AMPA and NMDA receptors are ionotropic glutamate receptors that make fundamental contributions to synaptic activity in the brain in different ways. Amin et al. show that their respective M4 segments, located on the periphery of their pore domains, contribute to their functional diversity.
Abstract
Ionotropic glutamate receptors (iGluRs), including AMPA receptor (AMPAR) and NMDA receptor (NMDAR) subtypes, are ligand-gated ion channels that mediate signaling at the majority of excitatory synapses in the nervous system. The iGluR pore domain is structurally and evolutionarily related to an inverted two-transmembrane K+ channel. Peripheral to the pore domain in eukaryotic iGluRs is an additional transmembrane helix, the M4 segment, which interacts with the pore domain of a neighboring subunit. In AMPARs, the integrity of the alignment of a specific face of M4 with the adjacent pore domain is essential for receptor oligomerization. In contrast to AMPARs, NMDARs are obligate heterotetramers composed of two GluN1 and typically two GluN2 subunits. Here, to address the function of the M4 segments in NMDARs, we carry out a tryptophan scan of M4 in GluN1 and GluN2A subunits. Unlike AMPARs, the M4 segments in NMDAR subunits makes only a limited contribution to their biogenesis. However, the M4 segments in both NMDAR subunits are critical for receptor activation, with mutations at some positions, most notably at the extreme extracellular end, completely halting the gating process. Furthermore, although the AMPAR M4 makes a minimal contribution to receptor desensitization, the NMDAR M4 segments have robust and subunit-specific effects on desensitization. These findings reveal that the functional roles of the M4 segments in AMPARs and NMDARs have diverged in the course of their evolution and that the M4 segments in NMDARs may act as a transduction pathway for receptor modulation at synapses.
Introduction
Ionotropic glutamate receptors (iGluR) are ligand-gated ion channels that mediate fast excitatory synaptic transmission in the mammalian nervous system. AMPA and NMDA receptors (AMPAR and NMDAR, respectively) are the two main postsynaptic iGluR subtypes. These receptors carry out different functions (Traynelis et al., 2010). AMPARs mediate fast synaptic currents and participate in rapidly translating presynaptic glutamate release into a postsynaptic signal. AMPARs also show strong desensitization in the continual presence of glutamate. NMDARs mediate a slower synaptic component impacting the electrical and biochemical activity of the postsynaptic neuron. The basis for these differences between AMPARs and NMDARs is incompletely understood (Mayer, 2016; Plested, 2016).
iGluRs are members of the pore loop superfamily of ion channels, which includes two-transmembrane K+ channels, voltage-gated K+, Na+, and Ca2+ channels, and transient receptor potential channels. This superfamily is defined by a pore domain, consisting of two membrane-spanning segments joined by a nonmembrane-spanning pore loop. Upon tetramerization, the pore domains from individual subunits form the ion channel. In iGluRs, the pore domain consists of the M1 and M3 transmembrane segments and an M2 pore loop (Sobolevsky et al., 2009; Chen et al., 2014; Karakas and Furukawa, 2014; Huettner, 2015). Homologous to TM2 in two-transmembrane K+ channels, M3 contains elements of the activation gate (Chang and Kuo, 2008; Sobolevsky et al., 2009) and undergoes extensive rearrangement upon pore opening (Jones et al., 2002; Sobolevsky et al., 2002).
In addition to a pore domain, all eukaryotic iGluRs have an additional transmembrane helix, the M4 segment (Fig. 1). For AMPARs, the M4 segment is critical to the transition from dimers to tetramers (Salussolia et al., 2011, 2013; Gan et al., 2015), in part by overcoming energy constraints imposed by other domains (Gan et al., 2016). In contrast to AMPARs, NMDARs are obligate heterotetramers composed of two GluN1 and typically two GluN2 subunits (Traynelis et al., 2010; Glasgow et al., 2015). Any role of the M4 segments in NMDAR assembly is uncertain (Meddows et al., 2001; Cao et al., 2011). In contrast, the NMDAR M4 segments may contribute to receptor gating. Deletion of the NMDAR M4 segments resulted in nonfunctional channels that could be rescued by coexpression of the M4 segments (Schorge and Colquhoun, 2003). Furthermore, the NMDAR M4 segments contain sites for ethanol modulation (Honse et al., 2004; Ren et al., 2012) and elements important for pore opening (Talukder et al., 2010). Highlighting the possible significance of the M4 segments to NMDAR function, numerous de novo missense mutations identified in the M4 segments of NMDAR subunits are implicated in neurological disorders (Lemke et al., 2013; Hamdan et al., 2014; Yuan et al., 2015; Hardingham and Do, 2016; Chen et al., 2017). Still, the functional role of the M4 segments in NMDARs and their relation to that in AMPARs is unknown.
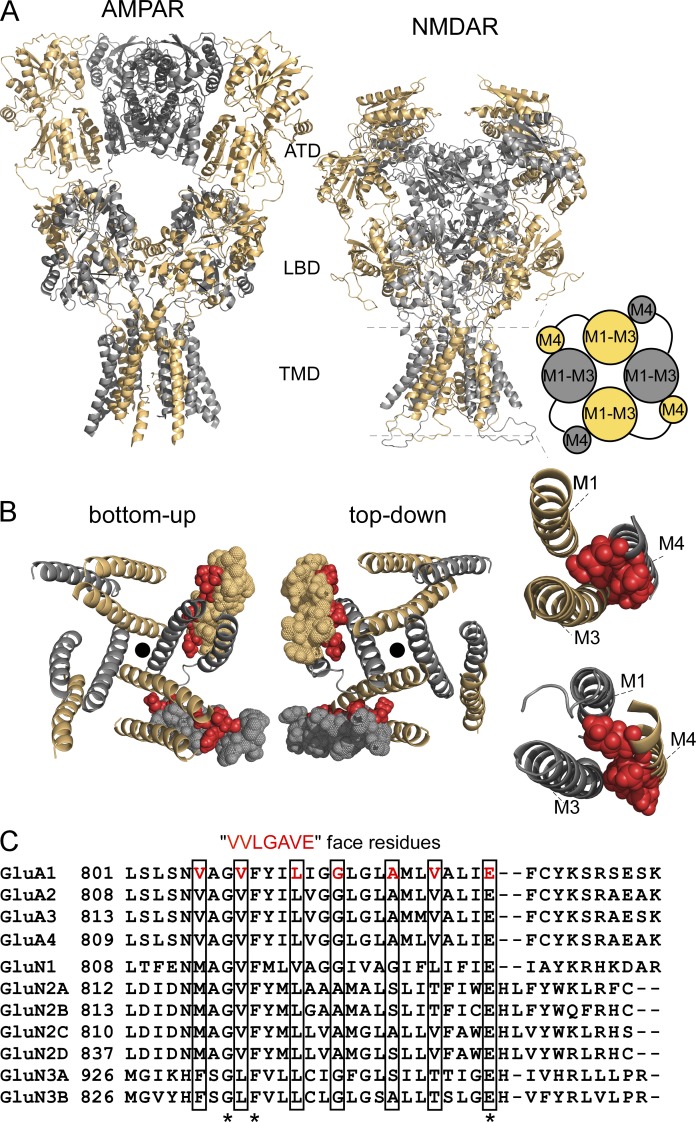
Figure 1.
Structural features of AMPAR and NMDAR TMDs. (A) Comparison of AMPARs (GluA2, 3KG2, Sobolevsky et al., 2009) and NMDARs (model structure based on 4TLM of GluN1/GluN2B, see Materials and methods) lacking the intracellular CTD. Subunits are colored light orange (GluA2 A and C, GluN1) and gray 60% (GluA2 B and D, GluN2A). For iGluRs, individual subunits as well as the oligomeric complexes are composed of four highly modular domains. Two of these domains are positioned on the extracellular side of the membrane: the amino-terminal domain (ATD) and the LBD. The TMD spans the lipid bilayer and forms the ion channel; in both receptor subtypes at the level of the TMD, the M4 segment of one subunit is associated with the pore domain or ion channel core (M1–M3) of a neighboring subunit (cartoon, right). The fourth domain is the intracellular CTD. (B) View of the TMD for the model NMDAR structure from either the intracellular (bottom-up view) or extracellular (top-down view) side illustrating the association of M4 with an adjacent pore domain. For clarity, one GluN1 and one GluN2A M4 segment are represented as spheres, with VVLGAVE face positions highlighted in red. The center of the pore is indicated by a black dot. (far right) Illustration showing that residues occupying the VVLGAVE face in NMDAR subunits are aligned quite closely, mainly with the M3 segment of an adjacent subunit. The S1-M1 linker of the same subunit is also positioned closely to the extracellular portion of its M4 segments (not depicted). (C) Alignment of the M4 segments and residues on the N- and C-terminal sides for rat AMPAR and NMDAR subunits. Only three residues, a glycine (G), phenylalanine (F), and a glutamate (E), are completely conserved across all subunits (asterisks). Still, residues occupying the VVLGAVE face (boxed) tend to have comparable noncharged (valine [V], leucine [L], methionine [M]) or small (glycine [G], alanine [A], serine [S]) side chains with the exception of a threonine (T) at the VVLGAVE position.
Structurally, the M4 segment of one subunit interfaces with the pore domain of a neighboring subunit (Fig. 1; Sobolevsky et al., 2009; Chen et al., 2014; Karakas and Furukawa, 2014). A specific face of the AMPAR M4 segment, termed VVLGAVE for the residues occupying it (Fig. 1, B and C), interacts with the pore domain, mainly the M3 transmembrane segment (Sobolevsky et al., 2009). The integrity of the entire extend of the VVLGAVE face is required for receptor tetramerization (Salussolia et al., 2013).
Here, we make a direct comparison of the roles of the M4 segments in AMPAR and NMDAR function by contrasting the effects of a tryptophan scan in both subtypes. In AMPARs, such a scan identified the importance of the VVLGAVE face to receptor tetramerization (Salussolia et al., 2011, 2013). In contrast, we find that the M4 segments in NMDARs make a highly significant contribution to receptor gating, while any contribution to biogenesis (i.e., assembly and/or trafficking) is limited. Notably, the M4 segments in both iGluR subtypes are split structurally in terms of their functional impact. For AMPARs, although the M4 segment contributes to receptor assembly, this role is dramatically reduced at the extreme extracellular end of M4. For NMDARs, the extreme extracellular ends of the M4s in both GluN1 and GluN2A also show a functional distinction, having the most dramatic impact on receptor gating possibly reflecting their interaction with their own S1-M1 (Ogden et al., 2017).
Materials and methods
Molecular biology and cell culture
All manipulations were made in rat GluN1 (GluN1a; NCBI Protein database accession no. P35439), GluN2A (Q00959), or GluA2 (P19491) subunits. In all instances, numbering included the signal peptide (GluN1, 18 residues; GluN2A, 19 residues; GluA2, 21 residues). In previous publications, we typically used numbering for the mature protein. Individual mutations were generated via QuikChange site-directed mutagenesis (Agilent Technologies) with XL1-Blue super-competent cells.
pHmystik-tagged constructs
pHmystik-pRK7 was a gift from M. Aurousseau and D. Bowie (Aurousseau, 2015). pHmystik generates a monomeric fluorescent protein that fluoresces in the blue-green spectrum. Purified pHmystik-pRK7 DNA was isolated via Mini Prep (QIAGEN) and confirmed by sequencing. Nhe1 restriction enzyme cloning sites were introduced via site-directed mutagenesis into GluN1 and GluN2A after the fourth amino acid of the mature protein (not including signal peptide). Nhe1 restriction enzyme sites were introduced at the 5′ and 3′ ends of the pHmystik insert in the pRK7 backbone. pHmystik-5′,3′ Nhe1, GluN1-Nhe1, and GluN2A-Nhe1 constructs were digested with Nhe1 in the presence of BSA, dephosphorylated with rSAP (NEB M0371S) and then separated on 0.8% agarose gel (1× TAE). Digested pHmystik and linearized GluN1 and GluN2A were extracted via Zymoclean gel-purification (Zymo D4001S). Purified pHmystik was inserted into the vector, with either GluN1 or GluN2A, using Roche rapid DNA ligation kit (04898117001; Roche). Ligation reaction of pHmystik-GluN1 and pHmystik-GluN2A was transformed into XL1-Blue super-competent cells (Agilent Technologies), and isolated colonies were picked for Mini Prep. Purified DNA of desired constructs were confirmed by DNA sequencing.
