Abstract
Parkinson’s disease is caused by a complex interplay of genetic and environmental factors. Although a number of independent molecular pathways and processes have been associated with familial Parkinson’s disease, a common mechanism underlying especially sporadic Parkinson’s disease is still largely unknown. In order to gain further insight into the etiology of Parkinson’s disease, we here conducted genetic network and literature analyses to integrate the top-ranked findings from thirteen published genome-wide association studies of Parkinson’s disease (involving 13.094 cases and 47.148 controls) and other genes implicated in (familial) Parkinson’s disease, into a molecular interaction landscape. The molecular Parkinson’s disease landscape harbors four main biological processes—oxidative stress response, endosomal-lysosomal functioning, endoplasmic reticulum stress response, and immune response activation—that interact with each other and regulate dopaminergic neuron function and death, the pathological hallmark of Parkinson’s disease. Interestingly, lipids and lipoproteins are functionally involved in and influenced by all these processes, and affect dopaminergic neuron-specific signaling cascades. Furthermore, we validate the Parkinson’s disease -lipid relationship by genome-wide association studies data-based polygenic risk score analyses that indicate a shared genetic risk between lipid/lipoprotein traits and Parkinson’s disease. Taken together, our findings provide novel insights into the molecular pathways underlying the etiology of (sporadic) Parkinson’s disease and highlight a key role for lipids and lipoproteins in Parkinson’s disease pathogenesis, providing important clues for the development of disease-modifying treatments of Parkinson’s disease.
Molecular interactions: the importance of fats
Lipids and lipoproteins play a central role in four key biological processes underlying Parkinson’s disease (PD). Using bioinformatics and other extensive analyses of previously published data, Geert Poelmans, Cornelius Klemann and colleagues in The Netherlands, Germany and the USA have mapped the interactions of proteins that are encoded by genes associated with both familial and sporadic forms of PD. They identify the oxidative stress response, lysosomal function, endoplasmic reticulum stress response and immune response activation as the main mechanisms leading to the death of dopaminergic neurons. Lipid signaling is implicated in all four of these processes and the authors find a link between the levels of particular lipids and lipoproteins and the risk of PD. These findings suggest that compounds that regulate lipid or lipoprotein levels offer a potential new treatment strategy for PD.
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disease, with an estimated prevalence of 0.3%, affecting 1–2% of people over 60 years of age.1, 2 The pathological hallmark of PD is loss of dopaminergic (DA) neurons in the substantia nigra (SN), and the presence of protein aggregates (i.e., Lewy bodies) involving synuclein alpha (SNCA) in the residual DA neurons.3 A number of biological processes that contribute to the pathogenesis of PD have been identified, including defects in mitochondrial function,4 oxidative stress,5 and protein aggregation.6–8 However, detailed insights into the molecular mechanisms underlying these processes, and how they interact with each other, are essentially lacking. In many studies exploring PD pathogenesis, familial PD genes served as starting point. Thus far, at least eighteen genetic loci for familial PD have been found, and twelve familial PD candidate genes have been identified (ATP13A2, DJ-1, DNAJC6, EIF4G1, FBXO7, LRRK2, PARK2, PINK1, PLA2G6, SNCA, SYNJ1, and VPS35) (refs 9, 10). However, as a mutation in one of these familial genes is found in only 5–10% of the cases, PD should be considered a predominantly sporadic disease,11, 12 with both genetic and environmental contributing risk factors. In recent years, 15 genome-wide association studies (GWASs) have investigated genetic risk factors for sporadic PD13–26 but the functional coupling of the proteins encoded by the GWAS-identified candidate genes to PD pathophysiology is often not clear. In the present study, we aimed to identify the core mechanisms underlying PD pathogenesis by using bioinformatics and extensive literature analyses to integrate (1) the genes corresponding to the top-ranked single-nucleotide polymorphisms (SNPs) found in published GWASs of sporadic PD, and (2) other PD candidate genes (e.g., familial PD genes) into a protein interaction landscape. This molecular landscape allowed us to identify the specific biological processes that are key in PD pathogenesis and provides clues for the development of novel PD treatment strategies.
Results
Selected PD GWAS genes and genetic network enrichment analysis
Thirteen of the fifteen published PD GWASs met our inclusion criteria (Supplementary Table 1) and were used to select a total of 451 PD GWAS candidate genes based on SNPs with p < 0.0001 (Supplementary Table 2). Of the five most significantly enriched ingenuity pathway analysis (IPA) networks (Supplementary Table 3), the network with the highest enrichment score (p = 1.00E-44) and the highest number of PD GWAS candidate gene-encoded proteins (28 proteins) served as the starting point for the building of the molecular landscape (Supplementary Fig. 1).
The molecular landscape of PD
Guided by the most significantly enriched genetic network and extensive literature searches, we built a molecular landscape consisting of 260 interacting proteins (i.e., encoded by approximately 58% of the 451 PD GWAS genes, Supplementary Table 2), 128 proteins implicated in PD through other evidence (Supplementary Table 4) and 49 proteins that have not been directly linked to PD (yet) but have multiple functional interactions within the landscape (Supplementary Table 4). Approximately one in three landscape proteins are implicated in PD etiology through at least two types of evidence.
