Abstract
Prenatal inorganic arsenic (iAs) exposure is associated with health effects evident at birth and later in life. An understanding of the relationship between prenatal iAs exposure and alterations in the neonatal metabolome could reveal critical molecular modifications, potentially underpinning disease etiologies. In this study, nuclear magnetic resonance (NMR) spectroscopy-based metabolomic analysis was used to identify metabolites in neonate cord serum associated with prenatal iAs exposure in participants from the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort, in Gómez Palacio, Mexico. Through multivariable linear regression, ten cord serum metabolites were identified as significantly associated with total urinary iAs and/or iAs metabolites, measured as %iAs, %monomethylated arsenicals (MMAs), and % dimethylated arsenicals (DMAs). A total of 17 metabolites were identified as significantly associated with total iAs and/or iAs metabolites in cord serum. These metabolites are indicative of changes in important biochemical pathways such as vitamin metabolism, the citric acid (TCA) cycle, and amino acid metabolism. These data highlight that maternal biotransformation of iAs and neonatal levels of iAs and its metabolites are associated with differences in neonate cord metabolomic profiles. The results demonstrate the potential utility of metabolites as biomarkers/indicators of in utero environmental exposure.
Graphical abstract
INTRODUCTION
Millions of individuals worldwide are exposed to levels of inorganic arsenic (iAs) in their drinking water that have been linked to chronic diseases such as precancerous skin lesions, cardiovascular disease, peripheral vascular disease, diabetes mellitus, hypertension, and cancers of the urinary bladder, skin, lung, and liver.1–3 Exposure to iAs during periods of increased susceptibility, including in utero and early childhood, is of particular concern. Early life iAs exposure is associated with both short- and long-term adverse health effects including increased risk for preterm birth, low birth weight, infant mortality, childhood neurological impairments, susceptibility to infectious disease, and chronic disease development later in life, including cancer.4
A precise mechanistic basis for the relationship between prenatal iAs exposure and disease has not been elucidated and is likely multifactorial. Potential mechanisms for iAs-associated diseases include the induction of oxidative stress, enzyme inhibition, interference with DNA repair, chromosomal aberrations, perturbation of cell signaling pathways, and epigenetic instability.5 Specific to prenatal iAs exposure, previous studies have highlighted changes in fetal gene expression,6–8 fetal leukocyte DNA methylation,7–10 and increased protein signaling of inflammatory mediators in cord serum.11 In spite of these advances, much remains unknown about the biochemical mechanisms underlying the health effects associated with prenatal iAs exposure.
One factor known to influence the susceptibility to iAs-associated diseases is an individual’s efficiency of iAs biotransformation/metabolism. Arsenic is biotransformed in humans to produce monomethylated arsenicals (MMAs) and dimethylated arsenicals (DMAs) that can exist in either a trivalent (+3) or pentavalent (+5) oxidation state. Specifically, six major arsenicals associated with iAs exposure have been detected in human urine, namely arsenite (iAsIII), arsenate (iAsV), monomethylarsonous acid (MMAIII), monomethylarsonic acid (MMAV), dimethylarsinous acid (DMAIII), and dimethylarsinic acid (DMAV).12,13 High urinary proportions of MMAs/total arsenic and increased MMAs/DMAs are likely indicators of inefficient iAs biotransformation. Inefficient biotransformation (or methylation) of iAs, as indicated by higher proportions of MMAs in urine, has been associated with the development of several adverse outcomes in humans including urinary bladder cancer, nonmelanoma skin cancers, carotid atherosclerosis, and chromosomal aberrations (reviewed in ref 14). In recent work, we have shown that elevated levels and proportions of MMAs in maternal urine are associated with decreases in placental weight and fetal birth weight.15
There are an increasing number of system-wide methodologies available to provide insight into mechanisms underlying environmentally mediated disease, including metabolomics. Metabolomics involves the analysis of the low molecular weight metabolites in cells, tissues, and biological fluids, providing a profile of the biochemistry and end result of multiple enzymatic processes, or the “metabotype,” of an individual. Metabolomic profiling has been used to study the impacts of nutritional, pharmaceutical, and environmental toxicant exposures.16 Metabolomic profiling is an ideal tool for biomarker assessment as it is efficient and relatively noninvasive, and it can convey information about disease/physiologic phenotypes.17,18 Metabolomic biomarkers identified in both maternal and fetal-health studies have been associated with fetal malformations, preterm delivery, premature rupture of membranes, gestational diabetes mellitus, preeclampsia, low birth weight, and hypoxic-ischemic encephalopathy - all representing adverse outcomes that are relevant to iAs exposure.19
Recent developments in metabolomic technologies and statistical methodologies have allowed for explorations for global changes in biological systems, predictive modeling of metabolomics alterations, and the use of metabolomics in clinical applications in regards to various exposures and/or disease states. Specifically, the use of nuclear magnetic resonance (NMR) spectroscopy to quantify and determine the metabotype of an individual or a population provides an excellent tool for detection of metabolites in ways that are nonbiased, easily quantifiable, require little to no separation - all allowing for the identification of novel compounds that are less tractable to GC-MS or LC-MS analysis.20 Additionally, a NMR-based metabolomic approach is particularly beneficial for human samples that may not be easily attainable or are limited in supply, as it is a nondestructive method that enables preservation of the sample and analytes for subsequent analyses.21 NMR spectroscopy, combined with multivariable statistical analysis, can be a powerful tool to detect metabolic changes induced by very low doses of an exposure based on over- or underexpression of endogenous molecules. Previous work has demonstrated the validity of NMR methods for populations exposed to iAs.22
We recently identified differences in the levels of 132 human urinary metabolites associated with iAs exposure in adults.23 Specifically, 59 metabolites that play a role in the tricarboxylic acid cycle and amino acid metabolism in diabetics were identified. Building upon our understanding of the relationship between iAs exposure and the metabolome, the aim of the present study was to identify a potential metabolomics signature of in utero iAs exposure in cord serum using samples from the Biomarkers of Exposure to ARsenic (BEAR) prospective pregnancy cohort in Gómez Palacio, Mexico. 15 Investigations of the metabolome of individuals exposed prenatally to iAs have not been previously investigated. The pregnant women in this cohort lived in areas with elevated iAs in their drinking water (up to 236 μg As/L, mean of 24.6 μg As/L).15 Several indicators of in utero iAs exposure were used, including the sum of iAsIII and iAsV in maternal urine (U-iAs), the sum of MMAIII and MMAV (U-MMAs), the sum of DMAIII and DMAV (U-DMAs), and the sum of U-iAs, U-MMAs, and U-DMAs as total As in maternal urine (U-tAs). As the efficiency of iAs metabolism has been linked to disease status, we set out to determine whether there was a relationship between metabolite patterns in cord serum and maternal iAs metabolism efficiency. Additionally, as a measurement of neonatal levels of iAs, we analyzed total cord serum arsenic (C-tAs) and/or iAs metabolites in cord serum (C-iAs, C-MMAs, and/or C-DMAs). Using a quantitative metabolomics approach coupled with multiple regression analyses, we identified metabolites altered in cord serum that are significantly associated with prenatal iAs exposure and/or percentages of its methylated metabolites.
