not implicated in the synthesis of C5 compounds. Therefore, CmLOX18 can be assumed to play an important role in C6 compound synthesis in melon fruits and might also be involved in JA biosynthesis in response to abiotic and biotic stresses.
Materials and Methods
Plant materials
Tomato Solanum lycopersicum L. cv. “Zhongshu 6” (WT) and the T0 and T1 transgenic tomato cultivars 35 S:: CmLOX18 were individually planted in pots (volume of 25 L, soil: peat: compost = 1:1:1) in a greenhouse (Shenyang Agricultural University, Shenyang, China) in spring and fall seasons, 2015 to obtain the fruits for analysis of volatile compounds. Flowers were hand pollinated and tagged on the day of bloom for each plant. Fruit from WT and the transgenic tomato 45 day after pollination (DAP) were equivalent in age to breaker fruit. The harvested flesh tissue was froze immediately in liquid nitrogen and then stored at −80 °C until use.
Heterologous expression and purification of CmLOX18 in yeast
Following the instructions provided by the manufacturer (Invitrogen, Paisely, UK), the CmLOX18 cDNAs were cloned in the pYES2.1 TOPO-TA vector for regulated protein expression in yeast. Furthermore, all the constructs were transformed into the Saccharomyces cerevisiae cell line INVSc1 for expression of recombinant protein. Time-course studies CmLOX18 gene expression in yeast was performed by harvesting an aliquot of cells at 0, 4, 8, and 24 hours after gala ctose induction.
The purification of recombinant LOX18 was performed according to the method described by Manriquez59.Protein purity was separated via a 12% Tris-glycine Coomassie brilliant blue stained SDS/PAGE.
Western blot analysis
Western blot analysis was performed to detect the recombinant CmLOX18 in yeast according to the method described by Patel et al.60. Total plant protein from ground leaves of transgenic tomato was extracted, quantified, separated and detected, by the method described by Mou et al.61 and NBT/BCIP buffer (Cat#00-2209, Zymed Lab., USA) system according to its manufacturer’s procedure.
Characterization of the recombinant CmLOX18
For pH optimum determination, glycin-HCl (pH 3.0, 3.5), CH3COOH-CH3COONa (pH 4.0, 4.5, 5.0, 5.5), Na2HPO4- Na2H2PO4 (pH 6.0, 6.5, 7.0, 7.5), and Tris-HCl (pH 8.0, 9.0) were used. LOX activities were determined according to the method described by Li et al.54. For the optimum temperature determination, 50 mM buffer was used for temperature range 20–45 °C increments of 5 °C. Each sample had three replicates. The kinetic parameters had been determined from a Michaelis-Menton plot in a range of substrate concentrations (linoleic acid and linolenic acid) between 15 and 350 μM.
According to the previous method17, we analyzed the CmLOX18 products. The reaction mixture of enzyme activity was stopped by adding 0.1 M HCl solution, n-hexane was then added to the mixture. The 9-HPOD and 13-HPOD isomers were analysed by a SP-HPLC using a Slica Ultrasphere column (Inertsil® SIL-100A, 250 × 4.6 mm, 5 μm particle size) with a solvent system of n-hexane/2-propanol/acetic acid (100:2:0.1, by volume) and a flow rate of 1.0 ml min−1. The enantiomer composition was analyzed by chiral phase HPLC (CP-HPLC) on a Chiralcel OB-H column (Diacel Chem. Industries, 4.6 × 250 mm, 5 μm particle size) with a solvent system of n-hexane/2-propanol/acetic acid (100:5:0.1, by volume) and a flow rate of 1.0 ml min−1. The absorbance at 234 nm (conjugated diene system of the hydroxy fatty acids) was recorded. Standards of 9- and 13-HPOD were purchased from Larodan (Malm, Sweden).
Subcellular localization analysis by transient expression of CmLOX18 in Arabidopsis protoplasts
Transient expression of GFP-tagged proteins CmLOX18 cDNAs was isolated and cloned into the vector pBI221-GFP to get the C termini of the corresponding coding regions fused in-frame to the amino terminal end of GFP. GFP-tagged proteins were expressed in A. thaliana cells using polyethylene glycol (PEG) (Bio-Rad) approach for transient transformation62. Subcellular localization analysis was performed according to the method described by Cao et al.65.
