Abstract
Shoot and inflorescence are central physiological and developmental tissues of plants. Flowering is one of the most important agronomic traits for improvement of crop yield. To analyze the vegetative to reproductive tissue transition in Jatropha curcas, gene expression profiles were generated from shoot and inflorescence tissues. RNA isolated from both tissues was sequenced using the Ilumina HiSeq 2500 platform. Differential gene expression analysis identified key biological processes associated with vegetative to reproductive tissue transition. The present data for J. curcas may inform the design of breeding strategies particularly with respect to reproductive tissue transition. The raw data of this study has been deposited in the NCBI's Sequence Read Archive (SRA) database with the accession number SRP090662.
Keywords: Biofuel, Differentially expressed genes, Flowering, Inflorescence, Shoot, Transcriptome
| Specifications | |
|---|---|
| Organism/cell line/tissue | Jatropha curcas |
| Sex | Inflorescence: Bisexual flowers |
| Sequencer or array type | Ilumina HiSeq 2500 platform |
| Data format | Raw: bam files |
| Experimental factors | Shoot and inflorescence |
| Experimental feature | n/a |
| Biospecimen IDs | Shoot: SAMN05827448, SAMN05827450, SAMN05827452 Inflorescence: SAMN05827449, SAMN05827451, SAMN05827453 |
| Sample source location | Bangi, Malaysia (2°55′09.0″N101°47′04.8″E). |
| Consent | n/a |
1. Direct link to deposited data
2. Introduction
Jatropha curcas L. is a sustainable feed stock for bio-energy productions. The oil extracted from seeds (40–58%) could be feasibly converted and/or incorporated into biodiesel and others products such as medicine, soap-making, lighting fuel and bio-pesticide industries [4]. In addition, the by-products from Jatropha oil extraction meet a wide array of uses; organic fertilizer, combustible fuel and biogas production. Irregularities in flowering and fruit set are among the major commercial problems affecting productivity of large-scale plantations [1]. The dynamics of flowering and fruit set begins at the floral development stage, initiated by phase transition. A substantial number of intercellular and intracellular signals and regulative mechanisms are involved in the vegetative-to-reproductive phase transition [5], [6], [7]. To date, no detailed molecular data are available for J. curcas to investigate this phenomenon. Using the transcriptomic data generated from shoot and inflorescence tissues of J. curcas, we investigated the molecular events underpinning shoot-to-inflorescence transition using the differential gene expression (DGE) analysis.
3. Experimental design, materials and methods
3.1. Plant material
Shoot and inflorescence tissues were collected from three different Jatropha curcas plants (Jcurcas1, Jcurcas2 and Jcurcas3) aged 2 years-old from Experimental Plot A, Universiti Kebangsaan Malaysia (UKM), Bangi (2°55′09.0″N101°47′04.8″E). The climate data of the experimental site from 2012 to 2015, indicated an average temperature of 25.6–28.8 °C and mean relative humidity of 95%. Annual rainfall during the growth period ranged from 1779.8–2630.1 mm. The soils are primarily Serdang series (Ultisols-Typic Paleudults); reddish brown and sandy clay loam. Shoot tissue was collected at 2.5 cm from the base of the peduncle. In presence of small leaves and corresponding petiole, the accessory tissues were excised. Inflorescence collected comprised of flowers, peduncle and pedicels (Fig. 1). All samples were immediately flash frozen in liquid nitrogen prior to RNA isolation.
Fig. 1.

Inflorescence (A) and shoot (B) tissues from Jatropha curcas used in the differential gene expression analysis.
3.2. RNA isolation, transcriptome assembly and analyses
Total RNA was isolated using a modified method of CTAB + silica column [8] and RNeasy Plant mini kit (Qiagen, Hilden, Germany). Quality and integrity of total RNA were determined prior to cDNA library preparation which followed the SureSelect Strand-Specific RNA Library Prep for NGS Workflow protocol (Agilent Technologies, USA). The fragmented genomic DNA was sequenced using Ilumina HiSeq 2500 (Yourgene Bioscience, Taiwan). For each library output, the raw fastq reads were trimmed using the Trim Galore package and aligned to Jatropha curcas genomic sequences from Kazusa's Jatropha Genome Database version r4.5 (ftp://ftp.kazusa.or.jp/pub/jatropha/). Alignment was performed using the STAR aligner (https://github.com/alexdobin/STAR). Cufflinks were used to produce a merged assembly. Unless stated, all computational applications were run using the default parameters. Prior to the DEG analysis, we normalized the number of reads that were mapped to each transcript into counts per million (CPM) using the weighted trimmed mean of M-values (TMM) method. To evaluate the number of differentially expressed genes between the shoot and inflorescence tissues, pairwise differential expression analysis using the Exact Test was performed as described in the BLAST2Go Pro 4.0 suite [3]. The differentially expressed genes were classified according to Gene ontologies (GO) annotations using WEGO, an online plotting tool [2].
