Abstract
Rab-interacting lysosomal protein (RILP) is a regulator of late stages of endocytosis. Recent work proved that depletion of RILP promotes migration of breast cancer cells in wound healing assay, whereas its overexpression influences re-arrangements of actin cytoskeleton. Here, we further characterized the role of RILP in cell migration by analyzing several aspects of this process. We showed that RILP is fundamental also for migration of lung cancer cells regulating cell velocity. RILP silencing did not affect Golgi apparatus nor microtubules reorientation during migration. However, both RILP over-expression and expression of its mutated form, RILPC33, impair cell adhesion and spreading. In conclusion, our results demonstrate that RILP has important regulatory roles in cell motility affecting migration velocity but also in cell adhesion and cell spreading.
Key words: RILP, cell migration, cell adhesion, microtubules, cell polarization
Introduction
Rab-interacting lysosomal protein (RILP) is a key regulatory protein of the endocytic pathway.1,2 RILP regulates late stages of endocytosis being the downstream effector for the small GTPases Rab7a and Rab34.1,3,4 In particular, GTP-bound Rab7a recruits on late endosomal and lysosomal membranes RILP, which in turn recruits the dynactin/p150Glued subunit of the dyneindynactin motor complex, responsible for transport of Rab7a-positive vesicles toward the minus end of microtubules.2 In fact, RILP and Rab7a control together lysosomal distribution and morphology, and are required for the proper degradation of a number of molecules inside lysosomes.1,3-5
Endocytosed molecules destined for degradation are sorted to degradative compartments, late endosomes and lysosomes, through multivesicular bodies (MVBs), endosomal organelles that contain multiple intraluminal vesicles (ILVs). Several proteins, belonging to different endosomal sorting complexes required for transport (ESCRTs), are responsible for sorting of proteins into (ILVs).6 RILP is fundamental for the biogenesis of MVBs.3,7,8 Indeed, RILP interacts and mediates the membrane recruitment of the mammalian counterpart of VPS22 and VPS36, two components of ESCRT-II.3,7,8
Increasing evidence proves a role for a number of Rab proteins in the regulation of different steps of cell migration, such as cell adhesion, Golgi complex reorientation, cytoskeleton rearrangements and trafficking of adhesion molecules.9-15 Alterations of migration play a key role in diseases such as, for instance, cancer.16 Notably, RILP has been associated with the suppression of invasion in prostate cancer cells.17,18 Moreover, it has been recently demonstrated that RILP expression is lower in highly invasive cells and that RILP silencing promotes migration and invasion of breast cancer cells, whereas RILP overexpression suppresses migration.19 Although it has been observed that RILP affects actin cytoskeleton by interacting with Ral guanine nucleotide dissociation stimulator (RalGDS), a regulator of RalA,19 how RILP affects cell motility and other aspects of cell migration has not been studied.
The aim of the present study was to better characterize the role of RILP in cell migration and we demonstrated that RILP affects migration velocity and regulates cell adhesion and spreading.
Materials and Methods
Cells and reagents
NCI H1299 cells (ATCC CRL-5803; human lung carcinoma) were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% FBS, 2 mM Lglutamine, 100 U/ml penicillin and 10 mg/ml streptomycin in 5% CO2 incubator at 37°C and confirmed to be contaminationfree. Chemicals were from Sigma-Aldrich. Tissue culture reagents were from Sigma- Aldrich (St. Louis, MO, USA), Gibco (Waltham, MA, USA), Lonza (Basel, Switzerland) and Biological Industries (Cromwell, CT, USA).
Plasmids and antibodies
PEGFP, pEGFP-RILP, pEGFP-RILPC33, pCDNA3_2XHA, pCDNA3_2XHARILP and pCDNA3_2XHA-RILP-C33 have been described previously.20-22 Rabbit polyclonal anti-HA (1:500, ab9110) and antigiantin (1:1000, ab24586) were from Abcam (Cambridge, UK). Mouse monoclonal antitubulin (1:500 for immunofluorescence analyses, 1:10000 for immunoblot analyses, T5168) was from Sigma-Aldrich. Rabbit anti-RILP polyclonal antibody (1:100) has been described previously.1 Secondary antibodies conjugated to fluorochromes (1:200) or horseradish peroxidase (HRP, 1:5000) were from Invitrogen (Carslbad, CA, USA) or GE Healthcare (Barrington, IL, USA).