35::CmLOX18 vector construction and tomato transformation
The full-length cDNA of CmLOX18 was cloned into the Gateway-compatible vector p ENTR D-TOPO (Invitrogen). The amplicon was transferred by LR recombination to the binary vector pB7FWG2.0, which harbours two 35 S Cauliflower mosaic vitus (CaMV) promoters, the marker gene for Spectinomycin (Spe), bialaphos acetyltransferase genes (Bar), and enhanced green fluorescent protein (Egfp) (Fig. S1), following the manufacturer’s instruction. According to the previous method63, binary vectors containing the expected insert were subsequently transformed into Agrobacterium tumefaciens EHA105 cells by electroporation.
PCR and Southern blotting
For PCR analysis, genomic DNA was extracted from transgenic lines and WT according to the manufacturer’s protocol (Tiangen Biotech Co.Ltd, Beijing, China). Froward primer TGTAGTGGTTGACGAT at + 768 and reverse primer TTGGAACTGACAGAAC at + 1085 of bar gene were used. For southern analysis, total DNA digested with HindШ restriction endonuclease, and electrophoresed in 0.8% agarose gels. Gels were transferred to nylon membranes and hybridized at 52 °C with DNA fragments labeled with a DIG High Prime DNA labeling kit (Roche, Switzerland). The blots were washed twice for 5 min at 65 °C with 40 min. NBT/BCIP was used for color detection.
RNA Isolation and Real-time RT-PCR analysis
Total RNA from leaves and fruit samples were performed according to the manufacturer’s protocol (Tiangen Biotech Co.Ltd, Beijing, China) as describe previously57. For measuring the mRNA level of CmLOX18, TomloxA-F, and LeHPL in tomato fruits, the gene-specific primers of Real-time quantitative PCR (qPCR) were listed in the Table S1. The reaction of quantitative PCR was performed for gene expression studies according to the method described by Tang et al.38.
Extraction and determination of volatiles
The volatiles of transgenic tomato fruit were exacted and detected according to the method described by Lewinsohn et al.64 and Tang et al.38 with a few modifications, in which an internal standard, namely 2-Octanone, (50 μL of, 59.5 mg/L, 0.5%, v/v, Aladdin Chemistry,China) was added to 10 mL supernatant of the juice for the determination of volatile matter content.
Statistical and image analysis
The data were analyzed by the analysis of variance (ANOVA) using the SPSS13.0 statistics program, and statistical significance of differences were calculated by a one-way ANOVA following Duncan’s multiple range tests for each experiment at a P < 0.05 level. Origin (version 8.0) was used to chart. Plants were sprayed with Egfp and kept in the dark for a few minutes prior to imaging. NightSHADE LB 985 (Berthold, BadWildbad, Germany) was used to image enhanced green fluorescent protein.
Electronic supplementary material
Acknowledgements
This work was supported by National Natural Science Foundation of China (31471868).
Author Contributions
C.Z., and H.Q. designed the study. C.Z. and S.C. contributed to acquisition of data. C.Z., H.Q. and S.C. contributed to analysis and interpretation of data. C.Z. and S.C. contributed to draftng of the manuscript. L.J., Q.C., Q.X. and Y.J. contributed to critical revision of the manuscript for important intellectual content. C.Z. and S.C. contributed to statistical analysis. H.Q., C.Z. and S.C. obtained funding. C.Z., S.C., L.J., Q.C., Q.X. and Y.J. contributed to administrative, technical, or material support. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Chong Zhang, Songxiao Cao and Yazhong Jin contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02559-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Defilippi BG, Manríquez D, Luengwilai K, González-Agüero M. Chapter 1 Aroma Volatiles: Biosynthesis and Mechanisms of Modulation During Fruit Ripening. Adv. Bot. Res. 2009;50:1–37. doi: 10.1016/S0065-2296(08)00801-X. [DOI] [Google Scholar]
- 2.And JCB, Grimm CC. Identification of volatile compounds in cantaloupe at various developmental stages using solid phase microextraction. J. Agric. Food. Chem. 2001;49:1345–1352. doi: 10.1021/jf0005768. [DOI] [PubMed] [Google Scholar]
- 3.Dirinck P, De Pooter H, Schamp N. Aroma development in ripening fruits. In: Flavor chemistry: trends and developments. Teranishi R and Buttery R (editors) American Chemical Society (ACS) Symposium series. 1989;388:23–34. [Google Scholar]
