Abstract
Background
The mesenchymal-epithelial transition factor (MET) receptor can be overexpressed in solid tumors, including small cell lung cancer (SCLC). However, the molecular mechanism regulating MET stability and turnover in SCLC remains undefined. One potential mechanism of MET regulation involves the C-terminus of Hsp70-interacting protein (CHIP), which targets heat shock protein 90-interacting proteins for ubiquitination and proteasomal degradation. In the present study, we investigated the functional effects of CHIP expression on MET regulation and the control of SCLC cell apoptosis and invasion.
Methods
To evaluate the expression of CHIP and c-Met, which is a protein that in humans is encoded by the MET gene (the MET proto-oncogene), we examined the expression pattern of c-Met and CHIP in SCLC cell lines by western blotting. To investigate whether CHIP overexpression reduced cell proliferation and invasive activity in SCLC cell lines, we transfected cells with CHIP and performed a cell viability assay and cellular apoptosis assays.
Results
We found an inverse relationship between the expression of CHIP and MET in SCLC cell lines (n=5). CHIP destabilized the endogenous MET receptor in SCLC cell lines, indicating an essential role for CHIP in the regulation of MET degradation. In addition, CHIP inhibited MET-dependent pathways, and invasion, cell growth, and apoptosis were reduced by CHIP overexpression in SCLC cell lines.
Conclusion
CHIP is capable of regulating SCLC cell apoptosis and invasion by inhibiting MET-mediated cytoskeletal and cell survival pathways in NCI-H69 cells. CHIP suppresses MET-dependent signaling, and regulates MET-mediated SCLC motility.
Keywords: Lung neoplasms, Small cell lung carcinoma, C-terminus of Hsp70-interacting protein, Mesenchymal-epithelial transition factor
Introduction
Lung cancer is one of the most common fatal malignancies in the developed world [1]. There are 2 main histological types of lung cancer: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [2]. SCLC is different from NSCLC in that SCLC grows more rapidly, spreads more easily, and is more responsive to chemotherapy and radiotherapy [2]. Although SCLC responds well to chemotherapy and radiation therapy, the long-term outcome of this disease remains unsatisfactory due to early recurrence, chemoradioresistance, and metastatic disease [2].
Mesenchymal-epithelial transition factor (MET) is a protein that is encoded by the MET proto-oncogene, and is also known as c-Met and hepatocyte growth factor (HGF) receptor [3,4]. Aberrantly active MET triggers angiogenesis, tumor growth, and metastasis [5,6]. Ma et al. [5] analyzed c-Met mutations in SCLC, including novel juxtamembrane domain mutations regulating cytoskeletal functions. Moreover, Maulik et al. [6] reported that the c-Met/HGF pathway is functional in SCLC, and may represent a useful target for novel therapies. Recent data indicate that the proteasomal degradation of MET occurs following acute HGF-induced MET endocytosis through binding to ubiquitin ligases [7].
The C-terminus of Hsp70-interacting protein (CHIP) is a well-described U-box-type E3 ubiquitin ligase that induces the ubiquitination and proteasomal degradation of its substrates, which include glucocorticoid receptors, c-Raf kinase, ErbB, and other oncogenic proteins [2,8–10]. The upregulation of CHIP has been found to inhibit tumor growth and metastasis, and its levels were negatively correlated with malignancy in human breast and gastric cancer [11]. However, the exact mechanism of the antitumor effect of CHIP on SCLC has not been identified. In this study, we identified CHIP as a physiologically negative regulator of the MET receptor that functions through a mechanism involving CHIP-mediated ubiquitination and proteasomal degradation. We also aimed to examine the role of CHIP in the regulation of MET, which may be a therapeutic target for SCLC.
Methods
1) Reagents and antibodies
The following antibodies and reagents were used in this study: anti-CHIP (SC-33264), protein kinase B (Akt) 1/2 (sc-1619), extracellular signal-regulated kinase (ERK) 1/2 (sc-94), pERK1/2 (sc-7383), focal adhesion kinase (FAK, sc-1688), pFAK (sc-16663), and green fluorescent protein (GFP, sc-9996) antibodies (sc-1055; Santa Cruz Biotechnology, Santa Cruz, CA, USA); protein A/G plus agarose beads (sc-2003, Santa Cruz Biotechnology); MG132 (c2211; Sigma, Indianapolis, IN, USA); anti-Myc (2278), pAkt1/2 (4060), p-paxillin (2541), paxillin (2542), and ubiquitin antibodies (3936; Cell Signaling, Boston, MA, USA); 10× lysis buffer (9803, Cell Signaling); horseradish peroxidase-conjugated secondary antibodies and SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL, USA); anti-Met antibody (ab51067; Abcam, Cambridge, UK); the Bradford Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA, USA); fetal bovine serum (FBS; HyClone Laboratories, Logan, UT, USA); and the Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA, USA).