Transfection and RNAi
Transfection was performed using Metafectene Pro from Biontex or Lipofectamine 2000 from Invitrogen as indicated by the manufacturers. Cells were analyzed after 24 h of transfection. For RNA interference, small interfering RNAs (siRNAs) were purchased from MWGBiotech. Transfection of cells with siRNA was performed using RNAiMAX from Invitrogen following the manufacturer’s instructions. RILP siRNA efficiency in silencing was reported previously:22 sense sequence 5’-GAUCAAGGCCAAGAUGUUATT- 3’ and antisense sequence 5’-UAACAUCUUGGCCUUGAUCTT- 3’. As a negative control we used a control RNA: sense sequence 5’-ACUUCGAGCGUGCAUGGCUTT- 3’ and antisense sequence 5’-AGCCAUGCACGCUCGAAGUTT-3’.
Wound-healing assay
Confluent monolayers of control or RILP-depleted NCI H1299 cells were subjected to wound-healing assay as previously described.13 Cells migrating toward the wound were imaged every 30 min over a 8 h time period with a 20X objective on an Olympus Fluoview 1000 IX-81 inverted confocal laser scanning microscope. Cell nuclei were tracked by using the Manual Tracking plugin of ImageJ software (National Institutes of Health) and cell migration parameters were calculated by using the Chemotaxis and Migration Tool software (Ibidi).
Cell adhesion assay
Cells transfected with various expression plasmids or siRNA were subjected to cell adhesion assay as described,23 after checking transfection efficiency. Briefly, cells were trypsinized and seeded in equal number into 96-well plates coated previously with 20 μg/mL fibronectin. Cells were incubated for different times, then washed with PBS and fixed. Cells were imaged with a 10X objective on a IncuCyte Zoom System.
Cell spreading assay
Cells were seeded onto fibronectin-coated coverslips (BD Biosciences) and after 30 minutes were fixed with 3% paraformaldehyde, permeabilized with 0.25% saponin in PBS and stained with anti-HA antibody, Rhodamine-conjugated phalloidin and Hoeschst 33258. Samples were then observed using an Olympus FluoView FV1000 microscope. Cell areas were calculated by using ImageJ software.
Golgi reorientation measurements
Confluent monolayers of control and RILP-depleted cells were subjected to wound-healing assay. After a 3-h incubation at 37°C under 5% CO2 cells were permeabilized with 0,25% saponin in PBS, fixed with 3% paraformaldehyde and stained with antigiantin antibody, Rhodamine-conjugated phalloidin and Hoeschst 33258. Cells were observed with a 63X PlanApo NA 1.42 objective on an Olympus FluoView FV1000 microscope with the FV1000 software. Cells with the Golgi complex localized at the side of the cell facing the wound were considered polarized.
Live imaging
NCI H1299 cells were grown on MatTek glass-bottomed dishes and incubated in DMEM without phenol red at 37°C under 5% CO2. Live cells were observed with a 63X PlanApo NA 1.42 objective on an Olympus IX-71 microscope equipped with a CSU22 spinning-disk confocal unit (Yokogawa), an Ixon EMCCD camera (Andor) and the Andor iQ1.8 software.
Western blot experiments
NCI H1299 cells were lysed with lysis buffer (25 mM Hepes, 125 mM potassium acetate, 2,5 mM magnesium acetate, 5 mM EGTA, 1 mM DTT, 0,5% Igepal CA-630 (NP-40), pH 7,2) plus protease inhibitor cocktail (Roche). Lysates were loaded onto SDS-PAGE gels, transferred onto polyvinylidene fluoride (PVDF) membrane from Millipore and probed with the specific primary antibodies diluted in 2% blotting grade non-fat dry milk (Bio-Rad) in PBS followed by HRP-conjugated secondary antibodies. Proteins were visualized by using the ECL system (GE Healthcare).
Statistical analysis
Data were statistically analyzed by using Student’s t-test (GraphPad Prism4 software). Statistical significance is indicated as follows: *P<0.05, **P<0.01 and ***P<0.001. Assays were performed at least in triplicate, error bars represent SEM.