- 4.Song J, Forney CF. Flavour volatile production and regulation in fruit. Can J Plant Sc i/Rev Can Phytotechi. 2008;88:537–550. doi: 10.4141/CJPS07170. [DOI] [Google Scholar]
- 5.Baldwin EA, Scott JW, Shewmaker CK, Schuch W. Flavor trivia and tomato aroma: biochemistry and possible mechanisms for control of important aroma components. Hort. sci. 2000;5:1013–1022. [Google Scholar]
- 6.Contreras C, Beaudry R. Lipoxygenase-associated apple volatiles and their relationship with aroma perception during ripening. Postharv Biol Technol. 2013;82:28–38. doi: 10.1016/j.postharvbio.2013.02.006. [DOI] [Google Scholar]
- 7.Rowan DD, Allen JM, Fielder S, Hunt MB. Biosynthesis of straight-chain ester volatiles in red delicious and granny smithapples using deuterium-labelled precursors. J. Agric. Food Chem. 1999;47:2553–562. doi: 10.1021/jf9809028. [DOI] [PubMed] [Google Scholar]
- 8.Fellman JK, Miller TW, Mattinson DS, Mattheis JP. Factors that influence biosynthesis of volatile flavor compound in apple fruits. Hort Sci. 2000;35:1026–1033. [Google Scholar]
- 9.Li M, Dunwell JM, Qiao X, Liu X, Zhang SL. Characterization of the lipoxygenase (LOX) gene family in the Chinese white pear (Pyrus bretschneideri) and comparison with other members of the Rosaceae. BMC Genomics. 2014;15:1–12. doi: 10.1186/1471-2164-15-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreou A, Feussner I. Lipoxygenases-Structure and reaction mechanism. Phytochemistry. 2009;70:1504–1510. doi: 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Feussner I, Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 12.Siedow JN. Plant lipoxygenase—structure and function. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:145–188. doi: 10.1146/annurev.pp.42.060191.001045. [DOI] [Google Scholar]
- 13.Hu T, Zeng H, Hu Z, Qv X, Chen G. Overexpression of the Tomato 13-Lipoxygenase Gene TomloxD Increases Generation of Endogenous Jasmonic Acid and Resistance to Cladosporium fulvum and High Temperature. Plant Mol. Biol. Rep. 2013;31:1141–1149. doi: 10.1007/s11105-013-0581-4. [DOI] [Google Scholar]
- 14.Kim ES, et al. Dual positional specificity and expression of non-traditional lipoxygenase induced by wounding and methyl jasmonate in maize seedings. Plant Mol Biol. 2003;52:1203–1213. doi: 10.1023/B:PLAN.0000004331.94803.b0. [DOI] [PubMed] [Google Scholar]
- 15.Veronico P, et al. A novel lipoxygenase in pea roots. Its function in wounding and biotic stress. Plant Physiol. 2006;141:1045–1055. doi: 10.1104/pp.106.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Allmann S, Wu J, Baldwin IT. Comparsions of LIP-OXYGENASES3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 2008;146:904–915. doi: 10.1104/pp.107.109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, Han B. Differential expression pattern of an acidic 9/13-lipoxygenase in flower opening and senescence and in leaf response to phloem feeders in the tea plant. BMC Plant Biol. 2010;10:1–15. doi: 10.1186/1471-2229-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemchenko A, Kunze S, Feussner I, Kolomiets M. Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J. Exp. Bot. 2006;57:3767–79. doi: 10.1093/jxb/erl137. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, et al. Disruption of a maize 9-lipoxygenae results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant. Microbe. In. 2007;20:922–933. doi: 10.1094/MPMI-20-8-0922. [DOI] [PubMed] [Google Scholar]
- 20.Gao X, Stumpe M, Feussner I, Kolomiets M. A novel plastidial lipoxygenase of maize (Zea mays) ZmLOX6 encodes for a fatty acid hydroperoxide lyase and is uniquely regulated by phytohormones and pathogen infection. Planta. 2008;227:491–503. doi: 10.1007/s00425-007-0634-8. [DOI] [PubMed] [Google Scholar]
- 21.Hwang IS, Hwang BK. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2010;152:948–967. doi: 10.1104/pp.109.147827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rancé I, Fournier J, Esquerré-Tugayé MT. The incompatible interaction between phytophthora parasitica var. nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Pro. Natl Acad. Sci.USA. 1998;95:6554–6559. doi: 10.1073/pnas.95.11.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang FC, Schwab W. Cloning and characterization of a 9-lipoxygenase gene induced by pathogen attack from nicotiana benthamiana, for biotechnological application. BMC Biotechnolog. 2010;11:1–15. doi: 10.1186/1472-6750-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang FC, Studart-Witkowski C, Schwab W. Overexpression of hydroperoxide lyase gene in Nicotiana benthamiana using a viral vector system. Plant Biotechnol J. 2010;8:783–95. doi: 10.1111/j.1467-7652.2010.00508.x. [DOI] [PubMed] [Google Scholar]