2) Cell lines and transient transfection
SCLC cell lines (NCI-H69, NCI-H82, NCI-H209, NCI-H345, and NCI-H526) were purchased from the America Type Culture Collection (Rockville, MD, USA). Cell lines were maintained in RPMI 1640 (Hyclone Laboratories, Logan, UT, USA) supplemented with 10% FBS (Hyclone Laboratories), 100 U/mL of penicillin, 100 μg/mL of streptomycin, and 2 mM glutamine in a humidified atmosphere of 5% CO2 and 95% air at 37°C. For transfection, cells were seeded and grown to a confluence of 80%, after which they were transfected using the Lipofectamine Plus reagent (Invitrogen) according to the manufacturer’s guidelines. When indicated, transfected cells were treated with MG132 or an equivalent amount of vehicle (dimethy lsulfoxide) for the indicated time periods.
3) Plasmid construction and small interfering RNA constructs
DNA fragments encoding full-length CHIP were amplified by polymerase chain reaction (PCR) and subcloned into pcDNA3 (Invitrogen) and pAcGFP1-N1 (Clontech, Mountain View, CA, USA). Truncated forms of CHIP (tetratricopeptide repeat [TPR] deletion and U-box deletion) were amplified by PCR and subcloned into pAcGFP1-N1 (Clontech). Similarly, MET was cloned into pcDNA3.1/myc-His A (Invitrogen). For knockdown studies, lung cancer cell lines were transfected with either a CHIP small interfering RNA (siRNA, sc-43555) or negative control siRNA (sc-37007, Santa Cruz Biotechnology).
4) Cell lysis, immunoprecipitation, and immunoblotting
Cells were lysed in a buffer (Cell Signaling) containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mg/mL of leupeptin, and 1 mM phenylmethane sulfonyl fluoride. Protein concentrations were determined using a Bradford assay kit (Bio-Rad Laboratories). Lysates (50 μg per lane) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride membranes, and probed with the indicated antibodies. All primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies and visualized using enhanced SuperSignal West Pico chemiluminescent substrate. The band density was quantified by using the Bio-Rad Gel Documentation System. Immunoprecipitation experiments were performed by the addition of 15 μL of protein A/G agarose to cell lysates at 4°C for 1 hour on a rotating platform. After several washes, the bound proteins were subjected to SDS-PAGE and detected by immunoblotting.
5) Cell viability assay
The Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) was used to evaluate cell viability. CCK-8 is a sensitive, nonradioactive colorimetric analysis that can be used to detect the number of living cells in order to measure cell proliferation. For the assay, the CCK-8 reagent was added directly to the culture medium. The detailed procedure for measuring cell viability was as follows. First, cells were transfected with pcDNA-CHIP or a vector and then incubated for 48 hours, after which 10 μL of CCK-8 solution was added and incubated at 37°C for 4 hours. Finally, the absorbance at 450 nm was measured using a microplate reader.
6) Cellular apoptosis assay
An annexin-V-fluorescein isothiocyanate apoptosis detection kit (Trevigen, Gaithersburg, MD, USA) was used to assess the binding of annexin-V to phosphatidylserine on the outside of cells, which is an indicator of apoptosis. To study the effects of CHIP overexpression on apoptosis, NCI-H69 cells were first transfected with CHIP for 48 hours. Next, the cells were washed twice with pre-chilled phosphate-buffered saline (4°C) and harvested. Approximately 1×105 cells were resuspended in a solution made up of 89 μL of binding buffer, 1 μL of annexin-V, and 10 μL of propidium iodide, and then incubated in the dark for 15 minutes. The cell suspension was then centrifuged at 5,000 rpm for 5 minutes, and the resulting cell pellet was resuspended with 400 μL of binding buffer and subsequently analyzed by flow cytometry.
7) Invasion assay
The invasive potential of NCI-H69 cells was tested using Matrigel invasion chambers (24-well format, 8-μm pore size; BD Biosciences, San Jose, CA, USA) as described previously. Briefly, 150,000 cells suspended in RPMI 1640 containing 1% FBS were transferred to the insert chamber. In the lower chamber, RPMI 1640 containing 10% FBS served as the source of chemoattractant. After 24 hours, the cells on the upper surface of the filter were wiped away with a cotton swab. The cells on the lower surface of the filters were then fixed with methanol for 10 minutes, incubated with Giemsa stain for 3 hours, and counted. For each replication, cells in 3 randomly selected fields were counted. The invading cells were quantified by dissolving the stained cells in 10% acetic acid (100–200 μL/well) followed by transferring a consistent amount of the dye-solute mixture to a 96-well plate for colorimetric optical density readings at 560 nm.