MATERIALS AND METHODS
Subcohort Selection
The BEAR cohort has been described previously.15 Briefly, women were recruited prior to the time of delivery from the time frame of August 2011 to March 2012 at the General Hospital of Gómez Palacio. All procedures associated with this study were approved by the Institutional Review Boards of Universidad Juárez del Estado de Durango (UJED), Gómez Palacio, Durango, Mexico, and the University of North Carolina at Chapel Hill (UNC), Chapel Hill, North Carolina, U.S.A. In the present study, a subset of 50 cord serum samples was selected from the larger BEAR cohort that spanned a range of iAs exposure where drinking water had variable levels of iAs. These samples have been previously described as they were prioritized for other molecular/mechanistic investigations, including assessment of iAs-associated microRNA expression,24 DNA methylation,8 gene expression,8,24 and protein expression.25
Methods for the measurements of iAs levels in this cohort have been previously described.15 Briefly, the assessment of the levels of iAs in drinking water included samples from the study participants’ home of their primary source of drinking water that was collected by the research team within 4 weeks of delivery. Maternal spot urine samples were collected at the time of delivery, immediately transferred to cryovials, placed in liquid nitrogen, and stored at −80 °C until further processing.
Arsenical levels in maternal urine have been used as indicators of prenatal iAs exposure.15,26 Concentrations of U-iAs, U-MMAs, and U-DMAs were determined by HG-AAS with cryotrapping (CT) as described previously.27–29 The LODs for U-iAs, U-MMAs, and U-DMAs were 0.2, 0.1, and 0.1 μg As/L, respectively. U-tAs was determined by summing U-iAs, U-MMAs, and U-DMAs. To account for differences in water intake/differential hydration, concentrations of U-iAs, U-MMAs, and U-DMAs in each urine sample were adjusted using the following equation: iAs × (mean SG-1)/(individual SG-1) as previously described.30 The efficiency of iAs biotransformation was determined by calculating the proportions of the individual arsenicals iAs, MMAs, and DMAs relative to total urinary As for each study subject. Metabolism efficiency for iAs in maternal urine was determined by calculating the percentage of the individual metabolites out of the total (U-% iAs, U-%MMAs, and U-%DMAs).
Cord blood was collected immediately after newborn delivery using an anticoagulant-free vacutainer tube. Following clot formation, the tube was centrifuged at 177.1 g-force, and the serum was collected and stored at −80 °C until aliquots were shipped on dry ice to UNC-Chapel Hill, NC, where they were immediately stored at −80 °C. Concentrations of cord serum arsenic included the measurement of cord serum-analyzed iAs (C-iAs), MMAs (C-MMAs), and DMAs (C-DMAs) and were determined using HG-CT-ICP-MS as described previously.28,31 The LODs for C-iAs, C-MMAs, and C-DMAs were 1.45, 0.06, and 0.12 pg As/mL, respectively. C-tAs was determined by summing C-iAs, C-MMAs, and C-DMAs. The percentages of the individual metabolites were calculated relative to the total (C-%iAs, C-%MMAs, and C-% DMAs).
A certified standard reference material, Arsenic Species in Frozen Human Urine (SRM 2669; National Institute of Standards and Technology), was used to ensure accuracy of iAs speciation analysis. Here, aliquots of SRM were diluted in urine or serum from an unexposed subject and analyzed by HG-CT-AAS and HG-CT-ICP-MS, respectively. The concentrations of iAs species in SRM in urine (after deduction of background iAs) ranged from 88% to 105% of the certified values (105% for iAs, 88% for MMAs, 99% for DMAs); the concentrations of iAs species in SRM in serum ranged from 85% to 90% of the certified value (85% for iAs, 90% for MMAs, and 87% for DMAs).
1H NMR Spectroscopy
The 50 cord serum samples were analyzed using nuclear magnetic resonance (NMR) spectroscopy to identify neonatal metabolites that were associated with maternal urinary arsenicals. Sample preparation, data acquisition, and data analysis followed procedures previously described.32–35 Each of the 50 samples were processed individually, whereby 150 μL of serum was prepared by adding 50 μL of 0.9% NaCl solution containing 4 mM sodium formate (Chemical Shift Indicator) and 0.2% NaN3 (to inhibit bacterial growth) in D2O. Each 200 μL sample was then vortexed for 30 s and transferred into 3 mm NMR tubes (Bruker Bio-Spin, Germany).
1H NMR spectra of serum samples were acquired on a Bruker Avance III 950 MHz NMR spectrometer (located at the David H. Murdock Research Institute at Kannapolis, NC, USA) using a 5 mm cryogenically cooled ATMA inverse probe and ambient temperature of 25 °C. A CPMG pulse sequence with presaturation (cpmgpr1d) was used for data acquisition. For each sample, 256 transients were collected into 32k data points using a spectral width of 19.5 kHz (20.5 ppm), 2 s relaxation delay, 400 μs fixed echo time, loop for T2 filter (l4) = 80, and an acquisition time of 1.678 s per FID. The water resonance was suppressed using resonance irradiation during the relaxation delay. NMR spectra were processed using TopSpin software (Bruker, Germany). Spectra were zero filled and Fourier-transformed after exponential multiplication with a line broadening factor of 0.5. Phase and baseline of the spectra were manually corrected for each spectrum. Spectra were referenced internally to the formate signal. The quality of each NMR spectrum was assessed for the level of noise and alignment of identified markers. Using a quality control (QC) procedure described by Chan and co-workers,36 pooled samples were prepared using aliquots of each study sample. Principal component analysis was performed to determine that sufficient separation of the data occurred. These QC samples were processed identically to each of the study samples. This was done by adding in small aliquots of each study sample by combining 12 μL aliquots into eppendorf tubes
Metabolites were identified, and the relative metabolite concentrations were determined prior to data analysis. This deconvolution of NMR peaks into metabolites and relative quantification of these identified metabolites are advantageous over the traditional binning method in pattern recognition methods and identifying statistically significant changes of metabolites.37,38 A targeted profiling approach was carried out to identify the metabolites. Metabolites were modeled using peak centers and splitting patterns of peaks (J-coupling) from the NMR spectra; signals were matched by the analyst to the 36 metabolites identified using the library in the Chenomx NMR Suite 8.1 Professional software (Chenomx, Edmonton, Alberta, Canada). This software contains an internal library adjustment for increments in chemical shift based on pH variation.38 Concentration determinations for metabolites were established using relative integration of the analyte to the formate internal standard. The library of concentrations was developed to account for differences in integral values as related to the relaxation time of the signal.38
Statistical Analyses
Spearman rank correlations between maternal urinary and cord serum arsenicals were calculated. Multivariable linear regression was used to determine the relationship between U-tAs, U-%iAs, U-%MMAs, and U-% DMAs C-tAs, C-%iAs, C-%MMAs, or C-%DMAs and the Chenomx concentration-matched metabolites using SAS 9.4 (SAS Institute Inc., Cary, NC). Potential confounders were identified a priori based on their known or potential association with both the exposure (iAs or its methylated metabolites) and the outcome (cord serum metabolite levels). All models were adjusted for maternal age (continuous), gender of the infant (male/female), and gestational age (continuous). To improve model fit U-tAs and C-tAs were was natural log transformed. Regression assumptions of linearity and the homogeneity of residuals were evaluated by examination of appropriate residual plots. A false discovery rate (FDR) correction was calculated for all p-values to account for multiple comparisons, with significance set at the acceptable level of q < 0.20.23,39,40 The identified cord serum metabolites that were significantly associated with U-tAs and/or the indicators for iAs biotransformation/metabolism efficiency indicators (U-%iAs, U-%MMAs, and U-%DMAs) and for cord serum total arsenic (C-tAs) and/or percentages of iAs in cord serum (C-%iAs, C-% MMAs, and/or C-%DMAs) were then analyzed for enriched pathways, using MetaboAnalyst.41 Significance was set at p < 0.05 for all analyses. These analyses were carried out separately for each indicator of exposure for both maternal U-tAs and for iAs biotransformation/metabolism efficiency indicators (U-% iAs, U-%MMAs, and U-%DMAs) and for cord serum total arsenic (C-tAs) and/or percentages of iAs in cord serum (C-% iAs, C-%MMAs, and/or C-%DMAs).