Results
RILP affects cell velocity and accumulated distance of migrating cells
It has been recently shown that RILP depletion in MCF7 (breast adenocarcinoma) cells causes a faster closure of the wound in wound-healing assay demonstrating a role of RILP in cell migration.19 In order to better characterize this role of RILP and to investigate if this is restricted to breast cancer cells we performed a woundhealing assay on NCI H1299 (lung carcinoma) cells transfected with either control RNA or siRNA against RILP. Confluent monolayers of cells were scratched with a pipette tip and migrating cells were imaged at time intervals of 30 min over an 8-h time period. We confirmed the role of RILP in cell motility as in RILP-depleted NCI H1299 cells migrated faster than control cells (Figure 1A; Supplementary Movie 1), proving that the role of RILP in migration is not cell-specific as it affects cell migration both in breast and lung cancer cells. Then, we evaluated several migration parameters such as velocity, accumulated distance, Euclidean distance and directness in order to better understand the effect of RILP on cell motility (Figure 1 B-E). Interestingly, upon RILP-depletion, cells speeded up of about 21% compared to control cells (Figure 1B). In accordance with this data, we observed a similar increase in the accumulated distance (of about 24%), which is the total distance that cells travelled in a certain amount of time, whereas Euclidean distance and directness were not affected upon RILP silencing (Figure 1 C-E). Western blot analysis of control and RILPdepleted cells demonstrated a strong reduction of RILP abundance of about 90% upon silencing (Figure 1 F-G).
Figure 1.
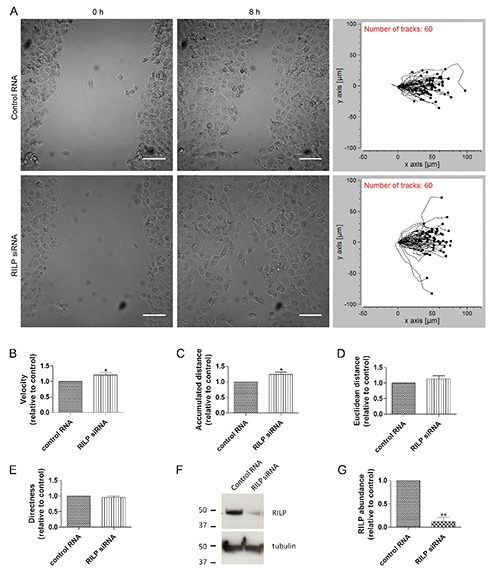
RILP silencing affects cell velocity and accumulated distance in migrating cells. NCI H1299 cells transfected with control RNA or with RILP siRNA were imaged every 30 min over an 8-h period during a wound-healing assay. A) Images refer to the initial time point (0 h) or after the closure of the wound (8 h); on the right panel the migratory tracks of 60 representative cells are shown for control and RILP-depleted cells; scale bars: 50 μm. Migration velocity (B), accumulated distance (C), euclidean distance (D) and directness (E) have been quantified and relative graphs are shown. Data represents the mean ± SEM of four independent experiments (n=60). F) Lysates of control and RILPdepleted NCI H1299 cells were subjected to western blot analysis using anti-RILP and anti-tubulin antibodies. G) Quantification of RILP abundance. The values represent the mean ± SEM of three independent experiments.
These data demonstrate that RILP regulates cell motility modulating cell velocity but not directness.
RILP depletion does not alter Golgi apparatus reorientation
Cell polarity is essential for directional migration and it is characterized by cytoskeleton rearrangements, positioning of the nucleus and reorientation of the Golgi complex and the microtubule-organizing center (MTOC) into the direction of the wound.24 Therefore, we evaluated a possible role of RILP on cell polarization by measuring the reorientation of the Golgi complex towards the leading edge. As suggested by the fact that directness was not affected by RILP silencing (Figure 1E), RILP depletion did not interfere with cell polarization and, in particular, with the Golgi apparatus reorientation (Figure 2A). In fact, about 80% of cells had the Golgi apparatus correctly oriented between the nucleus and the leading edge in both control and RILP-depleted cells (Figure 2B).
Figure 2.