8) Statistical analysis
Statistical significance was tested by the 2-tailed Student t-test, and p-values<0.05 were considered to indicate statistical significance.
Results
1) Inverse expression of MET and CHIP in SCLC cells
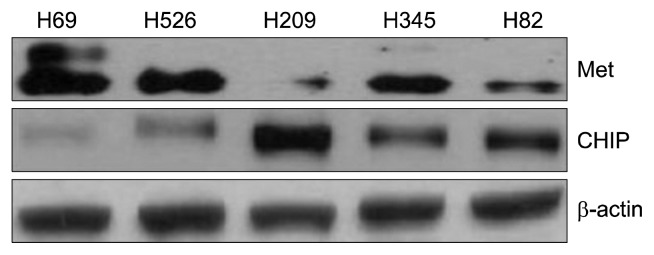
To evaluate the expression of CHIP and MET in this study, we examined the pattern of expression of MET and CHIP in SCLC cell lines using whole-cell lysates and western blotting. The level of actin expression in each sample served as a loading control. As shown in Fig. 1, MET expression was strongly detected in the NCI-H69 SCLC cell line. In contrast, moderate expression of MET was observed in the NCI-H526 and NCI-H345 cell lines, while the levels of MET in the NCI-H209 and NCI-H82 SCLC cell lines were low. Significant CHIP expression levels were detected in the NCI-H209 cell line, but weakly detected in the NCI-H69 cell line. Inversely, the NCI-H69 cell line had a high expression of MET, but low CHIP expression (Fig. 1).
Fig. 1.
MET and CHIP expression patterns in SCLC cell lines. MET (top panel) and CHIP (middle panel) expression patterns were examined using standard immunoblotting of whole cell lysates from the indicated lung cancer cell lines cultured under serum-containing conditions (10% fetal bovine serum): NCI-H69, NCI-H526, NCI-H209, NCI-H345, and NCI-H82. β-actin was included as a loading control (bottom panel). MET, mesenchymal-epithelial transition factor; CHIP, C-terminus of Hsp70-interacting protein; SCLC, small cell lung cancer.
2) CHIP inhibits invasion by downregulating MET-induced signaling proteins
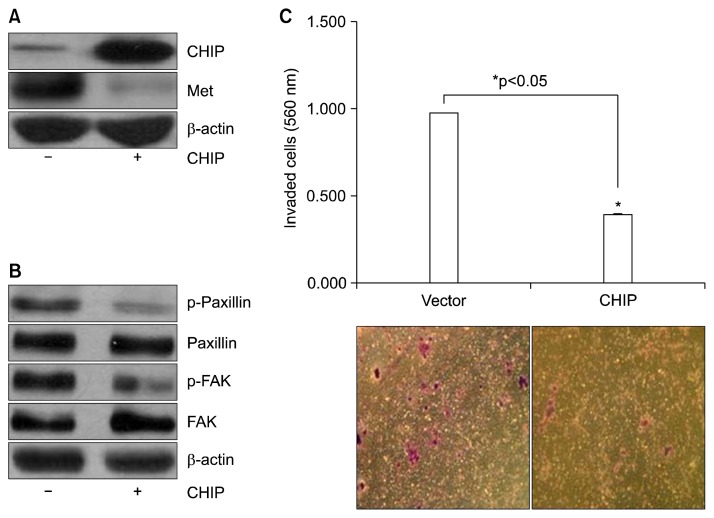
We asked whether CHIP overexpression would inhibit MET expression using transient transfection of pcDNA3-CHIP (+) and pcDNA3 (−) constructs in NCI-H69 cells. Consistent with our observations thus far, CHIP overexpression reduced MET expression levels (Fig. 2A). Using the cytoskeletal molecules paxillin and FAK, we next asked whether activation of paxillin and FAK was lower in NCI-H69 cells over-expressing CHIP. As shown in Fig. 2B, CHIP overexpression led to decreased phosphorylation of both paxillin and FAK. We also investigated the invasiveness of these cells, and found that CHIP overexpression significantly decreased the number of cells that were able to penetrate the Matrigel-coated membrane (Fig. 2C).
Fig. 2.
Effects of CHIP on the reduction of invasion and MET expression. (A) NCI-H69 cells were transfected with a pcDNA3 vector control (−) and pcDNA3-CHIP (+) for 48 hours and analyzed by immunoblotting using CHIP and MET antibodies. β-actin was used as a loading control. (B) NCI-H69 cell lysates were immunoblotted with anti-phospho-paxillin, anti-paxillin, phospho-FAK, and FAK antibodies. β-actin was used as a loading control. (C) CHIP-overexpressing and vector-transfected cells were evaluated for their ability to pass through membranes with 8-μm pores covered with Matrigel and gelatin (a mixture of proteins from basal membranes) to a lower chamber without fetal bovine serum. The invading cells were detected by colorimetric optical density readings at 560 nm and observed under a microscope. *p<0.05, statistically significant difference between the pcDNA3 vector-transfected and pcNDA3-CHIP-transfected cells. MET, mesenchymal-epithelial transition factor; CHIP, C-terminus of Hsp70-interacting protein; FAK, focal adhesion kinase.
