Sula and Wallace explore evidence supporting a role for a novel interaction motif in the gating of prokaryotic sodium channels.
Abstract
Voltage-gated sodium channels enable the translocation of sodium ions across cell membranes and play crucial roles in electrical signaling by initiating the action potential. In humans, mutations in sodium channels give rise to several neurological and cardiovascular diseases, and hence they are targets for pharmaceutical drug developments. Prokaryotic sodium channel crystal structures have provided detailed views of sodium channels, which by homology have suggested potentially important functionally related structural features in human sodium channels. A new crystal structure of a full-length prokaryotic channel, NavMs, in a conformation we proposed to represent the open, activated state, has revealed a novel interaction motif associated with channel opening. This motif is associated with disease when mutated in human sodium channels and plays an important and dynamic role in our new model for channel activation.
Introduction
The opening of eukaryotic sodium channels initiates the action potential in excitable tissues (Hille, 2001). The entire action potential functional cycle includes opening, closing, and inactivation of the sodium channels (Nav), and the closely coordinated functioning of potassium and calcium ion channels and sodium potassium ATPase pumps. These processes are essential in the functioning of all the closely related sodium channel isoforms found in different human tissues (central and peripheral nervous tissues, cardiac tissues, and muscle fibers). Mutations affecting the sodium channel functional cycles produce a range of channelopathies such as epilepsy, chronic pain, and ventricular fibrillations. As a result, these channels are important targets for pharmaceutical drug discovery.
A limited number of prokaryotes (mostly extremophiles) also possess sodium channels (Ren et al., 2001; Koishi et al., 2004); the biological roles of these channels are as yet unclear, but appear to be related to motility and chemotaxis, and may play a role in maintaining intracellular sodium ion concentrations and/or pH homeostasis (Ito et al., 2004). Although the quaternary structures of eukaryotic and prokaryotic sodium channels are different (the former are four-domain monomers that form pseudotetrameric structures, whereas the latter are single-domain monomers that associate to form true tetramers), their overall folds are similar, with each domain or monomer consisting of a voltage sensor (VS; transmembrane helices S1 to S4), selectivity filter (SF), and pore region (transmembrane helices S5 and S6; Ahern et al., 2016). There is considerable (∼20–35%) sequence identity between the transmembrane regions of prokaryotic and eukaryotic Navs, and consequently, they appear to have very similar secondary structures (Charalambous and Wallace, 2011). Hence, prokaryotic sodium channels, for which several crystal structures have been elucidated (Payandeh et al., 2011, 2012; McCusker et al., 2012; Zhang et al., 2012; Bagnéris et al., 2013, 2014; Shaya et al., 2014; Arrigoni et al., 2016; Naylor et al., 2016; Sula et al., 2017), tend to be good structural proxies for human sodium channels. This is indicated by the observations that several the prokaryotic sodium channels enable sodium flux with similar characteristics to human channels (Payandeh et al., 2011; Ulmschneider et al., 2013; DeCaen et al., 2014). One of the prokaryotic channels, NavMs, has also been shown to exhibit similar binding affinities and kinetics for eukaryotic channel blockers as human sodium channels (Bagnéris et al., 2014). Hence, the prokaryotic Nav structures were also anticipated to provide insight into structure–function relationships in eukaryotic channels.
A new crystal structure of the full-length prokaryotic sodium channel NavMs has recently been determined that provides new insight into the linkage between channel activation and opening. A striking feature of this structure is the presence of a novel “interaction motif” (IM), which we proposed is intimately associated with channel opening. Conserved residues in this motif are linked with mutations causing human diseases. Here, we discuss how residues from the S3 of the VS, the S4–S5 linker, and the S6 helix of the pore domain interact and the evidence that they play a dynamic role in the mechanism of channel gating.
Structures of sodium channels represent different functional states
Until recently, the prokaryotic sodium channel crystal structures that have been determined exhibited features associated with different phases of the activation/opening, closing, and inactivation cycle but did not reflect single identifiable functional states (Table 1). The first sodium channel crystal structure determined (Payandeh et al., 2011), NavAb, revealed a channel with a “preactivated” VS. It was similar but not identical to the expected “outward” facing fully activated form that had been identified by cross-linking experiments (DeCaen et al., 2009). Surprisingly, however, although the NavAb VS was similar to an activated form, it had a closed pore gate. This structure was rapidly followed by the structures of two “potentially inactivated” states (of NavAb [Payandeh et al., 2012] and of NavRh [Zhang et al., 2012]) and the pore-only structures of NavMs (McCusker et al., 2012; Bagnéris et al., 2013). Both the NavAb and NavRh structures had closed pore gates, preactivated VSs and collapsed SFs. The NavRh homologue did not exhibit sodium flux functioning, although the NavAb homologue did. Until very recently, the pore-only structures of NavMs were the only structures, which, based on HOLE calculations (Smart et al., 1996) of the radius of the pore gate being >4 Å, were proposed to be either partially (McCusker et al., 2012) or fully (Bagnéris et al., 2013) open pores, but because they did not have a VS, their activation state could not be identified. Interestingly, none of the aforementioned structures depicted the C-terminal domain (CTD) of the channel, although it was present in all of the constructs that were crystallized. This appears to be because the CTD is flexible and dynamic (Powl et al., 2010; Bagnéris et al., 2013) and hence was unordered within the crystal lattice. Later structures included a pore-only version of NavAe1 (Shaya et al., 2014; Arrigoni et al., 2016), which had a closed gate and again no VS but did depict the CTD, and a NavAb/Nav1.7DIV chimeric structure (Ahuja et al., 2015), which had an activated VS and a closed gate. A related structure was that of CavAb (Tang et al., 2016), a mutant of the NavAb sodium channel with a redesigned SF to make it selective for calcium ions rather than sodium ions. In all other regions, the CavAb sequence (and structure) was essentially the same as the native NavAb structure (preactivated VS, closed gate), except that its CTD domain was visible in this mutant construct, whereas it was absent in the wild-type NavAb structure. Because these latter structures (NavAe1, chimera, and CavAb) had all or part of their CTDs present, it became clear that in a structure with a closed gate, this region of the protein forms a coiled-coil/helical extension of the gate, with no pathway present for ion egress. A new structure of NavAb described as being in an “open” conformation has been produced by truncating all 40 residues of the CTD of the protein (Lenaeus et al., 2017). The structure of its pore and gate and the size of its interior hole are very similar to that of the NavMs pore-only open form, with a central kink in the S6 helix providing the means by which the gate appears to be opened. However, that construct is missing the entire CTD, as well as the all-important S6 residues that form the intracellular end of the gate, so it may not be an entirely suitable model for understanding the nature of gating.
