Abstract
Background
Anopheles sinensis is a dominant natural vector of Plasmodium vivax in China, Taiwan, Japan, and Korea. Recent genome sequencing of An. sinensis provides important insights into the genomic basis of vectorial capacity. However, the lack of a physical genome map with chromosome assignment and orientation of sequencing scaffolds hinders comparative analyses with other genomes to infer evolutionary changes relevant to the vector capacity.
Results
Here, a physical genome map for An. sinensis was constructed by assigning 52 scaffolds onto the chromosomes using fluorescence in situ hybridization (FISH). This chromosome-based genome assembly composes approximately 36% of the total An. sinensis genome. Comparisons of 3955 orthologous genes between An. sinensis and Anopheles gambiae identified 361 conserved synteny blocks and 267 inversions fixed between these two lineages. The rate of gene order reshuffling on the X chromosome is approximately 3.2 times higher than that on the autosomes.
Conclusions
The physical map will facilitate detailed genomic analysis of An. sinensis and contribute to understanding of the patterns and mechanisms of large-scale genome rearrangements in anopheline mosquitoes.
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-017-1888-7) contains supplementary material, which is available to authorized users.
Keywords: Chromosomal evolution, GRIMM, Inversion fixation, Fluorescence in situ hybridization, OrthoDB, Synteny blocks
Background
Anopheles sinensis is a member of the Anopheles hyrcanus group within the subfamily Anophelinae [1], which has a wide geographical distribution in Asia, mainly including Thailand, Malaysia, Indonesia, Singapore, Vietnam, China, Taiwan, Japan and Korea [2]. Within its range of distribution, An. sinensis has been historically considered as the most dominant and important natural vector of Plasmodium vivax in China, Taiwan, Japan, and Korea [3]. In China, An. sinensis is the most widespread vector of P. vivax, with a continuous range throughout 29 provinces and regions [4]. The recent re-emergence of vivax malaria, which started from 2001 in central China, appears in the areas where An. sinensis is the only vector [5] and further study reported high susceptibility of An. sinensis to P. vivax following artificial membrane feeding [6], suggesting that this species is responsible for the recent outbreaks of malaria. After being certified malaria-free, 204,428 and 300,000 cases of malaria in 2000 and 2001, respectively, were found in Korea [3], and three clusters of malaria cases were reported in Singapore in 2009 [7], all of which occurred in regions where An. sinensis was the predominant anopheline mosquito. As well as its role in malaria transmission, An. sinensis also plays a role in the transmission of a filarial worm, Brugia malayi, in China [8, 9].
Advances in next-generation sequencing (NGS) and assembly algorithms have rapidly promoted the analysis of genomes and comparative genomics in anopheline mosquitoes. The whole genome of An. sinensis was first published using the Roche/454 GS FLX sequencing approach with a Chinese laboratory strain and assembled into the 9595 scaffolds spanning 220.8 million base pairs (Mb) [10]. At almost the same time, the complete transcriptome of this species was obtained using the Illumina paired-end sequencing technology, and 38,504 unigenes were identified from another Chinese strain [11]. Later, the genome of a different strain of An. sinensis (‘SINENSIS’) was sequenced and assembled for comparative analyses by the 16 Anopheles mosquito genome project [12]. However, all these research efforts resulted in large numbers of scaffolds and contigs without chromosome assignment or orientations. The availability of a physical map for An. sinensis with scaffolds and contigs localized on the chromosomes will increase the quality of comparative genomic analyses with other mosquitoes that have chromosome-based genome assemblies, e.g. Anopheles gambiae. Such analyses will allow an exploration of the genomic basis of vectorial capacity and a study of the patterns of chromosome homology and rearrangements between species.
So far, physical maps have been developed for several Anopheles mosquito species including An. gambiae, Anopheles funestus, Anopheles stephensi, Anopheles atroparvus and Anopheles albimanus. These maps improved the draft genome assemblies and helped to understand the genome organization and evolution [13]. Anopheles gambiae and An. funestus represent two major African malaria vectors, while An. stephensi is a dominant vector in Asia. These species belong to the subgenus Cellia within the Series, Pyretophorus (An. gambiae), Myzomyia (An. funestus), and Neocellia (An. stephensi) [12]. Comparisons of the mapped genomes of An. funestus and An. stephensi with the An. gambiae genome have demonstrated that the X (sex) chromosome and the 2R arm are much more prone to rearrangement than the other chromosomal arms [14, 15].
Changes in gene order between An. gambiae and other species, including An. atroparvus and An. albimanus, demonstrated that the difference in the rate of evolution between the sex chromosome and autosomes is more than threefold [12]. A recent comparative genomic study between An. gambiae within genus Anopheles and Aedes aegypti in Culicinae also revealed that the sex-determining chromosome has a higher rate of genome rearrangements than autosomes [16]. However, whether fast evolution of the sex chromosome occurs in the majority of anophelines will not be clear until more species are investigated.
This study aimed to construct a physical map for An. sinensis by anchoring scaffold sequences onto the polytene chromosomes and to identify conserved synteny blocks and fixed inversions between An. sinensis and An. gambiae for exploring the patterns of chromosome evolution in Anopheles mosquitoes.