3) CHIP induces apoptosis and inhibits cell growth
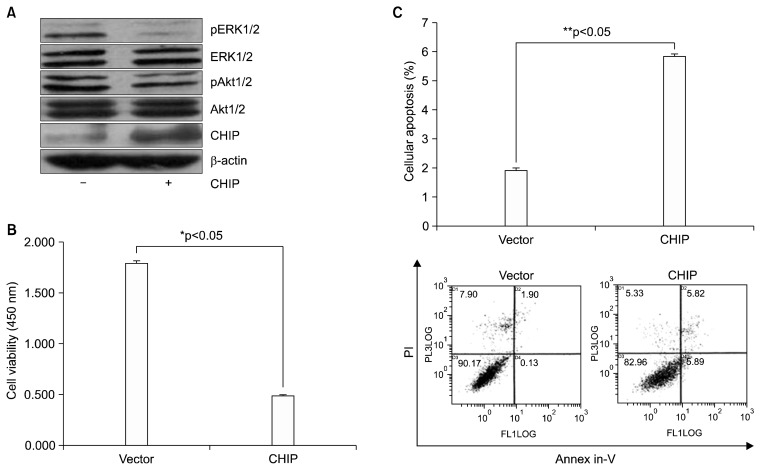
We next examined the effects of CHIP overexpression in NCI-H69 cells on the activation status of the phosphorylation of MET at tyrosine Y1349 downstream signaling pathways including Akt1/2 and ERK1/2. CHIP efficiently blocked the activation of both ERK1/2 and Akt1/2 (Fig. 3A). We also determined the effects of CHIP on the growth and apoptosis of NCI-H69 cells. As shown in Fig. 3B, the viability of NCI-H69 cells decreased following transfection with pcDNA3-CHIP for 48 hours compared to cells transfected with the pcDNA3 vector. To test the effect of CHIP on apoptosis, transfected cells were analyzed using annexin-V and propidium iodide staining and flow cytometry. Transfection of 5 μg of CHIP increased the level of apoptosis in NCI-H69 cell 3.02-fold compared to vector-transfected cells (5.83%±0.066% versus 1.93%±0.06%, p<0.0001) (Fig. 3C).
Fig. 3.
MET inhibition with CHIP overexpression in NCI-H69 lung cancer cells: the induction of apoptosis in vitro and inactivation of Akt and ERK signaling. (A) Cell lysates were immunoblotted using the indicated antibodies (pAkt1/2, Akt1/2, pERK1/2, ERK1/2, and CHIP; β-actin was used as a loading control). ‘−’=pcDNA3 vector, ‘+’=pcDNA3-CHIP. (B) Following transfection with a pcDNA3 vector and pcDNA3-CHIP, cell viability was evaluated by a CCK-8 assay. (C) The effects of CHIP were examined by flow cytometry using an annexin-V/PI cellular apoptosis assay. MET inhibition by CHIP induced significant cellular apoptosis in NCI-H69 cells (5.83%±0.066%, **p<0.001) compared with the vector control (1.93%±0.06%). The bar graph shows the mean values of the percentage of cells in early apoptosis (annexin-V +/PI +) from 3 independent experiments for each treatment condition. MET, mesenchymal-epithelial transition factor; CHIP, C-terminus of Hsp70-interacting protein; Akt, protein kinase B; ERK, extracellular signal-regulated kinase; CCK-8, Cell Counting Kit-8; PI, propidium iodide.