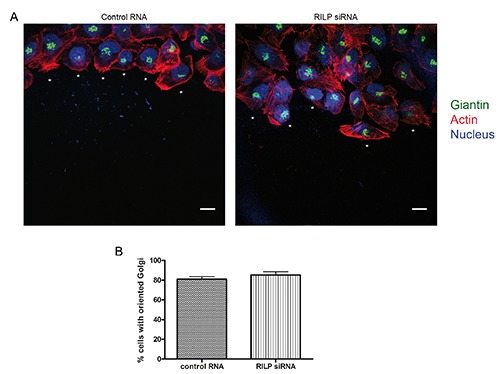
Golgi apparatus reorientation during migration is not affected in RILP-depleted cells. A) NCI H1299 cells transfected with either control RNA or siRNA against RILP were subjected to wound healing assay and stained after three hours with anti-giantin antibody (green), rhodamine-conjugated phalloidin (red) and Hoechst (blue); asterisks indicate a polarized cell with Golgi complex oriented into the direction of the wound; scale bars: 10 μm. B) Quantification of NCI H1299 cells transfected as described in (A) and showing oriented Golgi apparatus into the direction of the wound. Data represent the mean ± SEM of three independent experiments (n=50).
RILP does not influence microtubule cytoskeleton rearrangements
The reorganization of actin and microtubule cytoskeleton is fundamental during cell migration.25,26 Wang and coworkers demonstrated that RILP regulates the rearrangements of actin cytoskeleton19 and we confirmed this data in NCI H1299 cells expressing GFP-RILP (Supplementary Figure 1). However, nothing is known about the role of RILP on microtubules in migrating cells. Interestingly, RILP regulates microtubule minus-end directed transport of vesicles through the interaction with the dynactin/dynein complex.2,27 Therefore, we investigated whether RILP could affect microtubule organization during cell migration. As shown in Figure 3, control migrating cells showed oriented microtubules, and we did not detect any visible alteration of microtubule cytoskeleton in migrating cells silenced for RILP (Figure 3A-B). As RILP over-expression suppressed cell migration, 19 we investigated whether the expression of GFP-RILP could cause a defect in microtubule cytoskeleton rearrangements. However, the percentage of migrating cells with oriented microtubules was approximately 70% for both control and RILPoverexpressing cells (Figure 3 C-D). Moreover, RILP depletion did not affect tubulin filament dynamics (Supplementary Movie 2).
Figure 3.
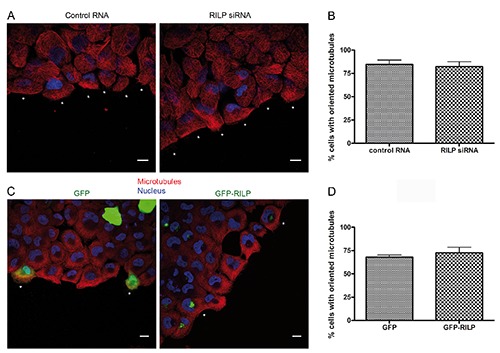
RILP does not regulate microtubule cytoskeleton rearrangements. A) NCI H1299 cells transfected with control RNA or RILP siRNA were subjected to a wound healing assay and fixed after three hours. Cells were stained with anti-tubulin (red) and Hoechst (blue); scale bars: 10 μm. B) Quantification of the percentage of cells with oriented tubulin in control and RILP-depleted cells after 3 h of migration. C) NCI H1299 cells expressing either GFP or GFP-RILP were fixed and stained 3 h after a wound healing assay with anti-tubulin (red) and Hoechst (blue); scale bars: 10 μm. D) Quantification of the percentage of cells with oriented tubulin in control and RILPoverexpressing cells after 3 h of migration. Data represent the mean ± SEM of three different experiments.
RILP modulates cell adhesion and Spreading
We further investigated the role of RILP in cell migration by analyzing the effect of RILP on cell adhesion and spreading. As Rab proteins control the trafficking of adhesion molecules28 and RILP interacts with Rab7a, we evaluated the effect of the expression of either exogenous RILP or its truncated form, RILP-C33, on cell adhesion and spreading. RILP-C33 retains the Rab7a-interacting domain but not the N-terminal half of the protein containing the domains responsible for the interaction with RalGDS and the dynein-dynactin complex. 1,2,19 Firstly, we evaluated a possible effect of RILP overexpression or silencing on cell adhesion. NCI H1299 cells transfected with GFP, GFP-RILP or GFP-RILP C33 were detached, suspended and replated on fibronectin-coated plates. Transfection efficiency was similar for the three samples. After 15, 30 and 60 min of incubation, unattached cells were removed by washing and attached cells were fixed and imaged (Figure 4A). Interestingly, the number of attached cells was decreased when GFPRILP or GFP-RILP C33 were expressed compared to GFP-expressing cells (Figure 4B). In particular, after 15 and 30 min, adhesion was decreased of about 35% or 45% upon expression of GFP-RILP or GFPRILP C33 compared to GFP-expressing cells, respectively. In contrast, cell adhesion was not affected by RILP depletion (Supplementary Figure 2). Given the role of RILP in cell adhesion, we studied the ability of control and RILP expressing cells to spread out on fibronectin-coated coverslips in 30 min by staining with rhodamine-phalloidin, and then we quantified the average cell area. Interestingly, overexpression of RILP or expression of the truncated RILP-C33 mutant determined a lower ability to spread compared to control cells (Figure 5A). In fact, the average cell area of control cells, both untransfected (data not shown) or transfected with the pCDNA3_2xHA vector was about 230 μm2, whereas HA-RILP and HA-RILP-C33 expressing cells had an average cell area of about 134 μm2. Therefore, expression of HA-RILP or of HA-RILP-C33 determined a decrease in the average area of about 42% (Figure 5B). Analysis of untransfected and transfected cells within the same sample produced similar results (data not shown). Moreover, we also tested the effect of RILP silencing on NCI H1299 cells but we did not observe any effect on cell spreading (Supplementary Figure 3).