3.3. Data description
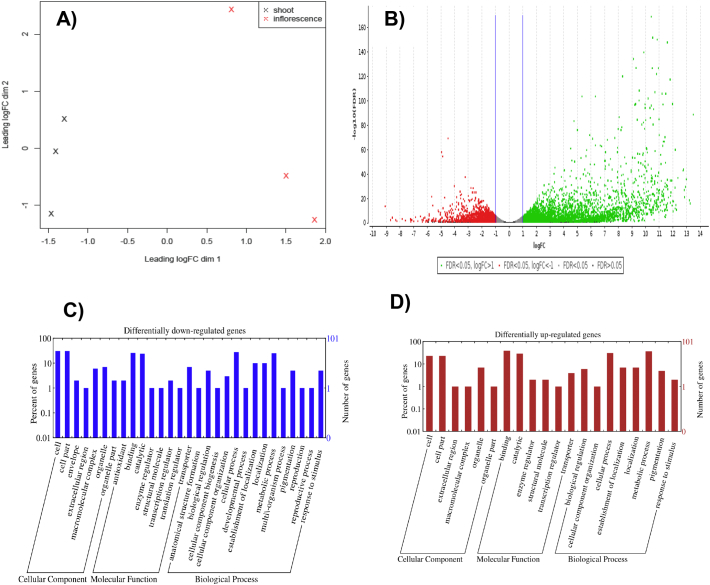
We obtained a total of 64,235 reads from 6 trancriptome assemblies, with the following biosample IDs: shoot (SAMN05827448, SAMN05827450, SAMN05827452) and inflorescence (SAMN05827449, SAMN05827451, SAMN05827453).The multidimensional scaling (MDS) plot assessing distance at log2-fold changes distinguished the shoot (black crosses) from the inflorescence (red crosses) samples in dimension 1. The tissue-specific clusters were in agreement with the biological replicates defined in this study (Fig. 2A). Differential expression analysis revealed a total of 6125 genes differentially expressed in the inflorescence relative to the shoot. The volcano plot shows up-regulated (FC > 1) and down-regulated (FC < 1) genes as green and red dots respectively. Among these genes, 4007 and 2118 number of genes were up- and down-regulated respectively at p < 0.05 (Fig. 2B). We selected the top 100 genes with FC > 4 (up-regulated) and FC < − 4 (down regulated) for GO annotation classifications. Cellular component terms showed similar distributions of associated gene numbers for both differentially up- and down-regulated genes with highest occurrence at cell, cell part and organelle. The molecular function term also displayed a similar trend with highest binding and catalytic activities. Biological process terms, however, showed differential distributions for down-regulated genes as compared to the up-regulated genes between shoot and inflorescence; structure formation, cellular component biogenesis, developmental process, establishment of localization, multi-organism process, reproduction and reproduction process (Fig. 2C and D). Our data highlighted several genes that may control the shoot-to-inflorescence tissue transition; gene IDs, fold change (FC) values and GO annotations of the selected candidates are given in Table 1. The present study is an important resource for mapping and marker assisted breeding of J. curcas yield-associated traits.
Fig. 2.
Differentially expressed gene analysis. A) Multidimensional plot of the shoot and inflorescence transcriptomes. B) Volcano plot shows up-regulated (FC > 1) and down-regulated (FC < 1) genes in green and red dots respectively. Gene ontology (GO) classification of the differentially expressed transcripts at FC > 4 (C) and FC < − 4 (D) are summarized: biological process, cellular component and molecular function.
Table 1.
Candidate genes identified from the differential gene expression analysis between the shoot and inflorescence tissues in Jatropha curcas Linn.
| Gene ID | Fold change | Annotation GO term |
|---|---|---|
| Jcr4S01319.50 | − 4.96 | Pathogenesis; extracellular region |
| Jcr4S00009.90 | − 4.69 | Clathrin adaptor complex; intracellular protein transport; vesicle-mediated transport |
| Jcr4S00083.20 | − 5.05 | Metal ion transport; metal ion binding |
| Jcr4S08020.10 | − 6.13 | Membrane; antiporter activity; drug transmembrane transport; drug transmembrane transporter activity |
| Jcr4S06192.20 | − 5.05 | Membrane; antiporter activity; drug transmembrane transport; drug transmembrane transporter activity |
| Jcr4S00009.90 | − 4.69 | Clathrin adaptor complex; intracellular protein transport; vesicle-mediated transport |
| Jcr4S10688.40 | − 4.92 | Membrane; seed development; protein storage vacuole |
| TCONS_00077217 | − 4.95 | Transporter activity; ATPase activity; outer membrane-bounded periplasmic space; ATP binding; cytochrome complex assembly |
Authors contributions
SS: performed the RNA isolation; NG: performed the bioinformatics analyses and wrote the manuscript; ZAMH, WR: conceived, coordinated and secured fundings for the project.
Acknowledgements
We thank Universiti Kebangsaan Malaysia (UKM) and Ministry of Higher Education, Malaysia (MOHE) for the financial support; Research University Grant AP-2012-008 and DIP-2012-019. Nisha Govender and Siju Senan were supported by UKM postdoctoral fellowships.
References
- 1.Tjeuw J., Slingerland M., Giller Ken. Relationships among Jatropha curcas seed yield and vegetative plant components under different management and cropping systems in Indonesia. Biomass Bioenergy. 2015;80:128. (e139) [Google Scholar]
- 2.Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunil N., Kumar V., Sujatha M., Rao G.R., Varaprasad K.S. Minimal descriptors for characterization and evaluation of Jatropha curcas L. germplasm for utilization in crop improvement. Biomass Bioenergy. 2013;48(1) (239e249) [Google Scholar]
- 5.Poethig R.S. Chapter 5: Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 2013;105:125–152. doi: 10.1016/B978-0-12-396968-2.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araki T. Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 2001;4:63–68. doi: 10.1016/s1369-5266(00)00137-0. [DOI] [PubMed] [Google Scholar]
- 7.Dong X., Jiang X., Qi G., Wang Q., Zhong M., Jin D., Hu J.Y. Genetic control of flowering time in woody plants: roses as an emerging model. Plant Diversity. 2017 doi: 10.1016/j.pld.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangha J.S., Gu K., Kaur J., Yin Z. An improved method for RNA isolation and cDNA library construction from immature seeds of Jatropha curcas L. BMC Res. Notes. 2010;3:126. doi: 10.1186/1756-0500-3-126. [DOI] [PMC free article] [PubMed] [Google Scholar]