Abstract
Aims
Darapladib, a potent inhibitor of lipoprotein-associated phospholipase A2 (Lp-PLA2), has not reduced risk of cardiovascular disease outcomes in recent randomized trials. We aimed to test whether Lp-PLA2 enzyme activity is causally relevant to coronary heart disease.
Methods
In 72,657 patients with coronary heart disease and 110,218 controls in 23 epidemiological studies, we genotyped five functional variants: four rare loss-of-function mutations (c.109+2T > C (rs142974898), Arg82His (rs144983904), Val279Phe (rs76863441), Gln287Ter (rs140020965)) and one common modest-impact variant (Val379Ala (rs1051931)) in PLA2G7, the gene encoding Lp-PLA2. We supplemented de-novo genotyping with information on a further 45,823 coronary heart disease patients and 88,680 controls in publicly available databases and other previous studies. We conducted a systematic review of randomized trials to compare effects of darapladib treatment on soluble Lp-PLA2 activity, conventional cardiovascular risk factors, and coronary heart disease risk with corresponding effects of Lp-PLA2-lowering alleles.
Results
Lp-PLA2 activity was decreased by 64% (p = 2.4 × 10–25) with carriage of any of the four loss-of-function variants, by 45% (p < 10–300) for every allele inherited at Val279Phe, and by 2.7% (p = 1.9 × 10–12) for every allele inherited at Val379Ala. Darapladib 160 mg once-daily reduced Lp-PLA2 activity by 65% (p < 10–300). Causal risk ratios for coronary heart disease per 65% lower Lp-PLA2 activity were: 0.95 (0.88–1.03) with Val279Phe; 0.92 (0.74–1.16) with carriage of any loss-of-function variant; 1.01 (0.68–1.51) with Val379Ala; and 0.95 (0.89–1.02) with darapladib treatment.
Conclusions
In a large-scale human genetic study, none of a series of Lp-PLA2-lowering alleles was related to coronary heart disease risk, suggesting that Lp-PLA2 is unlikely to be a causal risk factor.
Keywords: Human genetics, target validation, coronary heart disease, lipoprotein-associated phospholipase A2, darapladib
Introduction
Lipoprotein-associated phospholipase A2 (Lp-PLA2), an enzyme expressed by inflammatory cells in atherosclerotic plaques, is carried in the circulation bound predominantly to low-density lipoprotein (LDL).1,2 Lp-PLA2 (also called platelet-activating factor acetyl hydrolase) hydrolyses oxidized phospholipids to yield pro-inflammatory products implicated in endothelial dysfunction, plaque inflammation and formation of necrotic core in plaque.1 Observational3 and experimental studies in humans and animals have suggested that Lp-PLA2 could be a valid therapeutic target, postulating this enzyme to link oxidative modification of LDL and development of inflammatory responses to arterial intima.1 Previous studies have investigated genetic variants altering Lp-PLA2 function in relation to coronary heart disease (CHD) risk.4,5 However, these studies have generally yielded inconclusive, or conflicting results,4,5 perhaps due to limited statistical power and due to limited knowledge about variants altering Lp-PLA2 function (e.g. previous studies have been able to consider only one loss-of-function variant in PLA2G7, the gene encoding Lp-PLA2).
However, two phase 3 randomized trials of darapladib, a potent inhibitor of Lp-PLA2 activity, have not shown reductions in cardiovascular risk.6,7 These results could, at least in part, have been due to features of the trials. One of the phase 3 trials was restricted to patients recently hospitalized with acute coronary syndromes,6 yet many cardiovascular events occurring early after acute coronary syndromes may relate to thrombotic mechanisms and not be modifiable through Lp-PLA2 inhibition. Trials used statins as background therapy, so any Lp-PLA2 inhibition achieved with statins could have reduced any incremental benefits of darapladib. Trials could not assess the effects of prolonged Lp-PLA2 inhibition because they recorded only about 3–4 years of median follow-up.6,7
An alternative explanation is that darapladib did not reduce cardiovascular risk because Lp-PLA2 is not a causal risk factor in cardiovascular disease. We tested this possibility by investigating natural loss of Lp-PLA2 activity. Studies of Lp-PLA2-lowering alleles should complement randomized trials of darapladib because genotypes are fixed at conception, avoiding potential distorting effects of pre-existing disease and medication usage. Furthermore, Lp-PLA2-lowering alleles should produce lifelong, rather than shorter-term, Lp-PLA2 inhibition.
In over 260,000 participants of European, South Asian, or East Asian ancestries, we studied five functional variants in PLA2G7. We compared effects of Lp-PLA2-lowering alleles on soluble Lp-PLA2 activity, conventional cardiovascular risk factors and CHD risk with corresponding effects of darapladib, using results from randomized trials.