Figure 4.
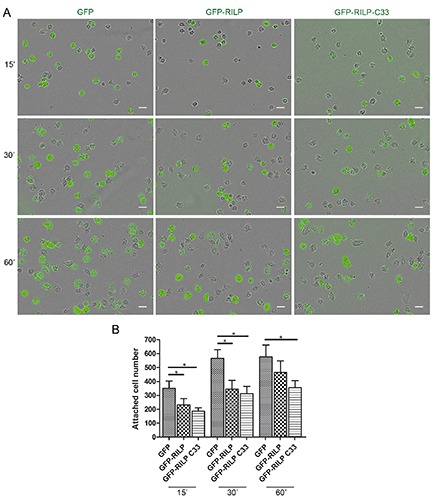
Overexpression of RILP alters cell adhesion. A) NCI H1299 cells expressing GFP, GFP-RILP or GFP-RILP-C33 were seeded in equal number on fibronectin-coated plates and left to adhere for the indicated time points; then, cells were washed with PBS and attached cells were fixed and imaged; scale bars: 50 μm. B) Quantification of the number of attached cells for the indicated samples. Data represent the mean ± SEM of three different experiments.
Figure 5.
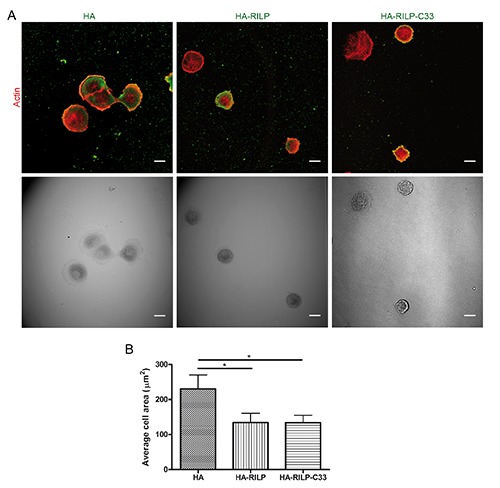
Overexpression of RILP alters cell spreading on fibronectin. A) NCI H1299 cells expressing either HA, HA-RILP or HA-RILP-C33 were plated on fibronectin-coated coverslips and left to adhere for 30 min; coverslips were then fixed and stained with rhodamine- conjugated phalloidin and anti-HA antibody. Confocal and relative transmission images are shown; scale bars: 10 μm. B) Quantification of the average area (in μm2) of NCI H1299 cells treated as described before. Data represent the mean ± SEM of at least three different experiments (n=50).
These data demonstrate that RILP influences cell adhesion and spreading on fibronectin and this might involve Rab7a but not RalGDS or dynactin as neither RalGDS, which has been linked to RILP-mediated role in migration and invasion of breast cancer cells, nor dynactin, which might modulate tubulin cytoskeleton rearrangements, can interact with RILP-C33.
Discussion
Cell migration is a process of extreme importance for cells and it consists of several integrated and highly regulated events such as cytoskeleton rearrangements, asymmetric polarization, cell adhesion and spreading.24 RILP regulates, together with Rab7a, late endocytic trafficking but recently its role in cell migration has been highlighted. 1-4,8,19 In fact, using wound-healing assay on a breast cancer cell line, it was demonstrated that RILP silencing fosters the closure of the wound,19 and therefore we decided to better characterize the role of RILP in cell migration.