Methods
Mosquito strains and chromosome preparation
The Wuxi laboratory strain (Jiangsu Institute of Parasitic Diseases, Wuxi, China) of An. sinensis was used in this study. Polytene chromosome preparations were made using salivary glands dissected from early fourth-instar larvae of An. sinensis as previously described [17]. Chromosomes with clear banding patterns were fixed in liquid nitrogen and dehydrated in 50, 70, 90 and 100% ethanol for in situ hybridization.
Fluorescence in situ hybridization
Genome sequences of the An. sinensis China strain were acquired from the database of Zhou et al. [10]. Polymerase chain reaction (PCR) primers for An. sinensis scaffolds were designed using the Primer3 Program [18]. PCR procedures were performed with genomic DNA of Anopheles lesteri extracted from live fourth-instar larvae with the DNeasy Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany) as templates. After PCR amplification, the PCR products were cut and purified from the agarose gel using a QIAquick Gel Extraction Kit (Qiagen GmbH, Hilden, Germany) and then labelled with either Cy3.5-AP3-dUTP or Cy5.5-AP3-dUTP (GE Healthcare UK Ltd. Chalfont St Giles, UK) using a Random Primed DNA Labelling Kit (Roche Applied Science, Penzberg, Germany). Following the in situ hybridization procedure performed using a previously described method [19], fluorescent signals were detected and recorded with a Zeiss LSM 710 laser scanning microscope (Carl Zeiss Microimaging GmbH, Oberkochen, Germany) and finally mapped to the cytogenetic map of An. sinensis [17].
Gene orthology, syntenic blocks and fixed inversion
OrthoDB was used to identify one-to-one orthologues from An. sinensis and An. gambiae and to determine their locations on the scaffolds [20]. The comparative positions of the orthologous genes from An. sinensis and An. gambiae were plotted using genoPlotR [21]. Synteny blocks for each pair of homologous chromosome arms between An. sinensis and An. gambiae were analysed from the database generated by OrthoDB (Additional file 1). Chromosomal regions containing two or more orthologous genes with the same order and orientations were defined as synteny blocks and numbered 1, 2, 3, etc. along the chromosomes. After obtaining the number of all synteny blocks, the inversion distances on homologous chromosome arms between An. sinensis and An. gambiae were estimated using the programs of Genome Rearrangements in Mouse and Man (GRIMM) [22].
Chromosome evolution in Anopheles mosquitoes
Chromosome evolution rates represented by inversions/Mb/MY were calculated as inversion number/mapped genome size/divergence time. To compare the evolution rates for each chromosomal arm in different species, previously published data was included for analysis [12]. To explore the fast evolution of sex chromosome, phylogenetic relationships of the 17 anopheline species were considered [12].
Results
A physical genome map of Anopheles sinensis
For physical mapping, scaffold sequences were acquired from the database of Zhou et al. [10]. Two pairs of PCR primers were designed from the start and the end of each scaffold. After amplification, the Cy3- and Cy5-labelled probes were hybridized to the polytene chromosomes of An. sinensis. Two examples of fluorescence in situ hybridization (FISH), with one clear signal in each, are presented in Fig. 1. A total of 104 clones were mapped to the polytene chromosomes of An. sinensis to determine the chromosomal locations of 52 scaffolds. The physical map and scaffold localizations of 52 An. sinensis scaffolds are summarized in Fig. 2 and Table 1, respectively. Of the 52 scaffolds, the orientations of 48 scaffolds could be determined, and four scaffolds have unique chromosome locations. This physical map includes 26 of the 30 largest scaffolds. The largest scaffold, AS2_scf7180000696055, with a size of 5,918,260 bp, was mapped to the regions 38C to 39C of the 3L chromosome and the second largest scaffold, AS2_scf7180000696060 (4,138,565 bp) was localized to the 22C-23B of the 2L arm of An. sinensis (Table 1). Although X is the shortest chromosome, it had the best mapping coverage among the five chromosomal arms, with eight mapped scaffolds from telomere to centromere, representing 13.42 Mb of genome. Chromosome 2R, 2L, 3R and 3L had 12, 8, 14 and 10 scaffolds, respectively (Table 2). The An. sinensis genome physical map composes 79.32 Mb, or 36%, of the total assembled (220.8 Mb) genome sequences (Table 2).
Fig. 1.
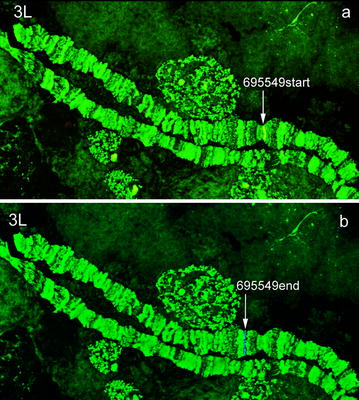
Fluorescence in situ hybridization of the genomic scaffold AS2_scf7180000695549 to the polytene chromosome 3L of An. sinensis. Probe 695549start is labelled with a red (Cy3.5) dye (a) and probe 695549end is labelled with a blue (Cy5.5) dye (b)
Fig. 2.