Methods
Study design
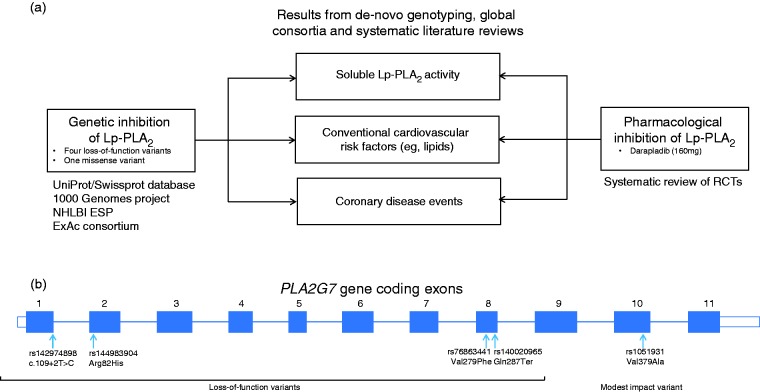
Figure 1 summarizes the study approach. Table 1 provides definitions and sources of data used. First, we identified four loss-of-function mutations and one missense variant in PLA2G7 suggested by previous experimental and bioinformatics studies, thereby developing an allelic series for Lp-PLA2 activity. Second, we assessed associations of these variants – both singly and in combination – with soluble Lp-PLA2 activity, conventional cardiovascular risk factors and CHD risk in people of European, South Asian or East Asian ancestries. Third, we compared associations of Lp-PLA2-lowering alleles with the aforementioned traits and CHD risk with the effects of darapladib treatment through a systematic review of randomized trials.
Figure 1.
Summary of study design. (a) Flow chart of study design. (b) Exonic structure of the PLA2G7 gene and location of variants used in this study. ExAc: Exome Aggregation consortium; NHLBI ESP: National Heart Lung and Blood Institute Exome Sequencing Project; Lp-PLA2: lipoprotein-associated phospholipase A2; RCT: randomized controlled trial; UniProt/Swissprot: manually annotated and reviewed section of the Universal Protein resource database.
Table 1.
Definitions and source of contributing data for the main study outcome.
| Lp-PLA2 assessment tool | Val279Phe, loss-of-function variant common in East Asians | Four loss-of-function variantsa, rare in Europeans and South Asians | Val379Ala, modest impact variant | Darapladib | ||||
|---|---|---|---|---|---|---|---|---|
| Data sources |
Systematic review: up to 12 East Asian studies |
De-novo genotyping and participant-level data: up to eight European or South Asian ancestry studies from the CHD Exome+ consortium19-26 De-novo genotyping and study-level data: up to 15 European ancestry studies from the MICAD Exome consortium38 and three European ancestry studies from the CHARGE Consortium17,29 Plus publicly available consortium data |
Systematic review: up to five randomized clinical trials6,7,39-41 from a systematic review | |||||
|
Endpoint
|
Number of studies and unique individuals contributing to analyses; n total or cases/controls
|
|||||||
| Coronary heart disease b | Seven East Asian studiesc | Eight European or South Asian ancestry studies from the CHD Exome+ consortium19-26 d | 15 European ancestry studies from the MICAD Exome consortium38 c | Eight European or South Asian ancestry studies from the CHD Exome+ consortium19-26 d | Eight European ancestry studies from the MICAD Exome consortium38 c | 14 European ancestry studies from the CARDIoGRAM consortium27 e | Four European or South Asian ancestry studies from the C4D consortium28 e | Two phase 3 randomized clinical trials of darapladib6,7 c |
| 10,088 cases 15,199 controls | 35,829 cases 44,948 controls | 35,533 cases 64,130 controls | 32,196 cases 41,464 controls | 14,976 cases 32,084 controls | 20,315 cases 58,419 controls | 15,420 cases 15,062 controls | 3364 cases 25,490 non-cases | |
| Lp-PLA2 activity f | 12 East Asian studiesc | One European ancestry study from the CHD Exome+ consortium25 d | One European ancestry study from the CHARGE consortium29 c | Two European ancestry studies from the CHD Exome+ consortium22,23,25 d | Three European ancestry studies from the CHARGE consortium17 c | Three phase 2 randomized clinical trials39-41 c | ||
| 8468 | 1240 | 8564 | 2173 | 11,662 | 854 | |||
Genetic variants
We defined loss-of-function variants as non-synonymous variants with in vitro or in vivo evidence demonstrating complete lack of Lp-PLA2 activity or sequence changes expected to abolish Lp-PLA2 function (e.g. nonsense variants or mutations in essential splice sites). We selected variants through a systematic search for loss-of-function variants using the UniProt database,8 the Exome Aggregation Consortium database (Cambridge, MA, USA; URL: http://exac.broadinstitute.org (accessed November 2014)),9 studies of site-directed mutagenesis10-12 and results from targeted gene sequencing.13 Among the full set of variants identified (Supplementary Material online Table 1), we selected the following variants that could be detected in the 1000 Genomes14 or the NHLBI Exome Sequencing Project15 projects (and, hence, potentially studied at the population level): the splice site mutation 109+2T>C (rs142974898); two non-synonymous variants – Arg82His (rs144983904) and Val279Phe (rs76863441); and the nonsense variant Gln287Ter (rs140020965). These loss-of-function variants are rare in European and South Asian ancestry populations, whereas carriage of 279Phe is common in East Asian ancestry populations and abolition of Lp-PLA2 activity is well documented.16 Additionally, we studied Val379Ala (rs1051931), a functional variant common in European ancestry populations, which lowers Lp-PLA2 activity only modestly,10,17 in contrast with the much stronger Lp-PLA2-lowering achieved by the loss-of-function variants described above.
