The majority of human rotavirus A are classified into two genetic groups, which have distinct genotype constellations of all the 11 gene segments encoding viral proteins (G, P, I, R, C, M, A, N, T, E and H) represented by prototype strains Wa and DS-1 respectively (Table 1) [1]. In nature, genotype constellations are stably maintained, and reassortment between them has rarely been described. However, recently in Japan and Thailand, the G1P[8] human rotavirus on the DS-1-like genotype constellation, in which the outer capsid proteins were replaced with those of the Wa-like virus with the DS-1-like genotype background, was reported [2], [3], [4], [5]. We also identified DS-1-like G1P[8] rotaviruses in the Philippines and genetically analysed them to elucidate their relatedness to previously reported rotaviruses and locally circulating rotaviruses to gain insights into their origin and evolutionary pathway.
Table 1.
Nucleotide sequence identity of DS-1-like G1P[8] strain TGO12-016 to other DS-1-like G1P[8] strains, G2P[4] and G1P[8] strains
| Strain and genotype (I to H) | Country (place) and collection date | Identity (%) to viral protein genes of strain TGO12-016 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | ||
| RVA/Human-wt/PHI/TGO12-016/2012/G1P[8] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Philippines (Palawan), August 2012 | 100a | 100a | 100a | 100a | 100a | 100a | 100a | 100a | 100a | 100a | 100a |
| RVA/Human-wt/PHI/TGO12-004/2012/G1P[8] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Philippines (Palawan), July 2012 | 100a | 100a | 100a | 100a | 100a | 95.4 | 100a | 99.9a | 100a | 100a | 100a |
| RVA/Human-wt/PHI/TGO12-012/2012/G1P[8] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Philippines (Palawan), November 2012 | 99.9a | 99.8a | 98.1 | 94.5 | 97.4 | 99.8a | 100a | 97.4 | 99.9a | ||
| RVA/Human-wt/PHI/TGO12-045/2012/G1P[8] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Philippines (Palawan), November 2012 | 99.8a | 99.9a | 97.8 | 94.3 | 97.4 | 99.8a | 100a | 96.9 | 99.7a | 100a | 100a |
| RVA/Human-wt/JPN/HC12016/2012/G1P[8] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Japan (Osaka), April 2012 | 99.1a | 99.9a | 97.9 | 94.5 | 97.3 | 99.5a | 99.6a | 97.0 | 99.7a | 99.4a | 99.8a |
| RVA/Human-wt/THA/PCB-180/2013/G1P[8] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Thailand (Phechaboon), 2013 | 98.6 | 98.9 | 98.1 | 94.4 | 97.3 | 99.5a | 99.7a | 96.9 | 99.5a | 99.2a | 99.5a |
| RVA/Human-wt/PHI/TGO12-003/2012/G2P[4] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
Philippines (Palawan), July 2012 | 65.8 | 83.9 | 99.6a | 99.7a | 99.5a | 86.3 | 98.0 | 99.6a | 97.1 | 98.1 | 98.5 |
| RVA/Human-tc/USA/DS-1/1976/G2P[4] I2-R2-C2-M2-A2-N2-T2-E2-H2 |
65.0 | 85.0 | 85.8 | 89.4 | 94.8 | 92.8 | 92.2 | 85.1 | 98.0 | 89.5 | 95.8 | |
| RVA/Human-tc/USA/Wa/1974/G1P[8] I1-R1-C1-M1-A1-N1-T1-E1-H1 |
91.4 | 88.7 | ||||||||||
| RVA/Human-wt/PHI/TGE12-045/2012/G1P[8] | Philippines (Leyte), September 2012 | 93.4 | 96.9 | |||||||||
| RVA/Human-wt/PHI/TGP12-005/2012/G1P[8] | Philippines (Manila), September 2012 | 93.5 | 97.0 | |||||||||
The nucleotide sequences determined in this study were deposited in the GenBank database under accession numbers KP007144-KP007211.
Two genes (NSP3 and NSP5) of TGO12-012 were not determined because of lack of sample.
Sequence identity of >99%.