- 25.Bell NJ, Creelman RA, Mullet JE. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen SA, et al. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 2013;74:59–73. doi: 10.1111/tpj.12101. [DOI] [PubMed] [Google Scholar]
- 27.Yang LJ, et al. Role of Tomato Lipoxygenase D in Wound-Induced Jasmonate Biosynthesis and Plant Immunity to Insect Herbivores. Plos Genetics. 2013;9:e1003964–e1003964. doi: 10.1371/journal.pgen.1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, et al. Using pseudo amino acid composition to predict protein subcellular location: approached with lyapunov index, bessel function, and chebyshev filter. Amino Acids. 2005;28:373–6. doi: 10.1007/s00726-005-0206-9. [DOI] [PubMed] [Google Scholar]
- 29.Cho K, et al. Cellular localization of dual positional specific maize lipoxygenase-1 in transgenic rice and calcium-mediated membrane association. Plant Sci. 2011;181:242–8. doi: 10.1016/j.plantsci.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, et al. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavour compounds. Plant Physiol. 2004;136:2641–2651. doi: 10.1104/pp.104.041608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen J, et al. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 2014;65:419–428. doi: 10.1093/jxb/ert382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leon J, et al. Lipoxygenase H1 gene silencing reveals a specific role in supplying fatty acid hydroperoxides for aliphatic aldehyde production. J Biol Chem. 2002;277:416–423. doi: 10.1074/jbc.M107763200. [DOI] [PubMed] [Google Scholar]
- 33.Farmaki T, et al. Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. J Exp Bot. 2007;58:555–568. doi: 10.1093/jxb/erl230. [DOI] [PubMed] [Google Scholar]
- 34.Feussner I, Kindl H. Particulate and soluble lipoxygenase isoenzymes: comparison of molecular and enzymatic properties. Planta. 1994;194:22–28. doi: 10.1007/BF00201030. [DOI] [Google Scholar]
- 35.Grayburn W, et al. Soybean leaves contain multiple lipoxygenases. Plant Physiol. 1991;95:1214–1218. doi: 10.1104/pp.95.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephenson L, Bunker T, Dubbs W, Grimes H. Specific soybean lipoxygenases localize to discrete subcellular compartments and their mRNAs are differentially regulated by source-sink status. Plant Physiol. 1998;1116:923–933. doi: 10.1104/pp.116.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, et al. The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L.) genome. Sci Hortic. 2014;170:94–102. doi: 10.1016/j.scienta.2014.03.005. [DOI] [Google Scholar]
- 38.Tang YF, Zhang C, Cao SX, Wang X, Qi HY. The Effect of CmLOXs on the Production of Volatile Organic Compounds in Four Aroma Types of Melon (Cucumis melo L.) Plos One. 2015;10:11. doi: 10.1371/journal.pone.0143567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes RK, et al. Probing a novel potato lipoxygenase with dual positional specificity reveals primary determinants of substrate binding and requirement for a surface hydrophobic loop and has implications for the role of lipoxygenase in the tuber. Eur J Biochem. 2001;353:345–355. doi: 10.1042/bj3530345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coffa G, Schneider C, Brash AR. A comprehensive model of positional and stereo control in lipoxygenases. Biochem. Bioph. Res. Co. 2005;338:87–92. doi: 10.1016/j.bbrc.2005.07.185. [DOI] [PubMed] [Google Scholar]
- 41.Veronico P, et al. A novel lipoxygenase in pea roots. Its function in wounding and biotic stress. Plant Physiol. 2006;141:1045–1055. doi: 10.1104/pp.106.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouet-Mayer MA, Bureau JM, Lauriere C. Identification and characterization of lipoxygenase isoforms in senescing carnation petals. Plant Physiol. 1992;98:971–978. doi: 10.1104/pp.98.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuchi-Mizutani M, et al. Molecular and functional characterization of a rose lipoxygenase cDNA related to flower senescence. Plant Sci. 2000;160:129–137. doi: 10.1016/S0168-9452(00)00373-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, et al. Effects of postharvest treatments on expression of three lipoxygenase genes in oriental melon (Cucumis melo var. makuwa Makino) Postharvest Biol Techol. 2015;110:229–238. doi: 10.1016/j.postharvbio.2015.08.024. [DOI] [Google Scholar]