Samples and data for genetic studies
We aimed to maximize study power and comprehensiveness by using the following complementary approaches to generate new data on, as well as to collate systematically existing relevant information about, the PLA2G7 variants mentioned above: (1) we conducted de-novo genotyping for 72,657 CHD patients and 110,218 controls (the majority of whom also had information available on some cardiovascular risk factors); (2) we accessed non-overlapping summary-level data from the only known global genetics consortium of CHD,18 yielding information on a further 35,735 CHD patients and 73,481 controls; (3) we conducted a systematic review (supplemented by provision of tabular data from each study investigator) of published East Asian CHD studies of Val279Phe because these studies were not represented in the global CHD consortium, yielding information on a further 10,088 CHD cases and 15,199 controls; (4) we accessed summary-level data from the largest available global genetics consortium on each of several relevant cardiovascular risk factors (e.g. Lp-PLA2 activity, conventional lipids, blood pressure), yielding information on 489,045 participants. Each of these sources of information is summarized below and in Table 1, with a key in Table 1’s legend denoting the level of data detail available for each source (e.g. individual-participant data versus tabular study-level results).
Coronary heart disease outcomes
For CHD outcomes, we had access to data for a total of 92,995 patients and 162,228 controls. For 182,875 of these participants (72,657 CHD patients, 110,218 controls), we did de-novo genotyping of the four loss-of-function variants (c.109 + 2T > C, Arg82His, Val279Phe, Gln287Ter) and Val379Ala using customized Exome arrays (Illumina, CA, USA) by technicians masked to the phenotypic status of the participants’ samples. For 35,829 CHD cases, 44,948 controls in eight studies, we had access to individual-participant data. The eight studies were: the Bangladesh Risk of Acute Vascular Events Study,19 Copenhagen City Heart Study,20 Copenhagen Ischemic Heart Disease/Copenhagen General Population Study,20 European Prospective Investigation into Cancer and Nutrition-Cardiovascular Disease Study (EPIC-CVD),21 MONICA Risk, Genetics, Archiving, and Monograph (MORGAM) study,22,23 Pakistan Risk of Myocardial Infarction Study,24 Pravastatin in elderly individuals at risk of vascular disease (PROSPER) trial25 and the West of Scotland Coronary Prevention Study26 (these eight studies are collectively called the ‘CHD Exome+ consortium’). For 15 additional studies (collectively called the ‘MICAD consortium’) we used similar genotyping methods to those described above but did not genotype c.109+2T>C and had access only to study-level data. We supplemented de-novo data on Val379Ala with non-overlapping consortium-level results from a further 35,735 CHD patients and 73,481 controls in the transatlantic Coronary Artery Disease Genome-wide Replication and Meta-analysis27 and Coronary Artery Disease Genetics28 consortia (Table 1). We obtained tabular data on Val279Phe from seven East Asian studies involving a total of 10,088 CHD cases and 15,199 controls, identified through systematic review (text and Table 5 in Supplementary Material online). About 90% of CHD patients in our genetic analysis had myocardial infarction or other major acute coronary events; the remainder had angiographic evidence alone (e.g. >50% coronary stenosis; Supplementary Tables 2 and 5).
Lp-PLA2 activity
For 13,835 participants, we had information on functional variants in PLA2G7 and Lp-PLA2 activity, using data from de-novo genotyping in MORGAM22,23 and PROSPER,25 supplemented by published data from the CHARGE Consortium (i.e. from the Atherosclerosis Risk in Communities study,29 Cardiovascular Health Study,17 Framingham Heart Study17 and Rotterdam study,17 and from 12 East Asian studies identified through the systematic review described above (Table 1; Supplementary Material text, Figure 1 and Tables 2 and 3).
Conventional cardiovascular risk factors
For 177,343 participants, we had information on functional variants in PLA2G7 and conventional cardiovascular risk factors and several other traits, including circulating concentrations of LDL-cholesterol, high-density lipoprotein (HDL)-cholesterol, triglycerides, glucose, insulin and C-reactive protein, and values of systolic and diastolic blood pressure, body-mass index and estimated glomerular filtration rate. Again, we supplemented data from our de-novo genotyping, with information from existing global genetics consortia (Table 1; Supplementary Tables 2 to 4).
Randomized trials of darapladib
To compare genetic associations with effects of pharmacological Lp-PLA2 inhibition, we conducted a systematic review to identify randomized placebo-controlled trials of darapladib that had reported on Lp-PLA2 activity, conventional risk factors and/or CHD events (Supplementary Material). CHD events in the trials were defined as fatal CHD, myocardial infarction or urgent revascularization, as recorded in STABILITY (Stabilization of Atherosclerotic Plaque by Initiation of Darapladib Therapy) and in SOLID-TIMI 52 (Stabilization of Plaque Using Darapladib-Thrombolysis in Myocardial Infarction 52).6,7 We pooled results across trials by fixed-effect inverse-variance weighted meta-analysis (Supplementary Figures 2 and 3; see Supplementary text for details of the methods used).