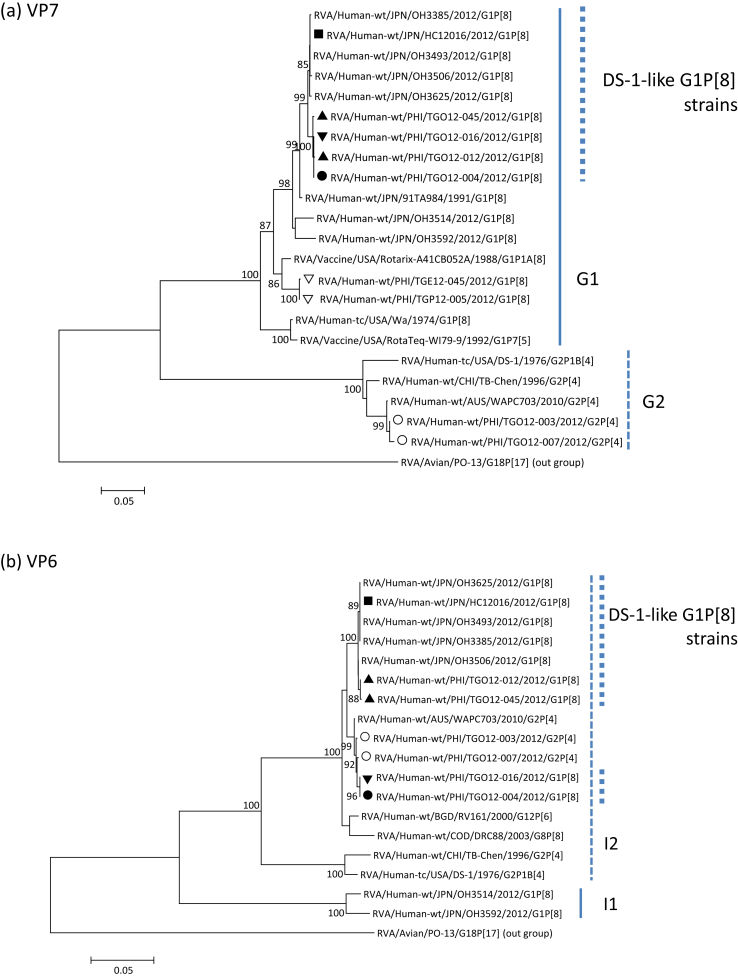
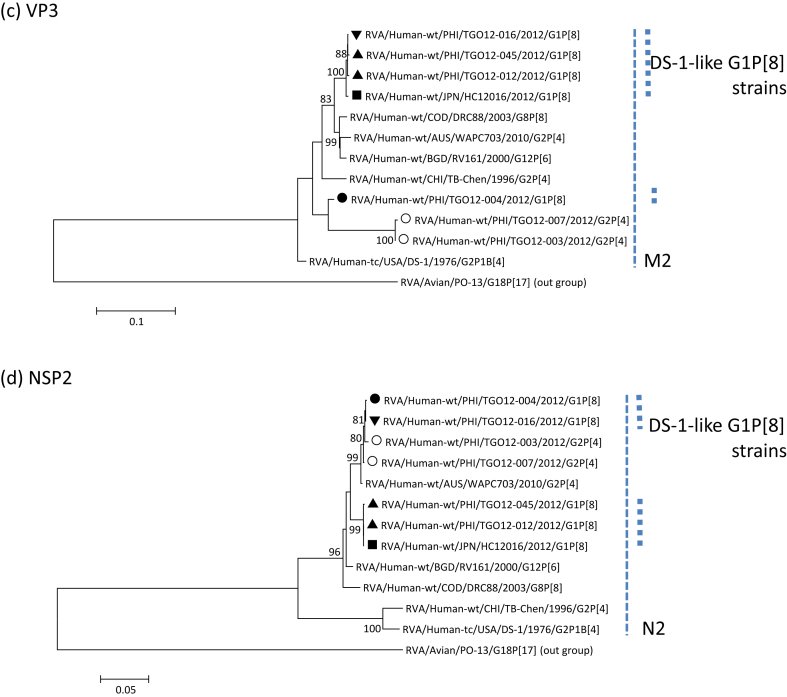
Stool specimens were collected from 45 children admitted with acute diarrhoea in Palawan, Philippines, in 2012. Rotaviruses were detected and genotyped by reverse transcriptase PCR [6]. Twenty-five specimens (56%) were rotavirus positive, including seven G1P[8] and 11 G2P[4]. Four G1P[8] and two G2P[4] specimens were subjected to direct sequencing for all gene segments. Sequence data were phylogenetically analysed (Fig. 1), and pairwise sequence identity was obtained. Our results revealed that all the rotavirus strains, including G1P[8], had a DS-1-like genotype constellation. Four DS-1-like G1P[8] strains were found to be genetically diverse (Table 1). Strain TGO12-016 had identical gene sequences to strain TGO12-004, except for the VP3 gene. Several genes of TGO12-016 (VP7, VP4, VP3, NSP1 and NSP3–NSP5) showed high sequence identities (>99%) to those of TGO12-045 and TGO12-012 (except for NSP3 and NSP5 genes), as well as to DS-1-like G1P[8] strains identified in other countries, whereas other genes, such as VP6, VP1, VP2 and NSP2, exhibited lower identities. Strains TGO12-012 and TGO12-045 were considered identical to strains HC12016 and PCB-180. VP6, VP1, VP2 and NSP2 genes of strains TGO12-016 and TGO12-004 showed >99% identity to those of G2P[4] strain TGO12-003, suggesting that these gene segments of DS-1-like G1P[8] strains might have been derived from a locally circulating G2P[4] strain. In contrast, none of the gene segments of strains TGO12-012 and TGO12-045 clustered with those of local G2P[4] strain TGO12-003.