Table 1. Conformational states of prokaryotic channel crystal structures.
| Parameter | NavMs | NavAb | NavAb | NavRh | NavMs (pore) | NavAe1 (pore) | CavAb |
|---|---|---|---|---|---|---|---|
| Apparent state | open/ activated | closed/ preactivated | closed/ inactivated | closed/ inactivated | open/- | closed/- | closed/ preactivated |
| VS | activated | preactivated | preactivated | preactivated | none | none | preactivated |
| Pore gate | open | closed | closed | closed | open | closed | closed |
| SF | open - Na ions | open - no ions | collapsed | collapsed | open - Na ions | open - no Na | open - Ca ions |
| CTD | visible - interaction | not visible | not visible | not visible | not visible | visible - coiled-coil | visible - coiled-coil |
| Conducts Na+ | yes | yes | yes | no | yes | no | NA |
A cryo-EM structure of the cockroach eukaryotic Nav has also recently been reported (Shen et al., 2017); like all the other Navs (except NavMs), this structure has a closed (pore) gate. Its four VSs exhibit different conformations ranging from “most” to “least” activated (as may well be expected for intermediate transition states), but as this “putative” channel is nonfunctional, it is difficult to relate this structure to a particular state in the opening/closing cycle.
The new full-length structure of the NavMs sodium channel
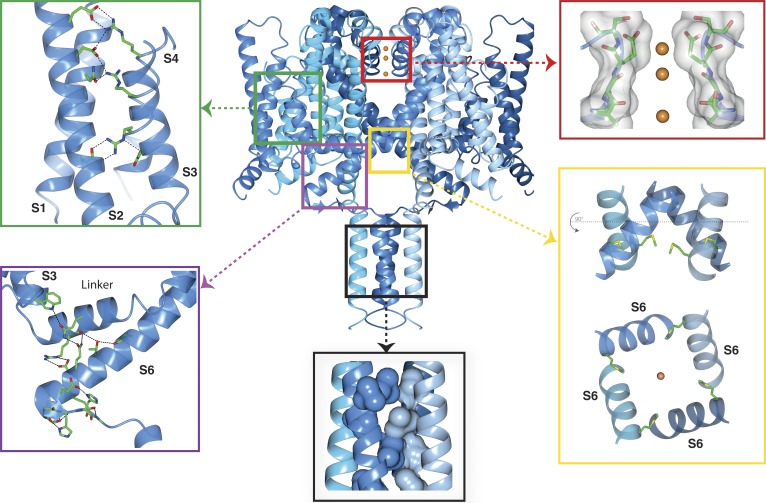
Very recently, a new high-resolution, complete crystal structure of the NavMs channel (Fig. 1; Sula et al., 2017) was determined that depicted for the first time the structure of a potentially open activated channel: it has a “outward facing” activated VS conformation joined via the S4–S5 linker to a pore domain with an open SF containing three sodium ions, a pore gate formed from the conjuncture of the C-terminal ends of the S6 helices, an extensively hydrogen bonded IM, and a four-stranded coiled-coil (displaying the typical knobs-into-holes motif associated with a coiled-coil sequence pattern) located at the distal end of the CTD, connecting the four monomers.
Figure 1.
Functional regions of the open activated full-length NavMs sodium channel. The four monomers of PDB ID 5HVX are depicted in ribbon motif in different shades of blue. The boxed regions point to expanded views of the VS, SF, pore gate, CTD, and IM. Top left (green box) inset: VS helices, showing the side chains of the canonical arginines of the S4 helix (indicated in the Fig. 4 sequence figure) and their partners in helices S1, S2, and S3. These are the ion pairings expected for an activated (“outwardly facing”) VS. The α-helical transmembrane helices are in cartoon ribbon depictions, whereas the top of the S4 helix, which is a 310 helix, is in worm depiction. The latter feature is consistent with the proposal that the transition from inactivated (fully α-helical) to activated states (310/α-helical) would involve changes in the helical parameters, as well as the arginine bonding partners (Villalba-Galea et al., 2008). Top right (red box) inset: The SF, with backbone and side chain atoms (only two subunits shown), is depicted as sticks overlaid in surface display mode; the three sodium ions are orange balls (radius not to scale, for ease of visualization). The closest residue to any of the ions is E178, which is adjacent to the top sodium ion, as previously seen in the NavMs pore structure (Naylor et al., 2016). All of the ion–polypeptide distances are too long to involve direct contacts and so must be via (mostly disordered) water molecules. Bottom right (yellow box) inset: (top) The open pore gate in S6 is depicted in ribbon mode with the side chains shown in stick mode for the Met222 residues, which correspond to the region previously identified as the closed channel gate constriction. The bottom panel is a 90° rotation view showing the end-on-view of the pore gate, again with the Met222 side chains shown. Bottom middle (black box) inset: The coiled-coil region of the CTD, with the hydrophobic side chains forming the knob-into-holes motif, shown in space-filling depiction (for clarity only the side chains from two of the helices are shown). Bottom left (purple box) inset: The IM showing the extensive network of hydrogen bonds and ion pairings (dashed black lines) involving both side chains and peptide backbone moieties. The S3, S4–S5 linker, and S6 helices are depicted in ribbon motif. The residues involved in the hydrogen-bonded network are shown as sticks in CPK coloring. This and the other graphics figures were created using CCP4Mg (McNicholas et al., 2011) software.