Statistical methods
We defined effect alleles as those associated with lower Lp-PLA2 activity and assumed an additive model. For participant-level data, we assessed associations of Lp-PLA2-lowering alleles with CHD using the genome-wide efficient mixed model analysis, an approach that models each genetic variant as a fixed-effect, but includes both fixed-effect and random-effects of genetic inheritance30 to account for population stratification and relatedness among participants (Supplementary Material). The four rare loss-of-function variants were tested jointly within each study by counting the number of loss-of-function alleles carried by each participant. Log odds ratios and standard errors were meta-analysed across studies using fixed-effect meta-analysis. For studies contributing only study-level data, we performed a similar test by conducting a combined burden test across studies using the R package seqMeta v1.2 (http://cran.r-project.org/web/packages/seqMeta/).
We calculated associations of Lp-PLA2-lowering alleles with soluble Lp-PLA2 activity and conventional risk factors using linear regression within each study, and then combined the regression coefficients using fixed-effect meta-analysis. When data were missing, we used information on rs1805018 as a proxy for Val279Phe and information on rs7756935 or rs3799277 as proxies for Val379Ala (Supplementary Material). To account for population stratification, we adjusted for the first principal component of ancestry (Supplementary Material). We calculated risk ratios for CHD with decrements in Lp-PLA2 activity, dividing the log transformed risk ratio and confidence interval (CI) by the effect on Lp-PLA2 activity of the instrument (i.e. the genetic variant).31 We investigated heterogeneity using the I2 statistic. We used Stata 13.1.
Results
Of the 261,950 total participants in this analysis, we studied 195,715 individuals of European ancestry, 34,221 individuals of South Asian ancestry and 32,014 individuals of East Asian ancestry. In people of European or South Asian ancestry without CHD, the frequency of alleles in PLA2G7 that lower Lp-PLA2 activity was 0.005% at c.109 + 2T > C, 0.04% at Arg82His, 0.04% at Val279Phe and 0.025% at Gln287Ter (i.e. in aggregate, 0.2% of the European or South Asian participants in the current study carried one of these loss-of-function alleles, although no one carried more than one of these variants), and about 80% at Val379Ala. In people of East Asian ancestry without CHD, the frequency of Val279Phe was about 15% and about 2% of the individuals were homozygous carriers of the 279Phe allele.
Soluble Lp-PLA2 activity
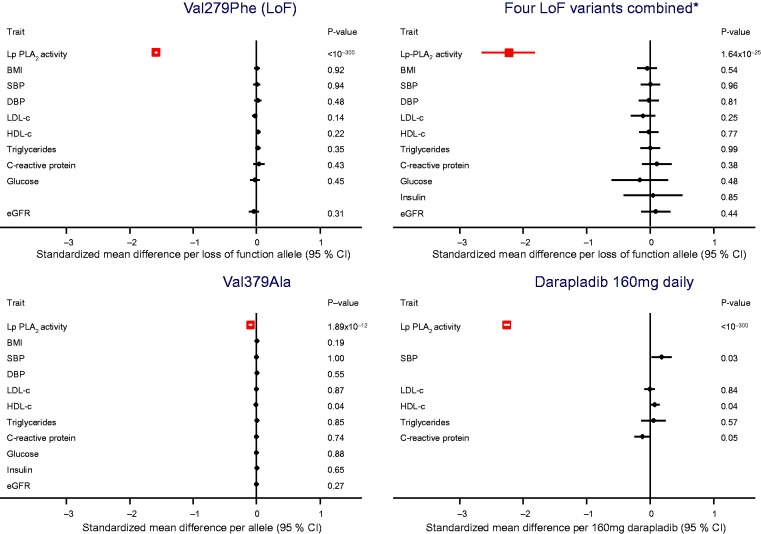
Compared with non-carriers, homozygote carriers of the 279Phe allele had 94% lower Lp-PLA2 activity (p < 10–300). For each 279Phe allele inherited, Lp-PLA2 activity decreased by 45% (1.59 SD, 95% CI: 1.61–1.57; p < 10–300). In Europeans who inherited any one of the four rare Lp-PLA2 loss-of-function alleles, Lp-PLA2 activity decreased by 64% (2.25 SD, 2.68–1.83; p = 1.6 × 10–25). For each 379Ala allele inherited, Lp-PLA2 activity decreased by 2.7% (0.096 SD, 0.122–0.069; p = 1.9 × 10–12). By comparison, 160 mg once-daily darapladib reduced Lp-PLA2 activity by 65% (2.26 SD, 2.31–2.21; p < 10–300). Study-level estimates are provided in Supplementary Figure 2.
Cardiovascular risk factors
None of the Lp-PLA2-related variants we studied was significantly associated with values of LDL-cholesterol, HDL-cholesterol, triglycerides, systolic or diastolic blood pressure, body-mass index, estimated glomerular filtration rate, glucose, insulin and C-reactive protein (Figure 2). By comparison, in previous randomized placebo-controlled trials, darapladib did not significantly affect concentrations of LDL-cholesterol or log triglycerides, but could have slightly increased systolic blood pressure and HDL-cholesterol values and slightly decreased C-reactive protein concentration (Figure 2).
Figure 2.
Mean per allele differences in Lp-PLA2 activity and cardiovascular risk factor levels by Lp-PLA2-lowering alleles or with darapladib 160 mg daily. To enable comparison of the magnitude of associations across several different markers, analyses were undertaken with standardized units of measurement for each marker. Associations are presented as per allele change in the biomarker expressed as standard deviations. Numbers of participants are provided in Table 1. Details of contributing studies are provided in Supplementary Material Tables 2 and 3 online.
