Abstract
Invasive amoebiasis is a life-threatening infection requiring immediate detection and treatment. However, diagnosis is challenging because conventional methods such as light microscopy and serology are unreliable. Molecular techniques are therefore considered the new diagnostic reference standard, but most of the developed assays are research tools and not widely available. Recently commercial multiplex PCR panels have been introduced which permit the simultaneous detection of multiple enteric pathogens including Entamoeba histolytica in stool samples. Our report demonstrates for the first time that these new assays might also serve as a rapid tool to diagnose amoebic liver abscess in patients with cystic focal liver lesions.
Keywords: Amoebiasis, amoebic liver abscess, diagnosis, Entamoeba histolytica, multiplex molecular panel
Introduction
Entamoeba histolytica is an important gastrointestinal pathogen of worldwide distribution, causing significant morbidity and mortality [1], [2]. The infection is highly endemic in nonindustrialized regions in Latin America, Africa and the Indian subcontinent, where it is associated with inadequate food and water hygiene and sanitation [3], [4]. In developed countries, the main risk groups include travellers, immigrants and men who have sex with men [5], [6]. E. histolytica inhabits the human colonic mucosa, where it causes a wide spectrum of pathology, from asymptomatic colonization to severe haemorrhagic colitis; as a result of its invasive capacity, the parasite is also able to cause metastatic extraintestinal infections, most frequently affecting the liver [5].
The discovery of several apathogenic doppelganger species, which are morphologically indistinguishable from E. histolytica, has led most of the established concepts regarding the epidemiology and pathophysiology of this pathogen to be questioned [4]. It has also led to questioning of traditional diagnostic methods such as microscopy, which is now considered obsolete by some experts [3], [7]. The dilemma was emphasized by the World Health Organization in 1997, which stated that all intestinal E. histolytica infections required diagnosis and treatment while carriers of apathogenic species should not receive therapy, highlighting the urgent need for new diagnostic techniques [1]. A major step was therefore the development of molecular methods that could detect E. histolytica–specific DNA in both intestinal and extraintestinal samples [8]. Until recently, however, this new approach was neither commercialized nor standardized and remained a research tool within reference laboratories. This has changed with the newly introduced multiplex molecular panels for enteric pathogens, some of which include E. histolytica, offering the detection of this pathogen by molecular methods within the routine microbiologic workup of stool samples [9]. Our reported case demonstrates for the first time that these new assays can also serve as a rapid tool to diagnose amoebic liver abscess (ALA) in patients with cystic focal liver lesions.
Patient and methods
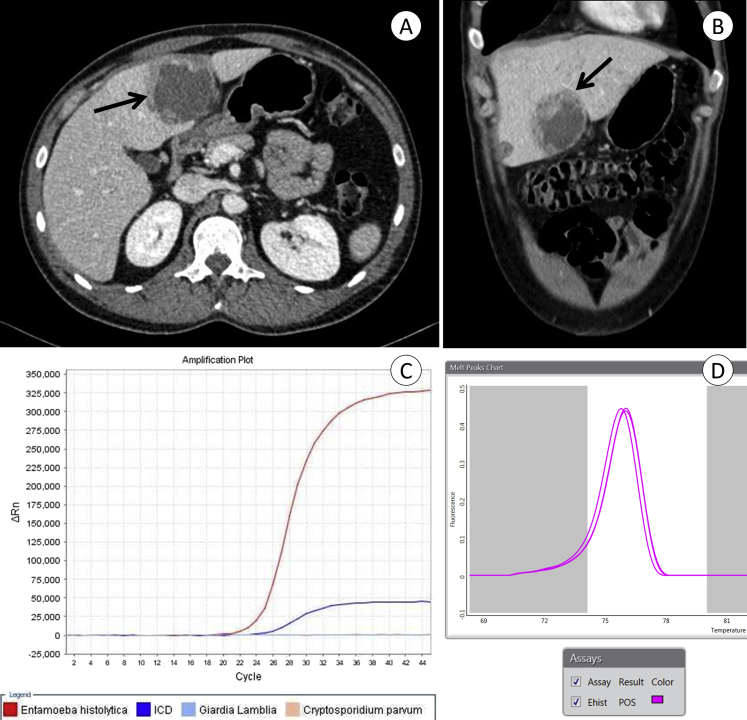