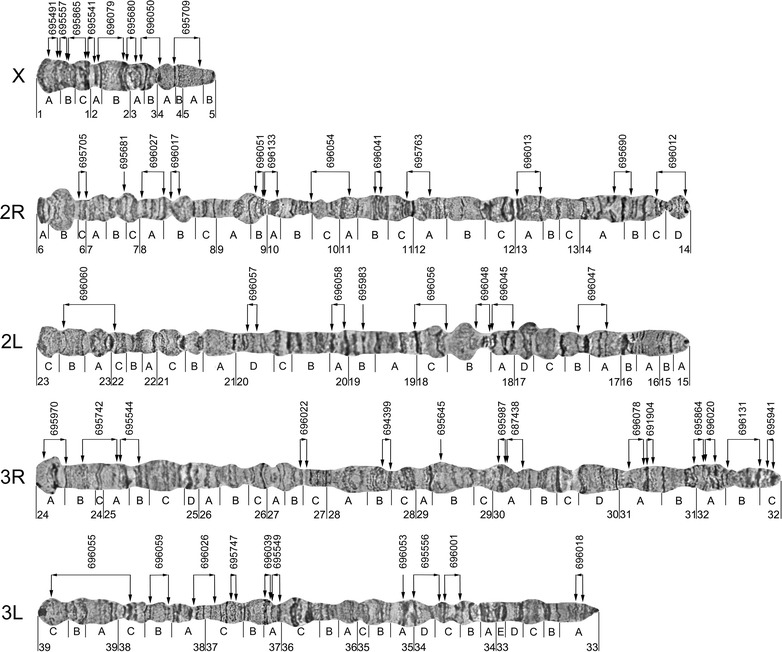
A physical map of the An. sinensis genome generated by FISH mapping on polytene chromosomes. Vertical arrows indicate the positions of the probes on chromosomes. Horizontal arrows show the orientations of scaffolds. The 6-digt numbers are the probe name and the corresponding scaffold name are listed in Table 1. The number and the A–D at the bottom of the map represent the division and subdivisions of the chromosome map
Table 1.
The localizations of 52 Anopheles sinensis scaffolds on the polytene chromosomes
| Clone name on the map | Scaffold name | Chromosome location in An. sinensis | Length (bp) | |
|---|---|---|---|---|
| 1 | 695491 | AS2_scf7180000695491 | X:1A | 730,280 |
| 2 | 695557 | AS2_scf7180000695557 | X:1A_B | 1,255,381 |
| 3 | 695865 | AS2_scf7180000695865 | X:1B_C | 2,684,564 |
| 4 | 695541 | AS2_scf7180000695541 | X:1C_2A | 841,721 |
| 5 | 696079 | AS2_scf7180000696079 | X:2A_B | 2,239,752 |
| 6 | 695680 | AS2_scf7180000695680 | X:2B_3A | 1,772,387 |
| 7 | 696050 | AS2_scf7180000696050 | X:3A_4A | 2,422,445 |
| 8 | 695709 | AS2_scf7180000695709 | X:4A_5A | 1,472,510 |
| 9 | 695705 | AS2_scf7180000695705 | 2R:6C | 3,132,144 |
| 10 | 695681 | AS2_scf7180000695681 | 2R:7B | 3,622,691 |
| 11 | 696027 | AS2_scf7180000696027 | 2R:8A | 1,402,797 |
| 12 | 696017 | AS2_scf7180000696017 | 2R:8B | 401,984 |
| 13 | 696051 | AS2_scf7180000696051 | 2R:9B | 1,646,812 |
| 14 | 696133 | AS2_scf7180000696133 | 2R:9B_10A | 2,071,771 |
| 15 | 696054 | AS2_scf7180000696054 | 2R:10C_11A | 2,075,225 |
| 16 | 696041 | AS2_scf7180000696041 | 2R:11B | 1,460,364 |
| 17 | 695763 | AS2_scf7180000695763 | 2R:11C_12A | 1,569,060 |
| 18 | 696013 | AS2_scf7180000696013 | 2R:13A | 1,443,628 |
| 19 | 695690 | AS2_scf7180000695690 | 2R:14A_B | 1,623,961 |
| 20 | 696012 | AS2_scf7180000696012 | 2R:14C_D | 1,036,301 |
| 21 | 696047 | AS2_scf7180000696047 | 2L:17A_B | 1,804,022 |
| 22 | 696045 | AS2_scf7180000696045 | 2L:18A | 1,433,044 |
| 23 | 696048 | AS2_scf7180000696048 | 2L:18B | 1,655,828 |
| 24 | 696056 | AS2_scf7180000696056 | 2L:18C | 2,410,210 |
| 25 | 695983 | AS2_scf7180000695983 | 2:19B | 1,798,621 |
| 26 | 696058 | AS2_scf7180000696058 | 2L:20A | 2,460,545 |
| 27 | 696057 | AS2_scf7180000696057 | 2L:20D | 3,056,258 |
| 28 | 696060 | AS2_scf7180000696060 | 2L:22C_23B | 4,138,565 |
| 29 | 695970 | AS2_scf7180000695970 | 3R:24A_B | 1,982,586 |
| 30 | 695742 | AS2_scf7180000695742 | 3R:24B_25A | 1,869,526 |
| 31 | 695544 | AS2_scf7180000695544 | 3R:25A_B | 1,280,009 |
| 32 | 696022 | AS2_scf7180000696022 | 3R:27B_C | 621,276 |
| 33 | 694399 | AS2_scf7180000694399 | 3R:28B | 146,261 |
| 34 | 695645 | AS2_scf7180000695645 | 3R:29B | 17,918 |
| 35 | 695987 | AS2_scf7180000695987 | 3R:30A | 525,874 |
| 36 | 687438 | AS2_scf7180000697438 | 3R:30A | 140,738 |
| 37 | 696078 | AS2_scf7180000696078 | 3R:31A | 229,900 |
| 38 | 691904 | AS2_scf7180000691904 | 3R:31A | 138,869 |
| 39 | 695864 | AS2_scf7180000695864 | 3R:32A | 170,159 |
| 40 | 696020 | AS2_scf7180000696020 | 3R:32A | 710,710 |
| 41 | 696131 | AS2_scf7180000696131 | 3R:32B | 2,359,367 |
| 42 | 695941 | AS2_scf7180000695941 | 3R:32C | 816,509 |
| 43 | 696018 | AS2_scf7180000696018 | 3L:33A | 515,011 |
| 44 | 696001 | AS2_scf7180000696001 | 3L:34C | 777,379 |
| 45 | 695556 | AS2_scf7180000695556 | 3L:34C_D | 1,591,401 |
| 46 | 696053 | AS2_scf7180000696053 | 3L:35A | 1,670,191 |
| 47 | 695549 | AS2_scf7180000695549 | 3L:37A | 814,231 |
| 48 | 696039 | AS2_scf7180000696039 | 3L:37A | 3,601,930 |
| 49 | 695747 | AS2_scf7180000695747 | 3L:37C | 566,275 |
| 50 | 696026 | AS2_scf7180000696026 | 3L:37C_38A | 1,091,046 |