*Carriage of any of the four loss-of-function variants c.109+2T>C, Arg82His, Val279Phe, Gln287Ter.
BMI: body-mass index; CI: confidence interval; DBP: diastolic blood pressure; eGFR: estimated glomerular filtration rate; HDL-c: high-density lipoprotein cholesterol; LDL: low-density lipoprotein cholesterol; LoF: loss-of-function; Lp-PLA2: lipoprotein associated phospholipase A2; SBP: systolic blood pressure
Clinical CHD outcomes
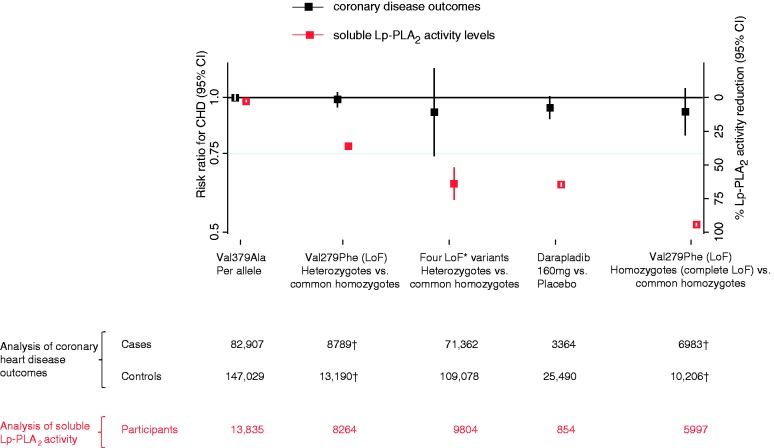
Compared with non-carriers, the odds ratio for CHD was 0.99 (0.95–1.03) in 279Phe heterozygotes, and 0.93 (0.82–1.05) in 279Phe homozygotes (i.e. nearly complete loss of Lp-PLA2 function: Figure 3). For each loss-of-function (279Phe) allele inherited, the odds ratio for CHD was 0.97 (0.91–1.02; I2 = 30%; pHeterogeneity = 0.2). In Europeans and South Asians who inherited one of the four rare Lp-PLA2-loss-of-function alleles, the odds ratio for CHD was 0.92 (0.74–1.16; I2 = 0%; pHeterogeneity = 0.8; Figure 3). For each 379Ala allele inherited, the odds ratio for CHD was 1.00 (0.98–1.02; I2 = 0.0%; pHeterogeneity = 0.5; Figure 3). Study-level results are provided in Supplementary Figure 3. In sensitivity analyses, odds ratios with each loss-of-function variant were similar to the odds ratio that combined information across the four loss-of-function variants we studied. There was no evidence of heterogeneity in odds ratios between European and South Asian ancestry populations (Supplementary Figure 4).
Figure 3.
Association of Lp-PLA2-lowering alleles with Lp-PLA2 activity and CHD risk. Spectrum of functional alleles in PLA2G7 and effects on Lp-PLA2 activity (red estimates) and coronary heart disease risk (black estimates); *Carriage of any of the four loss-of-function variants c.109+2T>C, Arg82His, Val279Phe, Gln287Ter. †One study did not provide tabular data to enable calculation of CHD odds ratios in heterozygotes or homozygotes. Hence, numbers are less than those presented for the per allele analysis in Table 2. CHD: coronary heart disease; CI: confidence interval; LoF: loss-of-function; Lp-PLA2: lipoprotein associated phospholipase A2.
Genetic risk ratios for CHD per 65% lower Lp-PLA2 activity (i.e. the reduction achievable with darapladib treatment) were: 0.95 (0.88–1.03) with Val279Phe in East Asians; and 0.92 (0.74–1.16) with carriage of any one of the four rare variants studied in Europeans and South Asians; and 1.01 (0.68–1.51) with Val379Ala (Table 2). By comparison, the risk ratio for CHD with darapladib treatment (i.e. also per 65% lower Lp-PLA2 activity) was 0.95 (0.89–1.02; Table 2).
Table 2.
Comparison on a common scale of human genetic and randomized trial evidence for Lp-PLA2 lowering and CHD.
| CHD patients | Controls | Risk ratio for CHD per 65% lower Lp-PLA2 activity (95% CI) | |
|---|---|---|---|
| Genetically lowered Lp-PLA2 | |||
| Val279Phe (East Asian LoF variant) | 10,088 | 15,199 | 0.95 (0.88–1.03) |
| Four loss-of-function variantsa | 71,362 | 109,078 | 0.92 (0.74–1.16) |
| Val379Ala | 82,907 | 147,029 | 1.01 (0.68–1.51) |
| Pharmacologically lowered Lp-PLA2 | |||
| Darapladib | 3364 | 25,490 | 0.95 (0.89–1.02) |
Further detail on the individual studies is provided in Supplementary Tables 2 and 3 online.
rs142974898 (c.109+2T>C), rs144983904 (Arg82His), rs76863441 (Val279Phe), rs140020965 (Gln287Ter); see also Figure 1(b) for further variant details.