A 34-year-old man from Santiago, Chile, was admitted with a 1-week history of progressive malaise, fever and upper abdominal pain. Two years before he had experienced a prolonged bout of travel-associated diarrhoea and abdominal pain after a trip to Colombia, which resolved spontaneously. At admission his temperature was 38.8°C; he was pale and had abdominal tenderness over the right upper quadrant. Laboratory examination revealed leukocytosis (12 300/mm3) and elevated values for C-reactive protein (37 mg/dL), erythrocyte sedimentation rate (87 mm/h), aspartate aminotransferase (28 U/L), alanine aminotransferase (48 U/L) and γ-glutamyl transferase (77 U/L). Abdominal ultrasound demonstrated a hypodense liver lesion of 5 cm diameter in the left lobe and computed tomographic scan a hypodense lesion in the IV-B segment of the left lobe, with contrast-enhancing walls compatible with abscess (Figs. 1A, B). The abscess was drained percutaneously, and the aspirated brownish fluid was tested by two commercial multiplex molecular panels, Rida®Gene Parasitic Stool Panel (R-Biopharm, Darmstadt, Germany) and FilmArray Gastrointestinal Panel (BioFire Diagnostics, Salt Lake City, UT, USA). Both assays amplified E. histolytica–specific nucleic acid, while other enteric pathogens were negative (Figs. 1C, D). The patient was also positive for E. histolytica IgG serum antibodies (E. histolytica IgG ELISA; R-Biopharm). Treatment with metronidazole and subsequent paromomycin was initiated, and the patient recuperated without complications.
Fig. 1.
Diagnosis of amoebic liver abscess by imaging and molecular techniques. Transverse (A) and sagittal (B) planes of contrast-enhanced computed tomographic scans showing single hypodense lesion of 5 × 6 cm, with well-defined lobulated margins and alternating hyper- and hypovascular peripheral halos (arrows). Amplification plot of Rida®Gene Parasitic Stool Panel (C) and melting curve of FilmArray Gastrointestinal Panel (D), indicating presence of Entamoeba histolytica–specific DNA in abscess aspirate material. ICD, internal control DNA.
Discussion
The differential diagnosis in patients with liver abscess is complex and includes a variety of bacterial, fungal and parasitic pathogens [10]. Imaging techniques are highly sensitive but are unable to differentiate the cause [5]. ALA is a frequent cause of liver abscess in tropical and developing countries, typically affecting the right liver lobe of men. As a life-threatening infection, it requires immediate diagnosis and treatment [11]. If untreated, it has a high fatality rate [2]. In countries of low endemicity, the diagnosis requires a high level of clinical suspicion because ALA might occur years after exposure [3]. Our patient most probably acquired the amoebic infection during his travels 2 years before, which is in accordance with observations from other imported cases [12].
Diagnosis of ALA is usually based on the detection of E. histolytica–specific serum antibodies [2], [5]. However, serology is unreliable during early stages (falsely negative) and in endemic regions with high background seroprevalence (falsely positive) [5], [8], [12]. The most powerful development to overcome the diagnostic problems of amoebiasis was the introduction of DNA-based tests, which are now considered the diagnostic reference standard [8]. Various PCR stool protocols have been developed, some of which were also successfully applied in ALA pus specimens [13], [14], [15], [16], [17]. Although most evaluations of molecular tests were performed in endemic regions, PCR proved to be a useful tool in the diagnosis of imported ALA cases in Spain and France [12], [18]. However, a comparative study demonstrated considerable performance differences using different in-house protocols [19]. Major disadvantages of molecular diagnostic methods for amoebiasis are the lack of standardization as well as a long hands-on time and high cost [8], [9]. The main obstacle for laboratories working under current quality standards is the absence of E. histolytica–specific molecular kits, which are licensed as in vitro diagnostics (IVD) because a commercial real-time PCR kit (Artus, Hamburg, Germany) has been discontinued [3].