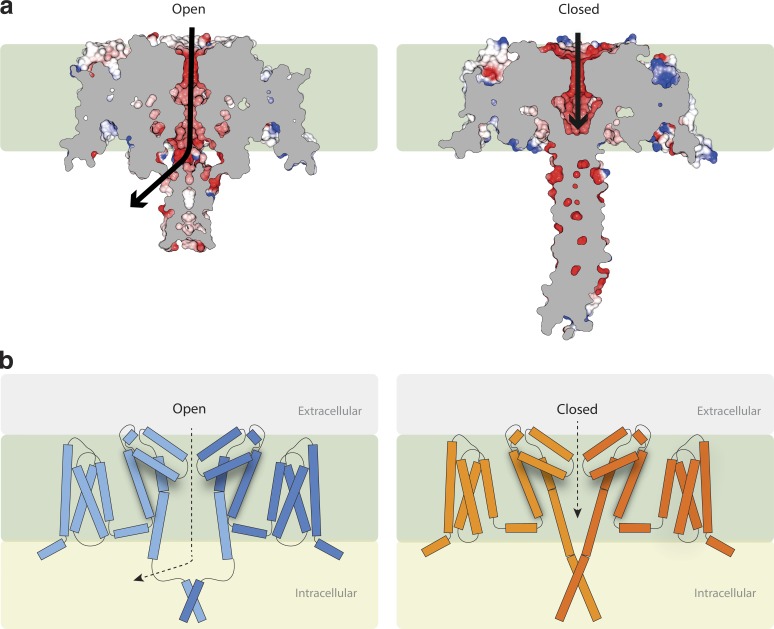
The gate has a minimum internal diameter of 4.0 Å (the narrowest region is adjacent to the Ile215 residues [Fig. 2]), as determined using HOLE software (Smart et al., 1996). This can be compared with the minimum diameter of <1.5 Å seen in the CavAb structure (Fig. 2) or 1.8 Å in the closed NavAb structure (Sula et al., 2017). The narrowest region of the NavAb structure (residue M221, equivalent to M222 in NavMs; Fig. 1, bottom right) corresponds to the region identified in human Navs (Oelstrom et al., 2014) as the hydrophobic constriction associated with a closed channel gate. The gate region of the full-length NavMs (Fig. 2) is much wider than that in the closed NavAb structures and is similar in dimensions to that of the NavMs pore structures, which have been shown to be compatible with passage of fully hydrated sodium ions (Ulmschneider et al., 2013; Naylor et al., 2016); hence, this structure is considered to have an open pore gate. It is notable that unlike eukaryotic sodium channels that have been proposed to passage large charged local anesthetics from the intracellular side, thereby blocking the pore (Qu et al., 1995), studies of NavMs (Bagnéris et al., 2014) have shown that the QX-314 does not block (or presumably enter) the pore, which may indicate prokaryotic channels are narrower than their eukaryotic relatives.
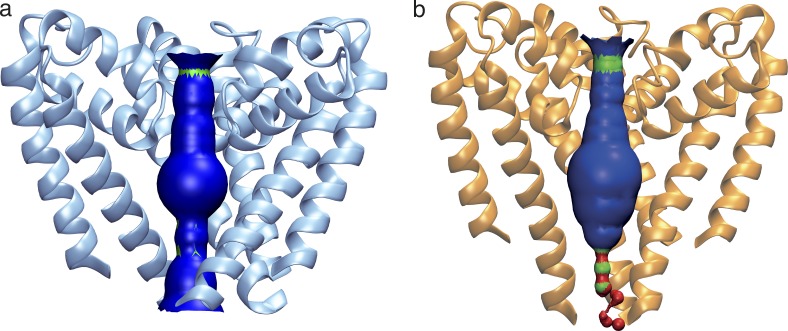
Figure 2.
Internal pore dimensions of open and closed transmembrane domains. (a) The open NavMs (in blue ribbon depiction). (b) The closed CavAb (in orange ribbon depiction). The inner surface dimensions as calculated using HOLE (Smart et al., 1996) are shown in dark blue, where the dimensions are sufficient to enable passage of fully hydrated sodium ions, in green where the dimensions are compatible with at least partially dehydrated ions, and red where the dimensions are too narrow to allow passage of sodium ions. Throughout the length of the pore from the extramembranous vestibule area, through the SF, into the bulbous hydrophobic cavity, which binds channel blocker compounds, until ∼14 Å from the intracellular surface, both open and closed structures are very similar and can accommodate sodium ions. However, near the exit of the pore, the closed pore narrows substantially, whereas the open pore retains a clear passage for the ions to exit the transmembrane region of the channel.
The CTD in this new open structure is very different than the CTDs visible in the closed NavAe1 (Arrigoni et al., 2016) and CavAb structures (Fig. 3, a and b): it consists of a helical extension of the S6 transmembrane helix at the proximal end and a short segment without any canonical secondary structure beginning at residue 234 and ending at residue 240; this is followed by a coiled-coil region that extends to the end of the structure. This type of structure is consistent with previous DEER-EPR spectroscopic studies (Bagnéris et al., 2013) on both detergent-solubilized and membrane-associated spin-labeled NavMs, thus indicating it is not an artifact of crystal packing. The role of the coiled-coil end of the CTD was previously indicated in studies where its removal resulted in a decrease in current density (Bagnéris et al., 2014). Although the coiled-coil was not essential for channel functioning, the decrease in current density likely arose from impaired assembly/enhanced disassembly of active channels, a result in line with studies of other prokaryotic orthologues (Mio et al., 2010; Powl et al., 2010) that had indicated the coiled-coil was important for stabilization of assembled channels.