Supplementary Figures 2 and 3 show all relevant protein interactions in the PD landscape that are functionally involved in four main biological processes:oxidative stress response, endosomal-lysosomal functioning, endoplasmic reticulum (ER) stress response, and neuron death and immune response. The Supplementary Information provides a detailed and referenced description of the evidence linking all the proteins in the landscape. In Supplementary Table 5, we have indicated in which process(es) each landscape protein exerts its main effect and where it is located in Supplementary Figs 2 and/or 3.
The above being said, we here give a succinct description of the four main biological processes and signaling cascades in the PD landscape that are depicted in Fig. 1. Central in the landscape is signaling involving lipoproteins—i.e., low-density lipoprotein (LDL), high-density lipoprotein (HDL) and very low-density lipoprotein (VLDL)—and their component lipids and metabolites (e.g., cholesterol, oxysterols, sphingolipids such as ceramide and sphingosine, and triglycerides). Lipid and lipoprotein signaling represents the ‘‘common denominator’’ that functionally integrates, regulates and is regulated by the four landscape processes (Fig. 1a–d). Either by themselves or in combination, deficits or impairments in any of these four processes—each composed of multiple signaling cascades—can contribute to the degeneration and ultimately death of DA neurons.
Fig. 1.
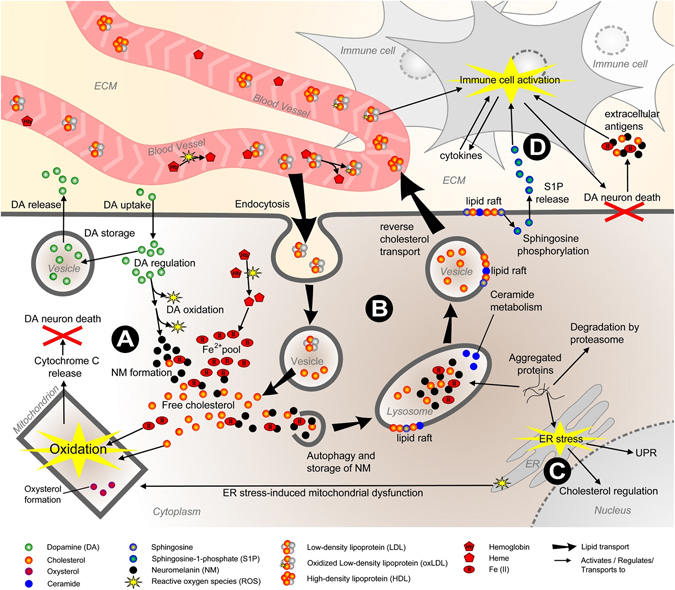
Overview of the molecular landscape of PD. The four main biological processes in the PD landscape—oxidative stress response (Fig. 1a), endosomal-lysosomal functioning (Fig. 1b), endoplasmic reticulum (ER) stress response (Fig. 1c), and neuron death and immune response (Fig. 1d)—are depicted. ECM extracellular matrix, ER endoplasmic reticulum, UPR unfolded protein response
First, deficits or impairments in dopamine synthesis and—linked to this—iron metabolism can cause an increased oxidative stress response (Fig. 1a). Dopamine can be either taken up through active transport or is newly synthesized in neurons and can subsequently be re-released (through vesicular exocytosis), degraded or (auto-)oxidized into neuromelanin (NM). Further, like erythrocytes (see below), SN DA neurons have a high-oxygen demand and express oxygen-carrying hemoglobin.
Through oxidation, cytotoxic heme is released from hemoglobin and then converted in DA neurons to ferrous iron, Fe(II). Fe(II) increases oxidative stress and together with free cholesterol—that is taken up by neurons through lipoproteins (see below)—induces mitochondrial oxysterol formation. In turn, this causes mitochondrial dysfunction and triggers the release of pro-apoptotic cytochrome c and, eventually, neuron death.
The second main landscape process centers around the (dys) regulation of endosomal-lysosomal functioning (Fig. 1b). Neuronal uptake of cholesterol occurs through the endosomal system, i.e., after neuronal uptake through vesicular endocytosis, LDL particles are processed into their composite parts: proteins, free cholesterol and other lipids. Free cholesterol and Fe(II) are bound in complexes by NM, which are then stored in lysosomes through autophagy. Hence, NM complex formation prevents the above described Fe(II)- and cholesterol-induced oxidative stress response. Moreover, their ageing-related increase in NM content and the associated increased demands on lysosomal function renders DA neurons particularly vulnerable to lysosomal defects. Other important lysosomal functions include the degradation of misfolded or aggregated proteins (such as pathological SNCA aggregates), the regulation of ceramide metabolism and reverse cholesterol transport, i.e., the vesicle-mediated transport and exocytosis of cholesterol into HDL particles in the bloodstream (and back to the liver). As such, a defect in any of these endosomal-lysosomal system components results in disturbed levels of lipids such as cholesterol and ceramide. In turn, these disturbed lipid levels affect membrane function in general and more specifically the functioning of so-called lipid rafts—microdomains of the vesicular, lysosomal, and plasma membrane containing high amounts of cholesterol and sphingolipids and crucial for membrane function—and hence processes such as autophagy, endo- and exocytosis. Deficient lysosomal function together with reduced degradation by the proteasome also leads to misfolded or aggregated protein formation.