Currently several multiplex PCR panels have been certified as IVD, allowing the simultaneous detection of multiple pathogens in stool samples, some of which also include protozoa such as Giardia lamblia, Cryptosporidium spp. and E. histolytica [9], [20]. Up to now data on the diagnostic efficacy and clinical usefulness of these multiplex kits in patients with suspected amoebiasis are limited. One study and literature review showed that the few published studies focussing on protozoa detection by enteric multiplex assays included only tiny numbers of E. histolytica–positive samples [21]. However, the authors acknowledged these tests as a robust alternative to traditional diagnosis of amoebiasis considering their diagnostic performance, rapid turnaround time and use in nonreference laboratories [21]. An exception might be the xTAG Gastrointestinal Pathogen Panel (Luminex Molecular Diagnostics, Toronto, Canada), which detected a high number of samples positive for E. histolytica that could not be confirmed by other techniques [22].
To our knowledge, no data on the use of commercial multiplex PCR panels in patients with liver abscess have been published. In our patient with a left lobe single lesion, both of the applied commercial kits confirmed the diagnosis of ALA by amplification of E. histolytica DNA from aspirated pus material within a short time. The FilmArray system gave faster results than the Rida®Gene test (1 hour vs. 3 hours) and required less hands-on time (5 minutes vs. 45 minutes). A possible advantage of Rida®Gene was the quantitative result, which revealed a high concentration of the E. histolytica–specific target (Cq value = 24), whereas the endpoint technique of the FilmArray platform only gives qualitative results. Although the multiplex design for enteric pathogens is not useful in aspirates of liver abscess, such panels represent the only option for IVD-licensed molecular testing for amoebiasis at present, and their usefulness and diagnostic performance for this indication should be further evaluated.
Our study suggests that multiplex PCR testing could permit a rapid and robust diagnosis of amoebiasis in patients with cystic focal liver lesions within routine microbiologic laboratories, overcoming the known diagnostic limitations of this neglected tropical disease.
Conflict of Interest
None declared.
References
- 1.World Health Organization Amoebiasis. Wkly Epidemiol Rec. 1997;72:97–100. [Google Scholar]
- 2.Stanley S.L., Jr. Amoebiasis. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 3.Fotedar R., Stark D., Beebe N., Marriott D., Ellis J., Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20:511–532. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ximénez C., Morán P., Rojas L., Valadez A., Gómez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Genet Evol. 2009;9:1023–1032. doi: 10.1016/j.meegid.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Haque R., Huston C.D., Hughes M., Houpt E., Petri W.A., Jr. Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 6.Hung C.C., Chang S.Y., Ji D.D. Entamoeba histolytica infection in men who have sex with men. Lancet Infect Dis. 2012;12:729–736. doi: 10.1016/S1473-3099(12)70147-0. [DOI] [PubMed] [Google Scholar]
- 7.Petri W.A., Jr., Singh U. Diagnosis and management of amebiasis. Clin Infect Dis. 1999;29:1117–1125. doi: 10.1086/313493. [DOI] [PubMed] [Google Scholar]
- 8.Stark D., Ellis J. Entamoeba. In: Liu D., editor. Molecular detection of human parasitic pathogens. CRC Press; Boca Raton, FL: 2013. pp. 63–76. [Google Scholar]