The metabolites identified in the present study were compared to prior population-based iAs-associated metabolomic studies. There were only three studies identified that investigated the metabolome in individuals exposed to iAs in human cohorts. Zhang et al. investigated iAs-related urinary changes in the metabolome in a cohort of 127 adult Chinese men.42 Another study investigated alterations in the metabolome of an occupationally exposed group of 389 men and a control group of 45 age-matched, nonexposed healthy men using NMR based metabolomics.22 Martin et al. investigated changes in the metabolome between individuals exposed to iAs and in 35 diabetic/nondiabetics.23
RESULTS
All maternal and birth characteristics and levels of the exposure indicators for the subcohort selected for the metabolomics assessment and the parent BEAR cohort are detailed in Table 1. Arsenic was detected in the urine of all women. The mean level of U-tAs was 61.4 μg As/L (range: 6.2 μg As/L to 319.7 μg As/L). The mean urinary level of U-iAs was 3.4 μg As/L (range: 0.22 μg As/L to 23.0 μg As/L). The mean level of U-MMAs was 3.8 μg As/L (range: 0.14 μg As/L to 18.2 μg As/L). The mean level of U-DMAs was 54.1 μg As/L (range: 5.3 μg As/L to 292.5 μg As/L). These correspond to proportions of U-tAs comprising 6.5% U-iAs, 6.5% U-MMAs, and 87.2% U-DMAs. The mean level of C-tAs was 0.61 pg As/mL (range: 0.048 to 2.98). The mean neonatal serum level of C-iAs was 0.055 pg As/mL (range: 0.0010 to 0.36). The mean level of C-MMAs was 0.10 pg As/mL (range: 0.000042 to 0.56). The mean level of C-DMAs was 0.45 pg As/mL (range: 0.047 to 2.70). This corresponds to proportions of C-tAs comprising 9.2% C-iAs, 15.8% C-MMAs, and 77.5% C-DMAs. The Spearman rank correlations of maternal urinary and cord serum arsenicals are presented in Table S2. U-tAs were significantly correlated with C-tAs (r = 0.84, p = < 0.0001). U-iAs were significantly correlated with C-iAs (r = 0.33, p = 0.02). U-MMAs were significantly correlated with C-MMAs (r = 0.85, p = < 0.0001). U-DMAs were significantly correlated with C-DMAs (r = 0.80, p = < 0.0001). U-%MMAs were significantly correlated with C-%MMAs (r = 0.45, p = 0.0013). In contrast, U-%iAs were not significantly correlated with C-%iAs (r = 0.015, p = 0.9), and U-%DMAs were not significantly correlated with C-%DMAs (r = 0.0047, p = 0.9).
Table 1.
Selected Characteristics of the NMR Metabolomics Subcohort and the BEAR Parent Cohort
| demographic characteristics | NMR subcohort (n = 50)
|
BEAR cohort (n = 200)
|
.
|
|---|---|---|---|
| mean, median [range] or N, (N%) | mean, median [range] or N, (N%) | p-value | |
| maternal age (years) | 24, 23 [18–39] | 24, 23[18–41] | 0.9 |
| maternal education | 0.5 | ||
| <high school | 13 (26) | 50 (25) | |
| high school | 25 (50) | 95 (48) | |
| >high school | 12 (24) | 54 (27) | |
| gestational age (weeks) | 39, 40 [34–41] | 39, 40 [34–42] | 0.9 |
| gender of infant | 0.5 | ||
| female | 27 (54) | 96 (48) | |
| male | 23 (46) | 104 (52) | |
| exposure indicators (μg As/L) | |||
| Dw-iAs | 53.9, 24.2 [<LODa–235.6] | 24.6, 13.0 [<LODa–236.0] | 0.001 |
| U-tAs | 61.4, 30.7 [6.2–319.7] | 37.5, 23.3 [4.3–319.7] | 0.001 |
| U-iAs | 3.4, 1.5 [0.22–23.0] | 2.1, 1.3 [0.14–23.0] | 0.001 |
| U-MMAs | 3.8, 1.3 [0.14–18.2] | 2.3, 1.4 [0.12–18.2] | 0.001 |
| U-DMAs | 54.1, 26.3 [5.3–292.5] | 33.1, 20.6 [1.4–292.5] | 0.001 |
| U-iAs (%) | 6.5, 5.3 [1.6–16.6] | 6.1, 5.3 [0.77–45.1] | 0.1 |
| U-MMAs (%) | 6.5, 5.9 [1.3–24.9] | 6.4, 6.0 [0.68–24.9] | 0.7 |
| U-DMAs (%) | 87.2, 88.2 [60.6–96.7] | 87.6, 88.5 [32.7–96.7] | 0.3 |
| C-tAs (μg/L) | 0.61, 0.322 [0.048–2.98] | 0.37, 0.41 [0.004–2.98] | 0.001 |
| C-iAs (μg/L) | 0.055, 0.010 [0.0010–.36] | 0.039, 0.013 [0.00085–0.50] | 0.001 |
| C-MMAs (μg/L) | 0.10, 0.050 [0.000042–0.56] | 0.064, 0.043 [0.000042–0.56] | 0.001 |
| C-DMAs (μg/L) | 0.45, 0.23 [0.047–2.70] | 0.26, 0.17 [0.0031–2.70] | 0.001 |
| C-iAs (%) | 9.2, 0.98 [0.088–69.5] | 10.9, 4.6 [0.088–69.5] | 0.5 |
| C-MMAs (%) | 15.8, 16.9 [0.088–33.5] | 16.7, 16.8 [0.032–51.8] | 0.3 |
| C-DMAs (%) | 77.5.0, 77.5 [21.6–97.8] | 72.3, 74.8 [21.6–97.8] | 0.04 |
Limit of detection (LOD) for iAs in drinking water is 0.456 mg As/L.