Firstly, we demonstrated that RILP silencing speed up the closure of the wound also in NCI H1299 lung cancer cells (Figure 1A; Supplementary Movie 1). This is interesting considering that alterations of migration are found in cancer cells and that also Rab7a regulates migration.14 In particular, Rab7a seems to favor or suppress cancer progression depending on the cell type or district of tumor.17,29,30 Indeed, Rab7a oncosuppressing role has been demonstrated in prostate cancer cells where its depletion affected lysosome localization, increasing tumor cell invasion.17,30 Conversely, Rab7a is upregulated in melanoma promoting cell proliferation.29 Thus, also alterations of RILP expression could impact on migration and invasive properties of cancer cells.
In RILP-depleted cells, velocity of cells increased compared to control cells (Figure 1B), however directness, which measures the ability of cells to move in a persistent manner toward the direction of migration, was not affected (Figure 1E). Therefore, the effect on the faster closure of the wound in NCI H1299 cells is the result of a higher velocity of cells. RILP does not seem to have a role on cell polarization, although other proteins involved in vesicular trafficking, such as Rab7b, have been shown to regulate this process.13 Indeed, RILP depletion did not affect the correct orientation of the Golgi complex (Figure 2). This result is in line with our data on directness (Figure 1E) as cell polarization is important for directional migration.
Another important event during migration is the reorganization of cytoskeletal components, in particular actin filaments and microtubules, that favor cell migration. 26,31 It has been demonstrated that RILP overexpression decreased the formation of stress actin fibers and cortical actin in breast cancer cells by interacting with RalGDS and acting indirectly on RalA.19 Here, we demonstrated that expression of GFP-RILP affects actin cytoskeleton reorganization also in lung cancer cells (Supplementary Figure 1). Dynein and dynactin are enriched at the leading edge during cell migration and their inhibition interferes with the reorientation of the microtubule network.2,32 As RILP interacts with the dynein-dynactin complex, we investigated whether modulation of RILP expression could alter microtubule rearrangements and we demonstrated that neither RILP silencing nor overexpression affect orientation or distribution of microtubules (Figure 3), suggesting that the role of RILP in cell migration does not involve the dynein-dynactin complex. In line with these data, we demonstrated that silencing of RILP does not affect microtubule dynamics through live imaging analysis (Supplementary Movie 2).
Cell adhesion to the extracellular matrix is another important step of cell migration regulated by numerous Rab GTPases. For instance, Rab7b controls cell adhesion and spreading by regulating actin filaments.13 Also, Rab1a knockdown inhibits adhesion and spreading on fibronectin, a component of the extracellular matrix, whereas Rab4 affects adhesion and spreading on vitronectin. 11,33 Furthermore, we recently demonstrated that Rab7a controls cell migration by influencing adhesion and spreading on fibronectin.14 Here, we demonstrated that also RILP plays a role in cell adhesion and spreading to fibronectin. Indeed, RILP overexpression decreased the adhesion and spreading on fibronectin compared to control cells (Figures 4 and 5). Moreover, expression of the truncated form of RILP, RILP-C33, which consists of the amino acids 217-401 containing the Rab7a binding domain, decreased cell adhesion and spreading similarly to the wild type protein. These data suggest that the role of RILP in cell adhesion is not mediated by the interaction with either dynactin or RalGDS, as the RILP domains responsible for these interactions are contained in the first 198 aminoacids, not present in RILP-C33.2,19 Notably, while RILP over-expression negatively affects these processes (Figures 4 and 5), its lack does not improve these steps (Supplementary Figures 2 and 3). This fact could be due to the presence of limiting factors that prevent the possibility of increasing adhesion and indicates that the increase velocity of cell migration in the woundhealing assay caused by RILP silencing (Figure 1; Supplementary Movie 1) is not due to changes in adhesion or spreading.