Cell culture and transfection
Human embryonic kidney 293 (HEK 293) or HEK 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% FBS, for 24 h before transfection. Non-tagged cDNA constructs were cotransfected into HEK 293 cells along with a separate pEGFP-Cl vector at a ratio of 4.5:4.5:1 (N1/N2/EGFP for NMDARs) or 9:1 (for AMPARs) using X-tremeGENE HP (Roche). pEGFP-Cl was not included in the transfection mix with GFP-tagged constructs, which were transfected at a 1:1 ratio. To improve cell survivability, HEK 293 cells transfected with NMDAR subunits were bathed in a media containing the NMDAR competitive antagonist APV (100 µM) and Mg2+ (100 µM; single-channel recording experiments) or the transfection mixture was replaced 4 h after transfection with fresh 5% FBS-DMEM culture media containing APV (100 µM) and Mg2+ (1 mM; whole-cell and imaging experiments). Cells transfected with AMPAR subunits were bathed in the competitive antagonist CNQX (10 µM). All experiments were performed 18–48 h after transfection.
Molecular modeling
The structural model for GluN1/GluN2B was modified from PDB 4TLM (Lee et al., 2014) by restoring the sequences of the N1 and N2B chains to the native Xenopus laevis sequences, containing residues 23–834 for N1 (chains A and C) and residues 26–839 for N2B (chains B and D). Specifically, mutations introduced for the crystallographic study were reverted to the native residues, and missing loop residues and missing atoms on other residues were added. These were all done by using MODELLER v9.10 (Šali and Blundell, 1993), except for one missing stretch (residues 482–502) in chain C, which was grafted from chain A.
After adding all hydrogens, the model was slightly refined by 5,000 steps of energy minimization with backbone fixed and 2 ns of Langevin dynamics with backbone constrained. The refinement procedure was performed in NAMD 2.9 (Phillips et al., 2005) using the CHARMM36 force field (Mackerell et al., 2004).
Fluorescence-detection size exclusion chromatography (FSEC)
For FSEC experiments, we used GluA2(Q) tagged with EGFP at the C terminus (Sobolevsky et al., 2009; Salussolia et al., 2013). In brief, transfected cells were rinsed with PBS, pelleted, and resuspended in 300 µl solubilization buffer containing 20 mM Tris-HCl, 200 mM NaCl (TBS), 1% DDM (Affymetrix), and 1 mM PMSF. Cells were lysed using Misonix Sonicator 3000 (2 min cycle of 30 s at power level 2 followed by 30 s resting) and rotated for 2 h at 4°C before ultracentrifugation (TLA110 rotor) at 70,000 rpm for 10 min. A fraction of supernatant (100 µl) was loaded onto a Superose 6 column (10/300 GL; GE Healthcare), preequilibrated with TBS buffer containing 0.05% DDM and run at a flow rate of 0.4 ml/min. The eluent from the Superose 6 column was passed through a fluorometer with the following settings: excitation, 475 nm; emission, 507 nm; time increment, 0.5 s; integration time, 1 s; recording time, 0–4,500 s. Chromatograms for a given transfection cycle were normalized to the tetramer peak (occurring between 1,700 and 2,000 s) for wild type collected during the same run and were analyzed over ∼1,700–2,400 s using the multi-peak fitting routine in Igor Pro (WaveMetrics; Salussolia et al., 2015). An area under the curve (AUC) was defined for the tetramer (AUCT), dimer (AUCD), and monomer (AUCM) fractions, from which we derived the %tetramer = 100 × AUCT/(AUCT + AUCD + AUCM) (see Gan et al., 2016).
Macroscopic current recordings
Macroscopic currents in the whole-cell mode or outside-out patches, isolated from HEK 293 cells, were recorded at room temperature (20–23°C) using an EPC-9 amplifier with PatchMaster software (HEKA), digitized at 10 kHz and low-pass filtered at 2.9 kHz (−3 dB) using an 8 pole low pass Bessel filter (Yelshansky et al., 2004). Patch microelectrodes were filled with our standard intracellular solution (mM): 140 KCl, 10 HEPES, and 1 BAPTA, pH 7.2 (KOH). Our standard extracellular solution consisted of (mM) 140 NaCl, 1 CaCl2, and 10 HEPES, pH 7.2 (NaOH). For experiments examining the number of channels expressing in whole-cell patches, 0.01 mM EDTA was added to our standard extracellular solution to approximate conditions used in single channels. Pipettes had resistances of 2–6 MΩ when filled with the pipette solution and measured in the standard Na+ external solution. We did not use series resistance compensation nor did we correct for junction potentials. Currents were measured within 15 min of going whole cell.
External solutions were applied using a piezo-driven double barrel application system. For NMDARs, one barrel contained the external solution +0.1 mM glycine, whereas the other barrel contained the same solution +1 mM glutamate. For AMPARs, we did not include glycine and 3 mM glutamate was applied instead of 1 mM. The open tip response (10–90% rise time) of the application system was between 400 and 600 µs. For display, NMDAR currents were digitally refiltered at 500 Hz and resampled at 1 kHz, and AMPARs at 1 kHz and at 2 kHz.
Assaying surface expression using whole-cell currents
We recorded whole-cell currents for each construct a minimum of two different transfection cycles (Salussolia et al., 2011). Constructs where we could not detect glutamate-activated currents were recorded on at least one additional transfection cycle. Wild type was recorded at minimum every other transfection cycle.
A caveat of using whole-cell currents to assay surface expression is a bias in cell selection: cells that express a high number of normal or high functioning NMDARs are more likely to die. Hence, when selecting cells for recording or imaging, we may be selecting cells that on average have fewer receptors expressed in the membrane. This issue will be most challenging for those constructs that show increased surface expression and/or show strongly enhanced gating properties. Most of our efforts focused on constructs that showed reduced current amplitudes.
Determining desensitization
To determine the extent and rate of desensitization, we applied glutamate at −70 mV for 100 ms to outside-out patches (AMPARs) or 2.5 s in the whole-cell mode (NMDARs). Percent desensitization (%des) was calculated from the ratio of peak (Ipeak) and steady-state (Iss) current amplitudes: %des = 100 × (1 − Iss/Ipeak). Time constants of desensitization (τdes) were determined by fitting the decaying phase of currents to either a single (AMPARs) or double (NMDARs) exponential function. In some instances when current amplitudes were small, we averaged three to five records.
Single-channel recordings
Single-channel recordings were performed in the on-cell configuration at steady-state using transfected HEK 293 cells at room temperature. The pipette solution mimicked extracellular conditions and contained (mM) 150 NaCl, 10 HEPES, 0.05 EDTA, 1 glutamate, and 0.1 glycine, pH 8.0 (NaOH). High pH and EDTA were used to minimize proton and divalent mediated inhibitory effects (Popescu and Auerbach, 2003). Recording pipettes were pulled from thick-wall borosilicate capillary glass (Sutter Instrument) and fire polished to final pipette resistances ranging from 5 to 27 MΩ. Cells were patched to resistances exceeding 1.5 GΩ. To elicit distinct inward current amplitudes, we held the electrode voltage at 100 mV. Currents were recorded using a patch clamp amplifier (Axopatch 200B; Molecular Devices), filtered at 10 kHz (four-pole Bessel filter), and digitized at 40 kHz (ITC-16 interfaced with PatchMaster). Experiments ran for ∼3–20 min to ensure a significant number of events for analysis.
Analysis of single-channel records was comparable with Talukder and Wollmuth (2011). In brief, recordings were exported from PatchMaster to QuB (http://www.qub.buffalo.edu). Processed data were idealized using the segmental k-means (SKM) algorithm in QuB with a dead time of 40 µs (Qin, 2004). From the idealization, we derived single-channel current amplitudes, mean closed time (MCT) and mean open time (MOT) as well as an equilibrium open probability (eq. Po). Eq. Po is the fractional occupancy of the open states in the entire single-channel recording, including long-lived closed states.
For recordings of wild-type GluN1/GluN2A, detection of single-channel patches was straightforward because this construct has a relatively high Po and recordings were made for a long duration (∼10,000–500,000 events). Certain constructs showed an extremely low Po and MOT. Although we did not detect any overlap in channel openings, we cannot rule out that these patches did not contain more than one channel. However, this issue is not critical to the present study because we did not do kinetic analysis on these patches.
Assaying surface expression using pH-sensitive GFP
HEK 293T cells were plated at a density of 1.2 × 105 on 35-mm glass-bottom dishes and were transfected as described in Cell culture and transfection section above. Samples were imaged 24–48 h after transfection using a Nikon Ti Eclipse microscope equipped with the Nikon TIRF slider. Excitation used the 488-nm line from a fiber-coupled argon Ion laser (Lasos). The fluorescence emission was collected with a Nikon 60× 1.45 NA oil-immersion TIRF objective and relayed to an iXon DU897 emCCD camera (Andor Technologies). The fluorescence emission was transmitted using a dual band 488/561 TIRF filter cube with a GFP band pass of 525/50 (Chroma). Cells were observed at 5-s intervals with 30–50 exposures collected for each field of view, which contained one to four fluorescent cells. Cells were imaged in either a bath solution at pH 7.4, consisting of (mM) 140 NaCl and 10 HEPES, pH 7.4 (NaOH); or a bath solution at pH 5.5, consisting of (mM) 140 NaCl and 30 Mes, pH 5.5 (HCl). These bathing solutions were exchanged using a continuous flow perfusion system.
Fluorescence intensity was quantified in ImageJ (National Institutes of Health; Schneider et al., 2012). Cells were selected for analysis that were at least 50% isolated from other cells. The fluorescence intensity (F) of a cell of interest was calculated for each frame: F = Fcell − Fbackgnd, where Fcell is the mean fluorescence of the cell, defined by a polygon circumscribing it, and Fbackgnd is the mean fluorescence of an acellular region directly adjacent to the cell of interest. For display and analysis, we normalized fluorescence intensity to a baseline fluorescence (Fo), which was the mean F just before the solution was changed from pH 7.4 to 5.5. The change in fluorescence (ΔF) was defined as ΔF = Fo − Ftest, where Ftest is the fluorescence intensity taken 15–30 s after the solution was switched from pH 7.4 to 5.5. We defined “detectable” surface expression as occurring when the fluorescence intensity decreased rapidly upon switching to pH 5.5 and returned to baseline with a return to pH 7.4. For some constructs, we detected both positive and negative (no detectable ΔF); in these instances, we averaged surface expression only for those cells that were positive.
Statistics
Data analysis was performed using Igor Pro, QuB, Excel, and MiniTab 17. All average values are presented as mean ± SEM. The number of replicates are indicated in the figure legend or in a table associated with the figure. In instances where we were only interested in whether outcomes were statistically different from that for wild type, we used an unpaired two-tailed Student’s t test to test for significant differences. Statistical significance was typically set at P < 0.05. In instances where we were interested in how constructs varied from each other, we used an ANOVA and followed with Tukey’s test (P < 0.05).
Online supplemental material
Three supplemental figures and two supplemental tables are included. Fig. S1 gives additional example FSEC chromatographs. Fig. S2 shows additional analysis of the interaction of the NMDAR M4 segments with other transmembrane segments. Fig. S3 shows control experiments with pHmystick constructs. Table S1 contrasts side chain properties of amino acids substituted in the GluA2 M4 segment. Table S2 shows single-channel data for positions that showed significant current potentiation, an issue we do not explore in the main text.
Results
Subtle mutations in the AMPAR M4 segment highlight its key role in receptor assembly
Tryptophan has a large and bulky side chain, which can disrupt transmembrane interactions (Soler-Llavina et al., 2006). For AMPARs, tryptophan substitutions of the M4 segment identified that a face (VVLGAVE) aligned with the pore domain (M1–M3) of an adjacent subunit (Fig. 1, B and C; Sobolevsky et al., 2009) disrupts receptor tetramerization (Salussolia et al., 2011, 2013). To further define the significance of the M4/ion channel core interactions to AMPAR function, we made more subtle mutations in the VVLGAVE face in the unedited GluA2 subunit tagged on the C terminus with EGFP (GluA2(Q)-EGFP). These subtle substitutions, rather than drastically changing the volume of the side chain as done by tryptophan, typically preserved the nature of the side chain while adding or removing a methyl group and hence changed the side chain volume in a more limited fashion (Table S1). Further, as we will show later, receptors containing these subtle substitutions still functioned, permitting us to test whether the VVLGAVE face has an impact on receptor gating, specifically desensitization. We quantified the effect of these subtle mutations on receptor assembly using FSEC (Fig. 2; Kawate and Gouaux, 2006; Salussolia et al., 2013).
