Abstract
Proprotein convertase subtilisin kexin 9 (PCSK9) is a member of the subtilisin serine protease family with an important role in cholesterol metabolism. PCSK9 expression is regulated by dietary cholesterol in mice and cellular sterol levels in cell culture via the sterol regulatory element binding protein transcription factors, and mutations in PCSK9 are associated with a form of autosomal dominant hypercholesterolemia. Overexpression of PCSK9 in mice leads to increased total and low-density lipoprotein (LDL) cholesterol levels because of a decrease in hepatic LDL receptor (LDLR) protein with normal mRNA levels. To study the mechanism, PCSK9 was overexpressed in human hepatoma cells, HepG2, by adenovirus. Overexpression of PCSK9 in HepG2 cells caused a decrease in whole-cell and cell-surface LDLR levels. PCSK9 overexpression had no effect on LDLR synthesis but caused a dramatic increase in the degradation of the mature LDLR and a lesser increase in the degradation of the precursor LDLR. In contrast, overexpression of a catalytically inactive mutant PCSK9 prevented the degradation of the mature LDLR; whereas increased degradation of the precursor LDLR still occurred. The PCSK9-induced degradation of the LDLR was not affected by inhibitors of the proteasome, lysosomal cysteine proteases, aspartic acid proteases, or metalloproteases. The PCSK9-induced degradation of the LDLR was shown to require transport out of the endoplasmic reticulum. These results indicate that overexpression of PCSK9 induces the degradation of the LDLR by a nonproteasomal mechanism in a post-endoplasmic reticulum compartment.
Keywords: autosomal dominant hypercholesterolemia, Narc-1, proprotein convertase subtilisin kexin 9, low-density lipoprotein receptor
Proprotein convertase subtilisin kexin 9 (PCSK9) is a member of the subtilisin serine protease family. The other eight mammalian subtilisin proteases, PCSK1–PCSK8 (also called PC1/3, PC2, furin, PC4, PC5/6, PACE4, PC7, and S1P/SKI-1) are proprotein convertases that process a wide variety of proteins in the secretory pathway and play roles in diverse biological processes (1–5). PCSK9 has been proposed to play a role in cholesterol metabolism. PCSK9 mRNA expression is down-regulated by dietary cholesterol feeding in mice (6), up-regulated by statins in HepG2 cells (7), and up-regulated in sterol regulatory element binding protein (SREBP) transgenic mice (6, 8), similar to the cholesterol biosynthetic enzymes and the low-density lipoprotein receptor (LDLR). Furthermore, PCSK9 missense mutations have been found to be associated with a form of autosomal dominant hypercholesterolemia (Hchola3) (9–11). PCSK9 may also play a role in determining LDL cholesterol levels in the general population, because single-nucleotide polymorphisms (SNPs) have been associated with cholesterol levels in a Japanese population (12).
Autosomal dominant hypercholesterolemias (ADHs) are monogenic diseases in which patients exhibit elevated total and LDL cholesterol levels, tendon xanthomas, and premature atherosclerosis (13, 14). The pathogenesis of ADHs and a recessive form, autosomal recessive hypercholesterolemia (ARH) (15), is due to defects in LDL uptake by the liver. ADH may be caused by LDLR mutations, which prevent LDL uptake, or by mutations in the protein on LDL, apolipoprotein B, which binds to the LDLR. ARH is caused by mutations in the ARH protein that are necessary for endocytosis of the LDLR–LDL complex via its interaction with clathrin. Therefore, if PCSK9 mutations are causative in Hchola3 families, it seems likely that PCSK9 plays a role in receptor-mediated LDL uptake.
Overexpression studies point to a role for PCSK9 in controlling LDLR levels and, hence, LDL uptake by the liver (16–18). Adenoviral-mediated overexpression of mouse or human PCSK9 for 3 or 4 days in mice results in elevated total and LDL cholesterol levels; this effect is not seen in LDLR knockout animals (16, 18). In addition, PCSK9 overexpression results in a severe reduction in hepatic LDLR protein, without affecting LDLR mRNA levels (16), SREBP protein levels, or SREBP protein nuclear to cytoplasmic ratio (18). These results indicate that PCSK9, either directly or indirectly, reduces LDLR protein levels by a posttranscriptional mechanism.
To better understand the mechanism by which overexpression of PCSK9 decreases LDLR protein levels, PCSK9 was overexpressed in HepG2 cells. These studies indicate that PCSK9 increases the degradation of the LDLR without affecting its synthesis. We also investigated the mechanism and location of the PCSK9-induced degradation of the LDLR, revealing a post-endoplasmic reticulum (ER) pathway. Overall, these studies provide information on the cellular function of PCSK9.
Methods
Materials. All cell-culture reagents were obtained from Invitrogen. HepG2 cells were maintained in DMEM supplemented with 10% FBS. Rabbit anti-mouse PCSK9 is described in ref. 16. Rabbit anti-human LDLR was from Research Diagnostics, and mouse anti-human Transferrin receptor was from Zymed. Mouse anti-human LDLR (clone IgG-C7) bioreactor supernatant was prepared from American Type Culture Collection hybridoma CRL-1691 by the Monoclonal Antibody Core Facility of The Rockefeller University. Goat anti-mouse IgG for LDLR immunoprecipitation was from Pierce or Jackson Immunochemicals. EZ-Link Sulfo-NHS-LC-Biotin and avidin-horseradish peroxidase (HRP) were from Pierce. MG132, lactacystin, ALLN, Suc-LLVY-AMC, and Z-RR-AMC were from Calbiochem. Ammonium chloride, chloroquine, E64d, brefeldin A (BFA), and nocodazole were from Sigma. Pepstatin and phosphoramidon were from Roche. Lipoprotein-deficient serum (LPDS) was made by ultracentrifugation by standard techniques.