Misfolded/aggregated proteins trigger the ER stress response (Fig. 1c), the third main landscape process, and subsequent activation of the protective unfolded protein response (UPR) as well as stimulation of cholesterol influx through upregulating the expression of key lipoprotein receptors. Prolonged ER stress that can no longer be counteracted by the UPR induces mitochondrial dysfunction, which eventually results in DA neuron death.
Lastly, apart from or in addition to dysregulated processes within DA neurons (as described above), DA neuron death can be the consequence of external factors such as an exaggerated immune response (Fig. 1d), the fourth landscape process. In this respect, immune cells are activated and attracted to damaged or already dying DA neurons by extracellular factors such as the sphingolipid-derived sphingosine-1-phosphate (S1P), triglyceride-rich extraneuronal VLDL particles (not shown), heme-oxidized LDL (oxLDL, see above), and various cytokines. Subsequently, the damaged/dying DA neurons are removed by the activated immune cells, an essentially normal and adequate response that is exaggerated in PD by DA neuron-specific antigens such as SNCA aggregates and NM complexes—released by dying DA neurons—creating a vicious cycle of DA neuron death and immune cell activation.
Polygenic risk score (PRS) analyses
Because our molecular landscape pointed towards an important role for lipids and lipoproteins in PD etiology (see above), we conducted PRS analyses using the tool PRSice,27 with GWAS data for the blood levels of various lipids and lipoproteins28 as base samples and meta-analytic PD GWAS data from the International Parkinson Disease Genomics Consortium (IPDGC)29 as target sample. We found statistically significant evidence (false discovery rate (FDR)-corrected p < 0.05) for a shared genetic etiology between the lipid traits ‘‘total cholesterol levels’’ and ‘‘total triglyceride levels’’ and PD, with the most predictive p-value threshold (p T) at 0.001 and 0.05, respectively (Fig. 2). In contrast, the lipoprotein traits ‘‘total HDL levels’’ and ‘‘total LDL levels’’ yielded no evidence for a shared genetic risk with PD (Fig. 2). For the various combinations of increased or decreased HDL and LDL levels, we found significant evidence for a shared genetic etiology between PD and the combined trait ‘‘increased HDL + increased LDL’’ (most predictive p T = 0.05) (Supplementary Fig. 4).
Fig. 2.
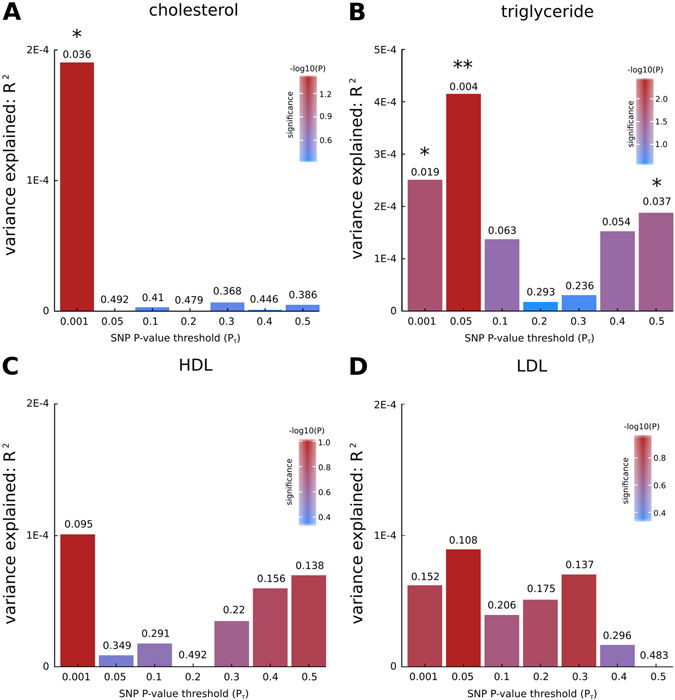
Bar plots from PRSice for shared genetic etiology between four lipid and lipoprotein traits (cholesterol, triglyceride, HDL, and LDL levels) and PD showing results at broad p-value thresholds (p T). The numbers above the bars indicate the p-values for shared genetic etiology, and these p-values were corrected using the false discovery rate (FDR) method; *denotes FDR-corrected p < 0.05, **denotes FDR-corrected p < 0.01
Discussion
In this study, we integrated the available genetic and expression data with data about environmental risk factors into a protein interaction landscape that reveals the main biological processes and signaling cascades that are affected in PD and occur in DA neurons and immune cells. Our PD landscape represents processes and cascades that are affected in both the monogenic, familial and the more prevalent polygenic, sporadic forms of PD. In this respect, the molecular landscape includes the ‘‘classic’’ processes and cascades known to be affected in PD that are based on the familial PD candidate genes (e.g., SNCA, PARK2, LRRK2): mitochondrial function, oxidative stress and protein aggregation. In addition, the landscape harbors more novel processes that have been less well studied in relation to PD pathogenesis yet, such as impairments in lysosomal function and immune response regulation. The landscape does not imply a ‘‘sequence of events’’ that leads to DA neuron loss, i.e., a number of (impaired) biological processes and cascades that occur in a temporally and/or spatially distinct order. Instead, deficits in any of the main landscape processes/cascades, either by themselves or in combination with deficits of other processes/cascades, may cause DA neurons to die. Moreover, an aging-related decline in the functionality and/or efficiency of landscape processes/cascades may play a role in PD onset and progression. For example, a gradual buildup of NM or aggregated proteins may disturb lysosomal function in DA neurons30 or an age-related decrease in the expression and activity of ER folding enzymes can compromise proper protein folding.31