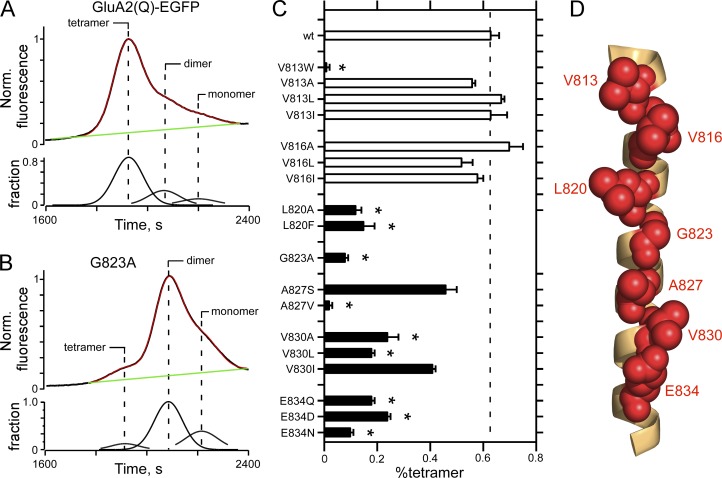
Figure 2.
FSEC to assay AMPAR oligomerization. (A and B) Raw FSEC chromatographs of wild-type GluA2(Q)-EGFP (A) or the same construct containing an alanine substitution at G823 (GluA2(Q)(G823A); B). For quantification, we normalized all chromatographs to the tetramer peak in the wild type for that transfection cycle. For the top panel in each plot, the black line is the original data, the green line the baseline, and the red line is the sum of the individual fits derived from a multi-peak fitting routine (see Materials and methods); the bottom panels show the fraction of the total chromatograph corresponding to, from left to right, tetramer, dimer, or monomer. (C) Mean (±SEM) of the %tetramer (see Materials and methods) for various substitutions of VVLGAVE face positions of the M4 segment in GluA2(Q). Black bars indicate values significantly different from wild type (P < 0.05, ANOVA); asterisks indicate values significantly different from subtle mutations at V813 and V816. Number of independent FSEC runs: wt, 16; V813W, 3; V813A, 4; V813L, 4; V813I, 3; V816A, 4; V816L, 3; V816I, 4; L820A, 4; L820F, 6; G823A, 5; A827S, 3; A827V, 4; V830A, 3; V830L, 3; V830I, 3; E834Q, 6; E834D, 7; E834N, 3. (D) GluA2 M4 segment with VVLGAVE positions shown in red space–filled balls.
On FSEC, wild-type GluA2 receptors predominantly give tetramers with a small fraction of dimers and monomers (Fig. 2 A). To quantify these results, we fit these curves with a polynomial to derive the tetramer, dimer, and monomer fraction (Fig. 2 A, bottom trace) from which we calculated the %tetramer (see Materials and methods; Salussolia et al., 2015; Gan et al., 2016). For GluA2(Q) homomers, this approach yielded a %tetramer of 63 ± 3%, n = 16 (mean ± SEM, n = number of independent runs), a result comparable with that published previously for homomeric AMPARs (Greger et al., 2003; Salussolia et al., 2013; Gan et al., 2016).
At the core of the VVLGAVE face is a GxxxG transmembrane–interacting motif (Moore et al., 2008). Substitution of alanine (A) for the glycine (G; G823) dramatically attenuates the extent of tetramerization (Fig. 2 B), reducing the %tetramer to 8 ± 1%, n = 5. Indeed, subtle substitutions at many of the positions in the VVLGAVE face, specifically from L820 inward, significantly attenuated tetramerization (Fig. S1 and Fig. 2 C, solid bars indicate significance relative to wild type). For example, changing side chains at the most intracellular position, a glutamate (E) at 834, with either a smaller, charged side chain (aspartate, D) or an uncharged side chain (glutamine, Q; or asparagine, N) in all instances significantly reduced the %tetramer. In contrast, subtle substitutions at the extreme extracellular positions, V813 and V816, had no significant effects on receptor assembly, though tryptophan substitutions at these same positions essentially eliminated tetramerization (for V813, Fig. 2 C; Salussolia et al., 2013). Thus, although all positions in the VVLGAVE face impact receptor assembly in AMPARs based on tryptophan substitutions, these actions based on subtle substitutions are more critical at positions intracellular to V816, encompassing L820 and inward.
Tryptophan scan of the NMDAR M4 segments indicates differences with AMPARs
Because of the robust effects of tryptophan substitutions in AMPARs (Salussolia et al., 2011), we performed a similar scan in NMDARs, substituting individual positions in the GluN1 and GluN2A M4 segments with tryptophan. As an initial means to assay the functional role of the M4 segments in NMDARs, we measured whole-cell currents for these constructs (Fig. 3 and Table 1) in a manner comparable with what was done previously (Salussolia et al., 2011) to facilitate direct comparisons (see Materials and methods for limitations of this approach).
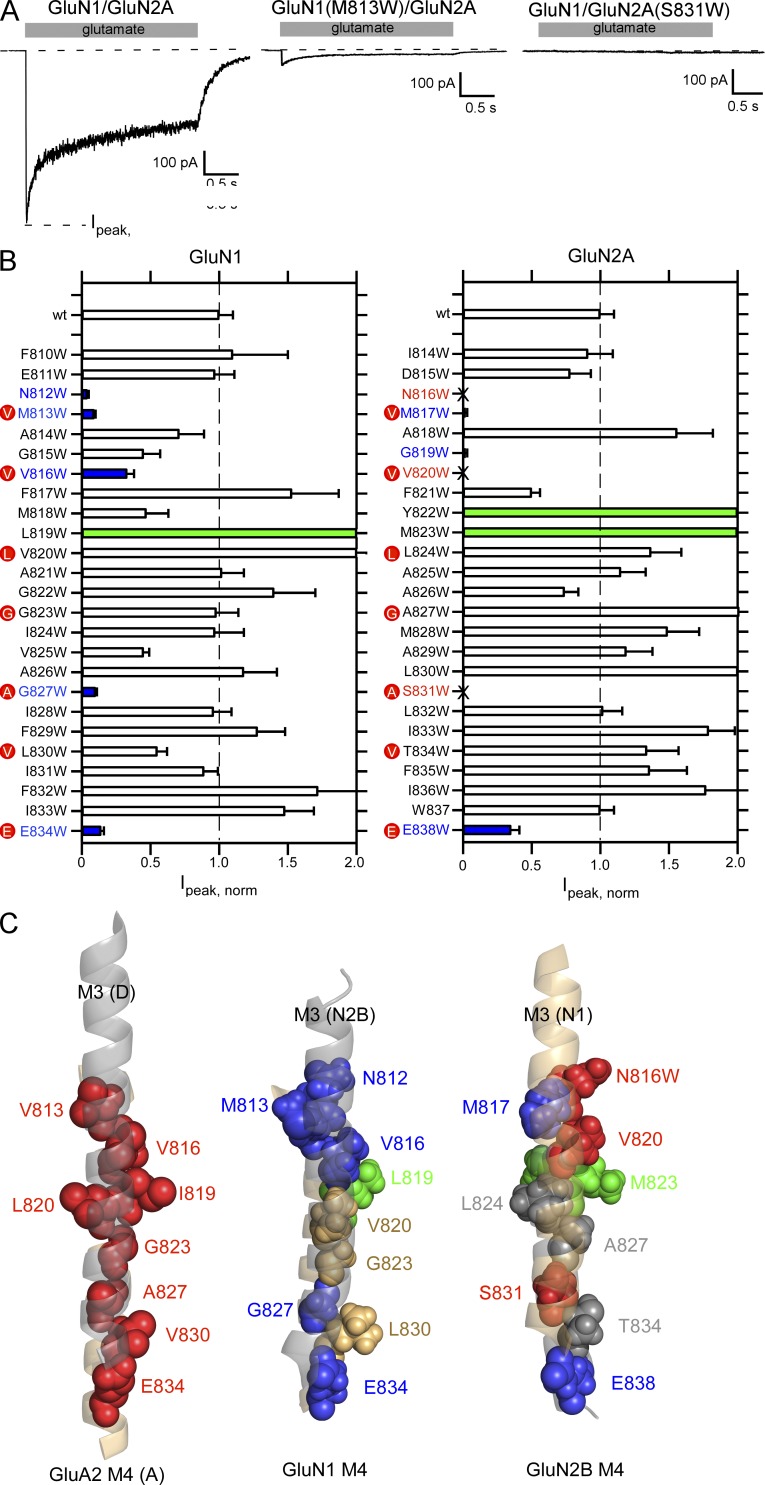
Figure 3.
Tryptophan mutagenesis scan of the M4 segments in the GluN1 or GluN2A subunits. (A) Representative whole-cell recordings of current through wild-type GluN1/GluN2A, GluN1(M813W)/GluN2A, or GluN1/GluN2A(S831W). Glutamate (1 mM, shaded box) was applied for 2.5 s. Cells were continuously bathed in glycine (0.1 mM). Holding potential, −70 mV. (B) Mean current amplitudes (±SEM) at −70 mV normalized to current amplitudes for wild-type GluN1/GluN2A (−750 ± 50, n = 18; raw values shown in Table 1). Positions that did not show detectable glutamate-activated currents are demarcated by an “X” (see Materials and methods). Positions that showed current amplitudes significantly less or greater than wild-type are colored blue and green, respectively (P < 0.05, two-tailed Student’s t test, unpaired). The red dots highlight positions homologous to the VVLGAVE face in AMPARs. (C) Orientation of the M3 segments (shadowed), viewed from the center of the pore, to the M4 segments in either AMPARs (left) or NMDARs either GluN2B M3 relative to GluN1 M4 (middle) or GluN1 M3 relative to GluN2B M4 (right). Positions that showed significant changes in current amplitudes are highlighted in blue (reduced), green (greater), or red (no detectable current). For AMPARs, only those positions that showed no detectable current (Salussolia et al., 2011) are indicated, which include the VVLGAVE positions as well as I819.
Table 1. Raw macroscopic peak current amplitudes, shown as normalized values in Fig. 3 B, for wild-type GluN1/GluN2A or GluN1/GluN2A containing tryptophan substitutions in either GluN1 or GluN2A M4 segments.
| Tryptophans in GluN1 subunit | Tryptophans in GluN2A subunit | ||||
|---|---|---|---|---|---|
| Construct | Ipeak | n | Construct | Ipeak | n |
| pA | pA | ||||
| N1/N2A | −750 ± 50 | 18 | |||
| F810W | −830 ± 290 | 4 | I814W | −680 ± 125 | 4 |
| E811W | −720 ± 95 | 4 | D815W | −580 ± 110 | 4 |
| N812W | −28 ± 6* | 4 | N816W | ND | 9 |
| M813W | −65 ± 9* | 5 | M817W | −17 ± 3* | 6 |
| A814W | −530 ± 130 | 4 | A818W | −1,160 ± 175 | 5 |
| G815W | −330 ± 90 | 4 | G819W | −11 ± 2* | 6 |
| V816W | −250 ± 30* | 4 | V820W | ND | 8 |
| F817W | −1,150 ± 240 | 5 | F821W | −370 ± 35 | 5 |
| M818W | −355 ± 110 | 6 | Y822W | −1,500 ± 120 | 4 |
| L819W | −2,010 ± 210^ | 4 | M823W | −2,090 ± 100^ | 5 |
| V820W | −1,830 ± 280 | 4 | L824W | −1,030 ± 150 | 4 |
| A821W | −760 ± 110 | 4 | A825W | −860 ± 120 | 4 |
| G822W | −1,050 ± 220 | 4 | A826W | −560 ± 65 | 5 |
| G823W | −730 ± 110 | 4 | A827W | −1,680 ± 205 | 5 |
| I824W | −730 ± 150 | 5 | M828W | −1,110 ± 150 | 5 |
| V825W | −330 ± 20 | 4 | A829W | −890 ± 130 | 6 |
| A826W | −880 ± 170 | 4 | L830W | −1,830 ± 470 | 5 |
| G827W | −78 ± 8* | 4 | S831W | ND | 9 |
| I828W | −720 ± 90 | 4 | L832W | −760 ± 90 | 5 |
| F829W | −960 ± 130 | 5 | I833W | −1,340 ± 100 | 4 |
| L830W | −410 ± 45 | 4 | T834W | −1,000 ± 160 | 4 |
| I831W | −670 ± 60 | 4 | F835W | −1,020 ± 190 | 4 |
| F832W | −1,290 ± 220 | 5 | I836W | −1,320 ± 160 | 5 |
| I833W | −1,110 ± 130 | 4 | W837 | −750 ± 50 | 18 |
| E834W | −100 ± 10* | 6 | E838W | −265 ± 40* | 6 |
Values shown are mean ± SEM. n indicates the number of whole-cell recordings. Peak current amplitudes were recorded in the whole-cell mode at −70 mV (e.g., Fig. 3 A). GluN1 constructs were co-expressed with wild-type GluN2A and vice-versa. Tagged values are significantly less (*) or greater (^) than wild-type (P < 0.05, two-tailed Student’s t test, unpaired). ND, no glutamate-activated currents detected. As done previously (Salussolia et al., 2011), constructs were tested on at least two different transfection cycles with wild type tested every other transfection cycle. Constructs that showed no detectable glutamate-activated current were tested on at least one additional transfection cycle.