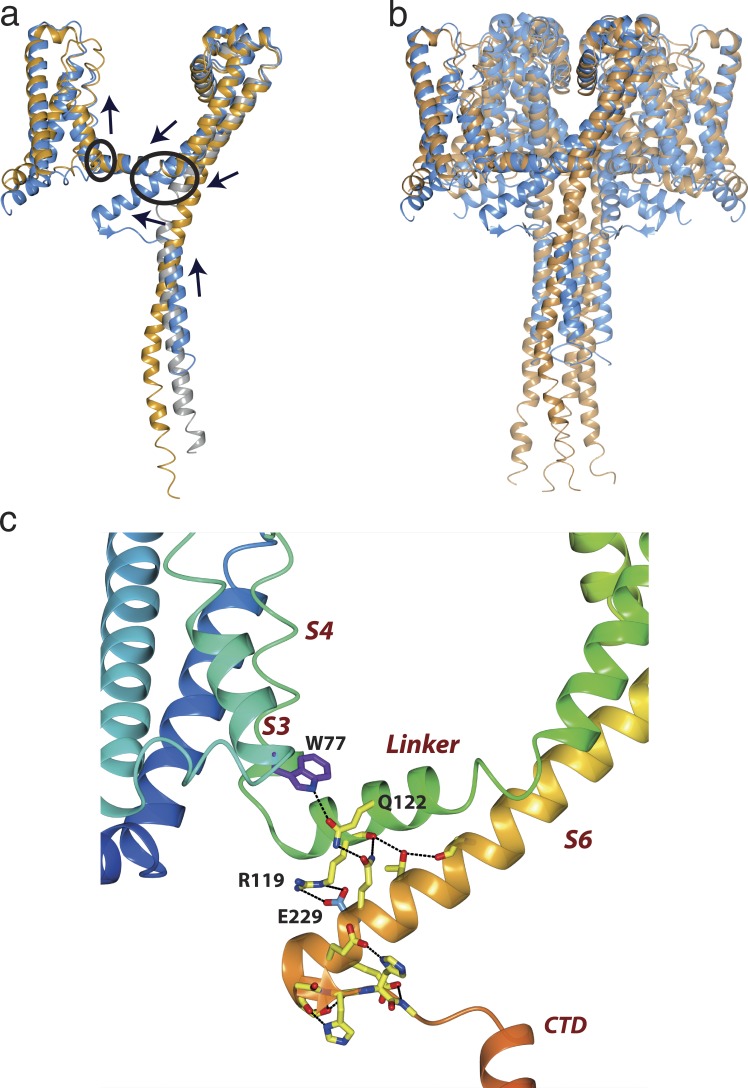
Figure 3.
Features of open and closed sodium channels. (a) Overlays of the NavMs (blue), CavAb (orange), and NavAe1 (gray) monomer structures, with black arrows indicating the relative directions of the concerted movements of the VS, S4–S5 linker, S6 pore helix, IM, and top of the CTD when transforming from the open to the closed conformation. (b) Overlays of the complete NavMs (blue) and CavAb (orange) tetrameric structures, showing the overall consequences of the open-to-closed conformational change. The extensions of the S6 helices in NavMs splay the “neck” region of the CTD in the direction of the S4–S5 linker, which then results in a bend angle of nearly 120° at the beginning of the region without canonical secondary structure. This region has been designated the IM of the open state. In the closed CavAb structure, the equivalent “neck” region at the top of the CTD is a helical extension of the narrower opening formed by its S6 helices. As a consequence of the different conformations at the tops of their CTDs, the distal ends of the CavAb CTD (formed of coiled-coils in both structures) protrude into the cytoplasm, further from the membrane interface than they do in NavMs. (c) Enlarged view of the NavMs IM, with the helices depicted in rainbow colored (N- to C-terminal direction) ribbon motif. The regions (S3, S4–S5 linker, S6, and CTD) comprising the IM are labeled in red italics. The side chain and backbone atoms that are involved in the extensive network of hydrogen bonds and ion pairs (dashed lines) are shown in stick motif. The conserved W77 is in purple, and E229 is in cyan.
A novel IM and its association with channel gating
The IM is a novel feature, as yet only seen in the complete open NavMs structure (Fig. 1, inset indicated by the purple box; and Fig. 3 c). It includes residues from different domains of the protein that form a conjunction of structures held together by an extensive network of hydrogen bonds and ion pairs involving both side chains and polypeptide backbones. Located at the N-terminal end of the S3 helix of the VS, the W77 side chain forms a hydrogen bond and hydrophobic stacking interaction with the Q122 side chain of the S4–S5 linker, which in turn forms a hydrogen bond with Q228 near the end of the S6 transmembrane helix (Fig. 3 c). At the other end of the motif, the three consecutive glutamates (E229, E230, and E231) form a salt bridge with R119 of the S4–S5 linker and hydrogen bonds with residues in the non-helical CTD region preceding the distal coiled-coil helix starting at I241. A further network of hydrogen bonds involves both side chains and backbone atoms from the region of the S6 helix closest to the membrane surface. As the IM either is located for the most part within the hydrophobic transmembrane region or else the interactions are buried within the interior of the folded domain, the energetic contributions of those hydrogen bonds are expected to be more substantial than if they were located in aqueous-exposed regions.
To produce the compact IM requires several coordinated shifts between different regions in the NavMs and NavAb structures (in the directions of the arrows in Fig. 3 a; Video 1): When the S4 VS helix moves to the fully outward-facing (up) position, the S4–S5 linker shifts outwards and toward the pore domain, which then acts as a lever to slightly translate the S6 helix within the plane of the membrane, enabling a small bend in the middle of this helix (seen previously in the NavMs pore crystal structures [McCusker et al., 2012; Bagnéris et al., 2013]), which produces the open gate. The resulting motion of the C-terminal end of S6 enables it to interact with the S3 helix and linker region, whereas the C-terminal end of the CTD moves upward toward the membrane, enabled by a disordering of the top of the CTD helix.