Construction of Adenovirus Expressing Catalytically Inactive Pcsk9. The creation of the mouse PCSK9 catalytic mutant, PCSK9S402A, is described in ref. 16. The PCSK9S402A ORF was inserted into the pAd5-CMV-NpA shuttling vector. Recombinant adenoviral particles containing the PCSK9S402A ORF under the control of a constitutive CMV promoter were created by ViraQuest (North Liberty, IA). Upon infection of multiple cell lines, PCSK9S402A produced only the pro-form of PCSK9. Control adenovirus (empty) and PCSK9 wild-type virus are described in ref. 16.
Adenoviral-Mediated Overexpression in Cell Culture. HepG2 cells were plated in collagen-coated six-well dishes at 250,000 cells per well. Twelve hours later, cells were infected with empty adenovirus, PCSK9 adenovirus, or PCSK9S402A adenovirus at a multiplicity of infection of 3,000 in DMEM containing 10% FBS. Twenty-four hours later, the medium was changed to DMEM containing 10% LPDS, and the cells were reinfected at the same titer. Twelve hours later, cells were processed for Western blotting, cell-surface biotinylation, endocytosis assay, or metabolic labeling studies.
Western Blotting. Preparation of cell lysates, determination of protein concentrations, and Western blotting were as described in ref. 16. Dilutions of antibodies used were as follows: anti-Pcsk9–583, 1:5,000; anti-LDLR, 1:1,000; and anti-transferrin receptor, 1:500. Secondary antibodies, goat anti-rabbit HRP and goat anti-mouse HRP, were used at 1:10,000. Avidin-conjugated HRP was used at 1:2,500. Band intensities were measured by using image pro plus.
Metabolic Labeling and LDLR Immunoprecipitation. Preformed immune complexes were made as described in ref. 19, except that 6 μg of IgG-C7 and 96 μg of goat anti-mouse IgG were used per sample. For metabolic labeling, cells were starved in DMEM without l-methionine and l-cysteine (DMEM*), pulse-labeled with DMEM* containing 200 μCi per well (1 Ci = 37 GBq) EXPRESS [35S] Protein Labeling Mix (NEN Life Sciences), and chased with DMEM* containing 1.5 mg/ml l-methionine and 0.5 mg/ml l-cysteine. At the end of the metabolic study, cells lysates were made and immunoprecipitated for the LDLR, and the mixture was subjected to sucrose gradient ultracentrifugation as described in ref. 19. Samples representing equal TCA precipitable counts of the original lysate were electrophoresed in 4–12% Tris-glycine gels, and gels were fixed in 25% methanol/10% acetic acid, washed in distilled water, treated with Autofluor (National Diagnostics, Atlanta), dried, and exposed to a phosphoimager screen or film. Band intensities were measured by using imagequant or image pro plus software.
Cell-Surface Biotinylation and Endocytosis Assay. For studies of cell-surface protein levels, cells were incubated in PBS containing 0.5 mg/ml Sulfo-NHS-Biotin at 4°C for 1 h. The reaction was quenched in 100 mM glycine in PBS, and the cells were washed five times in ice-cold Tris-buffered saline. Cells were lysed in LDLR lysis buffer (19) and immunoprecipitated for the LDLR. For endocytosis assays, cells were treated with monensin (20 μM) for various times before undergoing cell-surface biotinylation as described in ref. 20.
Metabolic Studies with Inhibitors. HepG2 cells were infected, starved, and pulsed as above. Cells were then chased in the presence of inhibitors at the following concentrations: ALLN, 100 μM; ammonium chloride, 10 mM; chloroquine, 75 μM; E64d, 10 μM; lactacystin, 10 μM; MG132, 20 μM; pepstatin, 50 μg/ml; phosphoramidon, 100 μM; BFA, 5 μg/ml; nocodazole, 20 μg/ml; and monensin, 0.5 μM. To show that the inhibitors were active, lysates from the experiments were mixed at 1:50 dilution in assay buffer (2.5 mM Hepes, pH 7.5/0.5 mM EDTA/0.05% Igepal/0.001% SDS) with or without 50 μM substrate. The substrate Suc-LLVY-AMC was used to assay proteasome activity, and the substrate Z-RR-AMC was used to assay cysteine protease activity. The reactions were incubated at 37°C for 1 h and then fluorescence due to the cleavage of the substrate was read in a microplate reader.
Results
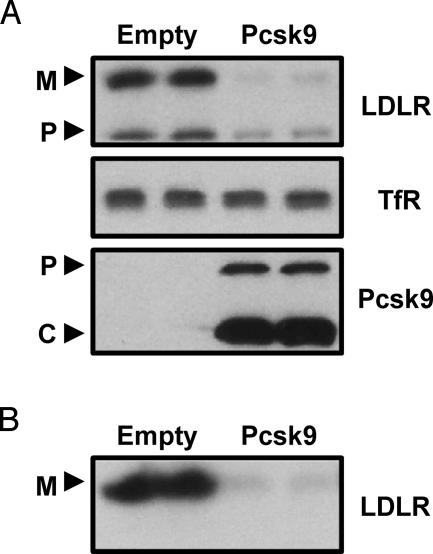
Overexpression of PCSK9 in HepG2 Cells Decreases LDLR Protein Levels. To determine the effects of PCSK9 on the LDLR in vitro, adenoviral-mediated overexpression of PCSK9 was used in HepG2 cells. As shown in Fig. 1A, overexpression of PCSK9 reduced whole-cell levels of both precursor LDLR (120 kDa) and mature LDLR (160 kDa). This effect was specific to the LDLR because PCSK9 overexpression had no effect on the levels of transferrin receptor (Fig. 1 A). To determine whether cell-surface levels of the LDLR were also affected by PCSK9 overexpression, cells were biotinylated, immunoprecipitated for the LDLR, and visualized with avidin-HRP. As shown in Fig. 1B, overexpression of PCSK9 also reduced cell-surface LDLR.