Lipid and lipoprotein signaling functionally integrates, regulates and is regulated by the key landscape processes and cascades. Any disturbance of these processes and cascades can (eventually) result in DA neuron death, which is further aggravated or initiated by an increased (auto-)immune response.32 The involvement of deficient lipid and lipoprotein signaling in PD pathophysiology is corroborated by a number of environmental risk factor studies. Increased plasma levels of total cholesterol are associated with a lower PD risk.33–35 Nevertheless, a recent meta-analysis did not show an effect of higher or lower dietary cholesterol intake on PD risk,36 suggesting that direct cholesterol intake through food may not play a major role in PD etiology. Further, low plasma levels of LDL are linked to a higher PD risk,37, 38 whereas high-plasma HDL and CSF oxysterol levels are associated with increased PD risk and duration.39, 40 In addition, the levels of oxidized LDL, oxysterols and sphingolipids are increased in the plasma of PD patients.41–43 Thus, PD patients have a lower LDL:HDL ratio that is associated with a lower risk of cardiovascular disease (CVD)44 and could at least to some extent explain why the PD population is indeed less susceptible to developing CVD.39 Apart from the observed dysregulated levels of cholesterol (metabolites) and cholesterol-containing lipoproteins in PD patients, lower serum levels of triglycerides—which are highly enriched in VLDL particles—associate with an increased PD risk.45, 46
Intriguingly, we found a significant overlap between the polygenic risk associated with total cholesterol and triglyceride levels and PD. We also identified a shared genetic etiology between the combined lipoprotein trait ‘‘increased HDL + increased LDL’’ and PD. To our knowledge, we are the first to find a shared genetic risk between quantitative traits and a neurodegenerative disease but these findings need to be replicated in larger data sets, especially for the target sample, i.e., the PD GWAS data set. Together, the epidemiological and our PRS analysis findings indicate that the link between specific lipid/lipoprotein traits and PD may be the result of both shared environmental and genetic risk factors.
Given the converging evidence for lipid and lipoprotein signaling playing a key role in PD etiology, compounds that modulate lipid/lipoprotein levels could represent effective novel PD treatments. In this respect, statins—inhibitors of peripheral cholesterol synthesis that are used to treat hypercholesterolemia and hence prevent cholesterol-associated CVD—have a neuroprotective effect in the rat brain,47 but their effect on PD risk remains unclear.48–55 Interestingly, the only published prospective study that has adjusted for baseline cholesterol levels before statin treatment has found that statin use is associated with a significantly higher PD risk,35 which is in keeping with the observation that higher total plasma cholesterol levels—which are lowered by statins—are protective against PD. Other signaling molecules from the landscape that affect cholesterol and lipoprotein levels are testosterone and vitamin D3. Caucasian male PD patients show significantly reduced testosterone levels,56–58 and free testosterone levels are positively correlated with LDL, HDL, and total cholesterol levels.59 Therefore, decreased testosterone levels may impact on several key PD landscape processes, as testosterone regulates the efflux of LDL and HDL to the circulation.60 Hence, testosterone could be used for treating PD in male patients and indeed, testosterone treatment has some modest beneficial effects in men with PD.61, 62 Deficiency of vitamin D3—which affects cholesterol metabolism through downregulating SREBF1 (ref. 63), the main transcriptional activator of lipid homeostasis and key landscape protein—has been consistently associated with an increased PD risk64 and its supplementation may stabilize PD symptoms.65
Lastly, a number of landscape proteins that both regulate lipid/lipoprotein signaling and landscape cascades involved in DA neuron death represent attractive (novel) drug targets for PD. Examples include HMOX1 that prevents oxidative stress by heme, PSAP and its receptor GPR37 that mediate ceramide metabolism, the immunity-related ICAM1 that is regulated by extracellular lipids and (oxidized) lipoproteins, and plasmin that regulates the degradation of extracellular SNCA and (lipo)proteins.
In conclusion, our integrated molecular landscape yields detailed insights into the mechanisms underlying PD pathogenesis, and highlights the involvement of deficient lipid and lipoprotein signaling. These findings warrant future rigorous perturbation experiments in PD cell and animal models that may eventually provide validated drug target ‘‘leads’’ for the development of novel disease-modifying PD treatments.