The structure of the compact but extensively hydrogen bonded IM then raises questions as to (a) whether the types of residues involved in these interactions are unique to NavMs (and hence the IM could simply arise as a result of a sequence variation within the prokaryotic channels) or whether there is a pattern of sequence similarities common to other orthologues and (b) what effects, if any, are there on function if the motif residue types are changed. The first residue of interest is the W77 of S3, a residue that has not previously been highlighted as being structurally essential to sodium channels. However, if we examine the sequences of all of the other prokaryotic sodium channels identified thus far, it is clear this residue type is completely conserved, despite the surrounding residues (which are not involved in the IM) being variable. This is suggestive of, but not evidence for, a conserved function. However, when this residue was changed (Sula et al., 2017) from tryptophan to other aromatic residues (either phenylalanine or tyrosine) that could not form the same types of interactions, the gating function was significantly impaired. Furthermore, the measured current density was abolished when the tryptophan was replaced by the nonaromatic residues alanine or methionine (although it was not possible to distinguish in these cases between lack of trafficking or lack of function). Nevertheless, these functional results on mutants are strong indications of the structural importance of the specific interactions formed by W77.
IM residues are conserved across phyla
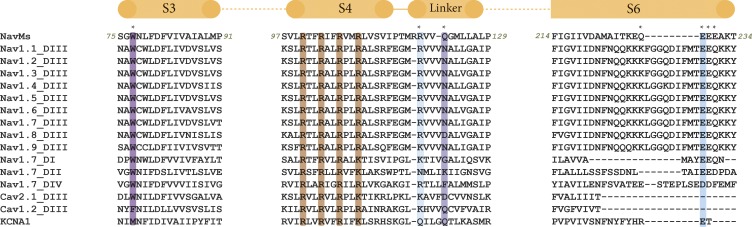
The W77 residue postulated above to have a potential role in stabilizing the open state is found in the equivalent position in all prokaryotic sodium channels that have been identified and sequenced thus far. This is not a singular uniquely conserved feature, as prokaryotic and eukaryotic sodium channels exhibit ∼20% sequence identities overall, but it is possibly noteworthy, as its level of conservation (identity) is even higher than many residues in the SFs of prokaryotic channels and it rivals or surpasses the conservation of the S4 arginines in the VSs. Furthermore, an equivalent tryptophan residue is also found in the S3 helix of all domains of all human sodium channels identified to date (Fig. 4), but not at equivalent locations in potassium channels (and only in some calcium channel domains). These observations also suggest it may be an important signature of sodium channel sequences.
Figure 4.
Alignments of sequences involved in the IM region. NavMs, domain III of all human Navs, all domains of the human Nav1.7 isoform, and other ion channels (calcium channels [one (Cav2.1) with, one (Cav1.2) without the conserved tryptophan], and the KCNA1 potassium channels) are listed. The locations of the helical regions in the NavMs structure are indicated by the orange bars at the top, and the numbers of the first and last residues of NavMs in each of the segments are noted in italics before/after each segment sequence. The residues of the IM where salt bridges/hydrogen bonds involving side chains are located are noted by the * above the sequence. The colored overlays indicate the conserved tryptophan in S3 (dark purple) and its partner residue (light purple), the E229 in S6 (cyan), and its partner R119 in the linker (light cyan), and for reference, the four VS arginines are in red.
A second interesting feature of the motif is the conserved E229 in S6, which forms a salt bridge with the conserved (or conservatively replaced) R119 in the linker region (Figs. 3 and 4). The E229 equivalent is found in both prokaryotes and at least one domain of all human Navs.
A role for corresponding residues in human sodium channels?
It is perhaps interesting to speculate whether the conserved tryptophan might have a role in human sodium channel functioning. It is striking that there are tryptophan residues at the equivalent locations in human channels, notably at the beginning of the transmembrane S3 helix in all domains of all nine human isoforms. This very high level of conservation is suggestive of a functional role for the tryptophan. Furthermore, although only three naturally occurring variants have yet been identified at the equivalent sites in human sodium channels, they are all associated with disease states: W190R in DI and W1284S in DIII of Nav1.1 are both single-site mutations (Fukuma et al., 2004; Depienne et al., 2009) linked to Dravet syndrome, a severe form of epilepsy beginning in infancy, and W1271C in DIII of Nav1.5 is the single-site variant (Kapplinger et al., 2010) linked with a form of Brugada syndrome, which is associated with increased risk of sudden cardiac death. Although no functional studies have yet linked these mutations to specific features of the opening/closing/inactivating cycle, the structure–function studies on the NavMs tryptophan and the human mutants associated with pathological traits suggest that the action potential cycle may be disrupted and may indicate the role of this residue in such a process. Of course this raises the issue of why more such mutants have not been isolated in the other human isoforms if this is a critical site. It might be hypothesized that this is because such mutations (especially if they occurred in more than one domain in a single channel), could completely disrupt the sodium channel–based electrical signaling, which could be fatal and hence no such variants would be detected in the population.
Likewise, when the residue corresponding to E229 (E1795 in hNav1.1 DIV) is mutated to K, this gives rise to a disease state (familial generalized epilepsy with febrile seizures; Li et al., 2010). The equivalent mutation in hNav1.6 DIII (E1483K) is associated with benign infantile seizures and paroxysmal dyskinesia (Gardella et al., 2016), again possibly suggestive of a function role in the action potential cycle.
Such speculations on functional roles for these residues will await both electrophysiological characterizations of the mutant human channels and further genetic screens. Nevertheless, they suggest that these IM residues could produce effects across the prokaryotic/eukaryotic divide, and thus provide an understanding of the molecular basis for several human diseases.