Fig. 1.
Overexpression of PCSK9 in HepG2 cells decreased whole-cell and cell-surface LDLR. (A) HepG2 cells were infected with control adenovirus (Empty) or PCSK9 adenovirus (Pcsk9), and whole-cell lysates were collected after 36 h. Cell lysates were subjected to Western blotting for LDLR, transferrin receptor (TfR), and PCSK9. M, mature LDLR; P, precursor LDLR; P, pro-PCSK9; C, cleaved PCSK9. (B) HepG2 cells were infected with empty adenovirus or PCSK9 adenovirus. After 36 h, cell-surface proteins were biotinylated at 4°C and lysates were collected and immunoprecipitated for the LDLR. Proteins were visualized with avidin-HRP. The results presented in this and other figures are representative of at least three experiments each.
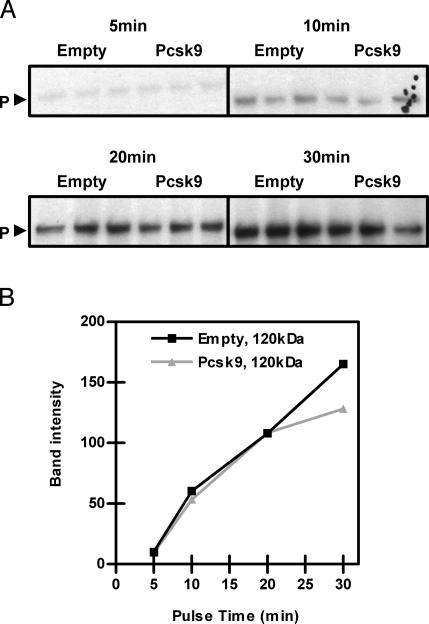
Overexpression of PCSK9 Has No Effect on LDLR Synthesis. Because overexpression of PCSK9 reduces LDLR protein with no effect on mRNA levels (16, 18), the effect of PCSK9 on LDLR protein synthesis was investigated to differentiate between effects at the level of translational yield or LDLR degradation. HepG2 cells infected with control adenovirus or PCSK9 adenovirus were pulsed with 35S-Met/Cys for 5, 10, 20, and 30 min. As shown in Fig. 2A, there were equivalent levels of labeled precursor LDLR in cells pulsed for 5, 10, and 20 min in control and PCSK9-overexpressing cells. These results indicate that PCSK9 overexpression had no effect on the synthesis of the LDLR (Fig. 2B). Of note, after 30 min of pulse, a decrease in the precursor LDLR was seen, suggesting an effect of PCSK9 on degradation of the LDLR.
Fig. 2.
Overexpression of PCSK9 did not affect synthesis of the LDLR. (A) HepG2 cells were infected with empty or PCSK9 adenovirus (Pcsk9). Cells were pulse-labeled with 35S-Met/Cys for 5, 10, 20, and 30 min and immunoprecipitated for the LDLR. P, precursor LDLR. (B) Quantification of the results in A; band intensities were quantified with image pro plus.
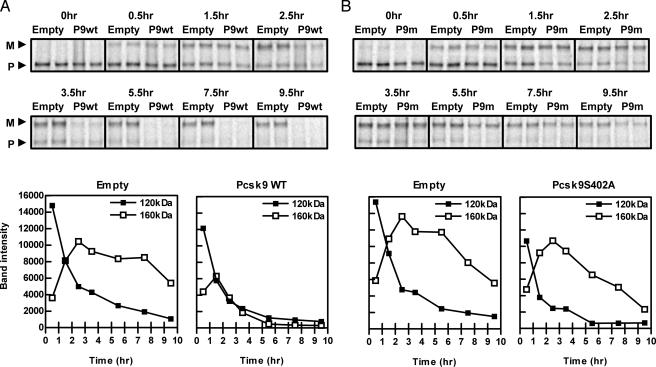
Overexpression of PCSK9 Accelerates the Degradation of the Mature LDLR. To investigate whether PCSK9 overexpression stimulated LDLR degradation, HepG2 cells infected with control or PCSK9 adenovirus were pulsed with 35S-Met/Cys for 30 min and chased for 0.5, 1.5, 2.5, 3.5, 5.5, 7.5, and 9.5 h. As shown in Fig. 3A, overexpression of PCSK9 caused rapid degradation of the mature LDLR. Of note, overexpression of PCSK9 also caused a small increase in the rate of degradation of the precursor LDLR.
Fig. 3.
Overexpression of PCSK9 accelerated the degradation of the mature LDLR in a catalytically dependent mechanism. (A) HepG2 cells were infected with empty or PCSK9 adenovirus (Pcsk9WT or P9wt). (B) HepG2 cells were infected with empty or PCSK9S402A adenovirus (Pcsk9S402A or P9m). Cells were pulse labeled with 35S-Met/Cys for 30 min, chased for 0, 0.5, 1.5, 2.5, 3.5, 5.5, 7.5, and 9.5 h, and immunoprecipitated for the LDLR. Band intensities were measured by using imagequant and graphed below. M, mature LDLR; P, precursor LDLR.