| 51 | 696059 | AS2_scf7180000696059 | 3L:38B | 2,208,365 |
| 52 | 696055 | AS2_scf7180000696055 | 3L:38C_39C | 5,918,260 |
Table 2.
Genome physical mapping information of Anopheles sinensis
| Chromosome arm | Scaffolds mapped (n) | Length (bp) | Total sequenced genome (%) |
|---|---|---|---|
| X | 8 | 13,419,040 | 6.07 |
| 2R | 12 | 17,382,848 | 7.87 |
| 2L | 8 | 18,757,093 | 8.50 |
| 3R | 14 | 11,009,702 | 4.99 |
| 3L | 10 | 18,754,089 | 8.49 |
| Total | 52 | 79,322,772 | 36.00 |
The physical map of An. sinensis presented in this study was compared with previous mapping data summarized in Table 3. Among mapped anopheline genomes, An. albiumanus had the most complete chromosomally anchored genome assembly covering 98.2% of the genome, followed by An. gambiae, An. stephensi and An. atroparvus [12, 23] (Table 3). Mapping of 52 scaffolds in An. sinensis and 103 scaffolds in An. funestus achieved similar portions of mapped genomes in both species (Table 3). Thus, the new genome map of An. sinensis can be used for exploration of chromosomal evolution in malaria mosquitoes.
Table 3.
Assembly and mapping metrics for anopheline genomes
| Species | Genome assembly | Total scaffolds | Mapped scaffolds | Scaffold N50, bp | Total length, Mb | Mapped length, Mb | Mapped, % |
|---|---|---|---|---|---|---|---|
| An. albimanus | AalbS1 | 204 | 40 | 18,068,499 | 170.5 | 167.4 | 98.2 |
| An. gambiae | AgamP4 | 8 | 5 | 49,364,325 | 264 | 273.1 | 84.3 |
| An. stephensi | AsteI2 | 23,371 | 86 | 1,591,355 | 221.3 | 137.14 | 62 |
| An. stephensi | AsteS1 | 1100 | 101 | 837,295 | 225 | 92.83 | 41 |
| An. atroparvus | AatrE1 | 1371 | 7 | 9,206,694 | 224.3 | 88.8 | 39.6 |
| An. sinensis | AsinC2 | 9592 | 52 | 814,231 | 220.8 | 79.3 | 35.9 |
| An. funestus | AfunF1 | 1392 | 103 | 671,960 | 225.2 | 79.0 | 35.1 |
Data for An. sinensis are from this study. Data for An. albimanus and An. stephensi AsteI2 are from Ref. [26] and Ref. [23], respectively. Data for An. gambiae are from Ref. [25] and https://www.vectorbase.org/organisms/anopheles-gambiae/pest/agamp4. Data for other species are from Ref. [12]
Synteny and gene order evolution in An. sinensis and An. gambiae
A total of 3955 one-to-one orthologues were identified from An. sinensis and An. gambiae using OrthoDB [20] (Additional file 1). The comparative positions of genes within mapped scaffolds based on orthology relationships were plotted on An. sinensis and An. gambiae chromosomes using genoPlotR [21] (Fig. 3). Physical mapping data were used to determine the orientations of scaffolds, and the default orientations were assigned to some scaffolds with only one probe. Figure 3 shows that the gene orders were reshuffled on five chromosome arms because of fixed inversions. The gene order changes on the X chromosome were more dramatic than those on the autosomes: 2R, 2L, 3R and 3L. The comparative chromosomal locations and orientations of 3955 orthologous genes were further used to determine the number of synteny blocks in the two species. Synteny blocks were defined as genomic regions containing at least two orthologous genes with the same order and orientation. A total of 364 synteny blocks have been identified between An. sinensis and An. gambiae (Table 4). The analysis revealed that the average length of 112 synteny blocks on the X chromosome (85,989 bp) is much smaller than those on the remaining chromosomes (237, 175; 239, 627; 197, 751 and 242, 299). Additionally, the largest synteny block on the X arm is only 766,489 bp, whereas the largest block on 2R is 1,796,395 bp, which is twice that on the X arm (Table 4). These results suggest that the sex X chromosome has smaller synteny blocks than the autosomes.