4) CHIP interacts with MET through its TPR domain
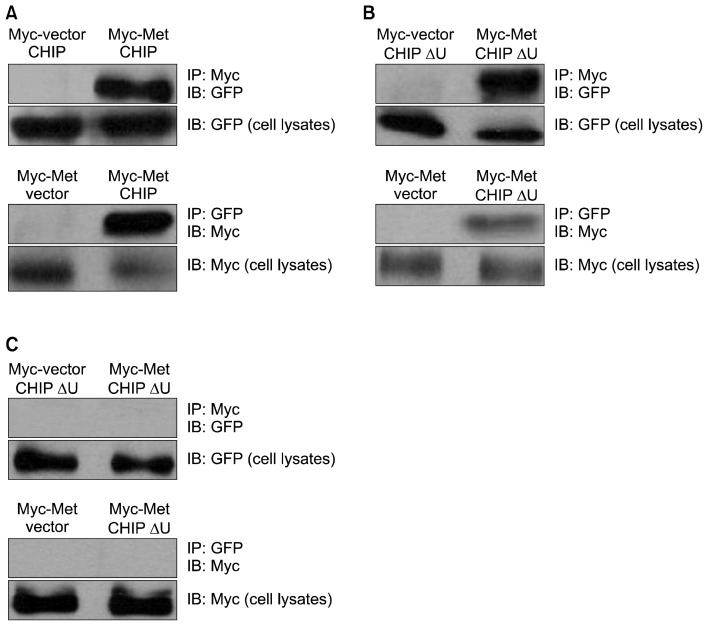
We sought to determine whether the U-box domain or the TPR domain is necessary for CHIP to interact with MET. To assess whether full-length CHIP could bind to MET, we co-transfected NCI-H69 cells with an expression vector for GFP-tagged full-length CHIP (pGFP-CHIP) and an expression vector coding for myc-tagged MET (pMyc-MET). Full-length CHIP co-immunoprecipitated with MET (Fig. 4A).
Fig. 4.
CHIP binds to MET using different mechanisms. (A) Co-immunoprecipitation of full-length CHIP and MET. (B) Co-immunoprecipitation of CHIP lacking the U-box domain (CHIPΔU) and MET. (C) Co-immunoprecipitation of CHIP lacking the TPR domain (CHIPΔT) and MET. MET, mesenchymal-epithelial transition factor; CHIP, C-terminus of Hsp70-interacting protein; TPR, tetratricopeptide repeat; GFP, green fluorescent protein; IP, immunoprecipitation analysis; IB, immunoblotting analysis.
To test whether a specific domain of CHIP is critical for MET binding, we co-transfected NCI-H69 cells with an expression vector for GFP-tagged CHIP lacking either the U-box domain (pGFP-CHIPΔU) or the TPR domain (pGFP-CHIPΔT), as well as expression vectors coding for myc-tagged MET (pMyc-MET). The U-box mutant was detected in the precipitated MET complex (Fig. 4B), but the TPR mutant was not (Fig. 4C), demonstrating that the TPR domain is required for the interaction of CHIP and MET.
5) Both the TPR and U-box domains are essential for CHIP-mediated MET degradation
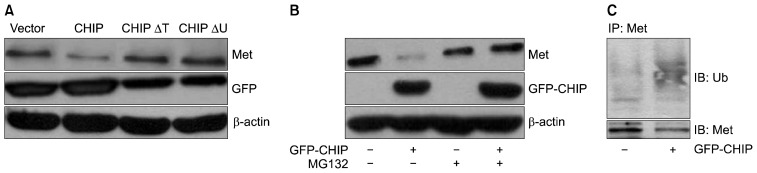
Given our findings that the N-terminal chaperone interaction (TPR) domain, but not the C-terminal U-box domain, of CHIP is essential for MET binding (Fig. 4B, C), we next examined whether these domains are required for CHIP-mediated MET degradation. To test this, we co-transfected NCI-H69 cells with Myc-MET and GFP-CHIP or CHIP mutant constructs of GFP-CHIP and GFP-CHIPΔU. Western blot analysis of cell lysates with anti-MET and anti-GFP showed that all of the constructs expressed similar amounts of protein (Fig. 5A). In contrast to wild-type CHIP (GFP-CHIP), which degraded MET, none of the CHIP mutants (GFP-CHIPΔT and GFP-CHIPΔU) exhibited decreased MET protein levels (Fig. 5A).
Fig. 5.
Both the TPR and U-box domains are required for MET degradation. (A) MET and GFP protein levels were determined by immunoblotting with GFP and MET antibodies, respectively. GFP was used as a control for efficient transfection and SDS-PAGE loading. (B) Cells were treated with MG132 and harvested. Lysates were analyzed by western blotting with antibodies for MET, GFP, and β-actin. (C) The cell lysates were immunoprecipitated using a polyclonal anti-MET antibody. The immunoprecipitation reactions were immunoblotted with a monoclonal anti-ubiquitin antibody (upper panel) and the corresponding cell lysates were probed using anti-MET. TPR, tetratricopeptide repeat; MET, mesenchymal-epithelial transition factor; GFP, green fluorescent protein; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; IP, immunoprecipitation analysis; IB, immunoblotting analysis; Ub, ubiquitination.