The PCSK9-Induced Degradation of the Mature LDLR Requires PCSK9 Serine Protease Activity. We next investigated whether the PCSK9 serine protease activity is necessary for LDLR degradation. A catalytically inactive PCSK9 was generated by mutating the catalytic-site residue serine 402 to alanine (PCSK9S402A) (16). Infection of HepG2 cells with an adenovirus expressing PCSK9S402A demonstrated an absence of intramolecular self-cleavage of PCSK9, indicating that this mutant is catalytically inactive (data not shown). HepG2 cells infected with control or PCSK9S402A adenovirus were pulsed with 35S-Met/Cys for 30 min and chased for 0, 0.5, 1.5, 2.5, 3.5, 5.5, 7.5, and 9.5 h. As shown in Fig. 3B, the PCSK9-induced degradation of the precursor LDLR still occurred in cells overexpressing PCSK9S402A. However, although there was less input into the mature LDLR pool, the PCSK9S402A catalytic mutant did not accelerate the degradation of the mature LDLR.
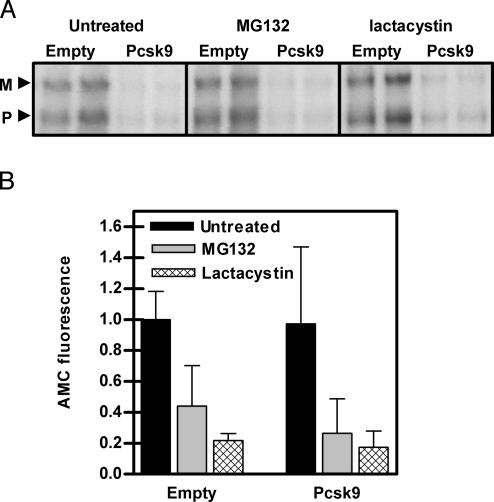
The PCSK9-Induced Degradation of the LDLR Does Not Depend on the Proteasome. The two major mediators of intracellular protein degradation are the proteasome and lysosome. To investigate whether the proteasome mediates PCSK9-induced degradation of the LDLR, HepG2 cells infected with control or PCSK9 adenovirus were pulsed with 35S-Met/Cys for 30 min and chased for 4 h in the absence or presence of the proteasome inhibitors MG132 and lactacystin. As shown in Fig. 4A, PCSK9 was still able to increase the degradation of the LDLR in the presence of MG132 and lactacystin, indicating that the PCSK9-induced degradation of the LDLR does not occur in the proteasome. To determine whether the proteasome was actually inhibited in these studies, cell lysates were incubated with the fluorescent proteasome substrate Suc-LLVY-AMC. As shown in Fig. 4B, the amount of fluorescence released by proteasome-mediated cleavage of the substrate was decreased in cells treated with MG132 and lactacystin.
Fig. 4.
PCSK9-induced degradation of the LDLR did not depend on the proteasome. (A) HepG2 cells were infected with empty or PCSK9 adenovirus (Pcsk9). Cells were pulse-labeled with 35S-Met/Cys for 30 min, chased for 4 h in the presence of the proteasome inhibitors MG132 and lactacystin, and immunoprecipitated for the LDLR. M, mature LDLR; P, precursor LDLR. (B) To measure proteasome activity in these cells, lysates were incubated with the proteasome substrate Suc-LLVY-AMC for 1 h. Fluorescence due to cleaved AMC by the proteasome was measured and compared to background fluorescence.
The PCSK9-Induced Degradation of the LDLR Does Not Depend on Various Classes of Lysosomal and Nonlysosomal Proteases. To investigate whether the PCSK9-induced degradation of the LDLR occurs in lysosomes, HepG2 cells infected with control or PCSK9 adenovirus were pulsed with 35S-Met/Cys for 30 min and chased for 4 h in the presence of the acidotropic agent ammonium chloride. This resulted in decreased conversion of precursor to mature LDLR, making interpretation of this experiment difficult. However, the decreased amount of mature LDLR that was made was protected from degradation by PCSK9 in the presence of ammonium chloride (Table 1), indicating that the degradation of the LDLR by PCSK9 may occur in a pH sensitive compartment like the lysosome. The major proteases in lysosomes are cysteine proteases of the cathepsin family. To investigate the role of this protease class, HepG2 cells infected with control or PCSK9 adenovirus were pulsed with 35S-Met/Cys for 30 min and chased for 4 h in the presence of the cysteine protease inhibitor E64d (Table 1). The PCSK9-induced degradation of the LDLR occurred in the presence of E64d, indicating that the effect was not mediated by cysteine proteases. That the cysteine proteases were actually inhibited in these experiments was demonstrated in cell lysates incubated with the cysteine protease substrate Z-RR-AMC, in which the amount of fluorescence was decreased by E64d treatment (data not shown). To determine whether other classes of proteases play a role in PCSK9-induced degradation of the LDLR, HepG2 cells infected with control or PCSK9 adenovirus were pulsed for 30 min and chased for 4 h in the presence of ALLN to inhibit calpain proteases, pepstatin to inhibit aspartic acid proteases, and phosphoramidon to inhibit metalloproteases. As shown in Table 1, PCSK9 was still able to increase degradation of the LDLR in the presence of all of these inhibitors.
Table 1. Pcsk9-induced degradation of the LDLR does not depend on various classes of proteases.
| Percent LDLR in Pcsk9 compared with empty
|
|||
|---|---|---|---|
| Inhibitor | Protease class | 120 kDa | 160 kDa |
| Untreated | NA | 27.7 | 15.2 |
| Ammonium chloride | NA | 18.8 | 84.0 |
| E64d | Cysteine | 26.4 | 19.9 |
| ALLN | Proteasome calpain | 17.1 | 14.7 |
| Pepstatin | Aspartic acid | 14.5 | 14.7 |
| Phosphoramidon | Metalloproteases | 22.3 | 14.4 |
NA, not applicable.