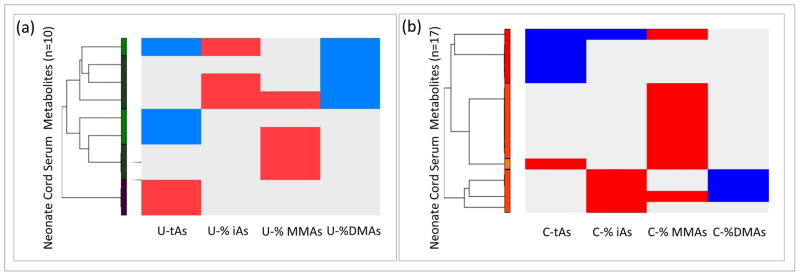
For every cord serum sample, 36 operator-matched metabolites were identified (Table S1). This method of analyte identification deconvolutes the entire metabolomic spectrum based on chemical shifts and coupling patterns and matches signals to a reference library of over 330 compounds. Multivariable linear regression models were used to analyze the linear relationships among total arsenic exposure (U-tAs) and/or iAs metabolism indicators (U-iAs, U-MMAs, U-DMAs, U-%iAs, U-%MMAs, U-%DMAs) analyzed separately as the independent variables and the 36 cord serum metabolites as the dependent variables (Table S1) across all subjects. Ten significant (p ≤ 0.05, q < 0.2) cord serum metabolites were identified from the multivariable regression analyses in relation to U-tAs and iAs metabolism efficiency indicators (U-%iAs, U-%MMAs, U-%DMAs) (Table 2). Specifically, of the 36 metabolites a total of five cord serum metabolite levels were found to be significantly (p ≤ 0.05, q < 0.2) associated with U-tAs (Table 2). For three of the metabolites there was a negative relationship between U-tAs and metabolite level. Specifically, there was a significant negative relationship between U-tAs and mean levels of isoleucine, with an estimated decrease of −0.003 mM for every one-unit change in ln U-tAs (p = 0.01). Similarly, there was an estimated decrease in mean levels of O-acetylcholine of −0.003 mM in relation to ln U-tAs (p = 0.04). There was an estimated decrease in mean tyrosine levels of −0.001 mM in relation to ln U-tAs (p = 0.03). In contrast, there was an estimated increase in mean levels of mannose of 0.005 mM in relation to ln U-tAs (p = 0.02) and mean levels of succinate of 0.002 mM (p = 0.05) (Table 2).
Table 2.
Beta and p-Values for the Cord Serum Metabolites Identified by Multivariable Linear Regression Modelsa as Having a Significant Association with Either Total Urinary iAs (ln U-tAs) or Percentages of Urinary iAs Metabolites (U-%iAs, U-%MMAs, U-%DMAs) and Associations with Either Total Cord Serum iAs (ln C-tAs) or Percentages of Cord Serum iAs Metabolites (C-% iAs, C-%MMAs, C-%DMAs)
| metabolite | maternal urinary arsenicals
|
|||
|---|---|---|---|---|
| U-tAs β (p-value) [FDR p-value] | U-%iAs β (p-value) [FDR p-value] | U-%MMAs β (p-value) [FDR p-value] | U-%DMAs β (p-value) [FDR p-value] | |
| glutamate | −0.01 (0.09) [0.14] | 0.005 (0.01)b [0.10] | 0.003 (0.1) [0.12] | −0.003 (0.03)b [0.066] |
| glycine | 0.005 (0.5) [0.5] | 0.004 (0.09) [0.22] | 0.004 (0.07) [0.11] | −0.003 (0.03)b [0.066] |
| methionine | −0.0009 (0.1) [0.14] | 0.0004 (0.05)b [0.16] | 0.0005 (0.01)b [0.05] | −0.0003 (0.01)b [0.066] |
| O-acetylcholine | −0.003 (0.04)b [0.10] | 0.0009 (0.05)b [0.16] | 0.0009 (0.06) [0.11] | −0.0006 (0.02)b [0.066] |
| isoleucine | −0.003 (0.01)b [0.10] | 0.02 (0.4) [0.57] | 0.0008 (0.01)b [0.05] | −0.0002 (0.3) [0.41] |
| taurine | −0.002 (0.3) [0.33] | 0.0007 (0.2) [0.33] | 0.001 (0.04)b [0.10] | −0.0006 (0.06) [0.11] |
| valine | −0.004 (0.3) [0.33] | −0.0003 (0.7) [0.77] | 0.002 (0.02)b [0.066] | −0.0007 (0.2) [0.31] |
| succinate | 0.002 (0.05)b [0.10] | 0 (0.9) [0.9] | −0.0001 (0.7) [0.77] | 0 (0.9) [0.90] |
| mannose | 0.005 (0.02)b [0.10] | −0.0004 (0.5) [0.62] | 0 (0.9) [0.90] | 0.01 (0.8) [0.88] |
| tyrosine | −0.001 (0.03)b [0.10] | 0.0002 (0.2) [0.33] | 0.0003 (0.1) [0.12] | 0.0002 (0.08) [0.18] |
|
| ||||
| metabolite | neonate cord serum arsenicals
|
|||
| C-tAs β (p-value) [FDR p-value] | C-%iAs β (p-value) [FDR p-value] | C-%MMAs β (p-value) [FDR p-value] | C-%DMAs β (p-value) [FDR p-value] | |
|
| ||||
| acetoacetate | 0.0003 (0.9) [0.9] | 0.0004 (0.02)b [0.1] | 0.0002 (0.01)b [0.04] | −0.0004 (0.01)b [0.05] |
| acetone | 0.003 (0.2) [0.4] | 0.0003 (0.03)b [0.1] | 0.0002 (0.5) [0.6] | −0.0004 (0.002)b [0.02] |
| betaine | −0.003(0.2) [0.4] | −0.0001 (0.7) [0.7] | 0.004 (0.004)b [0.01] | 0.0002 (0.2) [0.5] |
| ethanol | 0.02 (0.4) [0.7] | 0.004 (0.007)b [0.06] | −0.007 (0.09) [0.1] | −0.003 (0.06) [0.2] |
| glutamate | −0.02 (0.02)b [0.08] | −0.0005 (0.3) [0.6] | 0.002 (0.1) [0.1] | 0.0001 (0.7) [0.7] |
| glycerol | 0.004 (0.2) [0.4] | 0 (0.9) [0.9] | 0.002 (0.0003)b [0.1] | −0.0003 (0.1) [0.4] |
| glycine | 0.01 (0.1) [0.3] | −0.0004 (0.4) [0.7] | 0.005 (<0.001)b [0.001] | −0.0005 (0.3) [0.6] |
| isoleucine | −0.003 (0.03)b [0.08] | −0.0001 (0.5) [0.7] | 0.0001 (0.5) [0.6] | 0 (0.7) [0.7] |
| lactate | 0.02 (0.9) [0.9] | −0.005 (0.5) [0.7] | 0.04 (0.02)b [0.05] | −0.003 (0.7) [0.7] |
| mannose | 0.006 (0.02)b [0.08] | −0.0001 (0.3) [0.6] | 0.001 (0.001)b [0.005] | −0.0001 (0.6) [0.7] |
| methionine | −0.002 (0.02)b [0.08] | 0 (0.4) [0.6] | 0 (0.9) [0.9] | 0 (0.4) [0.6] |
| O-acetylcholine | −0.004 (0.03)b [0.08] | −0.0002 (0.1) [0.2] | 0.0001 (0.7) [0.7] | 0.0002 (0.1) [0.4] |
| pyruvate | 0.0005 (0.8) [0.9] | −0.0001 (0.6) [0.7] | 0.001 (0.001)b [0.005] | −0.0001 (0.4) [0.6] |
| serine | −0.0006 (0.8) [0.9] | 0 (0.8) [0.9] | 0.001 (0.027)b [0.05] | −0.0002 (0.4) [0.6] |
| taurine | −0.0005 (0.8) [0.9] | −0.0001 (0.4) [0.6] | 0.0009 (0.001)b [0.005] | −0.0001 (0.6) [0.7] |
| tyrosine | −0.002 (0.01)b [0.08] | −0.0001 (0.01)b [0.1] | 0.0002 (0.03)b [0.05] | 0.0001 (0.1) [0.4] |
| 3-hydroxybutyrate | 0.04 (0.7) [0.9] | 0.002 (0.007)b [0.06] | 0.001 (0.4) [0.5] | −0.002 (0.001)b [0.02] |
All models adjusted for maternal age, gender of the infant, and gestational age.