Methods
PD GWAS gene selection
The first step of our molecular landscape building approach66–68 is the selection of candidate genes based on GWAS SNPs and their corresponding p-values. All 15 PD GWASs published to date were considered. Criteria for inclusion were a publicly available GWAS discovery sample with all SNPs associated at p < 0.0001. From the GWASs for which these data were available, we then selected the SNPs that were associated with PD at p < 0.0001 to compile a list of associated genes. The selected genes either contained a SNP that was located within an exonic, intronic or untranslated region of the gene or were found within 100 kilobases (kb) downstream and upstream of the SNP. The latter was based on the fact that the vast majority of expression quantitative trait loci (eQTL) for a given gene are located within 100 kb downstream and/or upstream of a gene69–71 and because trait-associated SNPs are more likely to be eQTL.72 The chosen cutoff for association (p < 0.0001) is often employed to designate ‘‘suggestive’’ association and has been used in GWASs of multiple disorders.73–75 Subsequently, the literature was searched for additional (genetic) evidence linking the selected GWAS candidate genes to PD.
Genetic network enrichment analysis
To identify enriched protein networks in the PD GWAS candidate genes, a network analysis using the IPA software package (http://www.ingenuity.com) was performed with default parameters, i.e., the analysis used the so-called reference set of known genes and endogenous chemicals. This reference set is accessible through the ‘‘Ingenuity Knowledge Base’’, a repository of data from publicly accessible databases (e.g., BioGRID, IntAct) and data that are manually curated by Ingenuity through systematically reviewing published literature. In this respect, of all protein–protein interactions between the PD candidate proteins in the most significantly enriched IPA network shown in Supplementary Fig. 1 (see Results), we found approximately 4/5 through publicly accessible databases and 1/5 by IPA. In addition, only functional relationships that are corroborated by experimental evidence were included in the IPA networks. For each network, the Ingenuity software also generates an enrichment score that takes into account the number of eligible molecules/proteins in the network and its size, as well as the total number of network-eligible molecules analyzed and the total number of molecules in the Ingenuity Knowledge Base that could potentially be included in networks. This score is the negative logarithm of the right-tailed Fisher’s exact test result.
Molecular landscape building
Following the network enrichment analysis, the literature was extensively searched for the functions and interactions of all proteins encoded by the candidate genes implicated through PD GWASs as well as other PD candidates implicated via other evidence, including genetic association studies, messenger RNA/protein expression studies and/or functional studies. First, we used the UniProt Protein Knowledge Base (http://www.uniprot.org/uniprot)76 to gather basic information on the functions of all candidate genes and their encoded proteins. Subsequently and starting with the interactions in the most enriched genetic network, we used PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez) to search for all functional, experimental evidence-based interactions between all PD candidate genes/proteins. While building the landscape in this way, we also included genes/proteins and metabolites that have no known link with PD, but have multiple—i.e., at least two different—functional interactions with PD-implicated proteins. Based on all gathered information, we generated a protein interaction landscape. The figures depicting this landscape were made using the drawing program Serif DrawPlus version 4.0 (www.serif.com).
PRS analyses
Our molecular landscape pointed towards an important role for lipids and lipoproteins in PD etiology (see below). Therefore, we conducted PRS analyses using the tool PRSice,27 with summary statistics data from GWASs of blood levels of total cholesterol, total triglycerides, total HDL and total LDL as ‘‘base’’ samples (GWAS data for 188577 European-ancestry individuals from the general population)28 and summary statistics data from a meta-analytic PD GWAS by the IPDGC as ‘‘target’’ sample (GWAS data for 5333 PD cases and 12,019 healthy control subjects, all of European ancestry)29. Using the default settings in PRSice we calculated the shared genetic etiology between the four lipid/lipoprotein traits and PD at seven broad p-value thresholds (indicated by p T), which were used to select the SNPs from the base sample that were included in the PRS analysis, i.e., p T < 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5. As such, the seven p T thresholds led to the selection of all SNPs that were associated with the base lipid/lipoprotein phenotype at p < 0.001, 0.05, 0.1 etc.
The calculated p-values indicating the significance of a shared genetic etiology between each lipid/lipoprotein trait and PD were aggregated and corrected for multiple comparisons using the FDR method, incorporating potential dependencies between p-values.77
To calculate the FDR, we used the mafdr function in MATLAB (R2012a, The Mathworks, Natick, MA, USA) using the bootstrap selection method for the FDR parameter lambda. FDR was set to not lower p-values below uncorrected p-values, which would have occurred due to overall (relatively) low uncorrected p-values.