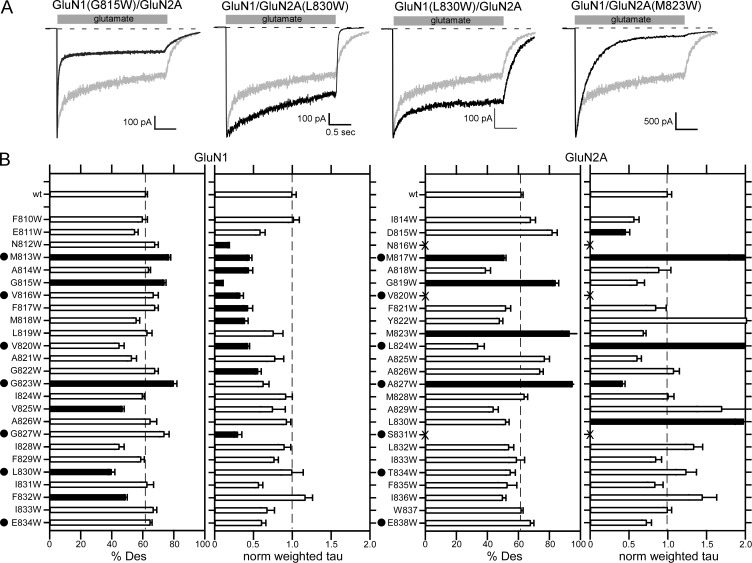
Wild-type GluN1/GluN2A shows robust glutamate-activated currents (Fig. 3 A, left; −750 ± 50 pA, n = 18; n = number of whole-cell recordings). Most other tryptophan-substituted constructs also showed robust glutamate-activated current, though certain constructs showed either significantly reduced peak current amplitudes (e.g., GluN1(M813W)/GluN2A; Fig. 3 A, middle) or no detectable glutamate-activated membrane current (e.g., GluN1/GluN2A(S831W); Fig. 3 A, right). To compare current amplitudes, we normalized current amplitudes to those for wild type (Fig. 3 B and Table 1). Constructs are highlighted as to whether they showed significantly reduced current amplitudes (blue), significantly greater current amplitudes (green) or no detectable current (X, red) relative to wild type (P < 0.05, t test).
This experiment suggests several initial conclusions. First, compared with similar experiments for AMPARs, where 9 out of 23 tested positions in the M4 segment showed no detectable current (Salussolia et al., 2011), the number of positions with no detectable current in NMDARs is notably less, only 3 (N816, V820, and S831, all within the GluN2A subunit) out of a total of 25 homologous positioned tested on either GluN1 or GluN2A. Second, when significantly reduced or no detectable current did occur in NMDAR subunits, it almost always occurred (4 of 5, GluN1; 4 of 6, GluN2A) at positions homologous to the AMPAR VVLGAVE face (red dots).
One possible explanation for the reduced number of positions showing no detectable current in NMDARs is that the tested constructs had substituted tryptophans in only two of the four subunits, whereas in homomeric AMPARs all four subunits possessed the substitution. In terms of transmembrane interactions, substitutions of M4 positions pointing toward M1 had no significant effect on current amplitudes (Fig. S2). In contrast, the majority of M4 positions that showed significant changes in current amplitudes are homologous to the AMPAR VVLGAVE and like the AMPAR VVLGAVE face point toward the M3 segments (Fig. S2). We therefore focused on positions homologous to the AMPAR VVLGAVE face and coexpressed GluN1 and GluN2A subunits with tryptophans substituted at homologous positions, corresponding to the VVLGAVE face, and again tested whole-cell current amplitudes (Table 2). We did not test two combinations in which any individual tryptophan substitution yielded no detectable current. Of the other five mutation pairs tested, three again showed high current amplitudes. The combination between the most extracellular and intracellular positions, GluN1(M813W)/GluN2A(M817W) and GluN1(E834W)/GluN2A(E838W), respectively, showed no detectable current. Thus, even when both subunits have tryptophans substituted for positions homologous to the AMPAR VVLGAVE face, any effect on whole cell is less notable than that for AMPARs.
Table 2. Macroscopic current amplitudes for GluN1/GluN2A containing tryptophan substitutions in the VVLGAVE face in the GluN1 and GluN2A M4 segments.
| Construct | Ipeak | n | Comments | |
|---|---|---|---|---|
| pA | ||||
| N1/N2A | −750 ± 50 | 18 | ||
| V | N1(M813W)/N2A(M817W) | ND | 9 | |
| V | N1(V816W)/N2A(V820W) | NT | - | N1/N2A(V820W) showed no current |
| L | N1(V820W)/N2A(L824W) | −340 ± 35 | 4 | |
| G | N1(G823W)/N2A(A827W) | −535 ± 60 | 4 | |
| A | N1(G827W)/N2A(S831W) | NT | - | N1/N2A(S831W) showed no current |
| V | N1(L830W)/N2A(T834W) | −570 ± 140 | 4 | |
| E | N1(E834W)/N2A(E838W) | ND | 6 |
Values shown are mean ± SEM. Currents recorded as in Fig. 1. n indicates the number of whole-cell recordings. None of the constructs that showed current were significantly different from wild type (P < 0.05, two-tailed Student’s t test, unpaired). NT, not tested. ND, no glutamate-activated currents detected.
In AMPAR subunits, the “VVLGAVE” face positions are aligned with the adjacent M3 segment (Fig. 3 C, left; Sobolevsky et al., 2009). For NMDAR M4 segments, the homologous positions are also generally aligned with the adjacent M3 segments (Fig. 3 C, middle and right). Positions L830 in GluN1 and T834 in GluN2A (VVLGAVE) appear oriented to the side of M3, possibly accounting for the lack of an effect at this position. However, G823 in GluN1 and A827 in GluN2A (VVLGAVE) as well as V820 in GluN1 and L824 in GluN2A (VVLGAVE) are oriented toward M3, yet show no change in function when both are substituted with tryptophan (Table 2). Three positions in NMDAR subunits, L819 in GluN1 and Y822 and M823 in GluN2A, which point away from the M3 segment, showed significant current potentiation when substituted with tryptophan (Fig. 3 B). The homologous position to L819 and M823 in AMPARs, I819, showed no detectable current when substituted with tryptophan (Salussolia et al., 2011).
In summary, assuming that a major effect of a tryptophan substitution is to disrupt transmembrane interactions, the interaction of the M4 segments in NMDAR subunits with other transmembrane segments affects receptor function, though this action is considerably less extensive than in AMPARs.
Many nonfunctional tryptophan-substituted NMDARs express robustly on the cell surface
For AMPARs, the lack of membrane currents for receptors containing tryptophan substitutions of the VVLGAVE face reflects an inability to tetramerize (Salussolia et al., 2011, 2013), thereby preventing surface expression. To discern the mechanistic basis of significantly reduced current amplitudes in tryptophan-substituted NMDARs, we initially assayed surface expression of these constructs. To do so, we tagged NMDAR subunits at the extracellular N-terminal end with a pH-sensitive GFP, pHmystik (Aurousseau, 2015). We used total internal reflection fluorescence (TIRF) to assay cellular fluorescence while changing the extracellular pH from 7.4 to 5.5. At pH 5.5, the fluorescence of surface-expressed pHmystik is nearly fully quenched in contrast to other pH-sensitive GFP constructs (Aurousseau, 2015).
Wild-type receptors, either pHmystik-GluN1/GluN2A (Fig. S3 A) or GluN1/pHmystik-GluN2A (Fig. 4 A), showed robust and rapid changes in fluorescence when the external solution was switched from pH 7.4 to 5.5. This fluorescence change was reversible upon switching back to pH 7.4. No rapid change in fluorescence occurred when pHmystik-GluN1 (−0.02 ± −0.02, n = 16) or pHmystik-GluN2A (−0.02 ± −0.01, n = 8) was expressed alone (Fig. S3 B), though a slow change did occur with very long exposures (>50 s) to pH 5.5 (Fig. S3 C). Thus, we used the rapid and reversible change in fluorescence (ΔF; e.g., points i and ii in Fig. 4 A) as an index of surface-expressed NMDARs.
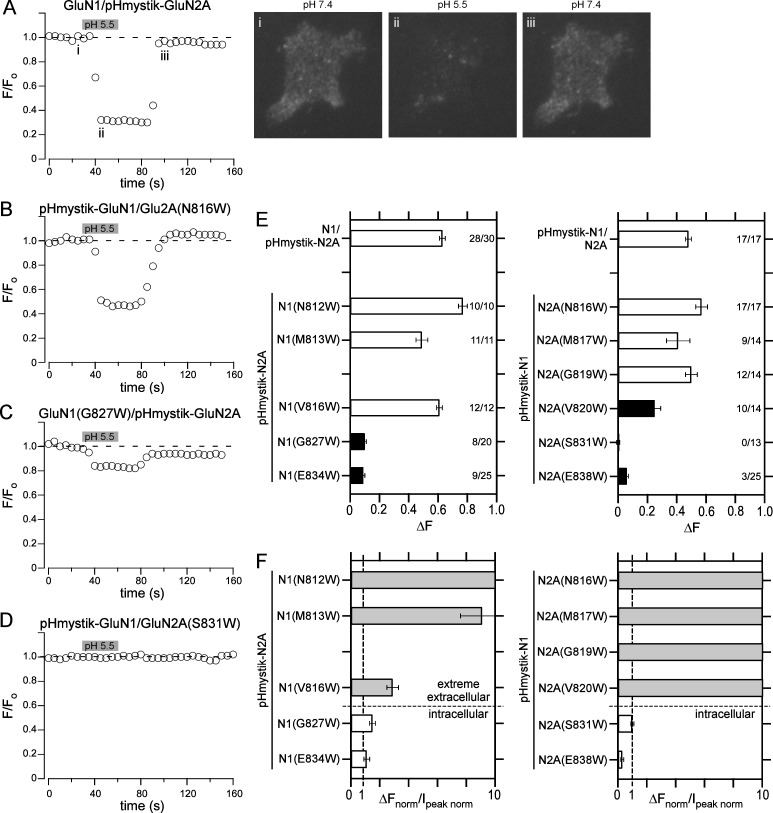
Figure 4.
Assaying surface expression of NMDARs using pHmystik, a pH-sensitive GFP. (A–D) Fluorescence time trace of GFP intensity as the extracellular bath solution pH was changed from 7.4 to 5.5 (gray bar, 30-s duration) and back again. Shown are HEK293T cells expressing GluN1/pHmystick-GluN2A (A), pHmystik-GluN1/GluN2A(N816W) (B), GluN1(G827)/pHmystick-GluN2A (C), or pHmystik-GluN1/GluN2A(S831W) (D). The representative cell images are from the time-points labeled in panel A; the images were taken at the approximate time points when the baseline fluorescence (Fo), corresponding to image (i), and the test fluorescence (Ftest), corresponding to image (ii), were measured as well as an image (iii) after return to pH 7.4. The change in fluorescence (ΔF = Fo − Ftest) was used as an index of surface expression. Sampling rate, 5 s. (E) Changes in cell fluorescence (±SEM) at low pH (ΔF, see Materials and methods) for positions in either GluN1 (left) or GluN2A (right). Solid bars indicate values significantly different from their respective wild type either N1/pHmystik-N2A (left) or pHmystik-N1/N2A (right; P < 0.05, t test). The numbers (far right in each plot) indicate the number of cells that showed detectable changes in surface expression relative to the total number of cells tested (see Materials and methods). Only cells that showed detectable changes in fluorescence were included in statistical analysis. (F) Relationship between surface expression, assayed by changes in fluorescence, and whole-cell current amplitudes. For comparison, values (±SEM) are normalized to their respective wild type, either for surface expression (ΔFnorm) or for peak whole-cell current amplitudes (Ipeak norm; Fig. 3 B). For this ratio of normalized values, a number close to unity implies a correlation between the two parameters, whereas a number much greater than unity indicates surface expression is much greater than expected from whole-cell currents.
Although both pHmystik-GluN1/GluN2A and GluN1/pHmystik-GluN2A showed robust surface expression, the expression was significantly greater for GluN1/pHmystik-GluN2A (0.63 ± 0.01, n = 17) than for pHmystik-GluN1/GluN2A (0.48 ± 0.01, n = 28). GluN1/pHmystik-GluN2A also showed considerably higher whole-cell current amplitudes (−2,050 ± 310 pA, n = 5) than pHmystik-GluN1/GluN2A (−930 ± 100 pA, n = 7).