FDR ≤ 0.1.
Three metabolites displayed levels that were significantly associated with U-%iAs including glutamate (β = 0.005; p = 0.01), methionine (β = 0.0004; p = 0.05), and O-acetylcholine (β = 0.0009; p = 0.05). The abundance of four metabolites was significantly associated with U-%MMAs including isoleucine (β = 0.0008; p = 0.01), methionine (β = 0.0005; p = 0.01), taurine (β = 0.001; p = 0.04), and valine (β = 0.002; p = 0.02). Additionally, four metabolites displayed levels that were significantly associated with U-%DMAs including glutamate (β = −0.003; p = 0.03), glycine (β = −0.003; p = 0.03), methionine (β = −0.0003; p = 0.01), and O-acetylcholine (β = −0.0006; p = 0.02). All metabolite levels displayed an increase in abundance in relation to U-%iAs and U-%MMAs, while in relation to U-%DMAs the metabolites were decreased as visualized in the heat map (Figure 1a). Methionine was the only metabolite with altered abundance that was common to U-%iAs, U-%MMAs, and U-%DMAs. Methionine levels displayed a positive significant association with U-%iAs and U-%MMAs and a negative association with U-%DMAs (Figure 2a). In addition to methionine, O-acetylcholine and glutamate displayed levels that were significantly associated with both U-%iAs and U-%DMAs.
Figure 1.
a: Hierarchical clustering of the neonate cord serum metabolites associated with total urinary arsenic (U-tAs) or the proportions arsenic methylated maternal urinary metabolites (U-%iAs, U-%MMAs, and U-%DMAs). b: Hierarchical clustering of the neonate cord serum metabolites associated with total cord serum arsenic (C-tAs) or the proportions arsenic methylated neonate serum metabolites (C-%iAs, C-%MMAs, and C-% DMAs). Blue represents a negative association, red represents a positive association, and gray represents no association.
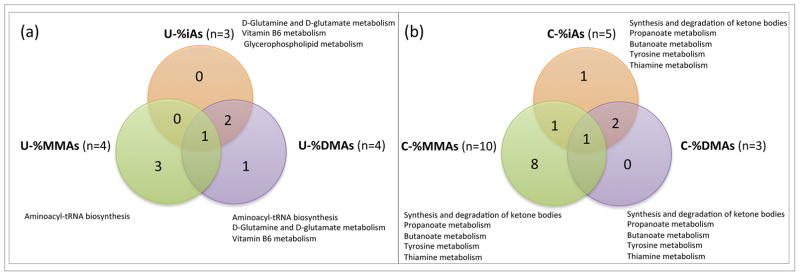
Figure 2.
a: Venn diagram demonstrating the number of neonate cord serum metabolites that are significantly associated with the proportions of the methylated maternal urinary arsenicals (U-%iAs, U-%MMAs, U-%DMAs). b: Venn diagram demonstrating the number of neonate cord serum metabolites that are significantly associated with the proportions of the neonate cord serum arsenicals (C-%iAs, C-%MMAs, C-%DMAs). Significant (p < 0.05) pathways enriched and shared among the methylated arsenicals are indicated.
Analyses of the 10 metabolites identified in relation to U-tAs and/or U-%iAs, U-%MMAs, and U-%DMAs resulted in the identification of several pathways (Table S3, Figure 2a). There were seven enriched pathways identified among the U-tAs-associated metabolites that are broadly involved in amino acid and energy metabolism, including the citrate cycle (TCA) enriched by succinate. There were three pathways that were enriched among the U-%iAs-associated metabolites. There were six pathways that were enriched among the U-%MMAs-associated metabolites, and six pathways were enriched among the U-%DMAs-associated metabolites identified that are all broadly involved in amino acid, energy, and vitamin metabolism. Two pathways were shared between U-%iAs and U-%DMAs, namely glutamine/D-glutamate metabolism and vitamin B6 metabolism. Aminoacyl-tRNA biosynthesis was common to both U-%MMAs and U-%DMAs-associated metabolites. There were no pathways shared between all tested arsenicals (Figure 2).
In a similar manner, multivariable linear regression models were used to analyze the linear relationships among total arsenic in cord serum (C-tAs) and/or iAs metabolites in cord serum (C-%iAs, C-%MMAs, C-%DMAs) as the independent variables and the levels of the 36 cord serum metabolites as the dependent variables (Table S1) across all subjects. Seventeen significant (p ≤ 0.05, q < 0.2) cord serum metabolites were identified from the multivariable regression analyses to display altered abundance in relation to C-tAs and iAs metabolites in neonate cord serum (C-%iAs, C-%MMAs, C-%DMAs) (Table 2). Specifically, of the 36 metabolites tested, a total of six cord serum metabolites were significantly (p ≤ 0.05, q < 0.2) associated with C-tAs (Table 2). Five of these had a negative relationship with C-tAs and include glutamate (β = −0.02; p = 0.02), isoleucine (β = −0.003; p = 0.03), methionine (β = −0.002; p = 0.02), O-acetylcholine (β = −0.004; p = 0.03), and tyrosine (β = −0.002; p = 0.01). Mannose displayed a positive relationship with C-tAs (β = 0.006; p = 0.02) (Table 2).
There were five metabolites with levels associated with C-% iAs. Among these, four displayed a positive relationship with C-%iAs, namely acetoacetate (β = 0.0004; p = 0.02), acetone (β = 0.0003; p = 0.03), ethanol (β = 0.004; p = 0.007), and 3-hydroxybutyrate (β = 0.002; p = 0.007) (Table 2). In contrast, tyrosine displayed a negative relationship with C-%iAs (β = −0.0001; p = 0.01). There were ten-cord serum metabolites with levels associated with C-%MMAs, all of which displayed a positive relationship. These include acetoacetate (β = 0.0002; p = 0.01), betaine (β = 0.004; p = 0.004), glycerol (β = 0.002; p = 0.0003), glycine (β = 0.0046; p = 0001), lactate (β = 0.04; p = 0.02), mannose (β = 0.001; p = 0.001), pyruvate (β = 0.0012; p = 0.0012), serine (β = 0.001; p = 0.02), taurine (β = 0.0009; p = 0.001), and tyrosine (β = 0.0002; p = 0.03). Three-cord serum metabolites displayed levels that were associated with neonate cord serum %DMAs, all having a negative association including acetoacetate (β = −0.0004; p = 0.01), acetone (β = −0.0004; p = 0.002), and tyrosine (β = 0–0.002; p = 0.001) (Table 2).