A proposed mechanism for gating
The new structure now enables us to make comparisons with other Nav structures that have both visible CTDs and closed gate conformations. The NavAe1 pore structure was the first to depict the full nature of the CTD of a closed gate (Shaya et al., 2014; Arrigoni et al., 2016): It consists of an essentially entirely helical structure, with the proximal end (designated the “neck”) consisting of individual helices and a distal end formed of the four-helix coiled-coil. The closed CavAb CTD is very similar (Tang et al., 2016); given that its structure (except in the engineered mutant SF, which makes it conduct calcium rather than sodium) is nearly identical to that of the NavAb orthologue, it provides a second very similar view of a closed CTD structure. In contrast, in the structure of the full-length NavMs, the region of the CTD just after the extended S6 helix that forms the gate is non-helical for a span of approximately seven residues (from Thr234 to Pro240), while the distal end of the CTD forms a four-helix bundle very similar to the NavAe1 and CavAB coiled-coils. This means the NavMs CTD is less helical overall. This observation corresponds well with the earlier synchrotron radiation circular dichroism (SRCD) spectroscopic studies that “dissected” the secondary structure of the NaChBac orthologue (Powl et al., 2010) and DEER experiments with NavMs (Bagnéris et al., 2013), which examined the CTD domain quaternary structure. Both of those studies identified approximately the top 10 residues of the CTDs as being non-helical. That prokaryotic Nav CTDs retain the helical bundle at the distal end in both open and closed structures is consistent with the suggestion that the CTD provides a means of stabilization of the monomers in the tetrameric (Mio et al., 2010; Powl et al., 2010), although it appears not to be required for initial assembly of the tetramer (Mio et al., 2010).
In the proposed open state NavMs channel, the gate at the end of S6 is sufficiently wide to enable the passage of ions to the cell interior, not directly through the entire length of the CTD (because of the tightly packed coiled-coil at the C-terminal end) but via a side exit (Fig. 5 a, left) near the intracellular membrane surface. In the closed CavAb and NavAe1 structures, the gate at the end of the S6 helix is too narrow to enable sodium passage to the cell interior by any pathway (Fig. 5 a, right). A striking and important difference between the open and closed structures (in addition to the size of the actual gate), is the conformation of the end of S6 and top of the CTD, included in what we have designated the IM. In the open channel structure, the S6 helix continues (in a more or less straight fashion) beyond the end of the membrane, to residue K233. Its structure is stabilized in this conformation because of its extensive network of hydrogen bonds and ion pairs involving the VS and S4–S5 linker. This creates an open pathway for ions that extends beyond the intracellular surface of the membrane. The ions exit not through the central axis (the coiled-coil of the CTD does not enable the straight-through passage of the ions), but via an opening to its side created by the IM structure (Fig. 5). The next seven residues beyond the end of the S6 helix have a well-defined structure, but that structure is not helical. This enables the CTD to change direction and ultimately accommodate the geometry of the coiled-coil distal region that is the point of association in the CTD between the individual monomers. In the closed gate structures of CavAb and NavAe1 pore, the S6 helix continues as an essentially straight helical structure from the inner membrane surface to the end of the CTD. The distal part of the helix also forms a tetramer-stabilizing coiled-coil helix bundle that is nearly identical to the one in the NavMs structure. However, in CavAb and NavAe1, both the top of the helix bundle and the distal coiled-coil sections are too tightly packed to enable the passage of an ion (Fig. 5 a, right), even if the gate had been wide enough. Notably, the CavAb channel (and hence the related NavAb channel, which is capable of conducting sodium ions) does contain a comparable sequence for nearly all of the key interaction domain residues, suggesting it could form an open gate in the same manner as NavMs. Why it does not do so in the crystal structure could be because the VS is in the preactivated state, where the slight shift in positions of the S3 and the S4-S5 linker suggest the relative alignment of the tryptophan with the CTD may not be sterically possible. It will be interesting to see whether in the future a fully open form of a complete construct of this orthologue can also be obtained.
Figure 5.
Gating and ion translocation. (a) Lateral (transmembrane) slice through the centers of the NavMs (left) and CavAb (right) structures. Slabs through the space-filling models show the pathway (black arrows) for ion egress in the open NavMs structure via the opening at the side of the CTD adjacent to the end of the intramembrane pore region and the blockage at the end of the CavAb structure, ending before the end of the transmembrane pore region, which prevents ions exiting the pore. (In both parts a and b of this figure, the hydrophobic region of the membrane is represented by the green background.) (b) New model for channel gating (a modification to the earlier Bagnéris et al. [2013] and Shaya et al. [2014] models), now depicting schematically the role of the IM in stabilizing the open conformation formed by the splayed bottoms of the S6 helices. Left: The activated (open) state. Right: The resting (closed) state. For simplicity, only two subunits are shown for each structure. The tubes represent helical structures, and the connecting lines are segments that are not helical. The distal ends of the CTDs in both cases act to stabilize the tetrameric structures.
Before the availability of this full-length structure, two very similar models were proposed for channel opening and closing (McCusker et al., 2012; Bagnéris et al., 2013; Shaya et al., 2014) based on crystallographic, functional, and spectroscopic data of pore-only structures. However, the assumptions made by both groups in the absence of the “missing link” structures of the full CTDs and the IM of the open state have proven to be remarkably prescient. Both suggested a coiled-coil (as now seen) at the distal end of the CTD would stabilize the tetramer and remain intact during the gating process and that the “neck” region at the proximal end of the CTD would change conformation to accommodate a splaying of the ends of the S6 pore helices. The coiled-coil would remain a constant feature, acting as the anchor tying together the monomers in the tetramer throughout the process of opening and closing. One of the models was been described as a “tethered oscillator” (Bagnéris et al., 2013). Such models are consistent with the newly available structures, with the exception that the now visible IM in the new structure reveals an additional possible feature of the mechanism, a means of energetically favoring and stabilizing the open state. This leads us now to propose an updated version of the gating model for a prokaryotic sodium channel (Fig. 5 b and Video 1). The NavMs full-length structure enables, for the first time, visualization of the process of opening and closing by morphing between open and closed structures (Video 1) as well as an examination of the roles of the different types of interactions stabilizing the IM in the open state.