Compared with wild-type constructs, surface expression for NMDARs containing tryptophan substitutions that were associated with reduced or no detectable glutamate-activated current fell into three categories (Fig. 4, B–E): (1) Surprisingly, despite showing either reduced or no whole-cell current, several positions in the extreme extracellular portion of the M4 (N1(N812W), N1(M813W), N1(V816W), N2A(N816W) [Fig. 4 B], N2A(M817W), N2A(G819W)) showed surface expression comparable with their respective wild-type construct (Fig. 4 E). In contrast, positions in the intracellular portion of the M4 showed (2) significantly reduced surface expression, including N1(G827W) (Fig. 4 C), N1(E834W), and N2A(E838W) or (3) no detectable surface expression, including N2A(S831W) (Fig. 4 D) and pHmystik-N1(E834W)/N2A(E838W) (−0.02 ± −0.04, n = 20).
Distinct features of NMDAR M4 extreme extracellular and intracellular portions
In contrast to AMPARs containing tryptophan substitutions in the VVLGAVE face, NMDARs containing comparable substitutions in the extreme extracellular portion of the M4s (at or near VVLGAVE) showed robust surface expression even when they displayed reduced or no detectable membrane currents (Fig. 3 B, Table 2, and Fig. 4 E). To define the relationship between surface expression as assayed by pHmystik (Fig. 4 E) and whole-cell current amplitudes (Fig. 3 B), we plotted a ratio of responses normalized to that for wild type (Fig. 4 F): surface expression normalized to the respective wild type (ΔFnorm) over whole-cell current amplitudes normalized to wild type (Ipeak norm; Fig. 3 B). A number close to unity indicates a good correspondence between surface expression and whole-cell current amplitudes; such a correspondence would suggest that the underlying single-channel activity of the construct is comparable with that for wild type. A ratio of normalized responses much greater than unity reflects that there is more surface expression than anticipated based on whole-cell currents, suggesting impaired channel function. Positions on the extreme extracellular portion of M4 in both GluN1 and GluN2A showed values much greater than unity, whereas intracellular positions showed values close or less than unity.
There are two conclusions from this plot of the ratio of normalized responses. First, tryptophan substitutions in the extreme extracellular portion of M4 still show robust surface expression (Fig. 4 E), much greater than that anticipated for whole-cell currents (Fig. 4 F). The most likely explanation for this is that these constructs can assemble and traffic to the membrane, like wild type, with the reduced or no detectable glutamate-activated currents probably caused by changes in receptor gating. Second, there is good correspondence between surface expression and whole-cell currents for intracellular positions (N1(G827), N1(E834), N2A(S831), N2A(E838); Fig. 4 F); because both surface expression and whole-cell currents show reduced or no expression, there appears some restriction for these constructs to get to the membrane, either a reduced receptor assembly and/or membrane trafficking. Thus, based on these considerations, there appears to be a dichotomy in the M4 segments in NMDARs with the extreme extracellular (N-terminal) and intracellular (C-terminal) portions taking on different functional roles.
Extreme extracellular M4 positions in NMDAR subunits impact receptor gating
The data in Fig. 4 based on tryptophan substitutions suggest that rather than participating in receptor assembly, as for AMPARs, the extreme extracellular portions of the M4 segments in NMDARs contribute to ion channel gating. To test this idea directly, we used single-channel recordings of tryptophan substitutions in either GluN1 or GluN2A that had significantly reduced whole-cell current amplitudes. We made these recordings in the on-cell mode at pH 8.0 and in the absence of divalents (see Materials and methods; Popescu and Auerbach, 2003; Talukder and Wollmuth, 2011).
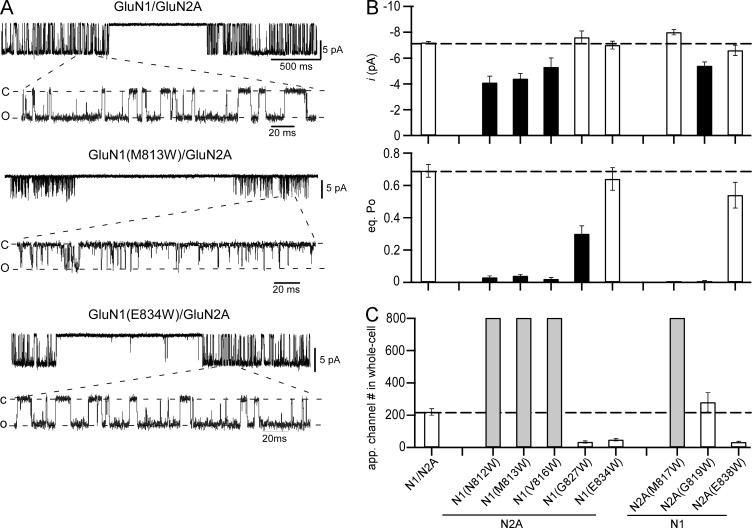
The single-channel profile for wild-type GluN1/GluN2A receptors, under the present recording conditions, showed robust activity (Fig. 5 A, top), with a mean single-channel current amplitude (i) around −7.2 pA (−7.2 ± 0.1 pA, n = 10) and an equilibrium open probability (eq. Po) of ∼0.7 (0.69 ± 0.04; Fig. 5 B and Table 3). Tryptophan substitutions in the extreme extracellular portion of M4, for example GluN1(M813W)/GluN2A (Fig. 5 A, middle), showed dramatic effects on receptor gating. These extracellular positions, N812, M813, and V816 in GluN1 and M817 and G819 in GluN2A, all showed significantly reduced eq. Po (Fig. 5 B and Table 3). The most consistent basis for this reduced open probability was a dramatic reduction in the MOT (Table 3). In wild type, MOT is ∼5.5 ms (5.5 ± 0.5 ms), whereas in these extracellular tryptophan substitutions it was reduced to <0.4 ms (Fig. 5 B and Table 3). For most of these constructs, there was also a trend for an increased MCT, though this was only consistently significant in the GluN2A positions (Table 3). Many of these same constructs also showed significantly reduced single-channel current amplitudes (Fig. 5 B and Table 3), though because of the extremely brief openings these amplitudes may reflect incomplete openings. In contrast, tryptophan substitutions of intracellular M4 positions, for example GluN1(E834W)/GluN2A (Fig. 5 A, bottom), had typically only weak or no effect on single-channel activity (Fig. 5 B and Table 3).
Figure 5.
Single-channel recordings of wild-type NMDARs and NMDARs with tryptophan substitutions that showed significantly reduced whole-cell current amplitudes. (A) Example single-channel recordings of GluN1/GluN2A, GluN1(M813W)/GluN2A, or GluN1(E834W)/GluN2A. Recordings were performed in the cell-attached configuration with a pipette potential of 100 mV. Downward deflections reflect inward currents. For each construct, the top half shows a low-resolution example (filtered at 1 kHz), and the bottom half shows a higher-resolution portion of the same record (filtered at 3 kHz). (B) Single-channel current amplitudes (top) and equilibrium open probability (eq. Po; bottom) shown as mean ± SEM (Table 3). Solid bars indicate values significantly different from wild type (P < 0.05, t test). (C) Approximate number of ion channels in the membrane mediating steady-state whole-cell current amplitudes (Table 4).
Table 3. Single-channel properties of wild-type GluN1/GluN2A or tryptophan-substituted GluN1 or GluN2A NMDARs where whole-cell currents were significantly changed.
| Construct | Total events (# of patches) | i | eq. Po | MCT | MOT |
|---|---|---|---|---|---|
| pA | ms | ms | |||
| N1/N2A | 1,531,680 (10) | −7.2 ± 0.1 | 0.69 ± 0.04 | 2.3 ± 0.2 | 5.5 ± 0.5 |
| N1(N812W) | 147,374 (5) | −4.1 ± 0.5* | 0.03 ± 0.01* | 31.8 ± 10.3* | 0.40 ± 0.07* |
| N1(M813W) | 484,988 (4) | −4.4 ± 0.4* | 0.04 ± 0.01* | 7.3 ± 1.5* | 0.28 ± 0.05* |
| N1(V816W) | 475,752 (4) | −5.3 ± 0.7* | 0.02 ± 0.01* | 7.2 ± 2.2 | 0.13 ± 0.01* |
| N1(G827W) | 218,498 (4) | −7.6 ± 0.5 | 0.30 ± 0.05* | 11.6 ± 1.9* | 4.7 ± 0.8 |
| N1(E834W) | 198,394 (4) | −7.0 ± 0.3 | 0.64 ± 0.07 | 3.8 ± 0.8 | 6.5 ± 0.6 |
| N2A(M817W) | 20,985 (5) | −8.0 ± 0.2 | 0.001 ± 0.0005* | 213 ± 73* | 0.20 ± 0.10* |
| N2A(G819W) | 27,892 (5) | −5.4 ± 0.3* | 0.005 ± 0.001* | 73.8 ± 7.1* | 0.33 ± 0.03* |
| N2A(E838W) | 81,550 (4) | −6.6 ± 0.4 | 0.54 ± 0.08 | 9.1 ± 3.4 | 8.8 ± 0.6* |
Values shown are mean ± SEM for single-channel current amplitude (i), equilibrium open probability (eq. Po), MCT, and MOT. Single-channel currents were recorded in the on-cell mode at approximately −100 mV and analyzed in QuB (see Materials and methods). Number of patches is in parenthesis to the right of total events. Eq. Po is the fractional occupancy of the open states in the entire single-channel recording, including long-lived closed states. All data were idealized and fit at a dead time of 40 µs. Asterisks indicate values significantly different from wild type (P < 0.05, t test).
The results in Figs. 4 F and 5 B suggest that the major effect of tryptophan substitutions at extreme extracellular M4 positions is to disrupt gating, whereas for intracellular positions it disrupts receptor biogenesis. To further test this idea, we estimated the number of ion channels in the membrane mediating whole-cell currents. To do so, we measured steady-state whole-cell current amplitudes under extracellular conditions that approximated those for which single-channel activities were recorded: no added divalents and 0.01 mM EDTA though at pH 7.2 (Table 4). As for peak current amplitudes (Fig. 3 B), steady-state current amplitudes measured under these conditions and relative to wild type were also significantly reduced (Table 4).
Table 4. Whole-cell current amplitudes in EDTA for wild-type GluN1/GluN2A or tryptophan-substituted GluN1 or GluN2A NMDARs where whole-cell currents were significantly reduced.
| Construct | Ipeak | Isteady-state | n | Estimated channel number (N) |
|---|---|---|---|---|
| pA | pA | |||
| N1/N2A | −1,170 ± 70 | −760 ± 50 | 13 | 220 ± 20 |
| N1(N812W) | −350 ± 40* | −96 ± 12* | 7 | 1,340 ± 390 |
| N1(M813W) | −590 ± 90 | −220 ± 30* | 7 | 1,810 ± 340 |
| N1(V816W) | −120 ± 20* | −80 ± 12* | 5 | 920 ± 210 |
| N1(G827W) | −150 ± 30* | −54 ± 13* | 5 | 34 ± 7 |
| N1(E834W) | −270 ± 40* | −150 ± 30* | 5 | 48 ± 7 |
| N2A(M817W) | −35 ± 4* | −27 ± 3* | 5 | 3,830 ± 330 |
| N2A(G819W) | −26 ± 6* | −5 ± 1* | 5 | 280 ± 60 |
| N2A(E838W) | −420 ± 50* | −230 ± 30* | 4 | 33 ± 5 |
Values shown are mean ± SEM. Tagged values are significantly less (*) or greater (^) than wild type (P < 0.05, t test). n indicates the number of whole-cell recordings. Currents were recorded in the whole-cell mode at a holding potential of −70 mV, as in Table 1, but with our standard extracellular solution (pH 7.2) also containing 0.01 mM EDTA. These conditions were aligned to those for on-cell single-channel recording except for the pH. The estimated channel number mediating the observed steady-state whole-cell current amplitudes (Isteady-state) was derived from: Isteady-state = N * Po * i, where N is the approximate number of channels on the membrane, Po is the equilibrium Popen, and i is single-channel current amplitude corrected for difference in membrane potential assuming Ohmic behavior (Table 3).