C-%iAs, C-%MMAs, and C-%DMAs shared one common metabolite, namely acetoacetate. C-%iAs and C-%MMAs shared an additional metabolite of tyrosine. C-%iAs and C-% DMAs shared two additional metabolites of acetone and 3-hydroxybutyrate (Figure 1b). There were a total of eight shared metabolites between the metabolites identified to be significantly associated with maternal exposure/biotransformation indicators and neonate cord measures of iAs and iAs metabolites; these include glutamate, glycine, isoleucine, mannose, methionine, O-acetylcholine, taurine, and tyrosine (Table 2).
Pathway-level analyses of the 16 metabolites identified an enrichment of several pathways associated with C-tAs and/or C-%iAs, C-%MMAs, and C-%DMAs (Table S3, Figure 2b). Specifically, two enriched pathways identified among the C-tAs-associated metabolites were aminoacyl-tRNA biosynthesis and D-glutamine and D-glutamate metabolism. There were four pathways enriched among the C-%iAs-associated metabolites broadly involved in lipid, carbohydrate, amino acid, and vitamin metabolism (Table S3, Figure 2b). There were 17 pathways enriched among the C-%MMAs-associated metabolites, and nine pathways enriched among the C-%DMAs-associated metabolites broadly involved in lipid, carbohydrate, amino acid, and vitamin metabolism, and translation (Table S3, Figure 2b). There were several shared broad pathways associated with the arsenic metabolism indicators. Specifically, metabolites associated with C-%iAs, C-%MMAs, and C-%DMAs were enriched for amino acid metabolism, carbohydrate metabolism, lipid metabolism, and metabolism of cofactors and vitamins. Metabolites associated with C-%MMAs and C-%DMAs were enriched for energy metabolism. Additionally there were shared pathways where metabolites associated with C-%iAs, C-% MMAs, and C-%DMAs were enriched for propanoate metabolism, synthesis and degradation of ketone bodies, tyrosine metabolism, and thiamine metabolism (Table S3; Figure 2b). Metabolites associated with C-%MMAs and C-% DMAs were enriched for butanoate metabolism.
As a secondary analysis, the metabolites identified here were compared to three other human iAs-associated metabolomics studies. Comparisons across studies revealed common metabolites changed in abundance in relation to iAs exposure (Table S1). Creatinine, glutamine, glycerol, glycine, lysine, methionine, proline, serine, succinate, taurine, and tyrosine were identified here as significantly associated with iAs and/or its metabolites and were previously identified in Martin et al.23 Glutamate was found to be altered here as well as in Dudka et al.22 Serine was identified as being significantly altered here and also in both Martin et al. and Zhang et al.18 Lastly, tyrosine was identified to be altered here and in both Dudka et al. and Martin et al.22,23
DISCUSSION
The relationship between prenatal iAs exposure and changes to the neonate metabolome in cord serum has not been previously explored. Metabolomics is a leading technology in functional genomics. Because the metabolome represents a biochemical endpoint of functional expression, where activity within the metabolome is associated with altered enzymatic changes that are relative to changes in the transcriptome and the proteome, it may represent an ultimate readout of the biochemical and physiological state of a cell.43 Thus, changes in the levels of metabolites in response to environmental contaminants may provide insight into specific genes/pathways associated with disease. In the present study, a NMR-based metabolomics assessment was used to identify metabolites in cord serum that were associated with levels of iAs and its major metabolites measured in both maternal urine and neonate cord serum. This pregnancy cohort is located in Gómez Palacio, Mexico where iAs contaminates the water and ranges up to 236 parts per billion.15 Of the metabolites that were analyzed, 10 were identified to be associated with total iAs or iAs metabolites assessed in maternal urine. In contrast, when analyzed in relation to iAs or iAs metabolites in cord serum, a total of 17 metabolites were identified. Interestingly, 11 metabolites were identified as common across a separate iAs-associated population23 and included creatinine, glutamine, glycerol, glycine, lysine, methionine, proline, serine, succinate, taurine, and tyrosine. These data support that prenatal iAs is associated with changes to the infant metabolome assessed in cord serum and a conservation of the metabolomic response in iAs-exposed individuals.44
Differences in iAs metabolism have been shown to be associated with varying disease risks in human populations. For example, in prior work we demonstrated that elevated levels and proportions of U-MMAs were associated with lower birthweight of infants in the BEAR cohort.15 In support of this, in the present study we found that the metabolomic patterns in neonate cord serum differed when analyzed in the context of levels of iAs or iAs metabolites in maternal urinary and/or neonate cord serum. In fact, methionine was the only common metabolite altered in relation to U-%iAs, U-%MMAs, and U-% DMAs. Here, methionine levels increased with higher proportions of U-%iAs and U-%MMAs and decreased in relation to higher proportions of U-%DMAs. This negative relationship between U-%DMAs and methionine is intuitive as methionine plays an important role in arsenic biotransformation and is converted to S-adenosylmethionine (SAM) by methionine adenosyltransferase. SAM serves as a methyl donor in the biotransformation of iAs to MMAs and MMAs to DMAs.12 Similarly, acetoacetate was the only shared metabolite identified in relation to C-%iAs, C-%MMAs, and C-%DMAs. The data from the present study suggest that these differences in iAs metabolism as indicated by the iAs arsenical proportions may also be associated with metabolomic differences in the neonate.45
When the metabolomic profiles were analyzed at the pathway level, interesting overlaps between previous findings from iAs-exposed adults emerged. In the present study, metabolites that are involved in the citrate cycle (TCA) were identified including succinate. Similarly, in prior work, TCA-associated metabolites were also identified to have altered abundance in relation to iAs.23 It has been suggested that the metabolism of iAs may impede the TCA cycle resulting in insufficient energy production.50 In addition to the TCA cycle, here we identified metabolites involved in vitamin B6 metabolism. In relation to this finding, vitamin B-dependent enzymes have been shown to influence metabolism of homocysteine, a known nutrient involved in the biotransformation of iAs, to cysteine in reactions.48 Furthermore, we identified glutamate and amino acid metabolism pathways as being significantly associated with iAs. Interestingly, glutamate is one of three amino acids that comprise glutathione, a major antioxidant involved in iAs biotransformation and a major defense against oxidative stress, including iAs-associated oxidative stress.46 Specifically, a reduction in glutamate can alter glutathione levels, and glutamate is also involved in the synthesis of homocysteine which is a cofactor involved in the methylation of iAs.47 The identified enriched pathways may provide insight into mechanisms by which prenatal iAs exposure influences cellular signaling.49
Several factors should be considered when interpreting the findings from the present study. While the sample size was relatively modest, this study is sufficiently powered to detect significant changes in metabolite levels as has been demonstrated in other studies.18,23 Given the age of the infants, it is currently unknown whether these changes in metabolite levels lead to disruption of biological pathways and whether they are related to health or disease early or later in life. Future research will allow for cross-cohort comparisons of the identified metabolites and the examination of relationships to health endpoints. The cross-population comparison of metabolites may be influenced by variation in demographic, disease status, and study design. Still, the overlap of some metabolites is indicative of the potential to identify a common metabolomic response in individuals exposed to iAs. This study has unique advantages including NMR-based assessment for sensitive detection of neonate cord metabolites and the identification of associations of neonate cord metabolites with efficiency maternal metabolism of iAs. These changes in metabolite abundance may suggest altered enzymatic function in the fetus. These results could inform future exposure assessment studies as well as clinical uses for iAs-associated diseases.