In genetic analysis, CHD was defined as myocardial infarction and other major coronary events (∼90% of cases) or angiographic stenosis only (∼10% of cases); see Supplementary Tables 2 and 3 for details. In the darapladib analysis CHD was defined as fatal coronary disease, non-fatal myocardial infarction or urgent revascularization for myocardial ischaemia.
Summary/tabular data available (by study).
Participant-level data available.
Meta-analysis data available.
See Supplementary Tables 2 and 3 for details on risk factor measurements.
BMI: body-mass index; C4D: Coronary Artery Disease Genetics consortium; CARDIoGRAM: Coronary ARtery DIsease Genome wide Replication and Meta-analysis; CHARGE: Cohorts for Heart and Aging Research in Genomic Epidemiology; CHD: coronary heart disease; CKDGen: Chronic Kidney Disease Genetics consortium; eGFR: estimated glomerular filtration rate; GIANT: Genetic Investigation of ANthropometric Traits consortium; GLGC: Global Lipids Genetics Consortium; ICBP: International Consortium for Blood Pressure; Lp-PLA2: lipoprotein associated phospholipase A2; MAGIC: Meta-Analyses of Glucose and Insulin-related traits Consortium; NA: data not available
Discussion
In a large-scale analysis of human genetic data, we tested whether Lp-PLA2 enzyme activity is causally relevant to CHD by studying five functional alleles that produce widely differing (i.e. small, moderate or large) degrees of reduction in Lp-PLA2 activity. We found that none was related to CHD risk, suggesting that Lp-PLA2 enzyme activity is unlikely to be causally relevant to CHD, a conclusion concordant with results from two phase 3 trials of a pharmacological Lp-PLA2 enzyme inhibitor.
Three features of our study merit comment. First, we studied almost 20 times more CHD patients than the previous largest study of loss-of-function PLA2G7 alleles, thereby providing the first robust genetic evaluation of effect sizes of Lp-PLA2 inhibition relevant to phase 3 trials such as relative risk reductions for CHD of 20%. For example, for the Val279Phe variant we had >99% power to detect a 20% risk reduction in CHD for a 65% genetic reduction in Lp-PLA2 activity (i.e. an effect on Lp-PLA2 activity similar to that achieved by darapladib).
Second, our study has provided the first investigation in CHD of a series of functional alleles that each reduce Lp-PLA2 function via different molecular mechanisms. Specifically, we studied five different Lp-PLA2-lowering alleles: three of the alleles were coding variants that produced different amino acid substitutions; two of the alleles produced protein truncations (one due to a nonsense mutation; the other due to a splice-site mutation). Because we observed null and broadly concordant findings for CHD risk across these alleles that each changed the enzyme in a different way (and to a different extent), we can more confidently conclude there is no material cause-and-effect relationship. By contrast, when the initial phase 3 trial of darapladib was launched in 2008, only two of the five alleles we studied had yet been identified: data on Val379Ala, a weak effect missense variant, were inconclusive because CHD studies were under-powered;32 data on Val279Phe, a loss-of-function variant, and CHD risk were sparse and restricted to East Asian populations.
A third feature was our study’s analysis of large-scale data from three different major ethnic groups: Europeans, South Asians and East Asians. This ethnic diversity enhanced the generalizability of our results.
Our study had potential limitations. To maximize comparability of CHD endpoints used in clinical trials with those used in human genetic studies, we restricted analysis of phase 3 darapladib trials to ‘major coronary events’ and principally focused in human genetic studies on the cognate endpoints of myocardial infarction or other major acute coronary events (which constituted ∼90% of the outcomes). Nevertheless, although the CHD definitions used in trials and genetic studies were similar, they were not identical.
It could be that cardioprotective benefits of Lp-PLA2 inhibition were obscured by pleiotropic effects of PLA2G7 variants; for example, 279Phe is known to produce a misfolded version of Lp-PLA2 not secreted by cells, prompting suggestions that its carriage could produce ‘off-target’ effects such as increased cell death.33,34 However, because we found null associations between four other functional alleles in PLA2G7 and CHD, each of which operates via a different molecular mechanism, it argues against this explanation. On the other hand, it is possible that darapladib may have additional effects beyond Lp-PLA2 inhibition. For example, darapladib may have had slight effects on CRP levels and systolic blood pressure, which we did not observe with the genetic variants.
Lifelong genetic reductions in Lp-PLA2 could result in compensatory responses that increase CHD risk. However, this explanation seems unlikely because it would require any such compensation to apply similarly across alleles that produce widely differing degrees of reduction in Lp-PLA2 activity. Furthermore, any such compensation could not operate through known cardiovascular mechanisms because we observed no associations between Lp-PLA2-lowering alleles and several established and emerging cardiovascular risk factors.
Soluble enzyme activity could be an imperfect indicator of the relevance of Lp-PLA2 to atherosclerotic plaques. However, for homozygote carriers of 279Phe, Lp-PLA2 activity should be almost abolished across all tissues. Finally, we studied life-long genetic reductions in Lp-PLA2 activity in relation to first-onset CHD outcomes rather than recurrent CHD, whereas darapladib trials studied recurrent coronary events in patients with stable or acute coronary disease.