- 9.Binnicker M.J. Multiplex molecular panels for diagnosis of gastrointestinal infection: performance, result interpretation, and cost-effectiveness. J Clin Microbiol. 2015;53:3723–3728. doi: 10.1128/JCM.02103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lardière-Deguelte S., Ragot E., Amroun K., Piardi T., Dokmak S., Bruno O. Hepatic abscess: diagnosis and management. J Visc Surg. 2015;152:231–243. doi: 10.1016/j.jviscsurg.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Salles J.M., Salles M.J., Moraes L.A., Silva M.C. Invasive amebiasis: an update on diagnosis and management. Expert Rev Anti Infect Ther. 2007;5:893–901. doi: 10.1586/14787210.5.5.893. [DOI] [PubMed] [Google Scholar]
- 12.Vallois D., Epelboin L., Touafek F., Magne D., Thellier M., Bricaire F. Amebic liver abscess diagnosed by polymerase chain reaction in 14 returning travelers. Am J Trop Med Hyg. 2012;87:1041–1045. doi: 10.4269/ajtmh.2012.12-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaman S., Khoo J., Ng S.W., Ahmed R., Khan M.A., Hussain R. Direct amplification of Entamoeba histolytica DNA from amoebic liver abscess pus using polymerase chain reaction. Parasitol Res. 2000;86:724–728. doi: 10.1007/pl00008558. [DOI] [PubMed] [Google Scholar]
- 14.Roy S., Kabir M., Mondal D., Ali I.K., Petri W.A., Jr., Haque R. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 2005;43:2168–2172. doi: 10.1128/JCM.43.5.2168-2172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderaro A., Gorrini C., Bommezzadri S., Piccolo G., Dettori G., Chezzi C. Entamoeba histolytica and Entamoeba dispar: comparison of two PCR assays for diagnosis in a non-endemic setting. Trans R Soc Trop Med Hyg. 2006;100:450–457. doi: 10.1016/j.trstmh.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Haque R., Kabir M., Noor Z., Rahman S.M., Mondal D., Alam F. Diagnosis of amebic liver abscess and amebic colitis by detection of Entamoeba histolytica DNA in blood, urine, and saliva by a real-time PCR assay. J Clin Microbiol. 2010;48:2798–2801. doi: 10.1128/JCM.00152-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zengzhu G., Bracha R., Nuchamowitz Y., Cheng I.W., Mirelman D. Analysis by enzyme-linked immunosorbent assay and PCR of human liver abscess aspirates from patients in China for Entamoeba histolytica. J Clin Microbiol. 1999;37:3034–3036. doi: 10.1128/jcm.37.9.3034-3036.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez-Cisneros M.J., Martin-Rabadan P., Menchen L., GarciaLechuz J.M., Fuentes I., Garate T. Autochthonous amebic liver abscess in Spain: an emerging disease? Case report and description of a PCR-based diagnostic test. Enferm Infecc Microbiol Clin. 2009;27:326–330. doi: 10.1016/j.eimc.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Qvarnstrom Y., James C., Xayavong M., Holloway B.P., Visvesvara G.S., Sriram R. Comparison of real-time PCR protocols for differential laboratory diagnosis of amebiasis. J Clin Microbiol. 2005;43:5491–5497. doi: 10.1128/JCM.43.11.5491-5497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zboromyrska Y., Vila J. Advanced PCR-based molecular diagnosis of gastrointestinal infections: challenges and opportunities. Expert Rev Mol Diagn. 2016;16:631–640. doi: 10.1586/14737159.2016.1167599. [DOI] [PubMed] [Google Scholar]
- 21.Laude A., Valot S., Desoubeaux G., Argy N., Nourrisson C., Pomares C. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin Microbiol Infect. 2016;22 doi: 10.1016/j.cmi.2015.10.019. 190.e1–8. [DOI] [PubMed] [Google Scholar]
- 22.Navidad J.F., Griswold D.J., Gradus M.S., Bhattacharyya S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J Clin Microbiol. 2013;51:3018–3024. doi: 10.1128/JCM.00896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]