6) CHIP interacts with endogenous MET to induce receptor ubiquitination and degradation
As described above, we identified a role for CHIP (possibly in association with chaperones) in the degradation of both exogenous and endogenous MET in NCI-H69 cells. Thus, we next decided to examine the ability of CHIP to promote the polyubiquitination of endogenous MET. Specifically, we examined the ubiquitination status of MET in NCI-H69 cells transfected with a vector control (pGFP: −) or a CHIP-expression construct (pGFP-CHIP: +). As shown in Fig. 5B, the MET protein levels were restored by MG132 treatment after CHIP overexpression. To determine whether CHIP promotes the polyubiquitination of endogenous MET, we also examined the ubiquitination status of MET in NCI-H69 cells transfected with a vector control (−) or a CHIP-expressing construct (+). The poly ubiquitinated MET exhibited a typical smear on the blot membrane, and overexpression of CHIP increased the smear intensity, suggesting elevated receptor polyubiquitination (Fig. 5C).
7) Knockdown of endogenous CHIP by siRNA increases MET levels in NCI-H69 cells
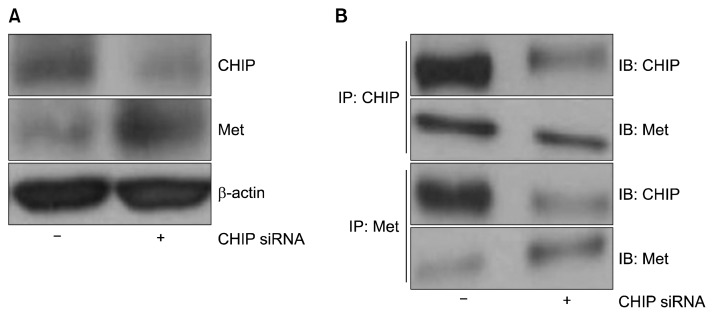
Next, we asked whether knockdown of endogenous CHIP protein by CHIP-siRNA could increase endogenous levels of MET. NCI-H69 cells were transfected with control siRNA (−) or CHIP siRNA (+), and immunoblotting analysis was performed using a MET-specific antibody. Consistent with our results thus far, we observed decreased CHIP expression and increased MET expression (Fig. 6A). To further investigate whether endogenous CHIP interacted with endogenous MET in NCI-H69 cells, we performed immunoprecipitation analysis using anti-MET and anti-CHIP antibodies. Our results showed that CHIP siRNA induced the dissociation of MET and CHIP (Fig. 6B).
Fig. 6.
Reduced CHIP expression enhances MET protein levels. (A) Following transfection with control siRNA and CHIP siRNA and incubation for 48 hours, cell lysates were prepared and the levels of CHIP and MET were determined by immunoblotting with anti-CHIP and anti-MET antibodies, respectively. (B) Cell lysates were precipitated with anti-MET and anti-CHIP antibodies, and the presence of conjugated MET in the immunocomplexes was detected by immunoblotting with anti-CHIP and anti-MET antibodies. In each case, representative results from 3 independent experiments are shown. CHIP, C-terminus of Hsp70-interacting protein; MET, mesenchymal-epithelial transition factor; siRNA, small interfering RNA; IP, immunoprecipitation analysis; IB, immunoblotting analysis.
Discussion
Previous studies have shown that MET is highly upregulated in a variety of human malignancies including SCLC, where it is involved in tumorigenesis, cell motility, scattering, invasion, and metastasis [12,13]. MET can activate Ras/mitogen-activated protein kinase, phosphatidylinositol 3-kinase, and the signal transducer and activator of transcription signaling pathway [14,15]. Thus, MET is a key factor influencing the events of tumor invasion and metastasis.
MET overexpression is an independent prognostic factor associated with adverse clinical outcomes. MET overexpression is also a prognostic factor in ovarian cancer and an effective target for the inhibition of peritoneal dissemination and invasion [16]. In addition, studies have also shown that MET overexpression in node-positive breast cancer can be used to identify patients with poor clinical outcomes independent of Her2/neu [17]. Moreover, it has been reported that blocking the MET/HGF pathway can inhibit the proliferation and invasion of multiple tumor cells [18]. A variety of small-molecule tyrosine kinase inhibitors and nucleic acid-directed gene silencing molecules targeting MET have been developed and have been shown to have direct effects on tumor cells [18,19]. In addition, both the physiological and pharmacological downregulation of MET have been linked to the induction of receptor ubiquitination, which targets the modified receptor for lysosomal and proteasomal degradation [20,21].
Ubiquitination and proteasomal degradation are critical components of protein quality control in cells. E3 ubiquitin ligases are widely thought to select target proteins for ubiquitination and thus commit the target protein to proteasomal degradation. CHIP is a U box-containing E3 ubiquitin ligase that binds to the independent TPR acceptor sites on Hsp90 and Hsp70 via its TPR domain, and mediates the ubiquitination of other Hsp90-chaperoned proteins [9,10].