Although this gating model is appropriate for prokaryotic sodium channel structures, it is important to remember, as previously noted (Sula et al., 2017), that as eukaryotic channels are monomeric and thus have only one CTD (which is not a coiled-coil), any similarities between prokaryotic and eukaryotic Navs in the distal end of the C-terminal region will not be found in eukaryotic Navs. Hence, although much of the IM could be common across phyla, the CTD coiled-coil feature will not be.
Conclusions and prospectives for the future studies
The recent crystal structure of the open, activated form of the NavMs sodium channel depicts a complete sodium channel, with all domains visible. Comparisons with closed structures of other sodium channel orthologues reveal previously unseen features associated with gating. In the context of the wide range of prokaryotic structures from different orthologues in different conformational states now available, it also provides a physical basis for understanding the molecular nature of functional transitions. The structure–function studies described have also suggested a potential molecular basis for several of the S3 tryptophan mutants that were previously identified as being associated with human diseases and for the conserved E229 residues also associated with human diseases. Additionally, sequence homology comparisons suggest that Trp77 may be a feature specifically associated with voltage-gated sodium channel members of the wider voltage-gated ion channel family, distinguishing them from potassium and some calcium channels.
The gallery of static “frozen” images now available as the result of crystallographic structures determined for prokaryotic sodium channels in different conformations and in different environments (detergent micelles, bicelles, all in the rigid environment of a crystal), provides intriguing opportunities for further structure–function speculations. Spectroscopic studies exploring the functional linkage between transmembrane voltage and the IM conformation of such channels when embedded in membranes could further add to our understanding of the dynamic nature of sodium channel conformations as hinted at by the static but detailed views produced by crystallographic studies described herein.
Finally, this structure of a prokaryotic sodium channel has stimulated new thoughts on structural features that may play important functional roles in the gating cycle of sodium channels. These, in turn, may give hints as to the molecular basis of some human sodium channelopathies. Future electrophysiology studies on these mutant human channels could provide further insight into the potential functional roles of the conserved residues thus far identified in the IM, providing a direct link between sequence, structure, function, and disease. Of particular interest would be comparison of the extent of the effects of mutations in single domains (as seen in the mutants thus far identified in human diseases) and mutations in all four domains (as is the case in the symmetric prokaryotic channels).
In summary, the new complete crystal structure of a prokaryotic sodium channel provides the starting point for understanding and exploring the functional relationships of newly revealed sets of interactions implicated in sodium channel gating.
Online supplemental material
Video 1 depicts morphing between NavMs (open pore, activated VS structure; PDB ID 5HVX; Sula et al., 2017) and CavAb (closed pore, preactivated VS structure; PDB ID 5KLB; Tang et al., 2016) structures.
Supplementary Material
Acknowledgments
We thank Jenny Booker (Birkbeck), Dr. Claire Naylor (formerly Birkbeck, now Molecular Dimensions), Prof. Paul DeCaen (Northwestern University), Prof. David Clapham (Harvard Medical School), and Prof. Martin Ulmschneider (Johns Hopkins) for stimulating discussions.
This work was supported by the UK Biotechnology and Biological Science Research Council (BBSRC) grant L006790 (to B.A. Wallace).
The authors declare no competing financial interests.
Author contributions: Both authors contributed to the analyses of the data and to the writing of the manuscript. B.A. Wallace produced the first draft of the manuscript, and A. Sula created the figures and video.
Lesley C. Anson served as editor.
Footnotes
Abbreviations used:
- CTD
- C-terminal domain
- IM
- interaction motif
- SF
- selectivity filter
- VS
- voltage sensor
References
- Ahern C.A., Payandeh J., Bosmans F., and Chanda B.. 2016. The hitchhiker’s guide to the voltage-gated sodium channel galaxy. J. Gen. Physiol. 147:1–24. 10.1085/jgp.201511492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja S., Mukund S., Deng L., Khakh K., Chang E., Ho H., Shriver S., Young C., Lin S., Johnson J.P. Jr., et al. . 2015. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 350:aac5464 10.1126/science.aac5464 [DOI] [PubMed] [Google Scholar]
- Arrigoni C., Rohaim A., Shaya D., Findeisen F., Stein R.A., Nurva S.R., Mishra S., Mchaourab H.S., and Minor D.L. Jr. 2016. Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell. 164:922–936. 10.1016/j.cell.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnéris C., DeCaen P.G., Hall B.A., Naylor C.E., Clapham D.E., Kay C.W.M., and Wallace B.A.. 2013. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat. Commun. 4:2465 10.1038/ncomms3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnéris C., DeCaen P.G., Naylor C.E., Pryde D.C., Nobeli I., Clapham D.E., and Wallace B.A.. 2014. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc. Natl. Acad. Sci. USA. 111:8428–8433. 10.1073/pnas.1406855111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous K., and Wallace B.A.. 2011. NaChBac: the long lost sodium channel ancestor. Biochemistry. 50:6742–6752. 10.1021/bi200942y [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Sharp E.M., Scheuer T., and Catterall W.A.. 2009. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl. Acad. Sci. USA. 106:22498–22503. 10.1073/pnas.0912307106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen P.G., Takahashi Y., Krulwich T.A., Ito M., and Clapham D.E.. 2014. Ionic selectivity and thermal adaptations within the voltage-gated sodium channel family of alkaliphilic Bacillus. eLife. 3:e04387 10.7554/eLife.04387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C., Trouillard O., Saint-Martin C., Gourfinkel-An I., Bouteiller D., Carpentier W., Keren B., Abert B., Gautier A., Baulac S., et al. . 2009. Spectrum of SCN1A gene mutations associated with Dravet syndrome: analysis of 333 patients. J. Med. Genet. 46:183–191. 10.1136/jmg.2008.062323 [DOI] [PubMed] [Google Scholar]