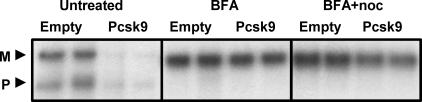
The PCSK9-Induced Degradation of the LDLR Requires Transport Out of the ER. To localize the cellular site of PCSK9-induced degradation of the LDLR, HepG2 cells infected with control or PCSK9 adenovirus were pulsed with 35S-Met/Cys for 30 min and chased for 4 h in the presence of BFA, which inhibits transport out of the ER. As shown in Fig. 5, PCSK9 was not able to degrade the LDLR in the presence of BFA. The single smaller LDLR band shown is described in refs. 21 and 22. Because BFA also collapses Golgi contents into the ER, cells were treated with BFA in the presence of nocodazole to prevent retrograde transport. As shown in Fig. 5, PCSK9 was also unable to promote the degradation of the LDLR in the presence of BFA and nocodazole. These results indicate that PCSK9-induced degradation of the LDLR occurs in a post-ER compartment.
Fig. 5.
PCSK9-induced degradation of the LDLR depends on exit from the ER. HepG2 cells were infected with empty or PCSK9 adenovirus (Pcsk9). Cells were pulse-labeled with 35S-Met/Cys for 30 min, chased for 4 h in the presence of BFA and BFA plus nocodazole (noc), and immunoprecipitated for the LDLR. M, mature LDLR; P, precursor LDLR.
Overexpression of PCSK9 Has No Effect on LDLR Endocytosis. ARH results from defective endocytosis of the LDLR due to the absence of ARH protein, which mediates LDLR binding to clathrin (15, 23). Because LDLR cell-surface levels are decreased by PCSK9 overexpression, it is possible that the rate of endocytosis is increased, targeting the LDLR for premature degradation. To investigate this possibility, cells were treated for variable lengths of time with monensin to inhibit LDLR recycling, while allowing endocytosis to continue (20). At each time point, cell-surface proteins were biotinylated and immunoprecipitated for the LDLR. The amount of biotinylated LDLR visualized with avidin-HRP at each time point represents the amount of LDLR remaining at the cell surface. As shown in Fig. 6, which is published as supporting information on the PNAS web site, infection with control and PCSK9 adenovirus both caused a reduction in cell-surface LDLR over the duration of monensin treatment. Furthermore, the rate of disappearance of the LDLR, and thus the rate of endocytosis, was identical in control and PCSK9-overexpressing cells.
Discussion
Overexpression of the subtilisin serine protease, PCSK9, reduces hepatic LDLR protein levels, with no change in LDLR mRNA levels (16, 18), indicating that PCSK9 may mediate posttranscriptional regulation of the LDLR. To investigate the mechanism of this effect, PCSK9 was overexpressed in HepG2 cells. Overexpression of PCSK9 did not have an effect on the incorporation of 35S-Met/Cys into the LDLR or on the conversion of the precursor to mature form of the LDLR, indicating that PCSK9 does not affect LDLR synthesis or maturation. In contrast, pulse–chase studies demonstrated that PCSK9 had a major effect on accelerating the degradation of the mature LDLR and that this effect required an active PCSK9 catalytic domain. Use of various inhibitors demonstrated that the PCSK9-induced degradation of the LDLR does not require the proteasome and occurs in a post-ER, pH-dependent compartment.
The LDLR is synthesized as a precursor, 120-kDa form in the ER. After its transport through the ER and Golgi, the LDLR acquires its mature carbohydrate residues and has an apparent molecular mass of 160 kDa (24). The mature LDLR is then transported from the Golgi to the cell surface, where it resides in clathrin-coated pits because of its interaction with ARH (25). The LDLR then undergoes endocytosis in the presence or absence of ligand, entering the endocytic recycling compartment. The pH change allows dissociation of the LDLR and ligand; the ligand moves to the lysosome to be degraded, and the LDLR recycles (24, 25). Despite an extensive list of mutations in the LDLR that affect the LDLR life cycle, very little is actually known about other proteins that play a role in and possibly regulate LDLR synthesis, maturation, trafficking to the cell surface, or degradation of receptor molecules originating from the secretory or the endocytic pathway.
Our data indicate that PCSK9 may play a role in regulating LDLR levels by inducing degradation of the mature LDLR in a PCSK9 catalytic dependent mechanism. PCSK9 is synthesized as a precursor, 72-kDa form in the ER and is trafficked to the Golgi, where it undergoes intramolecular self-cleavage to an active 63-kDa protease (26). Given this and the fact that the mature LDLR is found in the Golgi, it is therefore likely that the PCSK9 effect occurs in or is initiated in the Golgi or trans-Golgi complex. Our data in BFA-treated cells and data by Park et al. (18) indicating that PCSK9 can still degrade the LDLR in the absence of LDLR endocytosis localize the PCSK9 effect to the distal secretory pathway and support this hypothesis. These data also indicate that PCSK9 is probably not playing a role in the constitutive degradation of the LDLR that occurs at the end of the LDLR lifetime in the endocytic recycling compartment. To further localize the PCSK9 site of action, we attempted to use incubation of cells at 20°C (27) or in low concentrations of monensin (28) to determine whether the PCSK9 effect required transport out of the Golgi (data not shown). The results suggested that the PCSK9-induced LDLR degradation requires transport out of the Golgi, but this interpretation is complicated by the fact that PCSK9 in vitro is reported to be catalytically inactive below 25°C (29) and monensin inhibited the conversion of precursor to mature LDLR.