In summary, this is the first metabolomic assessment in neonate cord serum related to prenatal arsenic exposure in a human population. Our data suggest that prenatal arsenic exposure may be predictive of metabolomic alterations in neonate cord serum and that the biotransformation of arsenic may differentially influence the neonatal metabolome. The identified metabolites may provide insight into diseases associated with prenatal iAs exposure. These results provide more support for the need to assess metabolism of iAs as an etiologic factor underlying iAs-associated diseases. Future studies will need to consider the use of metabolomics as a useful method for determining biomarkers of iAs-associated diseases and/or provide insight into mechanisms that influence disease development.51
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institute of Environmental Health Sciences (P42-ES005948, R01-ES019315, U24 DK097193, and T32-ES07018).
Footnotes
ORCID
Rebecca C. Fry: 0000-0003-0899-9018
Notes
The authors declare no competing financial interest.
This material is available free of charge at The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.6b04374.
Tables S1–S3 (XLS)
References
- 1.Rahman MM, Ng JC, Naidu R. Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ Geochem Health. 2009;31(S1):189–200. doi: 10.1007/s10653-008-9235-0. [DOI] [PubMed] [Google Scholar]
- 2.Monographs, I. A. f. R. [accessed November 6, 2015];Arsenic, metals, fibres, and dusts volume, 100 C, A review of human carcinogens. http://monographs.iarc.fr/ENG/Monographs/vol100C/
- 3.Maull EA, Ahsan H, Edwards J, Longnecker MP, Navas-Acien A, Pi J, Silbergeld EK, Styblo M, Tseng CH, Thayer KA, Loomis D. Evaluation of the association between arsenic and diabetes: a National Toxicology Program workshop review. Environ Health Perspect. 2012;120(12):1658–70. doi: 10.1289/ehp.1104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, Suk WA. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect. 2013;121(3):295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey KA, Smith AH, Tokar EJ, Graziano JH, Kim KW, Navasumrit P, Ruchirawat M, Thiantanawat A, Suk WA, Fry RC. Mechanisms Underlying Latent Disease Risk Associated with Early-Life Arsenic Exposure: Current Research Trends and Scientific Gaps. Environ Health Perspect. 2016;124(2):170–5. doi: 10.1289/ehp.1409360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, Bhattacharya S, Kandjanapa K, Soontararuks S, Nookabkaew S, Mahidol C, Ruchirawat M, Samson LD. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007;3(11):e207. doi: 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fei DL, Koestler DC, Li Z, Giambelli C, Sanchez-Mejias A, Gosse JA, Marsit CJ, Karagas MR, Robbins DJ. Association between In Utero arsenic exposure, placental gene expression, and infant birth weight: a US birth cohort study. Environ Health. 2013;12:58. doi: 10.1186/1476-069X-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci. 2015;143(1):97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broberg K, Ahmed S, Engström K, Hossain MB, Jurkovic Mlakar S, Bottai M, Grandér M, Raqib R, Vahter M. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Origins Health Dis. 2014;5(4):288–98. doi: 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, Mahiuddin G, Mostofa G, Hsueh YM, Wright RO, Christiani DC. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect. 2012;120(7):1061–6. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed S, Mahabbat-e Khoda S, Rekha RS, Gardner RM, Ameer SS, Moore S, Ekström EC, Vahter M, Raqib R. Arsenic-associated oxidative stress, inflammation, and immune disruption in human placenta and cord blood. Environ Health Perspect. 2011;119(2):258–64. doi: 10.1289/ehp.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas DJ, Styblo M, Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicol Appl Pharmacol. 2001;176(2):127–44. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- 13.Vahter M. Mechanisms of arsenic biotransformation. Toxicology. 2002;181–182:211–7. doi: 10.1016/s0300-483x(02)00285-8. [DOI] [PubMed] [Google Scholar]
- 14.Tseng CH. Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2007;25(1):1–22. doi: 10.1080/10590500701201695. [DOI] [PubMed] [Google Scholar]
- 15.Laine JE, Bailey KA, Rubio-Andrade M, Olshan AF, Smeester L, Drobná Z, Herring AH, Stýblo M, García-Vargas GG, Fry RC. Maternal arsenic exposure, arsenic methylation efficiency, and birth outcomes in the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Mexico. Environ Health Perspect. 2015;123(2):186–92. doi: 10.1289/ehp.1307476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn WB. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys Biol. 2008;5(1):011001. doi: 10.1088/1478-3975/5/1/011001. [DOI] [PubMed] [Google Scholar]
- 17.Peng S, Yan L, Zhang J, Wang Z, Tian M, Shen H. An integrated metabonomics and transcriptomics approach to understanding metabolic pathway disturbance induced by perfluorooctanoic acid. J Pharm Biomed Anal. 2013;86:56–64. doi: 10.1016/j.jpba.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Yan L, Tian M, Huang Q, Peng S, Dong S, Shen H. The metabonomics of combined dietary exposure to phthalates and polychlorinated biphenyls in mice. J Pharm Biomed Anal. 2012;66:287–97. doi: 10.1016/j.jpba.2012.03.045. [DOI] [PubMed] [Google Scholar]
- 19.Fanos V, Atzori L, Makarenko K, Melis GB, Ferrazzi E. Metabolomics application in maternal-fetal medicine. BioMed Res Int. 2013;2013:720514. doi: 10.1155/2013/720514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart DS. Quantitative metabolomics using NMR. TrAC, Trends Anal Chem. 2008;27:228–237. [Google Scholar]
- 21.Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455(7216):1054–6. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 22.Dudka I, Kossowska B, Senhadri H, Latajka R, Hajek J, Andrzejak R, Antonowicz-Juchniewicz J, Gancarz R. Metabonomic analysis of serum of workers occupationally exposed to arsenic, cadmium and lead for biomarker research: a preliminary study. Environ Int. 2014;68:71–81. doi: 10.1016/j.envint.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Martin E, González-Horta C, Rager J, Bailey KA, Sánchez-Ramírez B, Ballinas-Casarrubias L, Ishida MC, Gutiérrez-Torres DS, Hernández Cerón R, Viniegra Morales D, Baeza Terrazas FA, Saunders RJ, Drobná Z, Mendez MA, Buse JB, Loomis D, Jia W, García-Vargas GG, Del Razo LM, Stýblo M, Fry R. Metabolomic characteristics of arsenic-associated diabetes in a prospective cohort in Chihuahua, Mexico. Toxicol Sci. 2015;144(2):338–46. doi: 10.1093/toxsci/kfu318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Drobná Z, Currier J, Douillet C, Olshan AF, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ Mol Mutagen. 2014;55(3):196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey KA, Laine J, Rager JE, Sebastian E, Olshan A, Smeester L, Drobná Z, Styblo M, Rubio-Andrade M, García-Vargas G, Fry RC. Prenatal arsenic exposure and shifts in the newborn proteome: interindividual differences in tumor necrosis factor (TNF)-responsive signaling. Toxicol Sci. 2014;139(2):328–37. doi: 10.1093/toxsci/kfu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes MF. Biomarkers of exposure: a case study with inorganic arsenic. Environ Health Perspect. 2006;114(11):1790–6. doi: 10.1289/ehp.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devesa V, Maria Del Razo L, Adair B, Drobna Z, Waters SB, Hughes MF, Styblo M, Thomas DJ. Comprehensive analysis of arsenic metabolites by pH-specific hydride generation atomic absorption spectrometry. J Anal At Spectrom. 2004;19(11):1460–1467. [Google Scholar]