Fig. 3.
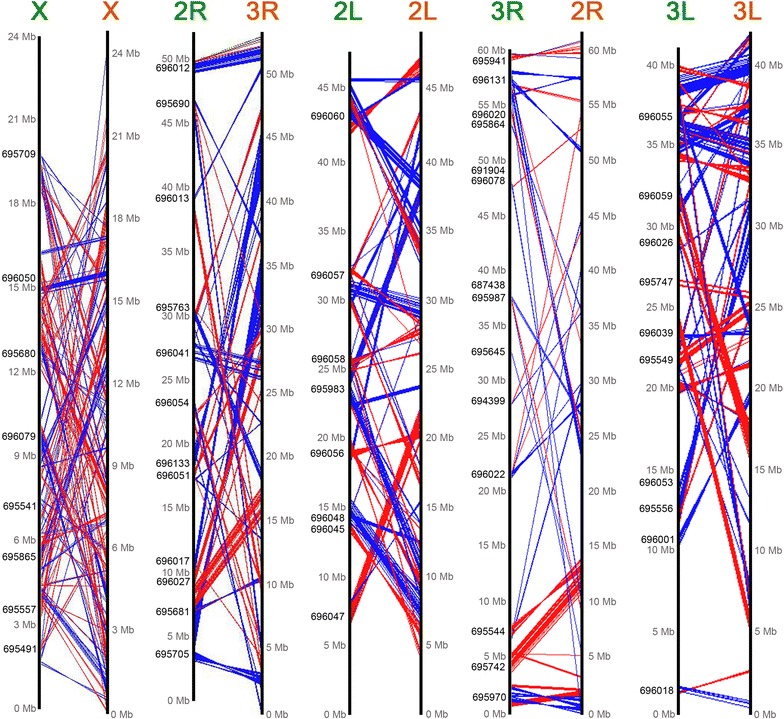
The relationship of the positions of orthologous genes between the An. sinensis and An. gambiae plotted with GenoPlotR. Orthologues with the same orientation in An. sinensis (left side) and An. gambiae (right side) are connected with red lines, and orthologues with the opposite orientation are connected with blue lines
Table 4.
The synteny blocks between An. sinensis and An. gambiae using An. sinensis as reference
| Chromosome | Number of synteny blocks (n) | The average length of synteny blocks (bp) | The largest synteny blocks (bp) | The smallest synteny blocks (bp) |
|---|---|---|---|---|
| X | 112 | 85,989 | 766,489 | 1676 |
| 2R | 68 | 237,175 | 1,796,395 | 1796 |
| 2L | 70 | 239,627 | 1,629,455 | 1514 |
| 3R | 47 | 197,751 | 1,709,848 | 3361 |
| 3L | 67 | 242,299 | 1,469,290 | 4748 |
| Total | 364 | 191,358 | 1,474,295 | 2619 |
To further analyse fixed inversions between An. sinensis and An. gambiae, we input the order of 361 synteny blocks (Additional file 2) into the Genome Rearrangements in Man and Mouse (GRIMM) program [22]. Table 5 shows that a minimum of 267 inversions were estimated between An. sinensis and An. gambiae. The sex chromosome exhibited a greater number of inversions (101), whereas the autosomes 2R, 2L, 3R and 3L had 42, 51, 33 and 40 inversions, respectively (Table 5). The total size of mapped scaffolds on each An. sinensis chromosome was used to calculate the density of inversions per megabase. Our data demonstrate that the inversion breaks per megabase on X chromosome is 7.527, which is approximately 3.2 times greater than the average density of inversions on autosomes (2.367) (Table 5). Among the autosomes, the inversion density between An. sinensis 3R and An. gambiae 2R is 2.997 inversions/Mb, which is higher than for the remaining autosomes. The 3L chromosome exhibits the lowest density of inversions (2.133 inversions/Mb). The most recent study of the chromosome evolution in Anopheles used the divergence time between An. atroparvus and An. gambiae of 58 MY [12], and An. atroparvus and An. sinensis belong to the subgenus Anopheles. The divergence time of 58 MY was used to calculate the number of inversions per magabase per million years (inversions/Mb/MY) (Table 5). For An. sinensis and An. gambiae, the rates of evolution were 0.130 for X and 0.042, 0.047, 0.052, 0.037 for 2R, 2L, 3R and 3L, respectively (Table 5).
Table 5.