Protein degradation in the secretory compartment is a known mechanism of protein quality control and, theoretically, may also be involved in the normal regulation of protein expression. The most well studied mechanism of protein degradation in the secretory compartment is ER-associated degradation (ERAD) (30–32). ERAD typically involves the retrotranslocation of luminal or transmembrane proteins across the ER membrane, followed by polyubiquitination and subsequent degradation by the proteasome. Our data do not support a PCSK9-induced degradation of the LDLR by ERAD because degradation was prevented in BFA-treated cells and was not sensitive to proteasome inhibitors. Thus, LDLR degradation by PCSK9 is different from that reported for class 2 LDLR mutants, which undergo proteasomal degradation (33). In the same study, wild-type LDLR was not affected by proteasome inhibitors.
Another mode of degradation from the secretory pathway involves targeting of proteins by an unknown mechanism from the Golgi to the endosomal system for lysosomal or vacuolar degradation (34). Furthermore, plasma membrane proteins may be ubiquitinated and then degraded by lysosomes or proteasomes (34). The data that localize the PCSK9-induced degradation of the LDLR between the ER and plasma membrane (18) and the data that ammonium chloride decreases PCSK9-induced degradation of the LDLR support a potential role for the lysosome in PCSK9-induced LDLR degradation. The lysosome contains proteases of mostly the cysteine class, but also aspartic acid, metalloprotease, and serine proteases (35, 36). Our data with E64d, pepstatin, and phosphoramidon indicate that the PCSK9-induced degradation of the LDLR does not depend on cysteine, aspartic acid, and metalloprotease families and does not provide support for lysosomal degradation. However, not all lysosomal proteases may be inhibited in our experiments, and it is still possible that the responsible proteases are found in lysosomes.
Regulation of LDLR levels has been mostly studied at a transcriptional level; however, evidence exists that the LDLR is also subjected to regulation at the level of mRNA stability (37–39), translation (40), protein degradation, and limited proteolysis. Regulation at the level of protein degradation has been suggested in the Zucker fatty rat (41), and in rats treated with endotoxin (42), glucagon (43), histamine (44), and coffee diterpenes (45, 46); however, the exact mechanisms are unknown. Furthermore, it is known that certain cell lines, such as the J774 macrophage cell line, have increased degradation rates of the LDLR (22, 47). Interestingly, the increased degradation in J774 cells was shown not to be affected by the cysteine protease inhibitors leupeptin and E64 but slowed by BFA treatment and incubation at 18°C (22). These results are similar to our data on the PCSK9-induced degradation of the LDLR. However, the mechanisms must be at least partially different in that ammonium chloride did not affect the increased degradation in J774 cells (22), and there is no evidence for PCSK9 expression in macrophages (K.N.M. and J.L.B., unpublished observations).
There is also evidence for regulation of the LDLR at the level of limited proteolysis. First, Begg et al. (48) demonstrated that phorbol ester treatment of HepG2 cells increased the LDLR degradation rate by inducing the release of a soluble LDLR. It does not appear, however, that PCSK9 is acting by this mechanism because no evidence for a soluble LDLR was found in the media of control or PCSK9-overexpressing cells (K.N.M. and J.L.B., unpublished results). Second, Kraemer et al. (49) demonstrated that treatment with the cAMP inducing agents isoproterenol and forskolin decreased LDLR levels in adipose cells concomitant with the appearance of a 90- to 95-kDa proteolytic product in the plasma membrane. Although we have not investigated plasma membrane fractions specifically, we have also not found evidence of a proteolytic LDLR cleavage product in PCSK9-overexpressing cells, although this still remains a possibility.
The existence of LDLR mutants with accelerated degradation also supports the notion of regulation at the level of protein degradation (50–55). These mutant LDLRs show either normal or only slightly slowed processing of precursor to mature forms and greatly accelerated degradation rates in the absence or presence of ligand, leading to decreased LDLR function and hypercholesterolemia. These mutations include missense mutations (50–52, 54), a small deletion (53), and a small duplication (55) all in the EGF precursor homology domain, indicating that this domain may be important for LDLR degradation. Of note, deletion of the entire EGF precursor homology domain only causes an increased degradation of the LDLR in the presence of ligand (56), indicating that the role of the EGF precursor homology domain is complex. It is possible that analysis of the accelerated degradation mutants may aid in the elucidation of the PCSK9 pathway, including the identification of other important members.
In summary, we have shown that overexpression of PCSK9 in HepG2 cells leads to accelerated degradation of the LDLR by a nonproteasomal mechanism in a post-ER, pH-sensitive compartment. This effect depends on an active PCSK9 catalytic domain as has been shown in vivo (18). However, it is still not certain whether PCSK9 is acting directly or through other mediators to degrade the LDLR in the Golgi or post-Golgi compartment. It is also not clear why PCSK9 overexpression in mice results in a similar phenotype to humans carrying PCSK9 missense mutations. The simplest interpretation is that these are gain-of-function mutations; however, it is still possible that overexpression of PCSK9 may result in the same phenotype as loss-of-function mutations.
Supplementary Material
Acknowledgments
We thank Meihui Pan, Sandy Simon, Sekhar Ramakrishnan, and Markus Stoffel for useful discussions. This work was supported by National Institutes of Health Medical Scientist Training Program Grant GM07739 (to K.N.M.) and National Institutes of Health Grants HL32435 (to J.L.B.) and HL58541 (to E.A.F.).