In our results, we observed an inverse relationship between CHIP and MET expression in several SCLC cell lines, including NCI-H69 cells. To determine whether the overexpression of CHIP plays a critical role in the growth and migration of SCLC cells, we also transfected cells with CHIP to downregulate the expression of MET, and subsequently analyzed the effect of MET downregulation on the phenotypes of SCLC cells. We found that CHIP was capable of specifically reducing MET expression levels in NCI-H69 cells. Additionally, the invasiveness and migration of SCLC cells was found to be strongly controlled by the cytoskeletal molecules paxillin and FAK, which are activated by MET regulation [6]. We found that that the phosphorylation of both paxillin and FAK was decreased by CHIP-mediated MET inhibition, which was associated with the reduction of invasiveness. Additionally, the auto-phosphorylation of MET on tyrosine Y1349 in its C-terminal tail provides a docking site for the recruitment of signal transducers [14,15,22]. In this way, phosphorylation of MET at tyrosine Y1349 is necessary for the activation of downstream signaling pathways including Akt1/2 and ERK1/2. In previous reports, the MET-mediated Akt1/2 and ERK1/2 pathways were shown to be the main pathways involved in the survival, proliferation, and differentiation of SCLC cells [23]. We found that overexpression of CHIP reduced the phosphorylation of Akt1/2 and ERK1/2 in NCI-H69 cells. The reduction of the phosphorylation and signaling output of Akt1/2 or ERK1/2 in the presence of CHIP overexpression is therefore consistent with this Met-induced degradation. Moreover, the overexpression of CHIP in NCI-H69 cells led to increased apoptosis.
The primary sequence and crystal structures of CHIP have revealed 3 domains: the N-terminal TPR domain, the C-terminal U-box domain, and the intervening charged domain [23,24]. U-box domains are structurally related to really interesting new gene (RING) finger domains, both of which confer E3 ubiquitin ligase activity [25]. The N-terminal TPR domain of CHIP has been shown to specifically bind to several chaperones, such as Hsp70 and Hsp90 [26]. Because it is known that CHIP interacts with Hsp90 or Hsc/Hsp70 through the TPR domain, our results suggest that a chaperone intermediate may be involved in CHIP-induced MET degradation [24]. The C-terminal U-box domain, which facilitates ubiquitin ligase activity, is required for CHIP-mediated degradation of MET as well as the TPR domain dose. The above experiments demonstrated that overexpression of CHIP promotes MET polyubiquitination and degradation in SCLC cells.
We previously showed that exogenous CHIP interacted with exogenous MET in HeLa cells in vitro, leading to the proteasomal degradation of CHIP [27]. In addition, we reported that CHIP affected MET proteasomal degradation and interacted with MET in NSCLC cell lines.
According to our results, CHIP may act as a tumor suppressor that inhibits tumor growth, motility, and invasion in SCLC. CHIP has also been suggested to play a role in other human malignancies. Kajiro et al. [28] reported that CHIP suppressed tumor progression by direct degradation of the oncogene SRC-3 in breast cancer cells, and Wang et al. [29] found that CHIP acted to downregulate the subunit protein of nuclear factor kappa-light-chain-enhancer of activated B cells and interfered with gastric tumorigenesis and angiogenesis.
Importantly, Morgan et al. [30] observed that the phosphorylation of tyrosine 1173 of EGFR is the tar get of erlotinib, which is confirmed by a significant survival benefit associated with gemcitabine in advanced pancreatic cancer. Wang et al. [11] suggested that CHIP might be a potential treatment target for pancreatic cancer, reporting that the phosphorylation of Tyr845 and Tyr1068 of EGFR could be regulated by CHIP, so this multitarget treatment may explain why CHIP enhanced the efficacy of erlotinib on pancreatic tumor growth and apoptosis; additionally, CHIP could also increase the apoptotic rate induced by erlotinib in pancreas-1 cells that present K-ras mutations.
Herein, we extended our results for SCLC cells in several significant ways. Specifically, we found that the TPR domain—but not the U-box domain—of CHIP is necessary for the interaction between CHIP and MET, and that full-length CHIP induces the degradation and ubiquitination of MET in SCLC cell lines.
In conclusion, the results of the present study indicate that the TPR domain of full-length CHIP is essential for MET regulation, and that CHIP is capable of regulating SCLC cell apoptosis and invasion by inhibiting MET-mediated cytoskeletal and cell survival pathways in NCI-H69 cells.
Acknowledgments
For the following study, a small cell lung cancer cell line was used, which the cell line was purchased of ones that were commercialized, hence we figured that it would not be an ethical issue and to be applicable of the IRB exemption.
This study was supported by a Grant of the Samsung Vein Clinic Network (Daejeon, Anyang, Cheongju, Cheonan; Fund no. KTCS04-069).