- 45.Halitschke, Baldwin. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003;36:794–807. doi: 10.1046/j.1365-313X.2003.01921.x. [DOI] [PubMed] [Google Scholar]
- 46.Halitschke R, Ziegler J, Keinänen M, Baldwin IT. Silencing of hydroperoxide lyase and allene oxide synthase reveals substrate and defense signaling crosstalk in Nicotiana attenuata. Plant J. 2004;40:35–46. doi: 10.1111/j.1365-313X.2004.02185.x. [DOI] [PubMed] [Google Scholar]
- 47.Chehab EW, et al. Distinct Roles of Jasmonates and Aldehydes in Plant-Defense Responses. Plos One. 2008;3:664–664. doi: 10.1371/journal.pone.0001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Froehlich JE, Itoh A, Howe GA. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 2001;125:306–317. doi: 10.1104/pp.125.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acosta IF, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 50.Podolyan A, White J, Jordan B, Winefield C. Identification of the lipoxygenase gene family from vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of sauvignon blanc. Funct Plant Biol. 2010;37:767–784. doi: 10.1071/FP09271. [DOI] [Google Scholar]
- 51.Padilla MN, Hernández ML, Sanz C, Martínez-Rivas JM. Functional characterization of two 13-lipoxygenase genes from olive fruit in relation to the biosynthesis of volatile compounds of virgin olive oil. J. Agric. Food Chem. 2009;57:9097–9107. doi: 10.1021/jf901777j. [DOI] [PubMed] [Google Scholar]
- 52.Bachmann AB, et al. Jasmonate-induced lipid peroxidation in barley leaves initiated by distinct 13-LOX forms of chloroplasts. Biological Chemistry. 2002;383:1645–1657. doi: 10.1515/BC.2002.185. [DOI] [PubMed] [Google Scholar]
- 53.Vörös K, et al. Characterization of a methyljasmonate-inducible lipoxygenase from barley (Hordeum vulgare cv. Salome) leaves. Eur. J. Biochem. 1998;251:36–44. doi: 10.1046/j.1432-1327.1998.2510036.x. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Qi HY, Liu Y, Guan XC, Liu Y. Effects of ethephon and 1-methylcyclopropene on fruit ripening and the biosynthesis of volatiles in oriental sweet melon (Cucumis melo var. makuwa Makino) J Horticultural Sci Biotechnol. 2011;86:517–526. doi: 10.1080/14620316.2011.11512798. [DOI] [Google Scholar]
- 55.Mariutto M, et al. The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 2011;11:29–29. doi: 10.1186/1471-2229-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gardner HW, Grove MJ, Salch YP. Enzymic pathway to ethyl vinyl ketone and 2-pentenal in soybean preparations. J. Agric. Food. Chem. 1996;44:882–886. doi: 10.1021/jf950509r. [DOI] [Google Scholar]
- 57.Heitz T, Bergey DR, Ryan CA. A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant physiol. 1997;114:1085–1093. doi: 10.1104/pp.114.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu JY, et al. Effects of abiotic stress and signal molecules on the expression of five 13-CmLOXs and enzyme activity in oriental sweet melon (Cucumis melo var. makuwa Makino) J Integr Agr. 2016;15:60345–7. [Google Scholar]
- 59.Manriquez D, et al. Two highly divergent alcohol dehydrogenases of melon exhibit fruit ripening-specific expression and distinct biochemical characteristics. Plant Mol Biol. 2006;61:675–685. doi: 10.1007/s11103-006-0040-9. [DOI] [PubMed] [Google Scholar]
- 60.Patel J, et al. Elastin-like polypeptide fusions enhance the accumulation of recombinant proteins in tobacco leaves. Transgenic Res. 2007;16:239–249. doi: 10.1007/s11248-006-9026-2. [DOI] [PubMed] [Google Scholar]
- 61.Mou ZL, Fan WH, Dong XN. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell. 2003;113:935–44. doi: 10.1016/S0092-8674(03)00429-X. [DOI] [PubMed] [Google Scholar]
- 62.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 63.Xu T, et al. Solanum lycopersicum iaa15 functions in the 2,4-dichlorophenoxyacetic acid herbicide mechanism of action by mediating abscisic acid signalling. J. Exp. Bot. 2015;66:13. doi: 10.1093/jxb/erv199. [DOI] [PubMed] [Google Scholar]
- 64.Lewinsohn E, et al. Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 2001;127:1256–65. doi: 10.1104/pp.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao S. X. et al. Heterologous Expression and Biochemical Characterization of Two Lipoxygenases in Oriental Melon, Cucumis melo var. makuwa Makino. PLoS ONE11(4), 1–20 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