Fixed inversions between An. sinensis and An. gambiae
| Chromosome name in An. sinensis | Size of mapped scaffolds in An. sinensis (Mb) | Inversions (GRIMM) | Inversions/Mb | Inversions/Mb/MY (divergence time 58 MY) |
|---|---|---|---|---|
| X | 13.419 | 101 | 7.527 | 0.130 |
| 2R | 17.382 | 42 | 2.416 | 0.042 |
| 2L | 18.757 | 51 | 2.719 | 0.047 |
| 3R | 11.010 | 33 | 2.997 | 0.052 |
| 3L | 18.754 | 40 | 2.133 | 0.037 |
| Total | 79.322 | 267 | 3.366 | 0.058 |
Rapid evolution of the sex chromosome in Anopheles mosquitoes
To understand the pattern of inversion fixations in malaria mosquitoes, the number of inversions/Mb/MY in our analysis was compared with the earlier published data [12] (Table 6). The results revealed that inversion rates on autosomes varied between An. gambiae and each of five Anopheles species. However, the density of fixed inversions on the X was consistently greater than that on autosomes (Table 6), suggesting the faster evolution of X chromosome in Anopheles mosquitoes. The ratio of the X chromosome evolution rate to the autosomal rate of rearrangements in An. sinensis and An. gambiae was also calculated and our data demonstrated that the X chromosome evolved approximately 3.2 times faster than autosomes. Our chromosomal evolution analysis data was added into the phylogenetic relationships of the 17 anopheline species constructed by Neafsey et al. [12] using the aligned protein sequences of 1085 single-copy orthologs. Figure 4 shows that the ratio of the X chromosome evolution to the autosomal rate of rearrangements varies among the Anopheles lineages with being higher in subgenera Anopheles and Nyssorhynchus and lower in genus Cellia.
Table 6.
The rates of inversion fixation between An. gambiae and other Anopheles species
| Species | X | 2R | 2L | 3R | 3L |
|---|---|---|---|---|---|
| An. albimanus–An. gambiae | 0.130 | 0.043 | 0.040 | 0.034 | 0.037 |
| An. atroparvus–An. gambiae | 0.124 | 0.036 | 0.035 | 0.025 | 0.041 |
| An. funestus–An. gambiae | 0.121 | 0.064 | 0.065 | 0.026 | 0.066 |
| An. sinensis–An. gambiae | 0.130 | 0.052 | 0.047 | 0.042 | 0.037 |
| An. stephensi–An. gambiae | 0.128 | 0.049 | 0.036 | 0.028 | 0.036 |
The rates represent the number of inversions per Mb per MY. Data for An. sinensis are from this study. Data for other species are from Ref. [12]
Fig. 4.
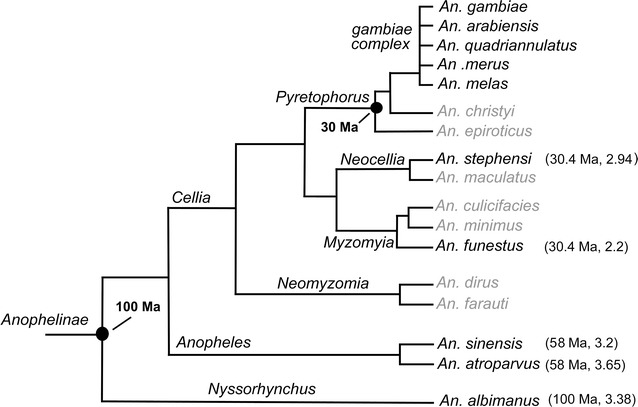
Reconstructed phylogenetic relationships of the 17 anopheline species and chromosomal evolution analysis from Ref. [12]. The aligned protein sequences of 1085 single-copy orthologs were used to construct the maximum likelihood molecular phylogeny. Chromosome evolution analysis was conducted between the species indicated with a dark font and An. gambiae. Comparative physical mapping has not been performed for the species marked with a grey font. Ma represents million years ago. The number in brackets after the divergence time is the ratio of the X chromosome evolution rate to the autosomal rate of rearrangements in each species compared with An. gambiae
Discussion
A physical map is a critical tool for improving a genome assembly and for studying chromosomal evolution
In this study, a physical map was constructed for an Asian malaria vector An. sinensis using fluorescence in situ hybridization (FISH) of DNA probes with polytene chromosomes. The physical mapping of An. sinensis placed 52 large scaffolds with total length of 79,322,722 bp from the genome database to the chromosomes (Fig. 2; Table 2). It accounted for approximately 36% of the total assembled (220.8 Mb) genome sequences of An. sinensis (Table 2).
So far, several genome maps have been developed for malaria mosquitoes and we compared the percentage of the physically mapped genome in An. sinensis with data from other species [12, 23]. Among mosquitoes, the African malaria vector An. gambiae was the first to have its genome sequenced [24]. More than 2000 BAC clones were originally placed onto the chromosomes for genome mapping and later, additional mapping added small scaffolds to the area around the centromeres, which resulted in ~84.3% of the An. gambiae genome assembly [25]. The physical map of An. albimanus initially placed ~76% of genome onto the chromosomes [12], while a more recent physical mapping effort reached the 98.2% coverage of the An. albimanus genome assembly [26], which is the most complete genome assembly to date. The genome of An. stephensi, a key vector of malaria throughout the Indian subcontinent and Middle East, has also been sequenced and assembled. A total of 86 scaffolds were in situ hybridized to the polytene chromosomes of An. stephensi, representing 62% of the genome assembly [23]. Anopheles atroparvus and An. funestus had mapped portions covering 39.6 and 35.1% of the total genome, respectively [12]. In this research, our new physical map for An. sinensis covers 35.9% of the genome, which is within the range of other Anopheles species (Table 3).