Author contributions: K.N.M., E.A.F., and J.L.B. designed research; K.N.M. performed research; E.A.F. contributed new reagents/analytic tools; K.N.M., E.A.F., and J.L.B. analyzed data; and K.N.M. and J.L.B. wrote the paper.
Abbreviations: ADH, autosomal dominant hypercholesterolemia; ARH, autosomal recessive hypercholesterolemia; BFA, brefeldin A; ER, endoplasmic reticulum; HRP, horseradish peroxidase; LDL, low-density lipoprotein; LDLR, LDL receptor; PCSK9, proprotein convertase subtilisin kexin 9.
References
- 1.Bergeron, F., Leduc, R. & Day, R. (2000) J. Mol. Endocrinol. 24, 1–22. [DOI] [PubMed] [Google Scholar]
- 2.Gensberg, K., Jan, S. & Matthews, G. M. (1998) Semin. Cell Dev. Biol. 9, 11–17. [DOI] [PubMed] [Google Scholar]
- 3.Seidah, N. G. & Chretien, M. (1999) Brain Res. 848, 45–62. [DOI] [PubMed] [Google Scholar]
- 4.Taylor, N. A., Van De Ven, W. J. & Creemers, J. W. (2003) FASEB J. 17, 1215–1227. [DOI] [PubMed] [Google Scholar]
- 5.Zhou, A., Webb, G., Zhu, X. & Steiner, D. F. (1999) J. Biol. Chem. 274, 20745–20748. [DOI] [PubMed] [Google Scholar]
- 6.Maxwell, K. N., Soccio, R. E., Duncan, E. M., Sehayek, E. & Breslow, J. L. (2003) J. Lipid Res. 44, 2109–2119. [DOI] [PubMed] [Google Scholar]
- 7.Dubuc, G., Chamberland, A., Wassef, H., Davignon, J., Seidah, N. G., Bernier, L. & Prat, A. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1454–1459. [DOI] [PubMed] [Google Scholar]
- 8.Horton, J. D., Shah, N. A., Warrington, J. A., Anderson, N. N., Park, S. W., Brown, M. S. & Goldstein, J. L. (2003) Proc. Natl. Acad. Sci. USA 100, 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abifadel, M., Varret, M., Rabes, J. P., Allard, D., Ouguerram, K., Devillers, M., Cruaud, C., Benjannet, S., Wickham, L., Erlich, D., et al. (2003) Nat. Genet. 34, 154–156. [DOI] [PubMed] [Google Scholar]
- 10.Timms, K. M., Wagner, S., Samuels, M. E., Forbey, K., Goldfine, H., Jammulapati, S., Skolnick, M. H., Hopkins, P. N., Hunt, S. C. & Shattuck, D. M. (2004) Hum. Genet. 114, 349–353. [DOI] [PubMed] [Google Scholar]
- 11.Leren, T. P. (2004) Clin. Genet. 65, 419–422. [DOI] [PubMed] [Google Scholar]
- 12.Shioji, K., Mannami, T., Kokubo, Y., Inamoto, N., Takagi, S., Goto, Y., Nonogi, H. & Iwai, N. (2004) J. Hum. Genet. 49, 109–114. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein, J. L., Hobbs, H. & Brown, M. S. (2001) in The Metabolic and Molecular Basis of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), Vol. 2, pp. 2863–2913. [Google Scholar]
- 14.Rader, D. J., Cohen, J. & Hobbs, H. H. (2003) J. Clin. Invest. 111, 1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen, J. C., Kimmel, M., Polanski, A. & Hobbs, H. H. (2003) Curr. Opin. Lipidol. 14, 121–127. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell, K. N. & Breslow, J. L. (2004) Proc. Natl. Acad. Sci. USA 101, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjannet, S., Rhainds, D., Essalmani, R., Mayne, J., Wickham, L., Jin, W., Asselin, M. C., Hamelin, J., Varret, M., Allard, D., et al. (2004) J. Biol. Chem. 279, 48865–48875. [DOI] [PubMed] [Google Scholar]
- 18.Park, S. W., Moon, Y. A. & Horton, J. D. (2004) J. Biol. Chem. 279, 50630–50638. [DOI] [PubMed] [Google Scholar]
- 19.Tolleshaug, H., Goldstein, J. L., Schneider, W. J. & Brown, M. S. (1982) Cell 30, 715–724. [DOI] [PubMed] [Google Scholar]
- 20.Michaely, P., Li, W. P., Anderson, R. G., Cohen, J. C. & Hobbs, H. H. (2004) J. Biol. Chem. 279, 34023–34031. [DOI] [PubMed] [Google Scholar]
- 21.Shite, S., Seguchi, T., Mizoguchi, H., Ono, M. & Kuwano, M. (1990) J. Biol. Chem. 265, 17385–17388. [PubMed] [Google Scholar]
- 22.Shite, S., Seguchi, T., Shimada, T., Ono, M. & Kuwano, M. (1990) Eur. J. Biochem. 191, 491–497. [DOI] [PubMed] [Google Scholar]
- 23.Soutar, A. K., Naoumova, R. P. & Traub, L. M. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 1963–1970. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein, J. L., Brown, M. S., Anderson, R. G., Russell, D. W. & Schneider, W. J. (1985) Annu. Rev. Cell Biol. 1, 1–39. [DOI] [PubMed] [Google Scholar]