In addition to calculating the shared genetic etiology between the four lipid/lipoprotein traits and PD, we performed similar analyses using four ‘‘combined lipoprotein traits’’. In order to do this, we first divided the HDL and LDL level summary statistics GWAS data into four groups of SNPs, i.e., all SNPs associated with (1) increased HDL levels, (2) increased LDL levels, (3) decreased HDL levels, and (4) decreased LDL levels (where ‘‘increased’’ and ‘‘decreased’’ refer to all SNPs that had an effect size-indicating beta > 0 and beta < 0, respectively). Subsequently, we conducted PRSice analyses with four combined data sets as base sample, i.e., (1) all SNPs increasing HDL and LDL levels, (2) all SNPs decreasing HDL and LDL levels, (3) all SNPs increasing HDL and decreasing LDL levels, and (4), all SNPs decreasing HDL and increasing LDL levels. Before the data sets were fed into PRSice, an equivalent, weighted p-value for each SNP in each of the four combined HDL/LDL data sets was calculated as:
Electronic supplementary material
Acknowledgements
We are particularly grateful to the International Parkinson Disease Genomics Consortium (IPDGC)—of which authors T. Gasser and M. Sharma are members—for providing us with the summary statistics GWAS data from the discovery sample of their meta-analytic GWAS of PD29 that we used as the target sample for our PRS analyses. J.E. Visser was supported by Stichting Parkinsonfonds, the Netherlands Organization for Scientific Research (NWO/ZonMw, VENI 916.12.167) and The Netherlands Brain Foundation (F2014(1)-16). T. Gasser and M. Sharma received funding through the EU Joint Programme—Neurodegenerative Disease Research (JPND) project “Courage-PD”. This project is supported through the following funding organizations under the aegis of JPND (www.jpnd.eu): France, the French National Research Agency; Germany, the German Bundesministerium für Bildung und Forschung; Israel, the Israeli Ministry of Health; Italy, the Italian Ministry of Health/Ministry of Education, Universities and Research; Luxembourg, the Luxembourgian National Research Fund; The Netherlands, the Netherlands Organization for Health Research and Development; Norway, the Research Council of Norway; Portugal, the Portuguese Foundation for Science and Technology; Spain, the Spanish National Institute of Health Carlos III; United Kingdom, the Medical Research Council. O. Isacson received funding from the National Institutes of Health grant R21NS084149, the Michael J Fox Foundation and the Consolidated Anti-Aging Foundation.
Author contributions
C.K. integrated the available data into the PD molecular landscape. C.K., G.M., J.V., and G.P. were involved in designing the study. C.K., G.M., O.I., T.G., J.V., G.P. have contributed to the interpretation of the results. M.S. and T.G. have provided the summary statistics GWAS data for the polygenic risk score analysis that was performed by M.M. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
J. E. Visser and G. Poelmans contributed equally to this work.
Electronic supplementary material
Supplementary Information accompanies the paper on the npj Parkinson’s Disease website (doi:10.1038/s41531-017-0015-3).
References
- 1.Nussbaum RL, Ellis CE. Alzheimer’s disease and Parkinson’s disease. N. Engl. J. Med. 2003;348:1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 2.de Lau LM, Breteler MM. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006;5:525–535. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- 3.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan BJ, Hoek S, Fon EA, Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends Biochem. Sci. 2015;40:200–210. doi: 10.1016/j.tibs.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Blesa J, Trigo-Damas I, Quiroga-Varela A, Jackson-Lewis VR. Oxidative stress and Parkinson’s disease. Front. Neuroanat. 2015;9:91. doi: 10.3389/fnana.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trojanowski JQ, Goedert M, Iwatsubo T, Lee VM. Fatal attractions: abnormal protein aggregation and neuron death in Parkinson’s disease and Lewy body dementia. Cell Death. Differ. 1998;5:832–837. doi: 10.1038/sj.cdd.4400432. [DOI] [PubMed] [Google Scholar]
- 7.Gundersen V. Protein aggregation in Parkinson’s disease. Acta. Neurol. Scand. Suppl. 2010;190:82–87. doi: 10.1111/j.1600-0404.2010.01382.x. [DOI] [PubMed] [Google Scholar]
- 8.Ebrahimi-Fakhari D, Wahlster L, McLean PJ. Protein degradation pathways in Parkinson’s disease: curse or blessing. Acta Neuropathol. 2012;124:153–172. doi: 10.1007/s00401-012-1004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonifati V. Genetics of Parkinson’s disease--state of the art, 2013. Parkinsonism. Relat. Disord. 2014;20:S23–S28. doi: 10.1016/S1353-8020(13)70009-9. [DOI] [PubMed] [Google Scholar]
- 10.Spatola M, Wider C. Genetics of Parkinson’s disease: the yield. Parkinsonism. Relat. Disord. 2014;20:S35–S38. doi: 10.1016/S1353-8020(13)70011-7. [DOI] [PubMed] [Google Scholar]