Fast evolution of the sex chromosome in Anopheles mosquitoes
The availability of the genome sequences and physical maps for Anopheles mosquitoes have promoted detailed analysis of the patterns of fixed inversions [12, 13]. In our study, 361 conserved synteny blocks and 267 fixed inversions were identified between An. sinensis and An. gambiae. Analysis of the density of inversions per Mb and the rate of chromosomal rearrangements in An. sinensis and An. gambiae suggested that fast evolution occurs on the sex chromosome. The earliest study of inversions on closely related species of the An. gambiae complex revealed that 5 of 10 inversions were on the X chromosome, providing the first evidence of fast evolution of sex chromosomes in Anopheles mosquitoes [27]. Several species belonging to different series within the subgenus Cellia have been extensively studied: An. gambiae (Pyretophorus), An. stephensi (Neocellia) and An. funestus (Myzomyia) [12]. The comparative analysis between An. funestus and An. gambiae as well as between An. stephensi and An. gambiae [14, 15] further demonstrated that the X chromosome evolved faster than the autosomes. The most recent analyses based on the genome assembly confirmed that the rate of evolution on X is approximately 2.2 times faster than the average autosomal rate for An. funestus and An. gambiae [12] or 2.94 times faster for An. stephensi and An. gambiae [23] (Fig. 4). Anopheles sinensis and An. atroparvus are members of the subgenus Anopheles, which is thought to have diverged from An. gambiae 58 MY ago [10, 12]. Previous studies have shown that the difference in the rate of evolution between the sex chromosome and autosomes is approximately 3.65 times in An. atroparvus and An. gambiae [12]. In this study, the density of inversions on the X chromosome is found to be 3.2 times greater than the average density of inversions on the autosomes between An. sinensis and An. gambiae (Fig. 4). These results suggest that the rapid evolution of sex chromosome is a common feature in Anopheles mosquitoes. The X chromosome rearrangements may play a role in speciation of malaria mosquitoes [14, 28]. Future genome studies can provide valuable information for dissecting the role of X chromosome inversions in speciation of malaria vectors.
Conclusions
This study constructed a physical genome map for an important malaria vector of P. vivax, An. sinensis, which is the most widely distributed vector in China, Korea, and Japan. This physical map includes 52 of the largest scaffolds from An. sinensis, spanning approximately 80 Mb of the 220 Mb, or approximately 36%, of the sequenced genome. The map coverage is similar to the mapped portion of An. funestus and An. atroparvus. By analysing the comparative positions of 3955 orthologous genes, 361 conserved synteny blocks and 267 fixed inversions between An. sinensis and An. gambiae were identified. The rate of evolution of the sex chromosome is approximately 3.2 times greater than the average autosomal rate of evolution. Thus, our comparative analysis in An. sinensis and An. gambiae inferred from physically mapped genome assemblies provided additional details for understanding chromosome evolution in malaria vectors.
Additional files
Additional file 1. Orthologous genes in An. sinensis and An. gambiae.
Additional file 2. The orders of synteny blocks for running GRIMM.
Authors’ contributions
AX conceived and designed the experiments, jointly performed data analysis and wrote the manuscript. YW, BC, JL, CW, and JW performed the experiments. DS contributed to data analysis. GZ, JT, and JC were responsible for mosquito strain collection. IVS analysed physical mapping data and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated during this study are included in this main paper and Additional files 1, 2.
Funding
The work was supported by a National Natural Science Foundation of China (31301877) and the Fundamental Research Funds for the Central Universities (KJQN201431) to AX. The comparative analysis of physical mapping data among Anopheles species was supported by the grant from the Russian Science Foundation No 15-14-20011 (to IVS). The funding agencies had no role in the design of the study, the collection, analysis, and interpretation of data or in writing the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-017-1888-7) contains supplementary material, which is available to authorized users.
Yun Wei and Biao Cheng contributed equally to this work
Contributor Information
Yun Wei, Email: 2015102097@njau.edu.cn.
Biao Cheng, Email: 2014102102@njau.edu.cn.
Guoding Zhu, Email: jipdzhu@hotmail.com.
Danyu Shen, Email: shendanyu@njau.edu.cn.
Jiangtao Liang, Email: jtliang@vt.edu.
Cong Wang, Email: 2015102098@njau.edu.cn.
Jing Wang, Email: 2016102093@njau.edu.cn.
Jianxia Tang, Email: tangjianxia78@163.com.
Jun Cao, Email: caojuncn@hotmail.com.
Igor V. Sharakhov, Email: igor@vt.edu
Ai Xia, Phone: +862584399675, Email: xiaai@njau.edu.cn.