The current data underscore the growing importance of human genetic approaches to enhance the efficiency of development of medicines by validating (or invalidating) novel drug targets.35-38 Specifically, despite beneficial effects of darapladib on surrogate markers (e.g. intravascular imaging) of coronary atherosclerosis in pre-clinical and clinical studies,39-41 these effects did not translate into reduced outcomes in the large phase 3 studies. Hence, human genetic studies may be useful in influencing prioritization of clinical outcome trials in the future.
Our results also illustrate how human genetic evidence can assist interpretation of observational epidemiological data. For example, we found that functional alleles in PLA2G7 do not alter levels of pro-atherogenic lipids (e.g. LDL-cholesterol), suggesting that such pro-atherogenic lipids do not mediate associations between Lp-PLA2 activity and CHD and supporting the need to adjust epidemiological associations of Lp-PLA2 activity with CHD risk for pro-atherogenic lipids (an approach which yields results consistent with non-causality).3
In summary, we found that none of a series of Lp-PLA2–lowering alleles was related to CHD risk, suggesting that Lp-PLA2 is unlikely to be a causal risk factor in CHD.
Supplementary Material
Acknowledgement
The work was conducted at the University of Cambridge.
Author contribution
ASB, DFF, JD, JMG contributed to the conception or design of the work. DFF, JMG and JD drafted the manuscript. ASB, JD, JMM, JMMH, JS, PG, PS, PW, SBu, SGT and SKap critically revised the manuscript. All the other authors contributed to the acquisition, analysis, or interpretation of data for the work. All gave final approval and agree to be accountable for all aspects of work ensuring integrity and accuracy. JMG, DFF, DS, JMMH, EDA, ASB and JD made equal contribution.
Declaration of conflicting interests
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Anders Malarstig and Maria Uria-Nickelsen are full time employees of Pfizer. Alex Thompson is a full-time employee of UCB. Since October 2015, Daniel Freitag has been a full time employee of Bayer. The funders had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work of the coordinating centre was funded by the UK Medical Research Council (G0800270), British Heart Foundation (SP/09/002), British Heart Foundation Cambridge Cardiovascular Centre of Excellence, UK National Institute for Health Research Cambridge Biomedical Research Centre, European Research Council (268834), European Commission Framework Programme 7 (HEALTH-F2-2012-279233). The Supplement includes a list provided by investigators of some of the funders of the component studies in this analysis.
References
- 1.Corson MA. Darapladib: An emerging therapy for atherosclerosis. Ther Adv Cardiovasc Dis 2010; 4: 241–248. [DOI] [PubMed] [Google Scholar]
- 2.Awan Z, Genest J. Inflammation modulation and cardiovascular disease prevention. Eur J Prev Cardiol 2015; 22: 719–733. [DOI] [PubMed] [Google Scholar]
- 3.Thompson A, Gao P, Orfei L, et al. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: Collaborative analysis of 32 prospective studies. Lancet 2010; 375: 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jang Y, Waterworth D, Lee JE, et al. Carriage of the V279F null allele within the gene encoding Lp-PLA2 is protective from coronary artery disease in South Korean males. PLoS One 2011; 6: e18208–e18208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada Y, Yoshida H, Ichihara S, et al. Correlations between plasma platelet-activating factor acetylhydrolase (PAF-AH) activity and PAF-AH genotype, age, and atherosclerosis in a Japanese population. Atherosclerosis 2000; 150: 209–216. [DOI] [PubMed] [Google Scholar]
- 6.O’Donoghue ML, Braunwald E, White HD, et al. Effect of darapladib on major coronary events after an acute coronary syndrome: The SOLID-TIMI 52 randomized clinical trial. JAMA 2014; 312: 1006–1015. [DOI] [PubMed] [Google Scholar]
- 7.White HD, Held C, Stewart R, et al. Darapladib for preventing ischemic events in stable coronary heart disease. N Engl J Med 2014; 370: 1702–1711. [DOI] [PubMed] [Google Scholar]
- 8.UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res 2015; 43: D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruse S, Mao XQ, Heinzmann A, et al. The Ile198Thr and Ala379Val variants of plasmatic PAF-acetylhydrolase impair catalytical activities and are associated with atopy and asthma. Am J Hum Genet 2000; 66: 1522–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tjoelker LW, Eberhardt C, Unger J, et al. Plasma platelet-activating factor acetylhydrolase is a secreted phospholipase A2 with a catalytic triad. J Biol Chem 1995; 270: 25481–25487. [DOI] [PubMed] [Google Scholar]
- 12.Yamada Y, Yokota M. Loss of activity of plasma platelet-activating factor acetylhydrolase due to a novel Gln281–>Arg mutation. Biochem Biophys Res Commun 1997; 236: 772–775. [DOI] [PubMed] [Google Scholar]