We estimated the approximate number of ion channels in the membrane mediating steady-state whole-cell current amplitudes (Isteady-state; Table 4) using the following relationship: Isteady-state = N * Po * i, where N is the approximate number of channels on the membrane, Po is the equilibrium open probability, and i is single-channel current amplitude (Table 3). For wild type, there was on average ∼200 channels on the membrane (Fig. 5 C and Table 4). For extreme extracellular positions in GluN1 (N812, M813, and V816) or GluN2A (M817 and G819), the estimated number of ion channels on the membrane was at least as high as wild type, if not considerably higher, indicating that reduced whole-cell currents is completely caused by a gating deficit rather than a disruption in receptor biogenesis. In contrast, for intracellular positions, N1(G827), N1(E834W), and N2A(E838W), the estimated number of ion channels was consistently lower than that for wild type.
In summary, these results strongly support the distinction between the extreme extracellular and intracellular portions of the NMDAR M4 segments. The extreme extracellular M4 in both GluN1 and GluN2A modifies receptor gating with little or no contribution to biogenesis, whereas the intracellular M4 appears to have a role in biogenesis, either assembly and/or trafficking to the membrane.
Recovery of function in pore-dead constructs
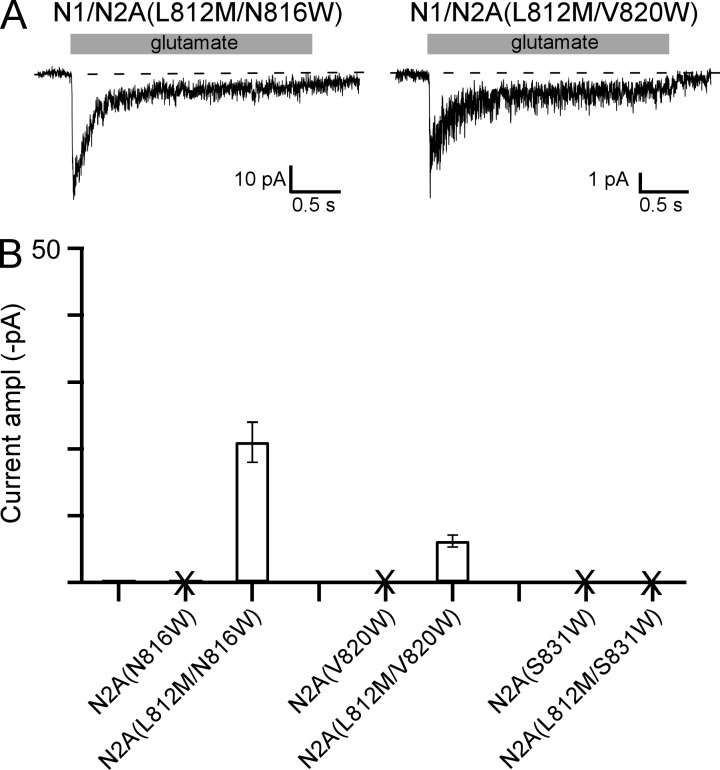
Tryptophan substitutions of three positions in the GluN2A subunit yielded receptors that showed no current (Fig. 3 B). Two of these positions, N816 and V820, are expressed on the membrane (Fig. 4 E) suggesting that they are “pore-dead”: they can assemble and traffic to the membrane, but agonists are unable to open the ion channel. Hence, the free energy generated by agonist binding is no longer able to overcome the stability of the closed conformation of the ion channel. To test this idea, we introduced in these backgrounds a missense mutation, GluN2A(L812M), that dramatically enhances receptor activation presumably by altering the energetics of the closed and open conformations (Yuan et al., 2014). Indeed, N816W and V820W, when combined with the L812M mutation, now showed detectable glutamate-activated currents (Fig. 6 A), though the amplitude remained significantly reduced relative to wild type (Fig. 6 B). In contrast, S831W, which does not express on the membrane (Fig. 4 F), still showed no detectable glutamate-activated current when combined with the L812M mutation, consistent with this position involved in biogenesis rather than gating.
Figure 6.
Recovery of function in “pore-dead” constructs. (A) Example whole-cell recordings, displayed as in Fig. 3 A, for NMDARs containing double mutations in the GluN2A subunit, L812M, and a tryptophan substituted at either N816W, V820W, or S831W. Receptors containing single tryptophan substitutions of N816, V820, or S831 showed no detectable glutamate-activated whole-cell currents (Fig. 3 B and Table 1). (B) Mean current amplitudes (±SEM) at −70 mV for single or double mutation constructs. Positions that did not show detectable glutamate-activated currents are demarcated by an “X.” Number of whole-cell current recordings for each construct: GluN1/GluN2A(N816W), 9; GluN1/GluN2A(L812M/N816W), 5; GluN1/GluN2A(V820W), 8; GluN1/GluN2A(L812M/V820W), 4; GluN1/GluN2A(S831W), 9; GluN1/GluN2A(L812M/S831W), 6.
The M4 segments in NMDAR subunits have strong effects on receptor desensitization
In the continual presence of glutamate, iGluRs can enter into a nonconducting, desensitized state. For non-NMDARs, the process of desensitization is fairly well understood being driven by rearrangements of the ligand-binding domain (LBD; Meyerson et al., 2016; Plested, 2016). In contrast to non-NMDARs, rearrangement of the LBD in NMDARs make only a limited contribution to desensitization (Borschel et al., 2011) and the energetics of domains in addition to the LBD strongly contribute to desensitization including the transmembrane domain (TMD; Krupp et al., 1998; Villarroel et al., 1998; Alsaloum et al., 2016). To directly contrast the contribution of the M4 segments to desensitization in iGluR subtypes, we characterized desensitization in these receptors, initially focusing on NMDARs containing tryptophan substitutions in the GluN1 or GluN2A M4 segment (Fig. 7).
Figure 7.
Impact of M4 substitutions on NMDAR desensitization. (A) Representative whole-cell recordings of current through wild-type GluN1/GluN2A (gray traces) or NMDARs containing a tryptophan substitution in the M4 segment (black traces). Currents were recorded as in Fig. 3. (B) Mean (±SEM) of the %Des or normalized weighted τ for wild-type GluN1/GluN2A or GluN1/GluN2A containing tryptophan substitutions in either the GluN1 (left) or GluN2A (right) M4 segments. Raw values and details of number of recordings made are shown in Table 5. Solid bars indicate values significantly different from wild type (P < 0.01, t test). We used a more stringent level of significance for this analysis to focus on only those positions with prominent effects on desensitization. Dots highlight positions homologous to the VVLGAVE face in AMPARs.
Wild-type GluN1/GluN2A (Fig. 7 A, gray traces) desensitize incompletely to ∼60% (62 ± 1%, n = 18) with a time course consisting of two components (weighted τ, 500 ± 20 ms). Tryptophan substitutions in the M4 segments of either the GluN1 or GluN2A subunit strongly altered receptor desensitization (Fig. 7 A and Table 5), increasing (e.g., GluN1(G815W)/GluN2A) or decreasing (GluN1/GluN2A(L830W)) the rate of desensitization as well as decreasing (e.g., GluN1(L830W)/GluN2A) or increasing (e.g., GluN1/GluN2A(M823W)) the extent of desensitization.
Table 5. Desensitization properties of wild-type GluN1/GluN2A or GluN1/GluN2A containing tryptophan substitutions in either GluN1 or GluN2A M4 segment.
| Tryptophans in GluN1 subunit | Tryptophans in Glu2A subunit | ||||||
|---|---|---|---|---|---|---|---|
| Construct | %des | τweighted | n | Construct | %des | τweighted | n |
| ms | ms | ||||||
| N1/N2A | 62 ± 1 | 500 ± 20 | 18 | 18 | |||
| F810W | 60 ± 3 | 510 ± 30 | 4 | I814W | 68 ± 3 | 290 ± 30 | 4 |
| E811W | 55 ± 2 | 300 ± 30 | 4 | D815W | 82 ± 3 | 230 ± 20* | 4 |
| N812W | 68 ± 2 | 88 ± 3* | 4 | N816W | – | – | |
| M813W | 77 ± 1^ | 220 ± 10* | 5 | M817W | 51 ± 1* | 990 ± 100^ | 4 |
| A814W | 64 ± 1 | 220 ± 25* | 4 | A818W | 39 ± 3 | 450 ± 70 | 4 |
| G815W | 74 ± 1^ | 48 ± 3* | 4 | G819W | 84 ± 2^ | 310 ± 40 | 5 |
| V816W | 67 ± 3 | 170 ± 20* | 4 | V820W | – | – | |
| F817W | 68 ± 2 | 220 ± 30* | 5 | F821W | 52 ± 3 | 420 ± 60 | 5 |
| M818W | 56 ± 2 | 200 ± 20* | 5 | Y822W | 48 ± 2 | 1,010 ± 145 | 4 |
| L819W | 63 ± 3 | 380 ± 60 | 4 | M823W | 93 ± 5^ | 350 ± 10 | 5 |
| V820W | 45 ± 3 | 210 ± 10* | 4 | L824W | 34 ± 4 | 1,350 ± 70^ | 4 |
| A821W | 53 ± 3 | 390 ± 50 | 4 | A825W | 77 ± 3 | 300 ± 20 | 4 |
| G822W | 68 ± 2 | 280 ± 20* | 4 | A826W | 74 ± 2 | 540 ± 30 | 5 |
| G823W | 80 ± 2^ | 310 ± 30 | 4 | A827W | 95 ± 1^ | 210 ± 10* | 5 |
| I824W | 60 ± 1 | 460 ± 30 | 5 | M828W | 64 ± 2 | 500 ± 30 | 5 |
| V825W | 47 ± 1* | 370 ± 80 | 4 | A829W | 44 ± 3 | 850 ± 160 | 5 |
| A826W | 65 ± 4 | 460 ± 20 | 4 | L830W | 52 ± 2 | 990 ± 40^ | 5 |
| G827W | 74 ± 3 | 150 ± 20* | 4 | S831W | – | – | |
| I828W | 45 ± 3 | 450 ± 40 | 4 | L832W | 54 ± 3 | 670 ± 50 | 5 |
| F829W | 59 ± 2 | 390 ± 20 | 5 | I833W | 59 ± 5 | 420 ± 30 | 4 |
| L830W | 40 ± 2* | 500 ± 70 | 4 | T834W | 55 ± 3 | 620 ± 60 | 4 |
| I831W | 63 ± 4 | 290 ± 25 | 4 | F835W | 53 ± 6 | 420 ± 50 | 4 |
| F832W | 49 ± 1* | 580 ± 40 | 5 | I836W | 50 ± 2 | 720 ± 90 | 5 |
| I833W | 67 ± 2 | 340 ± 40 | 4 | W837 | 62 ± 1 | 500 ± 20 | 18 |
| E834W | 65 ± 1 | 300 ± 20 | 6 | E838W | 68 ± 2 | 360 ± 30 | 5 |
Values shown are mean ± SEM. n indicates the number of whole-cell recordings. Tagged values are significantly less or slower (*) or greater or faster (^) than wild type (P < 0.01, t test).
Tryptophan substitutions of the M4 segments had strong effects in both the GluN1 (Fig. 7 B, left) and GluN2A (Fig. 7 B, right) subunits. There are several notable conclusions from these experiments. First, significant effects on desensitization occurred throughout both M4 segments, but such effects tended to be more limited at extreme intracellular positons, encompassing and C-terminal to G827 in GluN1 and S831 in GluN2A. Second, there is a strong subunit-specific difference in terms of the kinetics of desensitization: tryptophan substitutions in GluN1 consistently increase the rate of desensitization, whereas in GluN2A such substitutions preferentially decrease the rate of desensitization. Finally, substitutions at positions homologous to the AMPAR VVLGAVE face (Fig. 7 B, dots) consistently altered some aspect of NMDAR desensitization, suggesting that the interaction of M4 with the M3 segment impacts NMDAR desensitization.
The M4 segment in AMPARs has weak effects on receptor desensitization
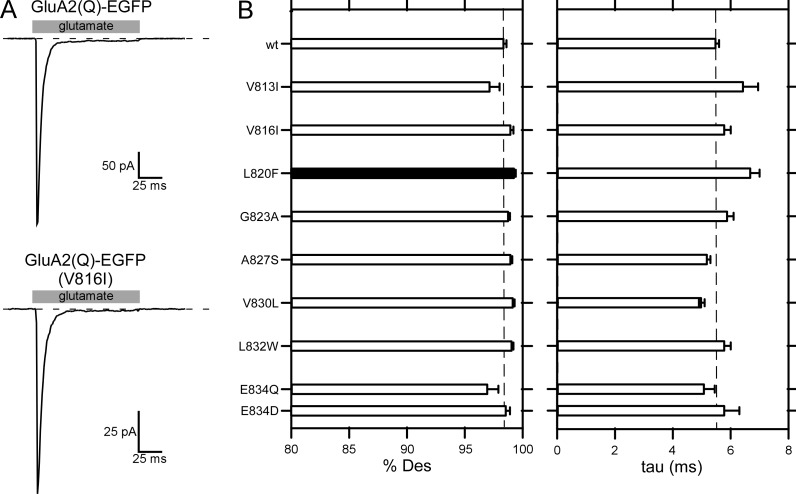
Many of the subtle substitutions of the AMPAR VVLGAVE face yielded tetramers (Fig. 2 C), and if they showed detectable tetramers, also showed membrane currents. We therefore characterized the effect of these subtle substitutions on AMPAR desensitization (Fig. 8). For these experiments, we used outside-out patches and fast agonist solution exchange. We did not test some constructs either because they showed extremely low whole-cell current amplitudes (e.g., L820A and E834N) or they had no effect on receptor assembly (e.g., V816A).