Fig. 1.
Phylogenetic dendrograms of rotavirus genes (a) VP7, (b) VP6, (c) VP3 and (d) NSP2 constructed by neighbour-joining methods with MEGA.5 program. Variation scale is described at bottom. Percent bootstrap support is indicated at each node (values <80% are omitted). Solid circle, triangle and inverted triangle indicate DS-1-like G1P[8] rotaviruses detected in Philippines; solid square indicates DS-1-like G1P[8] rotavirus in Japan; inverted open triangle indicates typical Wa-like rotaviruses in Philippines; open circle indicates typical DS-1-like rotaviruses in Philippines.
We identified three distinct types, suggesting that various reassortants were generated in Palawan. Six genes of all DS-1-like G1P[8] strains had high sequence identity, implying that they had possibly originated from the same rotavirus. Although genetic information was not sufficient because only a few rotavirus strains were analysed in the present study, two possibilities are conceivable for the diversity in DS-1-like G1P[8] viruses in the Philippines: first, various intergenogroup reassortants were generated in the Philippines, and a single strain has spread to other Asian countries; second, a single DS-1-like G1P[8] rotavirus strain was transmitted from any other Asian country to Palawan, where further reassortment occurred between this strain and local G2P[4] viruses. Although the origin is still not clear, intergenogroup reassortment has been more frequently reported in developing countries [7], [8], [9]. Therefore, it may be easily speculated that the DS-1-like G1P[8] rotavirus was generated in one of the Southeast Asian countries, including the Philippines, and was subsequently spread to other Asian countries. Continuous monitoring is necessary to identify the emergence and spread of novel reassortant viruses.
Acknowledgements
Supported in part by a Grant-in-Aid for Scientific Research (KAKENHI) (grant 23890016) and the Japan Initiative for Global Research Network on Infectious Diseases from the Ministries of Education, Culture, Sports, Science and Technology, Japan.
Conflict of Interest
None declared.
References
- 1.Matthijnssens J., Ciarlet M., Heiman E., Arijs I., Delbeke T., McDonald S.M. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008;82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto S.P., Kaida A., Kubo H., Iritani N. Gastroenteritis outbreaks caused by a DS-1-like G1P[8] rotavirus strain, Japan, 2012–2013. Emerg Infect Dis. 2014;20:1030–1033. doi: 10.3201/eid2006.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii Y., Nakagomi T., Nishimura N., Noguchi A., Miura S., Ito H. Spread and predominance in Japan of novel G1P[8] double-reassortant rotavirus strains possessing a DS-1-like genotype constellation typical of G2P[4] strains. Infect Genet Evol. 2014;28:426–433. doi: 10.1016/j.meegid.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Kuzuya M., Fujii R., Hamano M., Kida K., Mizoguchi Y., Kanadani T. Prevalence and molecular characterization of G1P[8] human rotaviruses possessing DS-1-like VP6, NSP4, and NSP5/6 in Japan. J Med Virol. 2014;86:1056–1064. doi: 10.1002/jmv.23746. [DOI] [PubMed] [Google Scholar]
- 5.Komoto S., Tacharoenmuang R., Guntapong R., Ide T., Haga K., Katayama K. Emergence and characterization of unusual DS-1-like G1P[8] rotavirus strains in children with diarrhea in Thailand. PLoS One. 2015;10:e0141739. doi: 10.1371/journal.pone.0141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iturriza-Gómara M., Kang G., Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31:259–265. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S., Kobayashi N. Whole-genomic analysis of rotavirus strains: current status and future prospects. Future Microbiol. 2011;6:1049–1065. doi: 10.2217/fmb.11.90. [DOI] [PubMed] [Google Scholar]
- 8.Ndze V.N., Esona M.D., Achidi E.A., Gonsu K.H., Dóró R., Marton S. Full genome characterization of human rotavirus A strains isolated in Cameroon, 2010–2011: diverse combinations of the G and P genes and lack of reassortment of the backbone genes. Infect Genet Evol. 2014;28:537–560. doi: 10.1016/j.meegid.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Komoto S., Tacharoenmuang R., Guntapong R., Ide T., Tsuji T., Yoshikawa T. Reassortment of human and animal rotavirus gene segments in emerging DS-1-like G1P[8] rotavirus strains. PLoS One. 2016;11:e0148416. doi: 10.1371/journal.pone.0148416. [DOI] [PMC free article] [PubMed] [Google Scholar]