Cell migration and cell adhesion and spreading are strictly related processes. In fact, for instance, alterations of molecules involved in adhesion, such as integrins, affect both adhesion and motility as a good balance between attachment and detachment is necessary for cell movement.34,35 However, few works have highlighted how alterations of a single protein could affect different aspects of cell migration through independent mechanisms.36 For instance, two different domains of E-cadherin are responsible for cadherin-mediated adhesion and cell motility.36
Wang and colleagues have previously shown that RILP regulates the invasion of breast cancer cells by interacting with RalGDS and, consequently, impairing RalA activity and actin filaments rearrangements.19 Therefore, the effect of RILP silencing on cell velocity could be mediated by RalGDS. However, RalGDS does not interact with the mutated form of RILP, RILP-C33 that affects adhesion and spreading. Therefore, we suggest that RILP could affect cell motility and cell adhesion and spreading through different mechanisms involving different players. Indeed, as RILP-C33 does not interact with the dynactin-dynein complex, and does not affect tubulin filaments rearrangements during migration (Figure 3), another RILP interactor could be responsible for the effect on cell adhesion and spreading mediated by RILP. RILP-C33 is able to interact with Rab7a, which has been recently shown to have a role in cell spreading, 14 therefore this small GTPase could be involved in the regulation of cell adhesion and spreading mediated by RILP.
The fact that Rab7a positively regulates migration while its effector RILP negatively regulates it gives rise to different hypothesis. Rab7a and RILP could regulate migration independently using different mechanisms and the migration outcome could be the result of the opposite contribution of the two proteins. Alternatively, as recently demonstrated for a newly discovered regulation mechanism of V-ATPase mediated by RILP,22,37 the interaction between Rab7a and RILP could modulate the availability of RILP (or of Rab7a) to regulate migration.
Altogether, our data demonstrate the importance of RILP in regulating cell velocity during migration but also in cell adhesion and spreading. Further work will be required to establish the exact molecular mechanism underlying these regulations, to identify possible other molecular players and to determine if the interaction with Rab7a is involved.
Acknowledgements
We thank Nikolai Engedal for his help with the IncuCyte Zoom System. We thank the NorMIC Oslo imaging platform, Department of Biosciences, University of Oslo and the 2HE-PONa3_00334 project for the Zeiss LSM700 confocal microscope.
Funding Statement
Funding: this work was supported by AIRC (Associazione Italiana per la Ricerca sul Cancro, Investigator Grant 2016 N. 19068 to CB), Telethon Italy (Grant GGP16037 to CB), the Norwegian Cancer Society (grants 5760850 to CP and 4604944 to OB), and the Research Council of Norway (grants 239903 to CP, 230779 to OB, and through its Centre of Excellence funding scheme, project number 179573).
References
- 1.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J 2001;20:683-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dyneindynactin motors. Curr Biol 2001;11: 1680-5. [DOI] [PubMed] [Google Scholar]
- 3.Progida C, Malerod L, Stuffers S, Brech A, Bucci C, Stenmark H. RILP is required for proper morphology and function of late endosomes. J Cell Sci 2007;120:3729-37. [DOI] [PubMed] [Google Scholar]
- 4.Wang T, Hong W. Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell 2002;13:4317-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitelli R, Santillo M, Lattero D, Chiariello M, Bifulco M, Bruni C, et al. Role of the small GTPase Rab7 in the late endocytic pathway. J Biol Chem 1997; 272:4391-7. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH. ESCRTs are everywhere. EMBO J 2015;34:2398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Progida C, Spinosa M, De Luca A, Bucci C. RILP interacts with the VPS22 component of the ESCRT-II complex. Biochem Biophys Res Commun 2006; 347:1074-9. [DOI] [PubMed] [Google Scholar]
- 8.Wang T, Hong W. RILP interacts with VPS22 and VPS36 of ESCRT-II and regulates their membrane recruitment. Biochem Biophys Res Commun 2006; 350:413-23. [DOI] [PubMed] [Google Scholar]