- 11.Thomas B, Beal MF. Parkinson’s disease. Hum. Mol. Genet. 2007;2:R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 12.Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maraganore DM, et al. High-resolution whole-genome association study of Parkinson disease. Am. J. Hum. Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung HC, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 15.Pankratz N, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 2009;124:593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latourelle JC, et al. Genomewide association study for onset age in Parkinson disease. BMC Med. Genet. 2009;10:98. doi: 10.1186/1471-2350-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 18.Simon-Sanchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards TL, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamza TH, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spencer CC, et al. Dissection of the genetics of Parkinson’s disease identifies an additional association 5’ of SNCA and multiple associated haplotypes at 17q21. Hum. Mol. Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saad M, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson’s disease in the European population. Hum. Mol. Genet. 2011;20:615–627. doi: 10.1093/hmg/ddq497. [DOI] [PubMed] [Google Scholar]
- 23.Simon-Sanchez J, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur. J. Hum. Genet. 2011;19:655–661. doi: 10.1038/ejhg.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Do CB, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, et al. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med. Genet. 2011;12:104. doi: 10.1186/1471-2350-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez DG, et al. Genome wide assessment of young onset Parkinson’s disease from Finland. PLoS ONE. 2012;7:e41859. doi: 10.1371/journal.pone.0041859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalls MA, et al. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isacson O. Lysosomes to combat Parkinson’s disease. Nat. Neurosci. 2015;18:792–793. doi: 10.1038/nn.4027. [DOI] [PubMed] [Google Scholar]
- 31.Salganik M, et al. The loss of glucose-regulated protein 78 (GRP78) during normal aging or from siRNA knockdown augments human alpha-synuclein (alpha-syn) toxicity to rat nigral neurons. Neurobiol. Aging. 2015;36:2213–2223. doi: 10.1016/j.neurobiolaging.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleidi M, Isacson O. Viral and inflammatory triggers of neurodegenerative diseases. Sci. Transl. Med. 2012;4:121ps3. doi: 10.1126/scitranslmed.3003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Lau LM, Koudstaal PJ, Hofman A, Breteler MM. Serum cholesterol levels and the risk of Parkinson’s disease. Am. J. Epidemiol. 2006;164:998–1002. doi: 10.1093/aje/kwj283. [DOI] [PubMed] [Google Scholar]
- 34.Miyake Y, et al. Case-control study of risk of Parkinson’s disease in relation to hypertension, hypercholesterolemia, and diabetes in Japan. J. Neurol. Sci. 2010;293:82–86. doi: 10.1016/j.jns.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Huang X, et al. Statins, plasma cholesterol, and risk of Parkinson’s disease: a prospective study. Mov. Disord. 2015;30:552–559. doi: 10.1002/mds.26152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang A, Lin Y, Wu Y, Zhang D. Macronutrients intake and risk of Parkinson’s disease: a meta-analysis. Geriatr. Gerontol. Int. 2015;15:606–616. doi: 10.1111/ggi.12321. [DOI] [PubMed] [Google Scholar]
- 37.Huang X, et al. Lower low-density lipoprotein cholesterol levels are associated with Parkinson’s disease. Mov. Disord. 2007;22:377–381. doi: 10.1002/mds.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, et al. cholesterol and increased risk of Parkinson’s disease: prospective results from Honolulu-Asia Aging Study. Mov. Disord. 2008;23:1013–1018. doi: 10.1002/mds.22013. [DOI] [PubMed] [Google Scholar]
- 39.Cassani E, et al. Cardiometabolic factors and disease duration in patients with Parkinson’s disease. Nutrition. 2013;29:1331–1335. doi: 10.1016/j.nut.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Bjorkhem I, et al. Oxysterols and Parkinson’s disease: evidence that levels of 24S-hydroxycholesterol in cerebrospinal fluid correlates with the duration of the disease. Neurosci. Lett. 2013;555:102–105. doi: 10.1016/j.neulet.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Andican G, et al. Plasma oxidative and inflammatory markers in patients with idiopathic Parkinson’s disease. Acta Neurol. Belg. 2012;112:155–159. doi: 10.1007/s13760-012-0015-3. [DOI] [PubMed] [Google Scholar]
- 42.Seet RC, et al. Oxidative damage in Parkinson disease: Measurement using accurate biomarkers. Free Radic. Biol. Med. 2010;48:560–566. doi: 10.1016/j.freeradbiomed.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 43.Mielke MM, et al. Plasma ceramide and glucosylceramide metabolism is altered in sporadic Parkinson’s disease and associated with cognitive impairment: a pilot study. PLoS ONE. 2013;8:e73094. doi: 10.1371/journal.pone.0073094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jukema JW, et al. LDL-C/HDL-C ratio in subjects with cardiovascular disease and a low HDL-C: results of the RADAR (Rosuvastatin and Atorvastatin in different dosages and reverse cholesterol transport) study. Curr. Med. Res. Opin. 2005;21:1865–1874. doi: 10.1185/030079905X74952. [DOI] [PubMed] [Google Scholar]
- 45.Wei Q, et al. Reduced serum levels of triglyceride, very low density lipoprotein cholesterol and apolipoprotein B in Parkinson’s disease patients. PLoS ONE. 2013;8:e75743. doi: 10.1371/journal.pone.0075743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saaksjarvi K, Knekt P, Mannisto S, Lyytinen J, Heliovaara M. Prospective study on the components of metabolic syndrome and the incidence of Parkinson’s disease. Parkinsonism. Relat. Disord. 2015;21:1148–1155. doi: 10.1016/j.parkreldis.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 47.He X, Jenner AM, Ong WY, Farooqui AA, Patel SC. Lovastatin modulates increased cholesterol and oxysterol levels and has a neuroprotective effect on rat hippocampal neurons after kainate injury. J. Neuropathol. Exp. Neurol. 2006;65:652–663. doi: 10.1097/01.jnen.0000225906.82428.69. [DOI] [PubMed] [Google Scholar]