Footnotes
Conflict of interest
No potential conflicts of interest relevant to this article are reported.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–96. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 3.Bottaro DP, Rubin JS, Faletto DL, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–4. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 4.Di Renzo MF, Narsimhan RP, Olivero M, et al. Expression of the Met/HGF receptor in normal and neoplastic human tissues. Oncogene. 1991;6:1997–2003. [PubMed] [Google Scholar]
- 5.Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–81. [PubMed] [Google Scholar]
- 6.Maulik G, Kijima T, Ma PC, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res. 2002;8:620–7. [PubMed] [Google Scholar]
- 7.Hammond DE, Urbe S, Vande Woude GF, Clague MJ. Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene. 2001;20:2761–70. doi: 10.1038/sj.onc.1204475. [DOI] [PubMed] [Google Scholar]
- 8.Connell P, Ballinger CA, Jiang J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–6. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 9.Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11:1569–77. doi: 10.1016/S0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhou P, Fernandes N, Dodge IL, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278:13829–37. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 11.Wang T, Yang J, Xu J, et al. CHIP is a novel tumor suppressor in pancreatic cancer through targeting EGFR. Oncotarget. 2014;5:1969–86. doi: 10.18632/oncotarget.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comoglio PM, Boccaccio C. Scatter factors and invasive growth. Semin Cancer Biol. 2001;11:153–65. doi: 10.1006/scbi.2000.0366. [DOI] [PubMed] [Google Scholar]
- 13.Kuba K, Matsumoto K, Date K, Shimura H, Tanaka M, Nakamura T. HGF/NK4, a four-kringle antagonist of hepatocyte growth factor, is an angiogenesis inhibitor that suppresses tumor growth and metastasis in mice. Cancer Res. 2000;60:6737–43. [PubMed] [Google Scholar]
- 14.Ma PC, Tretiakova MS, Nallasura V, Jagadeeswaran R, Husain AN, Salgia R. Downstream signalling and specific inhibition of c-MET/HGF pathway in small cell lung cancer: implications for tumour invasion. Br J Cancer. 2007;97:368–77. doi: 10.1038/sj.bjc.6603884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YH, Wei W, Xu H, Wang YY, Wu WX. Inducing effects of hepatocyte growth factor on the expression of vascular endothelial growth factor in human colorectal carcinoma cells through MEK and PI3K signaling pathways. Chin Med J (Engl) 2007;120:743–8. [PubMed] [Google Scholar]
- 16.Sawada K, Radjabi AR, Shinomiya N, et al. c-Met overexpression is a prognostic factor in ovarian cancer and an effective target for inhibition of peritoneal dissemination and invasion. Cancer Res. 2007;67:1670–9. doi: 10.1158/0008-5472.CAN-06-1147. [DOI] [PubMed] [Google Scholar]
- 17.Lengyel E, Prechtel D, Resau JH, et al. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer. 2005;113:678–82. doi: 10.1002/ijc.20598. [DOI] [PubMed] [Google Scholar]
- 18.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. The role of STATs in lung carcinogenesis: an emerging target for novel therapeutics. J Mol Med (Berl) 2007;85:427–36. doi: 10.1007/s00109-006-0152-3. [DOI] [PubMed] [Google Scholar]
- 19.Kim SJ, Johnson M, Koterba K, Herynk MH, Uehara H, Gallick GE. Reduced c-Met expression by an adenovirus expressing a c-Met ribozyme inhibits tumorigenic growth and lymph node metastases of PC3-LN4 prostate tumor cells in an orthotopic nude mouse model. Clin Cancer Res. 2003;9:5161–70. [PubMed] [Google Scholar]
- 20.Peschard P, Ishiyama N, Lin T, Lipkowitz S, Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J Biol Chem. 2004;279:29565–71. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 21.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 22.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–25. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Pashtan I, Tsutsumi S, Xu W, Neckers L. Cancer cells harboring MET gene amplification activate alternative signaling pathways to escape MET inhibition but remain sensitive to Hsp90 inhibitors. Cell Cycle. 2009;8:2050–6. doi: 10.4161/cc.8.13.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–8. doi: 10.1379/1466-1268(2003)008<0303:CALBTC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, Windheim M, Roe SM, et al. Chaperoned ubiq-uitylation: crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 26.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–5. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang KW, Lee JE, Kim SY, et al. The C-terminus of Hsp70-interacting protein promotes Met receptor degradation. J Thorac Oncol. 2011;6:679–87. doi: 10.1097/JTO.0b013e31820d9c7e. [DOI] [PubMed] [Google Scholar]
- 28.Kajiro M, Hirota R, Nakajima Y, et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol. 2009;11:312–9. doi: 10.1038/ncb1839. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Wu X, Zhang J, et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut. 2013;62:496–508. doi: 10.1136/gutjnl-2011-301522. [DOI] [PubMed] [Google Scholar]
- 30.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin Cancer Res. 2008;14:5142–9. doi: 10.1158/1078-0432.CCR-07-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]