- 9.Porther N, M.A. B. The role of endocytic Rab GTPases in regulation of growth factor signaling and the migration and invasion of tumor cells. Small GTPases 2015;6:135-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW, et al. Rab25 associates with alpha5beta1 integrin to promote invasive migration in 3D microenvironments. Dev Cell 2007; 13:496-510. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Yoo Y, Fan H, Kim E, Guan KL, Guan JL. Regulation of Integrin β 1 recycling to lipid rafts by Rab1a to promote cell migration. J Biol Chem 2010; 285:29398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawauchi T. Regulation of cell adhesion and migration in cortical neurons: Not only Rho but also Rab family small GTPases. Small GTPases 2011;2:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borg M, Bakke O, Progida C. A novel interaction between Rab7b and actomyosin reveals a dual role in intracellular transport and cell migration. J Cell Sci 2014;127:4927-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margiotta A, Progida C, Bakke O, Bucci C. Rab7a regulates cell migration through Rac1 and vimentin. Biochim Biophys Acta 2017;1864:367-81. [DOI] [PubMed] [Google Scholar]
- 15.Mendoza P, Ortiz R, Diaz J, Quest AF, Leyton L, Stupack D, et al. Rab5 activation promotes focal adhesion disassembly, migration and invasiveness in tumor cells. J Cell Sci 2013;126:3835-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedl P, Wolf K. Tumor-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003;3: 362-74. [DOI] [PubMed] [Google Scholar]
- 17.Steffan JJ, Williams BC, Welbourne T, Cardelli JA. HGF-induced invasion by prostate tumor cells requires anterograde lysosome trafficking and activity of Na+-H+ exchangers. J Cell Sci 2010;123:1151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steffan JJ, Cardelli JA. Thiazolidinediones induce Rab7-RILPMAPK- dependent juxtanuclear lysosome aggregation and reduce tumor cell invasion. Traffic 2010;11:274-86. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Zhou Y, Hu X, Chen W, Lin X, Sun L, et al. RILP suppresses invasion of breast cancer cells by modulating the activity of RalA through interaction with RalGDS. Cell Death Dis 2015;6: e1923. l [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colucci AMR, Spinosa MR, Bucci C. Espression, assay and functional properties of RILP. Methods Enzymol 2005;403:664-75. [DOI] [PubMed] [Google Scholar]
- 21.Colucci AMR, Campana MC, Bellopede M, Bucci C. The Rab-interacting lysosomal protein, a Rab7 and Rab34 effector, is capable of self-interaction. Biochem Biophys Res Commun 2005;334:128-33. [DOI] [PubMed] [Google Scholar]
- 22.De Luca M, Cogli L, Progida C, Nisi V, Pascolutti R, Sigismund S, et al. RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J Cell Sci 2014;127:2697-708. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda S, Kawamoto K, Miyamoto K, Tsuji A, Yuasa K. PCTK3/CDK18 regulates cell migration and adhesion by negatively modulating FAK activity. Sci Rep 2017;7:45545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci 2005;118:4917-9. [DOI] [PubMed] [Google Scholar]
- 25.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev 2014;94:235-63. [DOI] [PubMed] [Google Scholar]
- 26.Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol 2013; 29:471-99. [DOI] [PubMed] [Google Scholar]
- 27.Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci USA 1988;85:5946-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Franceschi N, Hamidi H, Alanko J, Sahgal P, Ivaska J. Integrin traffic - the update. J Cell Sci 2015;128:839-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso-Curbelo D, Riveiro-Falkenbach E, Perez-Guijarro E, Cifdaloz M, Karras P, Osterloh L, et al. RAB7 controls melanoma progression by exploiting a lineage-specific wiring of the endolysosomal pathway. Cancer Cell 2014;26:61-76. [DOI] [PubMed] [Google Scholar]
- 30.Steffan JJ, Dykes SS, Coleman DT, Adams LK, Rogers D, Carroll JL, et al. Supporting a role for the GTPase Rab7 in prostate cancer progression. PLoS One 2014;9:e87882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber F, Boire A, Lopez MP, Koenderink GH. Cytoskeletal crosstalk: when three different personalities team up. Curr Opin Cell Biol 2015;32:39-47. [DOI] [PubMed] [Google Scholar]
- 32.Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol 2003;163:1205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts M, Barry S, Woods A, van der Sluijs P, Norman J. PDGF-regulated rab4-dependent recycling of alphavbeta3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol 2001;11:1392-402. [DOI] [PubMed] [Google Scholar]
- 34.Truong H, Danen EH. Integrin switching modulates adhesion dynamics and cell migration. Cell Adh Migr 2009;3: 179-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawhney RS, Liu W, Brattain MG. A novel role of ERK5 in integrin-mediated cell adhesion and motility in cancer cells via Fak signaling. J Cell Physiol 2009;219:152-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Paradies NE, Fedor-Chaiken M, Brackenbury R. E-cadherin mediates adhesion and suppresses cell motility via distinct mechanisms. J Cell Sci 1997;110:345-56. [DOI] [PubMed] [Google Scholar]
- 37.De Luca M, Bucci C. A new V-ATPase regulatory mechanism mediated by the Rab interacting lysosomal protein (RILP). Commun Integr Biol 2014;7: e971572. [DOI] [PMC free article] [PubMed] [Google Scholar]