- 13.Song K, Nelson MR, Aponte J, et al. Sequencing of Lp-PLA2-encoding PLA2G7 gene in 2000 Europeans reveals several rare loss-of-function mutations. Pharmacogenomics J 2012; 12: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012; 337: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stafforini DM, Satoh K, Atkinson DL, et al. Platelet-activating factor acetylhydrolase deficiency. A missense mutation near the active site of an anti-inflammatory phospholipase. J Clin Invest 1996; 97: 2784–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grallert H, Dupuis J, Bis JC, et al. Eight genetic loci associated with variation in lipoprotein-associated phospholipase A2 mass and activity and coronary heart disease: Meta-analysis of genome-wide association studies from five community-based studies. Eur Heart J 2012; 33: 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015; 47: 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdhury R, Alam DS, Fakir, et al. The Bangladesh Risk of Acute Vascular Events (BRAVE) study: Objectives and design. Eur J Epidemiol 2015; 30: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamstrup PR, Tybjaerg-Hansen A, Steffensen R, et al. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA 2009; 301: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 21.Danesh J, Saracci R, Berglund G, et al. EPIC-Heart: The cardiovascular component of a prospective study of nutritional, lifestyle and biological factors in 520,000 middle-aged participants from 10 European countries. Eur J Epidemiol 2007; 22: 129–141. [DOI] [PubMed] [Google Scholar]
- 22.Evans A, Salomaa V, Kulathinal S, et al. MORGAM (an international pooling of cardiovascular cohorts). Int J Epidemiol 2005; 34: 21–27. [DOI] [PubMed] [Google Scholar]
- 23.Kulathinal S, Karvanen J, Saarela O, et al. Case-cohort design in practice – experiences from the MORGAM Project. Epidemiol Perspect Innov 2007; 4: 15–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleheen D, Zaidi M, Rasheed A, et al. The Pakistan Risk of Myocardial Infarction Study: A resource for the study of genetic, lifestyle and other determinants of myocardial infarction in South Asia. Eur J Epidemiol 2009; 24: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caslake MJ, Packard CJ, Robertson M, et al. Lipoprotein-associated phospholipase A(2), inflammatory biomarkers, and risk of cardiovascular disease in the Prospective Study of Pravastatin in the Elderly at Risk (PROSPER). Atherosclerosis 2010; 210: 28–34. [DOI] [PubMed] [Google Scholar]
- 26.Shepherd J, Cobbe SM, Ford I, et al. Prevention of coronary heart disease with pravastatin in men with hypercholesterolemia. West of Scotland Coronary Prevention Study Group. N Engl J Med 1995; 333: 1301–1307. [DOI] [PubMed] [Google Scholar]
- 27.Schunkert H, König IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011; 43: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011; 43: 339–344. [DOI] [PubMed] [Google Scholar]
- 29.Polfus LM, Gibbs RA, Boerwinkle E. Coronary heart disease and genetic variants with low phospholipase A2 activity. N Engl J Med 2015; 372: 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods 2014; 11: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer TM, Sterne JAC, Harbord RM, et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am J Epidemiol 2011; 173: 1392–1403. [DOI] [PubMed] [Google Scholar]
- 32.Casas JP, Ninio E, Panayiotou A, et al. PLA2G7 genotype, lipoprotein-associated phospholipase A2 activity, and coronary heart disease risk in 10 494 cases and 15 624 controls of European Ancestry. Circulation 2010; 121: 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem 2005; 74: 739–789. [DOI] [PubMed] [Google Scholar]
- 34.Zalewski A, Macphee C. Role of lipoprotein-associated phospholipase A2 in atherosclerosis: Biology, epidemiology, and possible therapeutic target. Arterioscler Thromb Vasc Biol 2005; 25: 923–931. [DOI] [PubMed] [Google Scholar]
- 35.Kamb A, Harper S, Stefansson K. Human genetics as a foundation for innovative drug development. Nat Biotechnol 2013; 31: 975–978. [DOI] [PubMed] [Google Scholar]
- 36.Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015; 47: 856–860. [DOI] [PubMed] [Google Scholar]
- 37.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov 2013; 12: 581–594. [DOI] [PubMed] [Google Scholar]
- 38.Stitziel NO, Won HH, Morrison AC, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med 2014; 371: 2072–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daida H, Iwase T, Yagi S, et al. Effect of darapladib on plasma lipoprotein-associated phospholipase A2 activity in Japanese dyslipidemic patients, with exploratory analysis of a PLA2G7 gene polymorphism of Val279Phe. Circ J 2013; 77: 1518–1525. [DOI] [PubMed] [Google Scholar]
- 40.Mohler ER, 3rd, Ballantyne CM, Davidson MH, et al. The effect of darapladib on plasma lipoprotein-associated phospholipase A2 activity and cardiovascular biomarkers in patients with stable coronary heart disease or coronary heart disease risk equivalent: The results of a multicenter, randomized, double-blind, placebo-controlled study. J Am Coll Cardiol 2008; 51: 1632–1641. [DOI] [PubMed] [Google Scholar]
- 41.Serruys PW, Garcá-Garcá HM, Buszman P, et al. Effects of the direct lipoprotein-associated phospholipase A(2) inhibitor darapladib on human coronary atherosclerotic plaque. Circulation 2008; 118: 1172–1182. [DOI] [PubMed] [Google Scholar]
- 42.Randall JC, Winkler TW, Kutalik Z, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet 2013; 9: e1003500–e1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011; 478: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013; 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dehghan A, Dupuis J, Barbalic M, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation 2011; 123: 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010; 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pattaro C, Köttgen A, Teumer A, et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 2012; 8: e1002584–e1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.