Figure 8.
Substitutions of the VVLGAVE face in GluA2 have no or only weak effects on receptor desensitization. (A) Example outside-out recordings from wild-type GluA2(Q)-EGFP (top trace) or V816I (bottom trace) in response to 100-ms pulses of glutamate. (B) Mean (±SEM) %Des (left) and rate of desensitization (right). Black bars indicate values significantly different from wild type (P < 0.05, t test). Many of the tested positions had strong effects on receptor tetramerization (Fig. 2). We also tested GluA2(L832W), a position on the side of the helix facing away from the pore domain. Number of outside-out patches for each construct: wt, 9; V813I, 4; V816I, 4; L820F, 5; G823A, 4; A827S, 4; V830L, 4; L832W, 4; E834Q, 4; E834D, 4; E834N, 3.
GluA2(Q)-EGFP showed rapid (5.5 ± 0.1 ms, n = 9) and strong (98.4 ± 0.1%Des) desensitization (Fig. 8, A and B) comparable with wild-type GluA2(Q) (Yelshansky et al., 2004). Receptors containing subtle substitutions of the VVLGAVE face, which often had dramatic effects on receptor assembly (Fig. 2), had no significant effect on desensitization with the exception of L820F, which showed a statistically significant but modest increase in %Des from ∼98.4% (wild type) to 99.1 ± 0.1%Des (Fig. 8 B). Thus, in contrast to what is observed for NMDARs (Fig. 7), the M4 segment in AMPARs, at least that component interacting with the M3 segment (VVLGAVE face), makes no notable contribution to receptor desensitization.
Discussion
In all iGluRs subtypes, an additional eukaryotic-specific transmembrane segment, the M4 segment, is associated with the pore domain of a neighboring subunit (Fig. 1). To test the functional role of the M4 segments to receptor function, we introduced tryptophans either in AMPAR (Salussolia et al., 2011, 2013) or NMDAR (present study) subunits. The M4 segments in AMPARs and NMDARs share similar sequences (Fig. 1 C) and structural arrangements (Figs. 1 and 3 C). Still, their functional roles are divergent. For AMPARs and based on tryptophan substitutions, the interaction along the entire extent of the M4 segment with the neighboring pore domain is required for receptor assembly. Even subtle mutations in the VVLGAVE face of AMPARs, except for two extreme extracellular positons, strongly disrupt tetramerization (Fig. 2). In contrast, and again based on tryptophan substitutions, the extracellular portion of the M4 segments in NMDAR subunits makes no obvious contribution to receptor assembly, but rather plays a dominant role in receptor gating, including generating “pore-dead” constructs—receptors that assemble and get to the cell surface efficiently but are completely unable to open their pore in response to agonist. Still, this distinction is not absolute: the M4 segment in AMPARs can impact receptor gating (Fig. 8; Terhag et al., 2010), and the extreme intracellular portion of the M4 in NMDARs can alter their surface expression (Fig. 4).
Impact of M4 segment on iGluR gating
In iGluRs, as well as other pore loop family members, the M3 transmembrane segment (homologous to TM2 or S6 in K+ channels) is the main pore-lining segment (Sobolevsky et al., 2009; Karakas and Furukawa, 2014; Lee et al., 2014). Surrounding the M3 segment are the outer transmembrane segments, M1 and M4. In NMDAR subunits, these outer transmembrane segment, M1 and M4, must be displaced for efficient pore opening to occur (Kazi et al., 2013) and contain missense mutations that alter receptor gating (Yuan et al., 2014; Chen et al., 2017; Ogden et al., 2017).
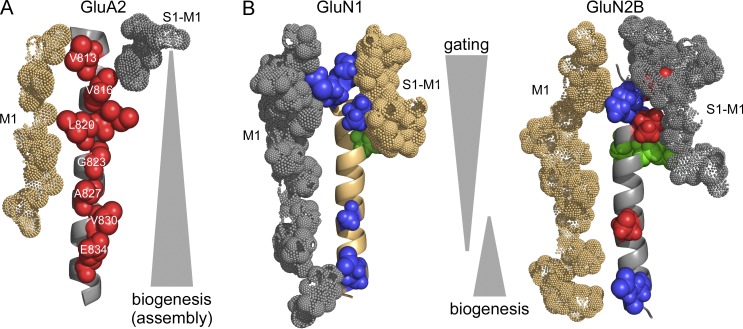
The GluN1 and GluN2A M4 segments, at least in the closed state, interact extensively with the M3 segments of GluN2A and GluN1, respectively (Figs. 1 and 3 C). In addition, the M4 segments interact with the respective adjacent subunits’ M1 segments (Figs. 1 B and 9). Notable is the strong interaction at the extracellular end of each M4 segment with the S1-M1 of the same subunit (Fig. 9 B), an interaction that is much less extensive in AMPARs (Fig. 9 A). This interaction may be part of the critical difference between AMPAR and NMDAR gating, as the S1-M1 of the same subunit most closely interacts with pore-dead residues identified in the tryptophan scan.
Figure 9.
Differential roles of M4 segments in AMPAR and NMDAR function. (A and B) The M4 segments, viewed from the center of the pore, in either AMPARs (A) or NMDARs (B) either GluN1 M4 (left) or GluN2B M4 (right). The M3 segment from the adjacent subunit (see Fig. 3 C) is hidden to emphasis other M4 interactions, including the S1-M1 LBD-TMD linker from the same subunit and the M1 transmembrane segment from the neighboring subunit. Positions that showed significant changes in current amplitudes are highlighted in blue (reduced), green (greater), or red (no detectable current). For AMPARs, only those positions that showed no detectable current (Salussolia et al., 2011) are indicated, which include the VVLGAVE positions as well as I819. Subunits are colored light orange (GluA2 A and C, GluN1) and gray 60% (GluA2 B and D, GluN2B). The M4 is represented as a cartoon, whereas the S1-M1 and the adjacent M1 are shown as dots.
A surprising feature is that tryptophan substitutions at extracellular positions, rather than destabilizing the closed state (i.e., reducing the MCT), as one might expect from disrupting transmembrane interactions observed in our closed state model, actually increased the MCT and at the same time also consistently decreased the MOT (Fig. 5 and Table 3). The lack of an available open state NMDAR structure makes it difficult to identify or hypothesize why this observation may occur. Further study will be needed to reveal how shifting interactions during channel opening facilitate efficient gating.
Differential contribution of the M4 segments to iGluR desensitization
For NMDARs, receptors containing tryptophan substitutions at 12 VVLGAVE face positions, 7 in GluN1 and 5 in GluN2A, showed whole-cell currents. Of these positions that showed current, 9 showed some form of altered desensitization, either a change in the extent and/or rate of desensitization (Fig. 7). Those that did not show any significant change, GluN1(L830W), GluN1(E834W), and GluN2A(E838W), are all located at the extreme intracellular end of the M4 segments, emphasizing that this region only has weak effects on NMDAR gating. In contrast, for AMPAR desensitization, substitutions at VVLGAVE positions had no effect on desensitization except for one L820 (VVLGAVE; Fig. 8), whereas these same substitutions had strong effects on receptor assembly (Fig. 2). These experiments highlight the differential roles of the M4 segments in iGluRs. In addition, these results also support the idea that desensitization of AMPARs and NMDARs are controlled by different processes, likely in different domains. For AMPARs, it is mainly regulated by the conformation of the LBD (Sun et al., 2002; Carbone and Plested, 2012), whereas for NMDARs, the TMD makes a more significant contribution (Krupp et al., 1998; Villarroel et al., 1998; Alsaloum et al., 2016; present study). Nevertheless, the structural basis for desensitization in NMDARs remains poorly defined.
For NMDARs, the two different subunits, GluN1 and GluN2A, also take on divergent roles. The GluN1 M4 seems to be highly important in regulating the rate of desensitization with 10 out 25 tryptophan substitutions drastically increasing the rate of desensitization and none decreasing the rate. Of these, nearly all fall within a tight seven-residue portion of extracellular region from N812 to M818 (Fig. 7 B). Interestingly, this region of the GluN1 M4 interacts strongly with a phenylalanine (F) previously identified to impact NMDAR desensitization (Alsaloum et al., 2016). In contrast, the interaction between similar regions in GluN2A is less notable; also, tryptophan substitutions in the GluN2A M4 often slowed the rate of desensitization. Still, the mechanistic and structural basis for the contribution of the GluN1 and GluN2A M4 segments to desensitization remains unknown.
Role of the M4 segments to receptor assembly
Except for the two valines located at the extreme extracellular end of M4, subtle substitutions in the VVLGAVE face in AMPARs can have profound effects on receptor tetramerization (Fig. 2 C), highlighting the important role of the M4 segment to AMPAR oligomerization. The exact mechanistic role of the AMPAR M4 segment in assembly however remains unclear because substitutions with both larger-sized as well as smaller-sized side chains disrupt tetramerization. Hence, factors in addition to steric hindrances between side chains such as van der Waals interactions may contribute to the role of the AMPAR M4 segment in receptor assembly.
Subtle substitutions at the two extreme extracellular positions in the AMPAR M4 segment, V813 and V816, had no significant effect on receptor oligomerization (Fig. 2 C) or receptor desensitization (Fig. 8). Interestingly, this extreme extracellular end corresponds to those positions in the GluN1 and GluN2A M4 segments that had extremely strong effects on receptor gating (Fig. 5) and also altered receptor desensitization (Fig. 7).
For NMDARs, the M4 segment plays an uneven role in receptor biogenesis. In contrast to AMPARs, the extracellular M4 of GluN1 and GluN2A makes no obvious contribution to receptor biogenesis. In addition, only two of the more extreme intracellular positions, GluN2A(S831) and the negative charges, GluN1(E834) and GluN2A(E838), contribute in some fashion to biogenesis, either receptor assembly or forward trafficking to the membrane. We were unable to test any contribution to receptor assembly using BN-PAGE or FSEC because the mammalian GluN1/GluN2A NMDAR has a strong tendency to dissociate into a dimer in detergents, making them difficult to be analyzed precisely.
Biology of the impact of the M4 segment on receptor gating
In iGluRs, the M4 segment is the most peripheral transmembrane segment, interacting extensively with phospholipids, and is attached to the intracellular C-terminal domain (CTD). NMDAR function is strongly modulated by lipids (Casado and Ascher, 1998; Korinek et al., 2015). Further, posttranslational modification of the CTDs in NMDAR (Salter and Kalia, 2004; Tu et al., 2010) and AMPAR (Lu and Roche, 2012) can alter their gating properties. How these factors mediate changes in receptor gating is unknown. One possibility is that the M4 segments act as transduction pathways for these effects, which may occur for non-NMDARs (Wilding et al., 2016). The lipid composition in contact with the M4 segment, for example, might alter the positioning or orientation of the M4 segments, which in turn alters how they interact with the pore domain, thereby impacting gating. The posttranslational status of the CTD might similarly impact the M4 positioning and orientation. Still, the importance of the M4 segment to these key biological pathways remains to be elucidated.
Supplementary Material
Acknowledgments
We thank Drs. Quan Gan and Kasper Hansen for helpful discussions and/or comments on the manuscript. We also thank Drs. Mark Aurousseau and Derek Bowie for their generous gift of pHmystik.
This work was supported by National Institutes of Health RO1 grants NS088479 (to L.P. Wollmuth), including a minority supplement (to J.B. Amin), MH081923 (to M.E. Bowen), NS093753 (to M.C. Regan), MH085926 and GM105730 (to H. Furukawa), and GM118091 (to H.-X. Zhou).
The authors declare no competing financial interests.
Author contributions: J.B. Amin and L.P. Wollmuth designed experiments, executed experiments, analyzed data, and wrote the paper. C.L. Salussolia and M.E. Bowen designed experiments, executed experiments, and wrote the paper. K. Chan and M.C. Regan executed experiments, analyzed data, and wrote the paper. J. Dai designed experiments, executed experiments, and analyzed data. H.-X. Zhou designed experiments and wrote the paper. H. Furukawa designed experiments and analyzed data.