- 28.Hernandez-Zavala A, Matousek T, Drobna Z, Paul DS, Walton F, Adair BM, Dĕdina J, Thomas DJ, Styblo M. Speciation analysis of arsenic in biological matrices by automated hydride generation-cryotrapping-atomic absorption spectrometry with multiple microflame quartz tube atomizer (multiatomizer) J Anal At Spectrom. 2008;23:342–351. doi: 10.1039/b706144g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Zavala A, Drobna Z, Styblo M, Thomas DJ. Analysis of arsenical metabolites in biological samples. Curr Protoc Toxicol. 2009;42:4.33.1–4.33.17. doi: 10.1002/0471140856.tx0433s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nermell B, Lindberg AL, Rahman M, Berglund M, Persson LA, El Arifeen S, Vahter M. Urinary arsenic concentration adjustment factors and malnutrition. Environ Res. 2008;106(2):212–8. doi: 10.1016/j.envres.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Matoušek T, Currier JM, Trojánková N, Saunders RJ, Ishida MC, González-Horta C, Musil S, Mester Z, Stýblo M, Dědina J. Selective hydride generation- cryotrapping- ICP-MS for arsenic speciation analysis at picogram levels: analysis of river and sea water reference materials and human bladder epithelial cells. J Anal At Spectrom. 2013;28(9):1456–1465. doi: 10.1039/C3JA50021G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Church RJ, Wu H, Mosedale M, Sumner SJ, Pathmasiri W, Kurtz CL, Pletcher MT, Eaddy JS, Pandher K, Singer M, Batheja A, Watkins PB, Adkins K, Harrill AH. A systems biology approach utilizing a mouse diversity panel identifies genetic differences influencing isoniazid-induced microvesicular steatosis. Toxicol Sci. 2014;140(2):481–92. doi: 10.1093/toxsci/kfu094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathmasiri W, Pratt KJ, Collier DN, Lutes LD, McRitchie S, Sumner SCJ. Integrating metabolomic signatures and psychosocial parameters in responsivity to an immersion treatment model for adolescent obesity. Metabolomics. 2012;8(6):1037–1051. [Google Scholar]
- 34.Sumner SC, Fennell TR, Snyder RW, Taylor GF, Lewin AH. Distribution of carbon-14 labeled C60 ([14C]C60) in the pregnant and in the lactating dam and the effect of C60 exposure on the biochemical profile of urine. J Appl Toxicol. 2010;30(4):354–6. doi: 10.1002/jat.1503. [DOI] [PubMed] [Google Scholar]
- 35.Tea I, Le Gall G, Kuster A, Guignard N, Alexandre-Gouabau MC, Darmaun D, Robins RJ. 1H-NMR-based metabolic profiling of maternal and umbilical cord blood indicates altered maternofoetal nutrient exchange in preterm infants. PLoS One. 2012;7(1):e29947. doi: 10.1371/journal.pone.0029947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan EC, Pasikanti KK, Nicholson JK. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat Protoc. 2011;6(10):1483–99. doi: 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- 37.Alonso A, Marsal S, Julià A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3:23. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78(13):4430–42. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 39.Sun Z, Asmann YW, Kalari KR, Bot B, Eckel-Passow JE, Baker TR, Carr JM, Khrebtukova I, Luo S, Zhang L, Schroth GP, Perez EA, Thompson EA. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PLoS One. 2011;6(2):e17490. doi: 10.1371/journal.pone.0017490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi Y, Li C, Miller C, George AL. Strategy for encoding and comparison of gene expression signatures. Genome Biol. 2007;8(7):R133. doi: 10.1186/gb-2007-8-7-r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6(6):743–60. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Shen H, Xu W, Xia Y, Barr DB, Mu X, Wang X, Liu L, Huang Q, Tian M. Urinary metabolomics revealed arsenic internal dose-related metabolic alterations: a proof-of-concept study in a Chinese male cohort. Environ Sci Technol. 2014;48(20):12265–74. doi: 10.1021/es503659w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbanczyk-Wochniak E, Luedemann A, Kopka J, Selbig J, Roessner-Tunali U, Willmitzer L, Fernie AR. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Rep. 2003;4(10):989–93. doi: 10.1038/sj.embor.embor944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Sevillano MA, Contreras-Acuña M, García-Barrera T, Navarro F, Gómez-Ariza JL. Metabolomic study in plasma, liver and kidney of mice exposed to inorganic arsenic based on mass spectrometry. Anal Bioanal Chem. 2014;406(5):1455–69. doi: 10.1007/s00216-013-7564-z. [DOI] [PubMed] [Google Scholar]
- 45.García-Sevillano MA, García-Barrera T, Navarro F, Montero-Lobato Z, Gómez-Ariza JL. Shotgun metabolomic approach based on mass spectrometry for hepatic mitochondria of mice under arsenic exposure. BioMetals. 2015;28(2):341–51. doi: 10.1007/s10534-015-9837-9. [DOI] [PubMed] [Google Scholar]
- 46.Flora SJ. Arsenic-induced oxidative stress and its reversibility. Free Radical Biol Med. 2011;51(2):257–81. doi: 10.1016/j.freeradbiomed.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Gamble MV, Liu X, Ahsan H, Pilsner R, Ilievski V, Slavkovich V, Parvez F, Levy D, Factor-Litvak P, Graziano JH. Folate, homocysteine, and arsenic metabolism in arsenic-exposed individuals in Bangladesh. Environ Health Perspect. 2005;113(12):1683–8. doi: 10.1289/ehp.8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selhub J. Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J Nutr Health Aging. 2002;6(1):39–42. [PubMed] [Google Scholar]
- 49.Zhao F, Liao Y, Jin Y, Li G, Lv X, Sun G. Effects of arsenite on glutamate metabolism in primary cultured astrocytes. Toxicol In Vitro. 2012;26(1):24–31. doi: 10.1016/j.tiv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Tseng CH. The potential biological mechanisms of arsenic-induced diabetes mellitus. Toxicol Appl Pharmacol. 2004;197(2):67–83. doi: 10.1016/j.taap.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Dunn WB, Wilson ID, Nicholls AW, Broadhurst D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis. 2012;4(18):2249–64. doi: 10.4155/bio.12.204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.