- 25.Gent, J. & Braakman, I. (2004) Cell. Mol. Life Sci. 61, 2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidah, N. G., Benjannet, S., Wickham, L., Marcinkiewicz, J., Jasmin, S. B., Stifani, S., Basak, A., Prat, A. & Chretien, M. (2003) Proc. Natl. Acad. Sci. USA 100, 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saraste, J., Palade, G. E. & Farquhar, M. G. (1986) Proc. Natl. Acad. Sci. USA 83, 6425–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa, S., Sakata, N., Ginsberg, H. N. & Dixon, J. L. (1992) J. Biol. Chem. 267, 22630–22638. [PubMed] [Google Scholar]
- 29.Naureckiene, S., Ma, L., Sreekumar, K., Purandare, U., Lo, C. F., Huang, Y., Chiang, L. W., Grenier, J. M., Ozenberger, B. A., Jacobsen, J. S., et al. (2003) Arch. Biochem. Biophys. 420, 55–67. [DOI] [PubMed] [Google Scholar]
- 30.Bonifacino, J. S. & Weissman, A. M. (1998) Annu. Rev. Cell Dev. Biol. 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sitia, R. & Braakman, I. (2003) Nature 426, 891–894. [DOI] [PubMed] [Google Scholar]
- 32.Trombetta, E. S. & Parodi, A. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 649–676. [DOI] [PubMed] [Google Scholar]
- 33.Li, Y., Lu, W., Schwartz, A. L. & Bu, G. (2004) J. Lipid Res. 45, 1084–1091. [DOI] [PubMed] [Google Scholar]
- 34.Arvan, P., Zhao, X., Ramos-Castaneda, J. & Chang, A. (2002) Traffic 3, 771–780. [DOI] [PubMed] [Google Scholar]
- 35.Pillay, C. S., Elliott, E. & Dennison, C. (2002) Biochem. J. 363, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turk, V., Turk, B. & Turk, D. (2001) EMBO J. 20, 4629–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, G. M., Vasa, M. Z. & Deeley, R. G. (1998) J. Lipid Res. 39, 1025–1032. [PubMed] [Google Scholar]
- 38.Nakahara, M., Fujii, H., Maloney, P. R., Shimizu, M. & Sato, R. (2002) J. Biol. Chem. 277, 37229–37234. [DOI] [PubMed] [Google Scholar]
- 39.Kong, W., Wei, J., Abidi, P., Lin, M., Inaba, S., Li, C., Wang, Y., Wang, Z., Si, S., Pan, H., et al. (2004) Nat. Med. 10, 1344–1351. [DOI] [PubMed] [Google Scholar]
- 40.Huang, Y., Ghosh, M. J. & Lopes-Virella, M. F. (1997) J. Lipid Res. 38, 110–120. [PubMed] [Google Scholar]
- 41.Liao, W., Angelin, B. & Rudling, M. (1997) Endocrinology 138, 3276–3282. [DOI] [PubMed] [Google Scholar]
- 42.Liao, W., Rudling, M. & Angelin, B. (1996) Biochem. J. 313, 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudling, M. & Angelin, B. (1993) J. Clin. Invest. 91, 2796–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao, W., Rudling, M. & Angelin, B. (1997) Endocrinology 138, 1863–1870. [DOI] [PubMed] [Google Scholar]
- 45.Rustan, A. C., Halvorsen, B., Huggett, A. C., Ranheim, T. & Drevon, C. A. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 2140–2149. [DOI] [PubMed] [Google Scholar]
- 46.de Roos, B. & Katan, M. B. (1999) Curr. Opin. Lipidol. 10, 41–45. [DOI] [PubMed] [Google Scholar]
- 47.Yoshimura, A., Seguchi, T., Yoshida, T., Shite, S., Waki, M. & Kuwano, M. (1988) J. Biol. Chem. 263, 11935–11942. [PubMed] [Google Scholar]
- 48.Begg, M. J., Sturrock, E. D. & van der Westhuyzen, D. R. (2004) Eur. J. Biochem. 271, 524–533. [DOI] [PubMed] [Google Scholar]
- 49.Kraemer, F. B., Natu, V., Singh-Bist, A., Patel, S., Komaromy, M. C., Medicherla, S., Azhar, S. & Sztalryd, C. (1996) J. Lipid Res. 37, 237–249. [PubMed] [Google Scholar]
- 50.Fourie, A. M., Coetzee, G. A., Gevers, W. & van der Westhuyzen, D. R. (1988) Biochem. J. 255, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leitersdorf, E., van der Westhuyzen, D. R., Coetzee, G. A. & Hobbs, H. H. (1989) J. Clin. Invest. 84, 954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyake, Y., Tajima, S., Funahashi, T., Yamamura, T. & Yamamoto, A. (1992) Eur. J. Biochem. 210, 1–7. [DOI] [PubMed] [Google Scholar]
- 53.van der Westhuyzen, D. R., Stein, M. L., Henderson, H. E., Marais, A. D., Fourie, A. M. & Coetzee, G. A. (1991) Biochem. J. 277, 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubinsztein, D. C., Coetzee, G. A., Marais, A. D., Leitersdorf, E., Seftel, H. C. & van der Westhuyzen, D. R. (1992) J. Lipid Res. 33, 1647–1655. [PubMed] [Google Scholar]
- 55.Bertolini, S., Patel, D. D., Coviello, D. A., Lelli, N., Ghisellini, M., Tiozzo, R., Masturzo, P., Elicio, N., Knight, B. L. & Calandra, S. (1994) J. Lipid Res. 35, 1422–1430. [PubMed] [Google Scholar]
- 56.Davis, C. G., Goldstein, J. L., Sudhof, T. C., Anderson, R. G., Russell, D. W. & Brown, M. S. (1987) Nature 326, 760–765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.