Angus C. Nairn served as editor.
Footnotes
Abbreviations used:
- AMPAR
- AMPA receptor
- CTD
- C-terminal domain
- FSEC
- fluorescence-detection size exclusion chromatography
- iGluR
- ionotropic glutamate receptor
- LBD
- ligand-binding domain
- MCT
- mean closed time
- MOT
- mean open time
- NMDAR
- NMDA receptor
- TMD
- transmembrane domain
References
- Alsaloum, M., Kazi R., Gan Q., Amin J., and Wollmuth L.P.. 2016. A molecular determinant of subtype-specific desensitization in ionotropic glutamate receptors. J. Neurosci. 36:2617–2622. 10.1523/JNEUROSCI.2667-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurousseau, M. 2015. Novel fluorescence-based methods to study the stoichiometric and surface expression properties of ionotropic glutamate receptors. McGill University. Available at: http://digitool.Library.McGill.CA:80/R/-?func=dbin-jump-full&object_id=135326&silo_library=GEN01
- Borschel, W.F., Murthy S.E., Kasperek E.M., and Popescu G.K.. 2011. NMDA receptor activation requires remodelling of intersubunit contacts within ligand-binding heterodimers. Nat. Commun. 2:498. 10.1038/ncomms1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J.Y., Qiu S., Zhang J., Wang J.J., Zhang X.M., and Luo J.H.. 2011. Transmembrane region of N-methyl-d-aspartate receptor (NMDAR) subunit is required for receptor subunit assembly. J. Biol. Chem. 286:27698–27705. 10.1074/jbc.M111.235333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone, A.L., and Plested A.J.. 2012. Coupled control of desensitization and gating by the ligand binding domain of glutamate receptors. Neuron. 74:845–857. 10.1016/j.neuron.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Casado, M., and Ascher P.. 1998. Opposite modulation of NMDA receptors by lysophospholipids and arachidonic acid: common features with mechanosensitivity. J. Physiol. 513:317–330. 10.1111/j.1469-7793.1998.317bb.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.R., and Kuo C.C.. 2008. The activation gate and gating mechanism of the NMDA receptor. J. Neurosci. 28:1546–1556. 10.1523/JNEUROSCI.3485-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Dürr K.L., and Gouaux E.. 2014. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science. 345:1021–1026. 10.1126/science.1258409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Tankovic A., Burger P.B., Kusumoto H., Traynelis S.F., and Yuan H.. 2017. Functional evaluation of a de novo GRIN2A mutation identified in a patient with profound global developmental delay and refractory epilepsy. Mol. Pharmacol. 91:317–330. 10.1124/mol.116.106781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Q., Salussolia C.L., and Wollmuth L.P.. 2015. Assembly of AMPA receptors: mechanisms and regulation. J. Physiol. 593:39–48. 10.1113/jphysiol.2014.273755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Q., Dai J., Zhou H.X., and Wollmuth L.P.. 2016. The transmembrane domain mediates tetramerization of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J. Biol. Chem. 291:6595–6606. 10.1074/jbc.M115.686246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow, N.G., Siegler Retchless B., and Johnson J.W.. 2015. Molecular bases of NMDA receptor subtype-dependent properties. J. Physiol. 593:83–95. 10.1113/jphysiol.2014.273763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger, I.H., Khatri L., Kong X., and Ziff E.B.. 2003. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 40:763–774. 10.1016/S0896-6273(03)00668-8 [DOI] [PubMed] [Google Scholar]
- Hamdan, F.F., Srour M., Capo-Chichi J.M., Daoud H., Nassif C., Patry L., Massicotte C., Ambalavanan A., Spiegelman D., Diallo O., et al. 2014. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 10:e1004772. 10.1371/journal.pgen.1004772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham, G.E., and Do K.Q.. 2016. Linking early-life NMDAR hypofunction and oxidative stress in schizophrenia pathogenesis. Nat. Rev. Neurosci. 17:125–134. 10.1038/nrn.2015.19 [DOI] [PubMed] [Google Scholar]
- Honse, Y., Ren H., Lipsky R.H., and Peoples R.W.. 2004. Sites in the fourth membrane-associated domain regulate alcohol sensitivity of the NMDA receptor. Neuropharmacology. 46:647–654. 10.1016/j.neuropharm.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Huettner, J.E. 2015. Glutamate receptor pores. J. Physiol. 593:49–59. 10.1113/jphysiol.2014.272724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.S., VanDongen H.M., and VanDongen A.M.. 2002. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J. Neurosci. 22:2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas, E., and Furukawa H.. 2014. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 344:992–997. 10.1126/science.1251915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate, T., and Gouaux E.. 2006. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 14:673–681. 10.1016/j.str.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Kazi, R., Gan Q., Talukder I., Markowitz M., Salussolia C.L., and Wollmuth L.P.. 2013. Asynchronous movements prior to pore opening in NMDA receptors. J. Neurosci. 33:12052–12066. 10.1523/JNEUROSCI.5780-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek, M., Vyklicky V., Borovska J., Lichnerova K., Kaniakova M., Krausova B., Krusek J., Balik A., Smejkalova T., Horak M., and Vyklicky L.. 2015. Cholesterol modulates open probability and desensitization of NMDA receptors. J. Physiol. 593:2279–2293. 10.1113/jphysiol.2014.288209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp, J.J., Vissel B., Heinemann S.F., and Westbrook G.L.. 1998. N-terminal domains in the NR2 subunit control desensitization of NMDA receptors. Neuron. 20:317–327. 10.1016/S0896-6273(00)80459-6 [DOI] [PubMed] [Google Scholar]
- Lee, C.H., Lü W., Michel J.C., Goehring A., Du J., Song X., and Gouaux E.. 2014. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 511:191–197. 10.1038/nature13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke, J.R., Lal D., Reinthaler E.M., Steiner I., Nothnagel M., Alber M., Geider K., Laube B., Schwake M., Finsterwalder K., et al. 2013. Mutations in GRIN2A cause idiopathic focal epilepsy with rolandic spikes. Nat. Genet. 45:1067–1072. 10.1038/ng.2728 [DOI] [PubMed] [Google Scholar]
- Lu, W., and Roche K.W.. 2012. Posttranslational regulation of AMPA receptor trafficking and function. Curr. Opin. Neurobiol. 22:470–479. 10.1016/j.conb.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell, A.D. Jr., Feig M., and Brooks C.L. III. 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415. 10.1002/jcc.20065 [DOI] [PubMed] [Google Scholar]
- Mayer, M.L. 2016. Structural biology of glutamate receptor ion channel complexes. Curr. Opin. Struct. Biol. 41:119–127. 10.1016/j.sbi.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Meddows, E., Le Bourdelles B., Grimwood S., Wafford K., Sandhu S., Whiting P., and McIlhinney R.A.. 2001. Identification of molecular determinants that are important in the assembly of N-methyl-d-aspartate receptors. J. Biol. Chem. 276:18795–18803. 10.1074/jbc.M101382200 [DOI] [PubMed] [Google Scholar]
- Meyerson, J.R., Chittori S., Merk A., Rao P., Han T.H., Serpe M., Mayer M.L., and Subramaniam S.. 2016. Structural basis of kainate subtype glutamate receptor desensitization. Nature. 537:567–571. 10.1038/nature19352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, D.T., Berger B.W., and DeGrado W.F.. 2008. Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 16:991–1001. 10.1016/j.str.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden, K.K., Chen W., Swanger S.A., McDaniel M.J., Fan L.Z., Hu C., Tankovic A., Kusumoto H., Kosobucki G.J., Schulien A.J., et al. 2017. Molecular mechanism of disease-associated mutations in the pre-M1 helix of NMDA receptors and potential rescue pharmacology. PLoS Genet. 13:e1006536. 10.1371/journal.pgen.1006536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., and Schulten K.. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested, A.J. 2016. Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat. Struct. Mol. Biol. 23:494–502. 10.1038/nsmb.3214 [DOI] [PubMed] [Google Scholar]
- Popescu, G., and Auerbach A.. 2003. Modal gating of NMDA receptors and the shape of their synaptic response. Nat. Neurosci. 6:476–483. [DOI] [PubMed] [Google Scholar]
- Qin, F. 2004. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys. J. 86:1488–1501. 10.1016/S0006-3495(04)74217-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, H., Zhao Y., Dwyer D.S., and Peoples R.W.. 2012. Interactions among positions in the third and fourth membrane-associated domains at the intersubunit interface of the N-methyl-d-aspartate receptor forming sites of alcohol action. J. Biol. Chem. 287:27302–27312. 10.1074/jbc.M111.338921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šali, A., and Blundell T.L.. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Salter, M.W., and Kalia L.V.. 2004. Src kinases: a hub for NMDA receptor regulation. Nat. Rev. Neurosci. 5:317–328. 10.1038/nrn1368 [DOI] [PubMed] [Google Scholar]
- Salussolia, C.L., Corrales A., Talukder I., Kazi R., Akgul G., Bowen M., and Wollmuth L.P.. 2011. Interaction of the M4 segment with other transmembrane segments is required for surface expression of mammalian α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J. Biol. Chem. 286:40205–40218. 10.1074/jbc.M111.268839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia, C.L., Gan Q., Kazi R., Singh P., Allopenna J., Furukawa H., and Wollmuth L.P.. 2013. A eukaryotic specific transmembrane segment is required for tetramerization in AMPA receptors. J. Neurosci. 33:9840–9845. 10.1523/JNEUROSCI.2626-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia, C.L., Gan Q., and Wollmuth L.P.. 2015. Assaying AMPA receptor oligomerization. In Ionotropic Glutamate Receptor Technologies. Popescu G.K., editor. Humana Press, New York. 3–14. [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorge, S., and Colquhoun D.. 2003. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J. Neurosci. 23:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky, A.I., Beck C., and Wollmuth L.P.. 2002. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 33:75–85. 10.1016/S0896-6273(01)00560-8 [DOI] [PubMed] [Google Scholar]
- Sobolevsky, A.I., Rosconi M.P., and Gouaux E.. 2009. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 462:745–756. 10.1038/nature08624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Llavina, G.J., Chang T.H., and Swartz K.J.. 2006. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker Kv channel. Neuron. 52:623–634. 10.1016/j.neuron.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Sun, Y., Olson R., Horning M., Armstrong N., Mayer M., and Gouaux E.. 2002. Mechanism of glutamate receptor desensitization. Nature. 417:245–253. 10.1038/417245a [DOI] [PubMed] [Google Scholar]
- Talukder, I., and Wollmuth L.P.. 2011. Local constraints in either the GluN1 or GluN2 subunit equally impair NMDA receptor pore opening. J. Gen. Physiol. 138:179–194. 10.1085/jgp.201110623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder, I., Borker P., and Wollmuth L.P.. 2010. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J. Neurosci. 30:11792–11804. 10.1523/JNEUROSCI.5382-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhag, J., Gottschling K., and Hollmann M.. 2010. The transmembrane domain C of AMPA receptors is critically involved in receptor function and modulation. Front. Mol. Neurosci. 3:117. 10.3389/fnmol.2010.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis, S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., and Dingledine R.. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62:405–496. 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, W., Xu X., Peng L., Zhong X., Zhang W., Soundarapandian M.M., Balel C., Wang M., Jia N., Zhang W., et al. 2010. DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell. 140:222–234. 10.1016/j.cell.2009.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroel, A., Regalado M.P., and Lerma J.. 1998. Glycine-independent NMDA receptor desensitization: localization of structural determinants. Neuron. 20:329–339. 10.1016/S0896-6273(00)80460-2 [DOI] [PubMed] [Google Scholar]
- Wilding, T.J., Lopez M.N., and Huettner J.E.. 2016. Chimeric glutamate receptor subunits reveal the transmembrane domain is sufficient for NMDA receptor pore properties but some positive allosteric modulators require additional domains. J. Neurosci. 36:8815–8825. 10.1523/JNEUROSCI.0345-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshansky, M.V., Sobolevsky A.I., Jatzke C., and Wollmuth L.P.. 2004. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J. Neurosci. 24:4728–4736. 10.1523/JNEUROSCI.0757-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H., Hansen K.B., Zhang J., Pierson T.M., Markello T.C., Fajardo K.V., Holloman C.M., Golas G., Adams D.R., Boerkoel C.F., et al. 2014. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat. Commun. 5:3251. 10.1038/ncomms4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H., Low C.M., Moody O.A., Jenkins A., and Traynelis S.F.. 2015. Ionotropic GABA and glutamate receptor mutations and human neurologic diseases. Mol. Pharmacol. 88:203–217. 10.1124/mol.115.097998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.