References
- 1.Harbach RE. The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull Entomol Res. 2004;94:537–553. doi: 10.1079/BER2004321. [DOI] [PubMed] [Google Scholar]
- 2.Hay SI, Sinka ME, Okara RM, Kabaria CW, Mbithi PM, Tago CC, et al. Developing global maps of the dominant anopheles vectors of human malaria. PLoS Med. 2010;7:e1000209. doi: 10.1371/journal.pmed.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ree HI. Studies on Anopheles sinensis, the vector species of vivax malaria in Korea. Korean J Parasitol. 2005;43:75–92. doi: 10.3347/kjp.2005.43.3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rueda LM, Pecor JE, Harrison BA. Updated distribution records for Anopheles vagus (Diptera: Culicidae) in the Republic of Philippines, and considerations regarding its secondary vector roles in Southeast Asia. Trop Biomed. 2011;28:181–187. [PubMed] [Google Scholar]
- 5.Zhou SS, Huang F, Wang JJ, Zhang SS, Su YP, Tang LH. Geographical, meteorological and vectorial factors related to malaria re-emergence in Huang-Huai River of central China. Malar J. 2010;9:337. doi: 10.1186/1475-2875-9-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu G, Xia H, Zhou H, Li J, Lu F, Liu Y, et al. Susceptibility of Anopheles sinensis to Plasmodium vivax in malarial outbreak areas of central China. Parasit Vectors. 2013;6:176. doi: 10.1186/1756-3305-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng LC, Lee KS, Tan CH, Ooi PL, Lam-Phua SG, Lin R, et al. Entomologic and molecular investigation into Plasmodium vivax transmission in Singapore, 2009. Malar J. 2010;9:305. doi: 10.1186/1475-2875-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin LZ, Xu JJ. Quantitative studies on the development of inoculated Brugia malayi microfilariae in Anopheles sinensis and Culex quinquefasciatus. Southeast Asian J Trop Med Public Health. 1990;21:418–423. [PubMed] [Google Scholar]
- 9.Li QJ, Duan JH, Hu GL, Yu LR, Yang WQ, Li LZ, et al. [Epidemiological characteristics and control of filariasis in Hunan Province](in Chinese) Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 1990;8:134–137. [PubMed] [Google Scholar]
- 10.Zhou D, Zhang D, Ding G, Shi L, Hou Q, Ye Y, et al. Genome sequence of Anopheles sinensis provides insight into genetics basis of mosquito competence for malaria parasites. BMC Genom. 2014;15:42. doi: 10.1186/1471-2164-15-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B, Zhang YJ, He Z, Li W, Si F, Tang Y, et al. De novo transcriptome sequencing and sequence analysis of the malaria vector Anopheles sinensis (Diptera: Culicidae) Parasit Vectors. 2014;7:314. doi: 10.1186/1756-3305-7-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Mosquito genomics. highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:e1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharakhov IV, Artemov GN, Sharakhova MV. Chromosome evolution in malaria mosquitoes inferred from physically mapped genome assemblies. J Bioinform Comput Biol. 2016;14:16300033. doi: 10.1142/S0219720016300033. [DOI] [PubMed] [Google Scholar]
- 14.Sharakhov IV, Serazin AC, Grushko OG, Dana A, Lobo N, Hillenmeyer ME, et al. Inversions and gene order shuffling in Anopheles gambiae and An. funestus. Science. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- 15.Xia A, Sharakhova MV, Leman SC, Tu ZJ, Bailey JA, Smith CD, et al. Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes. PLoS ONE. 2010;5:e10592. doi: 10.1371/journal.pone.0010592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timoshevskiy VA, Kinney NA, deBruyn BS, Mao CH, Tu ZJ, Severson DW, et al. Genomic composition and evolution of Aedes aegypti chromosomes revealed by the analysis of physically mapped supercontigs. BMC Biol. 2014;12:27. doi: 10.1186/1741-7007-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang J, Sharakhova MV, Lan Q, Zhu H, Sharakhov IV, Xia A. A standard cytogenetic map for Anopheles sinensis and chromosome arm homology between the subgenera Anopheles and Cellia. Med Vet Entomol. 2014;28(Suppl 1):26–32. doi: 10.1111/mve.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 19.Sharakhova MV, Xia A, McAlister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1093/jmedent/43.5.861. [DOI] [PubMed] [Google Scholar]
- 20.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guy L, Kultima JR, Andersson SG. GenoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Peery A, Hall AB, Sharma A, Chen XG, Waterhouse RM, et al. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol. 2014;15:459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 25.Sharakhova MV, Hammond MP, Lobo NF, Krzywinski J, Unger MF, Hillenmeyer ME, et al. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007;8:R5. doi: 10.1186/gb-2007-8-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artemov GN, Peery AN, JiangX TuZ, Stegniy VN, Sharakhova MV, et al. The physical genome mapping of Anopheles albimanus corrected scaffold misassemblies and identified inter-arm rearrangements in genus Anopheles. G3 (Bethesda) 2017;7:155–164. doi: 10.1534/g3.116.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coluzzi M, Sabatini A, Torre A, Di Deco MA. Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 28.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. doi: 10.1086/284701. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Orthologous genes in An. sinensis and An. gambiae.
Additional file 2. The orders of synteny blocks for running GRIMM.
Data Availability Statement
All data generated during this study are included in this main paper and Additional files 1, 2.