- 48.Wolozin B, et al. Simvastatin is associated with a reduced incidence of dementia and Parkinson’s disease. BMC Med. 2007;5:20. doi: 10.1186/1741-7015-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahner AD, Bronstein JM, Bordelon YM, Ritz B. Statin use and the risk of Parkinson disease. Neurology. 2008;70:1418–1422. doi: 10.1212/01.wnl.0000286942.14552.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X, Simon KC, Schwarzschild MA, Ascherio A. Prospective study of statin use and risk of Parkinson disease. Arch. Neurol. 2012;69:380–384. doi: 10.1001/archneurol.2011.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman B, Lahad A, Dresner Y, Vinker S. Long-term statin use and the risk of Parkinson’s disease. Am. J. Manag. Care. 2013;19:626–632. [PubMed] [Google Scholar]
- 52.Becker C, Jick SS, Meier CR. Use of statins and the risk of Parkinson’s disease: a retrospective case-control study in the UK. Drug Saf. 2008;31:399–407. doi: 10.2165/00002018-200831050-00004. [DOI] [PubMed] [Google Scholar]
- 53.Samii A, Carleton BC, Etminan M. Statin use and the risk of Parkinson disease: a nested case control study. J. Clin. Neurosci. 2008;15:1272–1273. doi: 10.1016/j.jocn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Hippisley-Cox J, Coupland C. Unintended effects of statins in men and women in England and Wales: population based cohort study using the QResearch database. BMJ. 2010;340:c2197. doi: 10.1136/bmj.c2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ritz B, et al. Statin use and Parkinson’s disease in Denmark. Mov. Disord. 2010;25:1210–1216. doi: 10.1002/mds.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ready RE, Friedman J, Grace J, Fernandez H. Testosterone deficiency and apathy in Parkinson’s disease: a pilot study. J. Neurol. Neurosurg. Psychiatry. 2004;75:1323–1326. doi: 10.1136/jnnp.2003.032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenangil G, Orken DN, Ur E, Forta H, Celik M. The relation of testosterone levels with fatigue and apathy in Parkinson’s disease. Clin. Neurol. Neurosurg. 2009;111:412–414. doi: 10.1016/j.clineuro.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 58.Brodacki B, et al. cGMP level in idiopathic Parkinson’s disease patients with and without cardiovascular disease - A pilot study. Parkinsonism. Relat. Disord. 2011;17:689–692. doi: 10.1016/j.parkreldis.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 59.Chock B, Lin TC, Li CS, Swislocki A. Plasma testosterone is associated with Framingham risk score. Aging Male. 2012;15:134–139. doi: 10.3109/13685538.2011.654369. [DOI] [PubMed] [Google Scholar]
- 60.Rubinow KB, et al. Acute sex steroid withdrawal increases cholesterol efflux capacity and HDL-associated clusterin in men. Steroids. 2012;77:454–460. doi: 10.1016/j.steroids.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okun MS, et al. Beneficial effects of testosterone replacement for the nonmotor symptoms of Parkinson disease. Arch. Neurol. 2002;59:1750–1753. doi: 10.1001/archneur.59.11.1750. [DOI] [PubMed] [Google Scholar]
- 62.Okun MS, et al. Testosterone therapy in men with Parkinson disease: results of the TEST-PD Study. Arch. Neurol. 2006;63:729–735. doi: 10.1001/archneur.63.5.729. [DOI] [PubMed] [Google Scholar]
- 63.Kong J, Li YC. Molecular mechanism of 1,25-dihydroxyvitamin D3 inhibition of adipogenesis in 3T3-L1 cells. Am. J. Physiol. Endocrinol. Metab. 2006;290:E916–E924. doi: 10.1152/ajpendo.00410.2005. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, et al. Vitamin D from different sources is inversely associated with Parkinson disease. Mov. Disord. 2015;30:560–566. doi: 10.1002/mds.26117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki M, et al. Randomized, double-blind, placebo-controlled trial of vitamin D supplementation in Parkinson disease. Am. J. Clin. Nutr. 2013;97:1004–1013. doi: 10.3945/ajcn.112.051664. [DOI] [PubMed] [Google Scholar]
- 66.Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am. J. Psychiatry. 2011;168:365–377. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- 67.Poelmans G, Franke B, Pauls DL, Glennon JC, Buitelaar JK. AKAPs integrate genetic findings for autism spectrum disorders. Transl. Psychiatry. 2013;3:e270. doi: 10.1038/tp.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van de Vondervoort I, et al. An integrated molecular landscape implicates the regulation of dendritic spine formation through insulin-related signalling in obsessive-compulsive disorder. J. Psychiatry Neurosci. 2016;41:140327. doi: 10.1503/jpn.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gherman A, Wang R, Avramopoulos D. Orientation, distance, regulation and function of neighbouring genes. Hum. Genomics. 2009;3:143–156. doi: 10.1186/1479-7364-3-2-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma D, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann. Hum. Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu W, et al. Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med. Genet. 2014;15:2. doi: 10.1186/1471-2350-15-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindstrom S, et al. Genome-wide association study identifies multiple loci associated with both mammographic density and breast cancer risk. Nat. Commun. 2014;5:5303. doi: 10.1038/ncomms6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.UniProt: a hub for protein information. Nucleic Acids Res. 43, (Database issue):D204-D212 (2015). [DOI] [PMC free article] [PubMed]
- 77.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57.1:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