- Fukuma G., Oguni H., Shirasaka Y., Watanabe K., Miyajima T., Yasumoto S., Ohfu M., Inoue T., Watanachai A., Kira R., et al. . 2004. Mutations of neuronal voltage-gated Na+ channel α1 subunit gene SCN1A in core severe myoclonic epilepsy in infancy (SMEI) and in borderline SMEI (SMEB). Epilepsia. 45:140–148. 10.1111/j.0013-9580.2004.15103.x [DOI] [PubMed] [Google Scholar]
- Gardella E., Becker F., Møller R.S., Schubert J., Lemke J.R., Larsen L.H., Eiberg H., Nothnagel M., Thiele H., Altmüller J., et al. . 2016. Benign infantile seizures and paroxysmal dyskinesia caused by an SCN8A mutation. Ann. Neurol. 79:428–436. 10.1002/ana.24580 [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. Sinauer Associates, Sunderland, Mass. [Google Scholar]
- Ito M., Xu H., Guffanti A.A., Wei Y., Zvi L., Clapham D.E., and Krulwich T.A.. 2004. The voltage-gated Na+ channel NaVBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc. Natl. Acad. Sci. USA. 101:10566–10571. 10.1073/pnas.0402692101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapplinger J.D., Tester D.J., Alders M., Benito B., Berthet M., Brugada J., Brugada P., Fressart V., Guerchicoff A., Harris-Kerr C., et al. . 2010. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 7:33–46. 10.1016/j.hrthm.2009.09.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koishi R., Xu H., Ren D., Navarro B., Spiller B.W., Shi Q., and Clapham D.E.. 2004. A superfamily of voltage-gated sodium channels in bacteria. J. Biol. Chem. 279:9532–9538. 10.1074/jbc.M313100200 [DOI] [PubMed] [Google Scholar]
- Lenaeus M.J., Gamal El-Din T.M., Ing C., Ramanadane K., Pomès R., Zheng N., and Catterall W.A.. 2017. Structures of closed and open states of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 114:E3051–E3060. 10.1073/pnas.1700761114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhang J., Guo J.-F., Yan X.-X., Xia K., and Tang B.-S.. 2010. Novel mutation of SCN1A in familial generalized epilepsy with febrile seizures plus. Neurosci. Lett. 480:211–214. 10.1016/j.neulet.2010.06.040 [DOI] [PubMed] [Google Scholar]
- McCusker E.C., Bagnéris C., Naylor C.E., Cole A.R., D’Avanzo N., Nichols C.G., and Wallace B.A.. 2012. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 3:1102 10.1038/ncomms2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas S., Potterton E., Wilson K.S., and Noble M.E.M.. 2011. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67:386–394. 10.1107/S0907444911007281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio K., Mio M., Arisaka F., Sato M., and Sato C.. 2010. The C-terminal coiled-coil of the bacterial voltage-gated sodium channel NaChBac is not essential for tetramer formation, but stabilizes subunit-to-subunit interactions. Prog. Biophys. Mol. Biol. 103:111–121. 10.1016/j.pbiomolbio.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Naylor C.E., Bagnéris C., DeCaen P.G., Sula A., Scaglione A., Clapham D.E., and Wallace B.A.. 2016. Molecular basis of ion permeability in a voltage-gated sodium channel. EMBO J. 35:820–830. 10.15252/embj.201593285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelstrom K., Goldschen-Ohm M.P., Holmgren M., and Chanda B.. 2014. Evolutionarily conserved intracellular gate of voltage-dependent sodium channels. Nat. Commun. 5:3420 10.1038/ncomms4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., and Catterall W.A.. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358. 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Gamal El-Din T.M., Scheuer T., Zheng N., and Catterall W.A.. 2012. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 486:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powl A.M., O’Reilly A.O., Miles A.J., and Wallace B.A.. 2010. Synchrotron radiation circular dichroism spectroscopy-defined structure of the C-terminal domain of NaChBac and its role in channel assembly. Proc. Natl. Acad. Sci. USA. 107:14064–14069. 10.1073/pnas.1001793107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Rogers J., Tanada T., Scheuer T., and Catterall W.A.. 1995. Molecular determinants of drug access to the receptor site for antiarrhythmic drugs in the cardiac Na+ channel. Proc. Natl. Acad. Sci. USA. 92:11839–11843. 10.1073/pnas.92.25.11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Navarro B., Xu H., Yue L., Shi Q., and Clapham D.E.. 2001. A prokaryotic voltage-gated sodium channel. Science. 294:2372–2375. 10.1126/science.1065635 [DOI] [PubMed] [Google Scholar]
- Shaya D., Findeisen F., Abderemane-Ali F., Arrigoni C., Wong S., Nurva S.R., Loussouarn G., and Minor D.L. Jr. 2014. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J. Mol. Biol. 426:467–483. 10.1016/j.jmb.2013.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhou Q., Pan X., Li Z., Wu J., and Yan N.. 2017. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 355:eaal4326 10.1126/science.aal4326 [DOI] [PubMed] [Google Scholar]
- Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., and Sansom M.S.P.. 1996. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14:354–360: 376. 10.1016/S0263-7855(97)00009-X [DOI] [PubMed] [Google Scholar]
- Sula A., Booker J., Ng L., Naylor C.E., DeCaen P.G., and Wallace B.A.. 2017. The complete structure of an activated open sodium channel. Nat. Commun. 8:14205 10.1038/ncomms14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Gamal El-Din T.M., Swanson T.M., Pryde D.C., Scheuer T., Zheng N., and Catterall W.A.. 2016. Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature. 537:117–121. 10.1038/nature19102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider M.B., Bagnéris C., McCusker E.C., DeCaen P.G., Delling M., Clapham D.E., Ulmschneider J.P., and Wallace B.A.. 2013. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc. Natl. Acad. Sci. USA. 110:6364–6369. 10.1073/pnas.1214667110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Galea C.A., Sandtner W., Starace D.M., and Bezanilla F.. 2008. S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. USA. 105:17600–17607. 10.1073/pnas.0807387105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., et al. . 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.